Abstract
The Targeting Protein for Xklp2 (TPX2) is a central regulator of spindle assembly in vertebrate cells. The absence or excess of TPX2 inhibits spindle formation. We have defined a TPX2 signature motif that is present once in vertebrate sequences but twice in plants. Plant TPX2 is predominantly nuclear during interphase and is actively exported before nuclear envelope breakdown to initiate prospindle assembly. It localizes to the spindle microtubules but not to the interdigitating polar microtubules during anaphase or to the phragmoplast as it is rapidly degraded during telophase. We characterized the Arabidopsis thaliana TPX2-targeting domains and show that the protein is able to rescue microtubule assembly in TPX2-depleted Xenopus laevis egg extracts. Injection of antibodies to TPX2 into living plant cells inhibits the onset of mitosis. These results demonstrate that plant TPX2 already functions before nuclear envelope breakdown. Thus, plants have adapted nuclear–cytoplasmic shuttling of TPX2 to maintain proper spindle assembly without centrosomes.
INTRODUCTION
Chromosome segregation is achieved through mitotic spindle activity in all eukaryotes. An early step of spindle assembly involves the nucleation of microtubules. In somatic animal cells, centrosomal microtubules form a prospindle, which may act as a basket to keep the chromosomes enclosed when the nuclear envelope breaks down. During prometaphase, these microtubules actively search for the chromosomes. Microtubules also nucleate from or in the vicinity of the chromosomes and align with the microtubules emanating from the centrosomes. Thus, the spindle is built up of two sets of antiparallel aligned microtubules (Hyman and Karsenti, 1996). In mature oocytes, centrosomes are absent and the meiotic spindle is formed solely by microtubule nucleation, sorting around the chromosomes and spindle bipolarization (Walczak et al., 1998). This activity can be simulated in Xenopus laevis egg extract around DNA-coated beads (Karsenti and Vernos, 2001; Carazo-Salas and Karsenti, 2003; Gruss and Vernos, 2004).
Higher plants are characterized by an acentrosomal spindle. A prospindle forms before nuclear envelope breakdown (NEB) by the convergence of aster-like microtubules nucleated at the nuclear envelope (Schmit et al., 1985; Stoppin et al., 1994; Canaday et al., 2000). It has been suggested that after NEB, the chromosome-based mechanism also takes place in higher plants (Lloyd and Chan, 2006).
In vertebrates, the chromosome-based mechanism relies on a gradient of active Ran GTPase around the chromosomes (Carazo-Salas et al., 2001; Hetzer et al., 2002; Caudron et al., 2005; Clarke and Zhang, 2008). One of the downstream effectors of this gradient is the Targeting Protein for Xklp2 (TPX2). TPX2 localizes to the nucleus during interphase and is released from importin-α and importin-β by active RanGTP at NEB. The activated TPX2 then induces microtubule nucleation at the kinetochores and around the chromosomes and binds to these microtubules, but not to the astral microtubules when present (Karsenti and Vernos, 2001; Gruss and Vernos, 2004; Tulu et al., 2006). Finally, microtubule motor proteins, stabilizers, and bundling proteins (microtubule-associated proteins [MAPs]) align and sort the microtubules to shape the spindle (Walczak et al., 1998). At the end of anaphase, TPX2 relocalizes to the spindle midzone. Thereafter, it is rapidly degraded (Stewart and Fang, 2005), although it is also claimed to be required for postmitotic nuclear envelope assembly (O'Brien and Wiese, 2006).
Downstream in the signaling pathway, TPX2 localizes the essential mitotic kinase Aurora A to the spindle microtubules. Aurora A is activated by TPX2 binding, and TPX2 is phosphorylated by Aurora A (Kufer et al., 2002; Bayliss et al., 2003). Aurora A functions in several aspects of cell division; among others, it activates microtubule nucleation from the centrosome (Ducat and Zheng, 2004; Brittle and Ohkura, 2005). The plus-end-directed kinesin Xklp2, which is involved in spindle pole stability, is also brought to the spindle microtubules by TPX2 (Walczak et al., 1998; Wittmann et al., 2000). Moreover, a short C-terminal domain of TPX2 plays an Eg5-dependent function in spindle pole segregation (Eckerdt et al., 2008).
Depending on stringency, depletion of TPX2 from Xenopus egg extracts or addition of TPX2 antibodies to these extracts causes effects that range from aberrant spindle poles to a complete block of spindle formation. Addition of excess TPX2 generates monopolar half-spindles with increased microtubule amounts or RanGTP-independent, ectopic asters (Wittmann et al., 2000; Gruss et al., 2001). In addition, TPX2 is essential for spindle formation in somatic cells, which possess centrosomes. Inhibition of TPX2 function in living HeLa cells by RNA interference or through the injection of antibodies causes the formation of two centrosome-based asters that do not interact with each other (Gruss et al., 2002).
Several of the genes involved in spindle formation have homologs in plants. Plants have a Ran GTPase and most factors that associate with it (Jeong et al., 2005; Zhao et al., 2006; Meier, 2007). Three Arabidopsis thaliana Aurora-like kinases (At AURORA1 to -3) were picked up in a large-scale green fluorescent protein (GFP)–tagging approach (van Damme et al., 2004; Demidov et al., 2005) and independently by homology with the animal and yeast Aurora kinases (Kawabe et al., 2005). All three proteins are able to phosphorylate histone H3, and At AURORA1 and -2 localize to the nucleus during interphase and the spindle during metaphase, thereby resembling the animal Aurora A–type kinases (Demidov et al., 2005; Kawabe et al., 2005).
Here, we characterize an Arabidopsis sequence that shares structural and functional similarities with animal TPX2. Therefore, it likely corresponds to the TPX2 ortholog. We characterized and used a signature sequence to reveal the relationships between TPX2 homologs. This sequence is repeated twice in all sequenced plant TPX2 proteins. Furthermore, Arabidopsis TPX2 has two nuclear localization signals (NLSs), a nuclear export signal (NES), and two microtubule binding domains. Structural analysis shows that At TPX2 may interact with Arabidopsis Aurora kinases. At TPX2 localizes to the nucleus during interphase and is exported through the activity of the NES at the G2/M transition to initiate the assembly of the prospindle. During mitosis, At TPX2 localizes to the spindle, but it does not interact with the microtubules located at the midzone during anaphase or telophase. However, truncated protein, lacking both NLSs, is cytoplasmic and binds cortical microtubules during interphase. At TPX2 is completely degraded at the end of anaphase. Functional analysis shows that At TPX2 plays a role in cell division. It can assemble microtubules in vitro and rescue aster formation in TPX2-depleted Xenopus egg extracts. Antibodies to TPX2 inhibit NEB and mitosis when microinjected into plant cells during late prophase. The entry into mitosis is probably arrested because the prospindle microtubules cannot be assembled when the interaction of antibodies with nuclear exported TPX2 inhibits its function. These results suggest that, in plants, TPX2 has adapted its behavior to play a key role in spindle formation. Moreover, if there is a perichromosomal spindle assembly mechanism analogous to the animal one, then it may not act independently from the perinuclear prospindle assembly mechanism.
RESULTS
Molecular Characterization of Plant TPX2 Homologs
Primary sequence comparisons were performed to determine the degree of identity/similarity between proteins homologous to human TPX2. A unique full-length gene sequence is present in the Arabidopsis genome (AT1G03780). Using the T-Coffee alignment tool, its sequence was compared with that of human TPX2 (Figure 1). The sequence is 758 amino acids long and shares <40% similarity with its animal counterparts (Table 1), including a PFAM 06886 targeting for Xklp2 that is considered a TPX2-specific domain (Figure 1). Except for Arabidopsis, all other plant TPX2 sequences found in data banks were incomplete and did not allow the establishment of identity/similarity rates for full-length proteins. Nevertheless, a self-comparison of the Arabidopsis peptide sequence revealed a duplicated region in the protein (Figure 1). Comparison of this duplication led us to a specific 26– to 28–amino acid motif that is conserved among eukaryotes (see Supplemental Figure 1 online). We defined this unique and specific TPX2 signature motif as R-x-R-[PAS]-x(3)-K-[STG]-x-[AS]-[ED]-x-E-x-E-[EM]-[LIFVM]-x(3,5)-[YFP]-K-F-[KR]-A (PROSITE format). The duplication was found in the 17 plant sequences analyzed (e.g., the moss Physcomitrella patens, the cicad Zamia fischeri, the monocot rice [Oryza sativa], and the dicot grape [Vitis vinifera]) but never in the animal sequences. Interestingly, using the signature sequence, we did not find any homologous proteins in insects, worms, or fungi, suggesting that they do not have a TPX2 gene at all. A relationship tree was established on the basis of these signature sequences (see Supplemental Figure 2 online; the alignment used to generate the tree is available in Supplemental Data Set 1 online). It showed that the vertebrate signature motif is closer to the second plant signature motif than the first one (Figure 1). In Arabidopsis, both copies of the signature are located in central positions (amino acids 330 to 356 and amino acids 430 to 456). The conservation of these specific amino acid sequences may reflect functional specificities, but as the signature is not known to have a predicted function, we first determined whether characteristic targeting domains are conserved in the At TPX2 sequence.
Figure 1.
T-Coffee Alignment of Arabidopsis and Human TPX2.
Conserved AURORA A binding sites are present in the N terminus (black boxes). TPX2 signature motifs are in gray boxes. The C-terminal PFAM-defined Xklp2 binding domain of TPX2 is underlined in black.
Table 1.
Percentage of Identity/Similarity of TPX2 between Species
| Species | Identity/Similarity with Arabidopsis |
|---|---|
| Oryza sativa | 38%/52% |
| Xenopus laevis | 22%/36% |
| Mus musculus | 23%/37% |
| Homo sapiens | 21%/39% |
In the search for putative NLSs in At TPX2, only one putative NLS motif (amino acids 60 to 66) was predicted using the NUCDISC program. This prediction did not overlap with any of the seven putative NLSs described in the Xenopus protein (Schatz et al., 2003). None of the other computer analysis software programs (Psort1 and Psort2, the Predict NLS server, etc.) identified any putative NLS. NLS124, -284, and -550, which are conserved between Xenopus, mouse, chicken, and human TPX2 (Schatz et al., 2003), were not conserved in Arabidopsis.
A large C-terminal domain of Xenopus TPX2 is involved in both Xklp2 targeting and microtubule binding. This region is more conserved among eukaryotes than the N-terminal part of the protein. Furthermore, two MAPs, WAVE-DAMPENED2 from Arabidopsis (WVD2; Perrin et al., 2007) and a 20.7-kD MAP of poplar (Populus sp; ABG57264) share homologies with the C-terminal domain of TPX2 (Rajangam et al., 2008), suggesting that a microtubule binding domain is present in At TPX2.
An Aurora kinase binding domain is present in the N-terminal part of animal TPX2 (Kufer et al., 2002; Bayliss et al., 2003). Sequence alignments of both human and Arabidopsis N termini are quite divergent, but hydrophobic cluster analysis revealed that the amino acids 1 to 42 region of human TPX2, which is functionally important for Aurora A binding, may correspond to the amino acids 14 to 65 region of the At TPX2 sequence (Figure 1). In order to address this question, we modeled the putative interaction between the At AURORA kinases and At TPX214-65 using the only available crystallographic model of human Aurora A/Hs TPX2 as a template (Figure 2A). Full-length At AURORA2 and Hs Aurora A share 65% identical and 78% similar residues with three gaps. The gaps consist of one deletion of one residue and one insertion of two residues in At AURORA2 (see Supplemental Figure 3 online). When we calculated identity/similarity based solely on the 38 amino acids from Hs Aurora A that make contact with Hs TPX2, identity with At AURORA2 rose to ∼74% and similarity rose to almost 95%. Thus, the plant model obtained by homology modeling is both accurate and very similar to the human Protein Data Bank model (entry 1OL5). The binding groove described by Bayliss et al. (2003) was almost identical in At AURORA2, and all of the major amino acids involved in both the recognition and binding of TPX2 were conserved (Figure 2B). When applied to At AURORA1 and -3, homology modeling suggested a putative binding for the former and excluded it for the latter. All of these data suggest that the N-terminal region of At TPX2 may interact with At AURORA1 and -2 and that At TPX2 may rescue Xl TPX2 deletion in Xenopus egg extract.
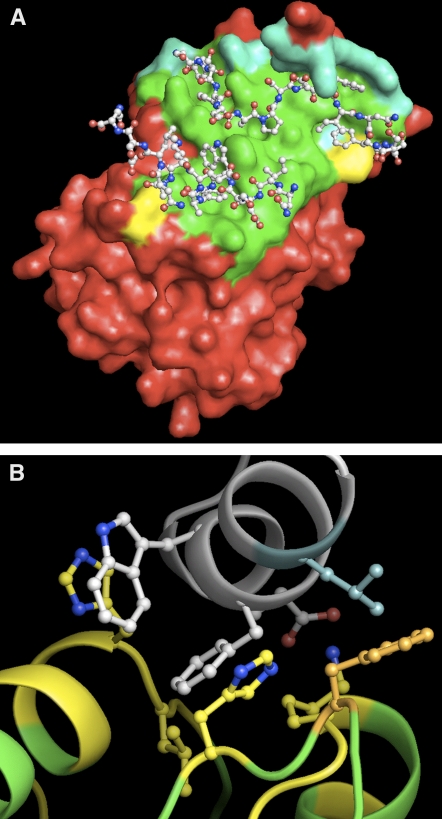
Figure 2.
Three-Dimensional Model of the Putative Interaction between the At AURORA Kinases and At TPX214-65 and Comparison with the Crystallographic Model of Human Aurora A/Hs TPX2.
(A) Crystallographic representation of the interaction between Hs Aurora A (colored residue surface) and Hs TPX2 (balls and sticks). The Aurora A residues not involved in the contact with Hs TPX2 (i.e., those whose accessible surface is not modified upon Hs TPX2 binding) are shown in red. The Aurora A residues involved in the interaction are shown in green, cyan, and yellow, according to their conservation in At AURORA2. The corresponding At AURORA2/At TPX2 putative interacting sequences completely agree with the human representation (green). Conserved replacements of amino acids are shown in cyan, and only two amino acids appear not to be conserved (yellow).
(B) Detailed ribbon view of the At AURORA2/At TPX2 model (Sali and Blundell, 1993) showing the amino acids closely involved in the interaction. At TPX2 is shown in white, At AURORA2 is shown in yellow or green depending on whether the residue is involved in the contact or not, respectively. The side chains of strongly interacting amino acids are shown as balls and sticks.
Full-Length At TPX2 Is Targeted to the Nucleus and Binds Microtubules
Antibodies raised against human TPX2 (Gruss et al., 2002) and the Arabidopsis TPX2 signature motif cross react with plant proteins (Figure 3A). A band of ∼65 kD was highlighted in Arabidopsis seedling, tobacco (Nicotiana tabacum BY-2) suspension culture, and Tradescantia virginiana flower bud total protein extracts. Anti-At TPX2 labeled a faint supplemental band of >100 kD in the Arabidopsis seedling extract. This latter labeling may correspond to native At TPX2. To ensure that these antibodies, which were used for immunolocalization and microinjection experiments, really recognize plant TPX2, we prepared nuclear extracts of Nicotiana benthamiana leaves infiltrated with Agrobacterium tumefaciens carrying an E-tag:GFP:At TPX2 construct. The transgenic protein was labeled using antibodies against human TPX2, GFP, and E-tag (Figure 3B), confirming the expected cross reactivity. Surprisingly, these bands remained <70 kD instead of the expected mass of ∼120 kD. These data strongly argue for a high lability of At TPX2 despite the use of wide-spectrum protease inhibitor mix. The cross reactivity with the two anti-TPX2 antibodies was finally demonstrated by inducing the expression of 6His-At TPX2 in the yeast Pichia pastoris (which has no endogenous TPX2) and comparing the labeling of total protein extracts with anti-6His, anti-human TPX2, and anti-Arabidopsis TPX2 (Figure 3C). Also here, common bands were labeled with all antibodies, but Pichia extract carrying the empty expression vector did not cross react with any of the antibodies. Despite the degradation that is enhanced by the induction of expression, a faint band corresponding to the suspected full-length protein could be seen (arrowhead), validating the further use of these antibodies. We then immunolabeled BY-2 cells using these two antibodies. In both samples, the nucleoplasm was labeled during interphase until late G2, the perinuclear region was labeled during early prophase, and the spindle was labeled during mitosis (Figures 3D to 3I; see Supplemental Figure 4 online). The phragmoplast, however, was not labeled (see Supplemental Figure 4 online). These results are consistent with the localization of TPX2 in human and Xenopus cells.
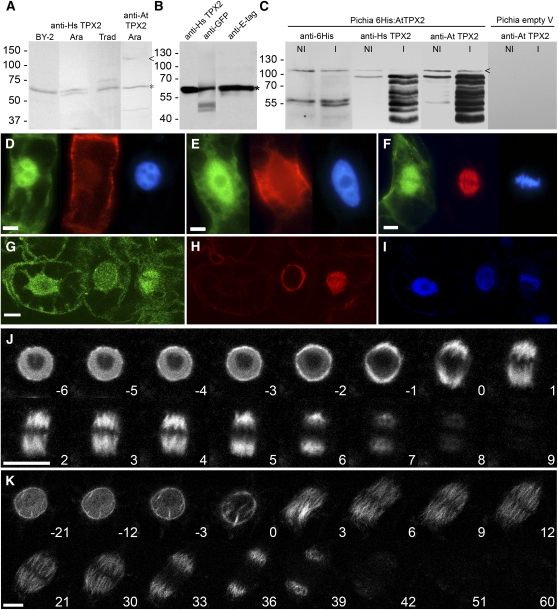
Figure 3.
Expression and Dynamics of Full-Length At TPX2 in Various Plant Cells.
(A) Protein gel blotting of total protein extracts of tobacco BY-2 suspension cells, T. virginiana flower buds (Trad), and Arabidopsis seedlings (Ara) probed with anti-human and anti-Arabidopsis TPX2 antibodies. In all samples, a band of ∼65 kD was labeled (asterisk). Full-length At TPX2 is marked by an arrowhead.
(B) Nuclear extract from N. benthamiana leaves that were infiltrated with P35S:E-tag:GFP:At TPX2. When probed with anti-human TPX2, anti-GFP, and anti-E-tag antibodies, the same band was labeled, indicating that anti-human TPX2 antibodies cross react with expressed TPX2 despite degradation of the protein, as suggested by the observed low molecular weight.
(C) Total protein extract of P. pastoris expressing 6His-At TPX2 and probed with anti-6His, anti-human TPX2, and anti-Arabidopsis TPX2 antibodies. A low expression was still observed in noninduced cells (NI) but was enhanced after induction (I). Extract from cells carrying only the empty vector did not cross react with anti-Arabidopsis TPX2 antibodies. Small amounts of native 6His-At TPX2 were detected with all three antibodies (arrowhead), confirming that the TPX2 antibodies cross react with At TPX2. The lower bands correspond to At TPX2 degradation products.
(D) to (F) Immunolabeling of BY-2 cells using anti-human TPX2 antibody (left cells), anti-tubulin antibody (middle cells), and DAPI chromatin staining (right cells). (D) shows interphase cells with cortical microtubules and predominantly nucleus-localized TPX2. (E) shows late G2 phase cells with preprophase band and nucleus-localized TPX2. (F) shows prometaphase cells with TPX2 decorating the spindle. Bars = 10 μm.
(G) to (I) Immunolabeling of BY-2 cells with anti-Arabidopsis TPX2 antibody (G), anti-tubulin antibody (H), and DAPI staining (I). TPX2 is intranuclear in interphase (left cells), perinuclear in prophase (middle cells), and localized to the spindle together with the microtubules after NEB (right cells). Bar = 10 μm.
(J) and (K) GFP-At TPX2 localization during cell division. (J) shows GPF-At TPX2 localization in Arabidopsis root cells from late G2 through mitosis and cytokinesis. (K) shows GFP-At TPX2 localization in BY-2 suspension culture cells. In both cells, the shift from the nuclear localization to the prospindle and the sudden breakdown of the protein at the end of anaphase are clearly visible. Time in minutes after NEB is indicated. Bars = 10 μm.
In order to observe the subcellular dynamics of At TPX2 during cell division, we expressed the full-length protein in fusion with GFP at its N terminus using various constructs, fluorescent markers, transformation methods, and plant model species, such as stably transformed Arabidopsis and BY-2 suspensions (Figures 3J and 3K) and bombarded BY-2 cells (see Supplemental Table 1 online). At TPX2 in Arabidopsis roots was located within 65% of interphase nuclei (see Supplemental Table 1 online). Thirty-five percent of nuclei were not labeled, suggesting that At TPX2 production and import are cell cycle–regulated. Similar nuclear localization was observed when wild-type or transgenic tobacco leaves were agroinfiltrated with P35S:GFP:At TPX2 constructs. Nuclear targeting of GFP-At TPX2 was not impaired by tubulin inhibitors (oryzalin or colchicine) or actin inhibitors (latrunculin B), indicating that the protein import was not cytoskeleton-dependent (see Supplemental Table 1 online). This suggests that At TPX2 possesses at least one nuclear localization signal. Furthermore, the overexpression of GFP:At TPX2 led to the labeling of cortical microtubules in bombarded BY-2 cells, agroinfiltrated N. benthamiana leaves, and polyethylene glycol–transfected Nicotiana plumbaginifolia protoplasts (see Supplemental Table 1 online). This confirms that, as in animal TPX2, At TPX2 presents a microtubule binding capability.
During G2 and early prophase in Arabidopsis root cells that express either GFP-At TPX2 (Figure 3J; see Supplemental Movie 1 online ) or both GFP-At TPX2 and the histone H2AX-monomeric red fluorescent protein (mRFP) (see Supplemental Movie 2 online), fluorescence progressively increased as chromatin condensed. Shortly before NEB, At TPX2 was exported within 5 min from the nucleus and organized into two polar crescents. After NEB, the forming spindle was progressively labeled. From anaphase onward, the labeling followed the shortening kinetochore fibers but marked neither the central spindle nor the phragmoplast microtubules before degradation (Figure 3J; see Supplemental Movies 1 and 2 online). In BY-2 lines expressing P35S:GFP:At TPX2, similar TPX2 dynamics were observed (Figure 3K; see Supplemental Figure 5 and Supplemental Movie 3 online). These data point to a role for At TPX2 in perinuclear microtubule and spindle assembly but preclude roles in phragmoplast assembly, nuclear envelope reformation, and cortical microtubule assembly at the onset of G1.
Characterization of the Domains Required for At TPX2 Localization in the Nucleus and on Microtubules
As nuclei and microtubules were labeled with GFP-At TPX2, we characterized the minimal domain(s) needed for nuclear localization (i.e., putative NLSs) and microtubule binding. To determine these sequences, we designed truncated P35S:GFP:At TPX2 constructs and expressed them in bombarded BY-2 cells (Figure 4).
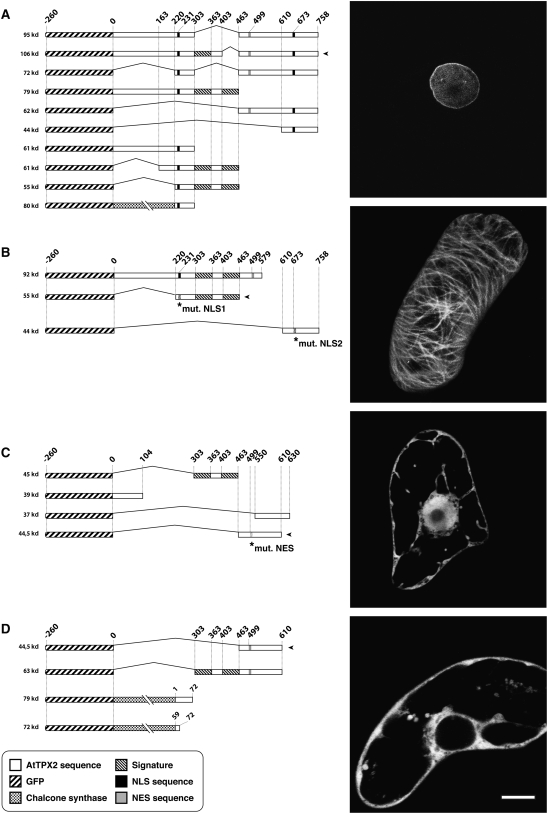
Figure 4.
Typical Expression Profiles of GFP:At TPX2 Domains in BY-2 Cells.
Tobacco BY-2 cells were bombarded with the constructs represented by the drawings at left. Based on the resulting TPX2 localization patterns, the various constructs were grouped, and representative images are shown at right (the construct used for illustration is marked by an arrowhead to its right). (A) shows exclusively nuclear targeting. (B) shows microtubule targeting. (C) shows diffuse nucleocytoplasmic location. (D) shows exclusively cytoplasmic location. Specific sequences are marked and explained at bottom. Bar = 10 μm.
Depending on the construct (see Supplemental Table 1 online), the fluorescence was distributed in four subcellular locations. An exclusive nuclear targeting (Figure 4A) demonstrated that an NLS activity was carried by sequences located between amino acids 220 and 303 and between amino acids 610 and 758. The amino acid mutations KR231NG and KR673NG abolished nuclear targeting, confirming the suspected NLS activity of these basic amino acids. The KR733NG mutation did not modify the nuclear targeting. Two domains corresponding to amino acids 220 to 463 and amino acids 684 to 758 were seen to preferentially target At TPX2 to the microtubule cytoskeleton (Figure 4B). The region from amino acids 220 to 303 seems necessary for this targeting, as the sequence strictly limited to the signatures (amino acids 303 to 463) was no longer specifically targeted. It revealed a diffuse nucleocytoplasmic pattern also obtained with short truncated proteins not sharing any specific targeting sequence and whose sizes were below the nuclear pore exclusion size (∼60 kD; Figure 4C). An exclusive cytoplasmic location (Figure 4D) suggested the presence of a NES activity. The widely accepted NES sequence L-x(2,3)-[LIVFM]-x(2,3)-L-x-[LI] (Bogerd et al., 1996) is present in At TPX2 and starts at amino acid position 499. When this LDIFDKLSL motif was replaced by SDIFDKHSL, the polypeptide was no longer exported from the nucleus (Figure 4D). As most of the protein export mechanisms are dependent on Crm1 exportin activity, we treated BY-2 cells stably expressing full-length GFP:At TPX2 and transiently expressing GFP:At TPX2463-610 with leptomycin B (Figure 5). After leptomycin B treatment, GFP-At TPX21-758 entirely relocalized to the nucleus (Figure 5A), and in GFP:At TPX2463-610–expressing cells, the protein, which was initially excluded, now also moved into the nucleus (Figure 5B). From all of these localization results, we can conclude that at least three domains may be involved in nuclear targeting, one in nuclear export and two in microtubule binding.
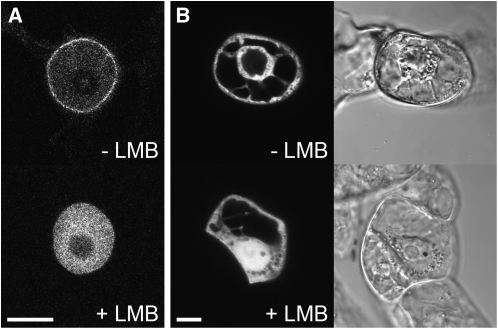
Figure 5.
Inhibition of Nuclear Export with Leptomycin B.
(A) Without leptomycin B (LMB), the GFP-At TPX2 localizes to the nucleus and the nuclear envelope of stably transformed BY-2 cells (top). With 100 nM leptomycin B, the protein accumulates in the nucleus and is completely absent from the cytoplasm (bottom). Bar = 10 μm.
(B) Transient expression of GFP:At TPX2463-610, which contains the NES sequence, in BY-2 cells observed 4 h after bombardment. Without leptomycin B, the protein is excluded from the nucleus due to the constant export against the constant inward diffusion (top). With 100 nM leptomycin B, the protein is no longer excluded from the nucleus (bottom). The cells are visualized with a CLSM (left) and DIC optics (right). Bar = 10 μm.
Arabidopsis TPX2 Induces Microtubule Assembly and Binds Importin-α in Xenopus Egg Extracts
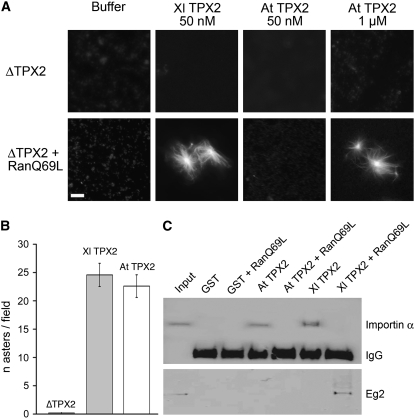
We investigated whether At TPX2 could stimulate microtubule nucleation through the Ran-dependent pathway in Xenopus egg extract (Figure 6). We depleted cytostatic factor–arrested (CSF) extract of endogenous TPX2 and reconstituted it with either 6His-At TPX2 or 6His-Xl TPX2 as a positive control. These extracts were then incubated in the presence or absence of RanQ69L-GTP (RanGTP; Gruss et al., 2001) and their ability to form microtubule asters was determined by microscopy. As previously demonstrated (Gruss et al., 2001), RanGTP did not promote microtubule aster formation in the absence of TPX2, and the addition of recombinant Xl TPX2 at endogenous levels (50 to 150 nM) restored this activity (Figure 6A). We found that, in the presence of RanGTP, 6His-At TPX2 could trigger microtubule assembly in TPX2-depleted extracts, although a higher concentration of protein was needed (1 μM; Figure 6A). We also tested the in vitro microtubule nucleation activity of At TPX2 by mixing 6His-At TPX2 with purified tubulin and GTP. Like Xl TPX2, 6His-At TPX2 promoted microtubule polymerization in vitro. In addition, the number of asters formed indicates a similar activity for both Xenopus and plant TPX2 proteins (Figure 6B).
Figure 6.
Arabidopsis TPX2 Rescues Ran-Dependent Aster Formation in TPX2-Depleted Xenopus Egg Extract.
(A) TPX2-depleted Xenopus egg extract was incubated with bacterially expressed Xl TPX2 or At TPX2 in the absence or presence of RanQ69L-GTP. Microtubule aster formation was analyzed by fluorescence microscopy. Bar = 5 μm.
(B) At TPX2 induces microtubule nucleation in vitro. Microtubule asters were assembled by mixing Xl TPX2 or At TPX2 with tubulin and GTP. The control treatment was without TPX2. The graph shows the average number of asters counted per field (number of fields, 10) and is representative of two independent experiments. Error bars represent se.
(C) Protein gel blot of anti-GST immunoprecipitation experiments from egg extracts containing GST-At TPX2 or GST-Xl TPX2. Both Xl TPX2 and At TPX2 bind importin-α (top blot) in the absence of RanGTP. However, only Xl TPX2 is able to bind Eg2 (bottom blot) in the presence of RanQ69L-GTP.
To investigate whether At TPX2 protein binds to importin α, as does the Xenopus protein (Gruss et al., 2001), we added purified, bacterially produced glutathione S-transferase (GST)–At TPX2 to a CSF Xenopus egg extract in the presence or absence of RanGTP and recovered it by immunoprecipitation with anti-GST antibodies. We found that importin-α coprecipitated with GST-At TPX2 and that this interaction was abolished when the extract was supplemented with RanGTP (Figure 6C). This suggests that At TPX2 has a similar importin binding regulation to its Xenopus counterpart. However, in contrast with Xl TPX2, GST-At TPX2 did not pull down Xenopus Aurora A (Eg2), whether RanGTP was present or not (Figure 6C). These results indicate that At TPX2 has conserved functions and can efficiently trigger microtubule assembly induced by RanGTP in the M-phase cytoplasm.
Microinjection of Anti-TPX2 Antibodies Blocks NEB and Prospindle Formation in Plant Cells
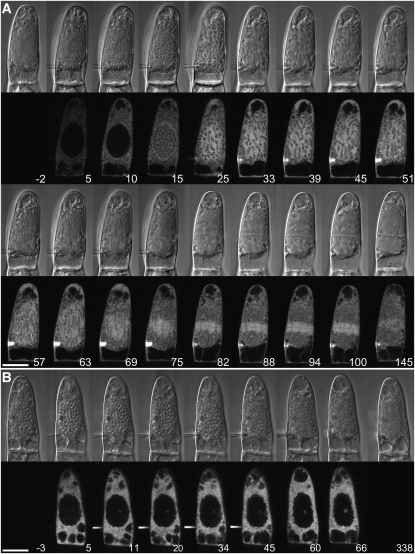
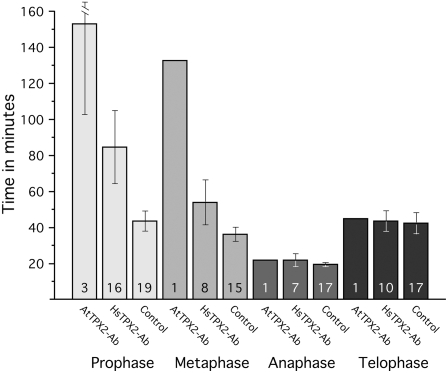
To decipher the function of At TPX2 in plant cells, both anti-human and anti-Arabidopsis TPX2 antibodies were injected into living stamen hair cells of T. virginiana plants. Microinjection of these antibodies mixed with 160-kD tetramethyl rhodamine isothiocyanate (TRITC)-dextran altered neither cell morphology nor cytoplasmic streaming in interphase cells. Cells that were injected with goat anti-mouse or goat anti-rabbit IgG (Figure 7A) or with just the TRITC-dextran (160 kD; see Supplemental Figure 6 online) progressed through cell division like noninjected stamen hair cells (Vos et al., 1999, 2000). On the other hand, microinjection of anti-TPX2 antibodies during prophase caused cell cycle arrest and increased prophase duration of cells that did not arrest (Figure 7B). Also, most of these arrested cells failed to form the typical brightly labeled halo around the nucleus, which coincides with the prospindle (see control cells in Figure 7A and Supplemental Figure 6 online at 10 to 15 min and at 35 to 45 min after injection, respectively). Two of eight cells that were injected during late prophase and still went through NEB were arrested in prometaphase, as was the cell that was injected with anti-At TPX2 antibody immediately after NEB (hence the increased metaphase transition times). Cells that were injected during metaphase, anaphase, or telophase were not affected (Figure 8). The prophase transition time for anti-Hs TPX2 injections during prophase was 85 ± 20 min (mean ± se) and was statistically different from the transition time for control injections (44 ± 5 min; two-sample independent t test, P = 0.04). The metaphase, anaphase, and telophase transition times for the anti-Hs TPX2 injections were not statistically different from those of control injections (P = 0.10, 0.32, and 0.95, respectively). Thus, plant TPX2 is essential for the assembly of the mitotic spindle during late prophase and early prometaphase.
Figure 7.
Microinjection of Antibodies to TPX2 Inhibits the Onset of Mitosis.
(A) Control injection. TRITC-goat anti-rabbit IgG was injected in a T. virginiana stamen hair cell during late prophase (as indicated by the condensed chromatin). The cell is visualized using DIC optics (top) and a CLSM (bottom). The cell progresses through mitosis and cytokinesis normally. Note the elevation of fluorescence around the nucleus just before NEB (10 to 15 min after injection). The needle concentration was 0.8 mg/mL. Time is in minutes after injection. Bar = 20 μm.
(B) Microinjection of antibodies to TPX2 in T. virginiana stamen hair cells inhibits NEB. No bright halo is formed, and after several hours the chromatin decondenses and the cell returns to interphase. In this example, the cell was injected with 0.2 mg/mL anti-Hs TPX2 antibody and 0.2 mg/mL TRITC-dextran (160 kD) as a control for cytoplasmic injection. The cytoplasmic concentration of IgG was ∼48 nM (assuming that 33% of the cell volume is cytoplasm). Time is in minutes after injection. Bar = 20 μm.
Figure 8.
Mean Mitosis Transition Times for Microinjections with Antibodies and Dextran.
Microinjection of antibodies to Hs TPX2 and At TPX2 caused prophase arrest or delay and metaphase arrest or delay when injected immediately after NEB (1× for anti-At TPX2 and 2× for anti-Hs TPX2). Injections later during metaphase had no effect, nor did injections during anaphase or telophase. The length of prophase in anti-Hs TPX2 antibody–injected cells was significantly longer (P = 0.4) than in control injections. In total, 5 cells were injected with anti-At TPX2 antibody, 32 cells were injected with anti-Hs TPX2 antibody, and 39 cells were injected with control solutions. The numbers in the bars refer to the number of data points for that particular transition time. Error bars represent se.
DISCUSSION
The data presented here describe the structural and functional characterization of At TPX2 and suggest that At TPX2 is the ortholog of vertebrate TPX2. The behavior of the plant TPX2 protein mostly matches that of its animal counterpart, particularly during mitosis, yet it also shows interesting plant-specific features during the onset of mitosis. With the identification of At TPX2, we characterized a more specific TPX2 signature motif than the PFAM Xklp2 binding sequence. Using the TPX2 signature in the search for homologs, we did not identify any fungal protein sequence, suggesting at least a high divergence in this phylum. Interestingly, the N-terminal domain of TPX2, which does not possess the signature, seems to be conserved in Caenorhabditis elegans (Özlü et al., 2005). This Ce TPX2–like (TPXL) protein possesses different functional specificities. However, both TPX2 and Ce TPXL-1 belong to a family of Aurora A–regulating proteins (Karsenti, 2005).
The MAP WVD2 protein described by Perrin et al. (2007) seems distantly related to TPX2 but does not carry the TPX2 signature motif. WVD2-like genes have a conserved C-terminal KLEEK domain that shares homology with the C-terminal domain of vertebrate TPX2, but they have no further similarity to TPX2. The protein localizes to the cortical microtubules during interphase and possibly to all other microtubule arrays of the plant (preprophase band, spindle, and phragmoplast). Overexpression caused the cortical microtubules to be randomly organized and resulted in right-handed root twisting (leftward slanting), a reduction of cell elongation, and reduced numbers of trichome branches (Yuen et al., 2003; Perrin et al., 2007). Knockdown of the gene caused an opposite root-twisting phenotype (Yuen et al., 2003). The presence in the WVD2/WDL protein family of a domain distantly related to the C-terminal TPX2 sequence reinforces the idea that proteins are built of functional modules (Karsenti, 2005): in both proteins, the C-terminal region binds or bundles microtubules.
In all interphase plant cells, TPX2 was primarily concentrated in the nucleus, but a moderate elevation was visible at the nuclear envelope (Figure 3). This distribution coincides with AURORA2 distribution (Kawabe et al., 2005) and could suggest a role for TPX2 on or just outside the nuclear envelope, maybe in nucleating microtubules that are involved in nuclear positioning. Our in vitro experiments established that purified At TPX2 is indeed able to induce microtubule polymerization (Figure 6B). Whether the protein is localized to the outside of or on the envelope, in addition to the nucleoplasmic labeling during interphase, remains an interesting question that can only be answered at the electron microscopic level.
Starting at ∼15 min before NEB in BY-2 cells and at 5 min before NEB in Arabidopsis root cells, TPX2 was exported from the nucleus and clearly localized to the crescent-shaped prospindle (Figure 3). TPX2 activity outside the nucleus, just like in vitro (Figure 6B), is likely to cause microtubule polymerization, which is essential for prospindle assembly. Prospindle accumulation of TPX2 coincides with the AURORA2 and γ-tubulin localizations (Liu et al., 1993; Kumagai et al., 2003; Kawabe et al., 2005). During prometaphase, At TPX2 associated with the spindle microtubules. Notably, TPX2 did not label the (nonkinetochore) polar microtubules during anaphase. The fluorescence decrease during telophase (see Supplemental Figures 4 and 5 online) suggests that At TPX2 is degraded in a cell cycle–dependent manner before the exit from mitosis, similar to Hs TPX2 and many APC/C substrates, such as cyclin B and Cdc20 (Stewart and Fang, 2005; Francis, 2007). The inability to be degraded, as described for human GFP:TPX2 (Stewart and Fang, 2005), was not observed in our GFP:At TPX2–expressing cell lines and plants.
Microinjection of anti-TPX2 antibodies into living plant cells showed that TPX2 indeed probably functions during late prophase and at the onset of mitosis in plants. The antibodies to human and Arabidopsis TPX2 interacted with plant TPX2 (Figure 3), and when injected they arrested or delayed the cell cycle in late prophase or prometaphase (Figures 7 and 8). The antibodies presumably bind the exported endogenous TPX2 and thereby prevent the protein from getting involved in prospindle assembly. This was supported by the absence of a bright halo around the nucleus, a common feature of late prophase cells injected with fluorescently labeled control IgGs and/or dextrans that is due to the elevated accessible volume in the prospindle (Figure 7; see Supplemental Figure 6 online). The antibodies to Hs TPX2 were shown earlier to cause aberrant mitotic spindles and chromosome missegregation when injected into human cells or added to Xenopus egg extracts (Gruss et al., 2002).
Unlike human and Xenopus TPX2, which do not possess a NES, plant TPX2 seems to play a role in spindle formation already before NEB. This is only possible if TPX2 is actively exported from the nucleus. NLS activity may control plant TPX2 nuclear import during interphase, and NES activity may be up-regulated just before NEB to allow for the massive export of TPX2 and the consequent microtubule nucleation and polymerization to form the prospindle. Indeed, evidence for a functional NES and cell cycle–regulated TPX2 export came from the subcellular localization of various At TPX2 truncations and treatments with leptomycin B. Truncations without an NLS but with the NES were excluded from the nucleus and were either cytoplasmic or microtubule-associated (Figure 4). When cells that express a fusion protein containing the NES domain (GFP-TPX2463-610) were treated with leptomycin B, the protein became evenly distributed between cytoplasm and nucleoplasm. Furthermore, leptomycin B treatment of interphase cells that expressed full-length GFP:At TPX2 caused the protein to further accumulate in the nucleus and abolished its elevated concentration at the nuclear envelope (Figure 5). The timing of TPX2 export coincided not only with the formation of the prospindle but, at least in tobacco BY-2 cells, also with the start of preprophase band breakdown (Vos et al., 2004). Interestingly, experimentally induced activation of KCBP, a calcium/calmodulin-regulated minus end–directed kinesin-14, results in premature NEB and metaphase arrest, but only during late prophase (Vos et al., 2000). Obviously, this period is critical for spindle formation.
Overexpression of TPX2 in plant cells led to an enrichment of prometaphase cells and cell death. Only moderately expressing lines with a 16–amino acid linker between the GFP and the TPX2 protein were viable (Figure 3). As in HeLa cells (Gruss et al., 2002), overexpression of TPX2 in plant cells probably causes spindle abnormalities. Overexpression of TPX2 in animal cells also allows the protein to interact with interphase microtubules. Similarly, in plants, overexpression induced by a strong constitutive promoter may overload cells with At TPX2 (see Supplemental Table 2 online). Under such conditions, if the At TPX2 nuclear import is rate-limited or restricted to a precise period of interphase, this overexpression may lead to the accumulation of the protein in the cytoplasm and cause (cortical) microtubule labeling. Our preliminary results show that TPX2 knockout in Arabidopsis is lethal; only plants that were heterozygous for the T-DNA insert could be obtained..
Xenopus TPX2 interacts with the mitotic kinase Aurora A and targets the kinesin Xklp2 to the microtubule minus ends (Brunet et al., 2004). The lack of interaction between At TPX2 and Xenopus Aurora A (Eg2; Figure 6C) is not surprising in light of the tight binding specificities of this interaction. Indeed, the N-terminal fragment of human TPX2 involved in the direct interaction with human Aurora A does not pull down Eg2 from a Xenopus egg extract. In addition, point mutations in the sequences involved in the interaction either in the kinase or in TPX2 impair complex formation (Bayliss et al., 2004). Although plant Aurora kinases diverge from the animal Aurora kinases (Kawabe et al., 2005) and At TPX2 does not pull down Xenopus Aurora A, the crystallographic model of human Aurora A/TPX2 (Bayliss et al., 2003) overlaps with the reconstructed model of At AURORA2/At TPX2 (Figure 2). Furthermore, At TPX2 can, at least partially, rescue microtubule assembly in depleted Xenopus extracts (Figure 6A). Therefore, we expect that At TPX2 interacts with At AURORA2 and possibly with At AURORA1.
Through evolution, various fundamental functions of At TPX2 have been conserved, like its ability through similar pathways to nucleate and bind microtubules, as described for vertebrate TPX2 (Brunet et al., 2004). Nevertheless, some unique features occur in the plant kingdom, perhaps linked to the absence of centrosomes, which play major roles during the G2/M transition, at the moment when the mitotic spindle is formed outside the still intact nucleus. The rapid export of TPX2 during late G2 seems essential for plant prospindle assembly around the nucleus, which replaces the role of the centrosomes in prospindle assembly in animal cells. After NEB, TPX2-dependent spindle fiber assembly may be necessary for kinetochore fiber stabilization and mitotic activity, as is also observed for its animal counterpart.
METHODS
Plant Materials, Bacteria, and Yeast
Tobacco (Nicotiana tabacum) BY-2 cells were grown according to Nagata et al. (1992). Nicotiana benthamiana and Tradescantia virginiana plants were grown in climate chambers under 16 h of light at 23°C and 6 h of darkness at 18°C. Growing stamen hairs were excised from young flower buds of ∼6 mm. Arabidopsis thaliana (ecotype Columbia) seeds were grown on MS0255 medium (Duchefa) supplemented with 0.5% agar under 14 h of light at 20.5°C and 10 h darkness at 17°C. Escherichia coli was grown under agitation in Luria-Bertani medium (Euromedex) at 37°C, and Agrobacterium tumefaciens was grown in Luria-Bertani medium supplemented with 10 mM MgSO4 at 28°C. The Pichia pastoris strain was grown as described by the supplier (Invitrogen).
Similarity Search and Alignments
The BLAST tool of the National Center for Biotechnology Information was used to determine potential TPX2 homologs in the Arabidopsis genome. ClustalW was used for identity/similarity evaluation. T-Coffee version 6.07, at the Website http://tcoffee.vital-it.ch, was used for alignment comparisons. Prosite pattern screening was performed on Myhits (myhits.isb-sib.ch), and hydrophobic cluster analysis (Eudes et al., 2007) was performed with Ressource Parisienne en Bioinformatique Structurale resources (bioserv.rpbs.jussieu.fr/RPBS/html/fr/T0_Home.html).
Bioinformatics
Three-dimensional structures of Aurora and TPX2 were built using Modeler (Sali and Blundell, 1993) with human Aurora A as a template (Protein Data Bank model entry 1OL5; Bayliss et al., 2003). Analysis was performed and illustrations were made with PyMOL (DeLano, 2002), and computation of accessible surfaces was done with the MSMS program version 2.5.7 (M.F. Sanner, unpublished data).
At TPX2 Cloning and Recombinant Protein Purification
At TPX2 cDNA was amplified by PCR from an Arabidopsis seed cDNA library (a kind gift from The Arabidopsis Information Resource) with the following primers: At TPX2 Fwd (5′-ggatccATGGAAGCAACGGCGGA-3′) and At TPX2 Rev (5′-gcggccgcCTATCTCATCTGACCAGCAGAGGCGC-3′). The lowercase letters correspond to BamHI and NotI restriction sites, respectively. Amplified DNA was inserted after digestion with BamHI and NotI either into pGEX6P-1 (Amersham Biosciences) or pHAT2 vector to generate GST and 6His fusion proteins, respectively.
GST- and 6His-At TPX2 were expressed in Escherichia coli BL21 (DE3) (Stratagene) and purified on glutathione–Sepharose 4B 5 (Amersham Bioscience) or TALON metal affinity resin (BD Biosciences Clontech), respectively, according to the manufacturers' instructions. The purified proteins were dialyzed in 20 mM HEPES, pH 7.7, 150 mM KCl, and 1 mM DTT. The purity and concentration of the purified proteins were determined on Coomassie Brilliant Blue–stained SDS-PAGE gels and Bradford (Bio-Rad) protein assay following the manufacturer's protocol.
GST-tagged Xenopus laevis TPX2 was prepared as described by Wittmann et al. (2000). The expression, purification, and loading with GTP of RanQ69L was as described by Brunet et al. (2004).
Construction of Recombinant Plasmids
A full-length At TPX2 coding sequence fragment was generated by PCR with 5′-acgcgtATGGAAGCAACGGCGGAGGAA-3′ and 5′-gcggccgcCTATTATCTCATCTGACCAGCAGA-3′ primers and subcloned using MluI and NotI restriction sites downstream of the eGFP gene and under the control of the cauliflower mosaic virus 35S promoter (P35S) into the plant expression vector pNEG-X1 (see Supplemental Figure 7 online). Amplified plasmid was used directly in bombardment experiments. For stable transformations, the plant expression cassette was excised from pNEG-X1 with HindIII and EcoRI and subcloned into pGreenII 0029 Ti vector (Hellens et al., 2000). A. tumefaciens GV3101-pSoup was then transformed with recombinant pGreenII and selected with kanamycin. Alternatively, At TPX2 was amplified with 5′-CACCATGGAAGCAACGGCGGAG-3′ and 5′-CTATCTCATCTGACCAGCAGAGGC-3′ primers and cloned into pENTR/SD/D-TOPO. From there the gene was placed into the P35S:5′-eGFP–containing plasmid pK7WGF2 (Plant System Biology, Vlaams Instituut voor Biotechnologie-Ghent University, Belgium) using Gateway recombination technology (Invitrogen). All primers used for modified TPX2 constructs are listed in Supplemental Table 3 online.
Transient and Stable Cell Transformations
For biolistic transfection, BY-2 cells were bombarded as described by Hunold et al. (1995) with modifications (Haas et al., 2005) and observed 4 h after bombardment.
Two-week-old N. benthamiana leaves were used for agroinfiltration as described previously (van der Hoorn et al., 2000; Goodin et al., 2002). Equal volumes of a P35S:p19 strain were added as a silencing suppressor (Voinnet et al., 2003). Leaves were examined by confocal laser scanning microscopy between 40 and 90 h after infiltration.
Stably expressing GFP:At TPX2 tobacco BY-2 cell lines were obtained according to the procedure of Geelen and Inzé (2001) using A. tumefaciens LBA440 carrying the P35S:GFP:At TPX2 Gateway plasmid. Arabidopsis ecotype Columbia plants with young flower buds were transformed with A. tumefaciens carrying the P35S:GFP:At TPX2 pGreenII plasmid by the floral dip technique as described by Bechtold et al. (1993). Seeds of these plants were sterilized and germinated onto Murashige and Skoog medium with kanamycin for selection. Plants showing GFP expression were transferred to the greenhouse for further growth and seed production by self-crossing. GFP-At TPX2/H2AX-mRFP plants were obtained by crossing kanamycin-resistant GFP-At TPX2 homozygote Arabidopsis with hygromycin-resistant H2AX-mRFP homozygote plants (generous gift of M.E. Chabouté, Institut de Biologie Moléculaire des Plantes). Double transformants were selected on medium containing both antibiotics.
At TPX2 transgenic BY-2 cells were incubated for 4 h in culture medium containing 100 nM leptomycin B (Sigma-Aldrich), 2 μM oryzalin (Sigma-Aldrich), 10 μM colchicin (Merck), or 10 μM latrunculin B (Calbiochem).
Immunoblotting
Total protein extracts were prepared from Arabidopsis seedlings, tobacco BY-2 cells, and young T. virginiana flower buds. Tissue was ground in liquid nitrogen and added to boiling standard SDS-PAGE loading buffer (Laemmli, 1970). The supernatant after centrifugation was put on SDS-PAGE gels. Protein extracts were also prepared from purified nuclei of N. benthamiana leaves that were agroinfiltrated with a 35S:E-tag:GFP:At TPX2 construct. Leaves were collected 2 d after agroinfiltration, and nuclei were prepared according to Carrington et al. (1991). They were then dissolved in SDS-PAGE loading buffer for protein extraction (see above). All samples were blotted after SDS-PAGE using anti-human TPX2, anti-Arabidopsis TPX2, anti-GFP, or anti-E-tag antibodies as described elsewhere (Evrard et al., 2002). The rabbit polyclonal anti-Arabidopsis TPX2 antibody was raised against the conserved TPX2 signature motif. For this purpose, a PCR fragment corresponding to At TPX2 amino acids 303 to 463 was generated using the primers 5′-AGCACGCGAGACCTATTCGTC-3′ and 5′-TATCTTTTTGTTCAAAGGTTT-3′. This fragment was subcloned into pBAD/TOPO ThioFusion expression vector (Invitrogen). Expression and purification were done as indicated by the kit supplier, followed by immunization as described previously (Evrard et al., 2002). For P. pastoris expression, the Invitrogen EasySelect expression kit was used as described by the supplier. The His-tagged-At TPX2 coding frame was amplified using 5′-gaattcAAAAATGTCTCATCATCATCATCATCATCTCGAGGCAACGGCGGAGGAATCAGTTAG-3′ and 5′-gcggccgcTATTATCTCATCTGACCAGCAGAGGC-3′ primers and properly subcloned with EcoRI and NotI enzymes into pPICZ-A vector. After 7 h of methanol induction, cells were directly lysed into SDS-PAGE loading buffer.
Immunolocalization
BY-2 cells were fixed and immunostained as described previously (Erhardt et al., 2002). Cells were incubated overnight with anti-human TPX2 or anti-Arabidopsis TPX2 and 1:6000 diluted anti-α tubulin antibodies (Sigma-Aldrich) at 4°C and then incubated with 1:300 diluted anti-rabbit or anti-mouse IgG secondary antibodies conjugated with Alexa Fluor 488 or 568, respectively (Molecular Probes), for 1 h at room temperature. DNA was stained using l μg/mL 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI).
Fluorescence Microscopy
Transformed tobacco leaves and cells were observed on a Zeiss LSM510 confocal laser scanning microscope (CLSM) equipped with argon and helium/neon lasers, standard filters, and a 63×, 1.2 numerical aperture (NA) C-Apochromat water-immersion lens or on a Zeiss Axiovert 200M microscope with an LSM5 Pascal CLSM system equipped with argon and helium/neon lasers, standard filters, and a 63×, 1.4 NA differential interference contrast (DIC) lens. Immunolabelings and Arabidopsis roots were observed on a Nikon TE2000 inverted microscope equipped with a 60× 1.45 NA total internal reflection fluorescence objective and a Photometrics CoolSnap digital CCD camera that was controlled by Metamorph version 6.2. Transmitted light reference images were captured using DIC optics. Images were processed with LSM510 version 2.8 (Zeiss), OsiriX PACS DICOM 3D workstation (http://homepage.mac.com/rossetantoine/osirix/), LSM Image Examiner version 3.5, AxioVision LE version 4.5, ImageJ 1.37v, and Adobe Photoshop CS version 8.0.
Xenopus Egg Extract Experiments and in Vitro Microtubule Nucleation Assay
CSF extracts were prepared as described previously (Murray, 1991). TPX2 immunodepletion was performed according to Wittmann et al. (2000). To study the functionality of At TPX2 protein, the indicated amounts of His-tagged At TPX2 or Xl TPX2 were added to 20 μL of TPX2-depleted CSF extract supplemented with 0.2 mg/mL rhodamine-labeled tubulin in the presence or absence of 20 μM RanQ69L-GTP. The mixture was incubated at 20°C for 20 to 30 min. Aliquots of 1 μL were fixed and squashed onto cover slips for examination by fluorescence microscopy. Pictures were taken at 40× magnification with identical illumination settings and processed identically with Adobe Photoshop.
TPX2 aster formation assays in vitro were done according to Brunet et al. (2004). In brief, 0.5 μM His-Xl TPX2 or His-At TPX2 was mixed with 10 μM tubulin containing 5% rhodamine-labeled tubulin and 1 mM GTP in BRB80 and incubated for 20 min at 25°C, as described previously (Brunet et al., 2004). Samples were fixed and squashed onto slides and observed as above. Microtubule asters from at least 10 randomly selected fields were counted.
Immunoprecipitation experiments were performed as described by Brunet et al. (2004). In brief, protein A–conjugated Dynabeads 280 (Dynal Biotech) were coated with anti-GST (6 μg/20 μL beads) in PBS with 0.1% Triton X-100 for at least 1 h at 4°C on a rotating wheel and washed thoroughly. GST-tagged Xl TPX2 or At TPX2 were incubated in CSF-egg extract for 20 min at 20°C before addition of the antibody-coated beads. After incubating the mixtures on ice for 30 min, beads were retrieved on a magnet and washed. Immunoprecipitated proteins were eluted from beads with SDS-PAGE sample buffer and then analyzed by protein gel blotting with the following primary antibodies: anti-GST (1:1000 dilution) and anti-importin-α (1:1000 dilution; a gift from I. Mattaj, European Molecular Biology Laboratory) antibodies and the monoclonal anti-Eg2 antibody 1C1 (a gift from C. Prigent, Centre National de la Recherche Scientifique, Université de Rennes 1). The secondary antibodies were goat anti-rabbit Alexa Fluor 680 (1:10,000 dilution; Invitrogen) and donkey anti-mouse IRDye 800 (1:10,000 dilution; Rockland). The blots were analyzed with the Odyssey infrared imaging system (Li-Cor).
Microinjection
To assess the role of TPX2, the protein function was inhibited by the microinjection of antibodies to human and Arabidopsis TPX2 (0.1 to 1.5 mg/mL in 5 mM HEPES, pH 7.0, 100 mM KCl, and 0.2 mg/mL 4.4-kD fluorescein isothiocyanate–dextran or 160 kD TRITC-dextran) into young T. virginiana stamen hair cells. As a control, cells were injected with 4.4-kD fluorescein isothiocyanate–dextran (0.2 mg/mL) or 160-kD TRITC-dextran (0.2 mg/mL) or with either dextran plus fluorescently labeled goat anti-rabbit or goat anti-mouse IgG conjugated to Alexa Fluor 448, 546, or 568 (0.1 to 0.7 mg/mL; Molecular Probes). Microinjection was performed with backfilled needles as described previously (Vos et al., 1999) on a Narishige micromanipulation system mounted on the Zeiss Axiovert 200M microscope mentioned above. Typically, microinjection gives an ∼1 to 2% volume increase in a 10- to 20-pL stamen hair cell. At 0.2 mg/mL IgG (150 kD), this gives a cytoplasmic concentration of 39 to 76 nM (assuming that the cytoplasm constitutes 33% of the total volume of a prophase cell and the nucleus is ∼5 pL). Injected cells were imaged on the Zeiss LSM5 Pascal CLSM system mentioned above, and mitosis transition time data were analyzed with Origin software (OriginLab).
Accession Numbers
The Arabidopsis Genome Initiative locus identifier for the TPX2 gene analyzed in this study is AT1G03780. The accession numbers of all other genes are listed in Supplemental Table 2 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of the TPX2 Signature Motifs.
Supplemental Figure 2. Phylogram of Eukaryote TPX2 Signatures.
Supplemental Figure 3. Sequence Alignment of Arabidopsis and Human Aurora and TPX2.
Supplemental Figure 4. At TPX2 Localization during Cell Division in Immunolabeled BY-2 Cells.
Supplemental Figure 5. Dynamics of At TPX2 during Cell Division.
Supplemental Figure 6. Microinjection of TRITC-Dextran.
Supplemental Figure 7. Map and Multiple Cloning Sites of pNEG-X1
Supplemental Table 1. Combined Results of Experiments Concerning GFP:At TPX2 Expression.
Supplemental Table 2. Accession Numbers Corresponding to Various Organism Sequences in Which a TPX2 Signature Is Present.
Supplemental Table 3. List of Primers Used in Generating Various At TPX2 Constructs.
Supplemental Movie 1. GFP-At TPX2 Localization during Cell Division in an Arabidopsis Root Cell.
Supplemental Movie 2. Simultaneous GFP-At TPX2 and Histone H2AX-mRFP Localization during Cell Division in an Arabidopsis Root Cell.
Supplemental Movie 3. GFP-At TPX2 Localization during Cell Division in a Tobacco BY-2 Suspension Culture Cell.
Supplemental Data Set 1. FASTA Text File of the Alignment Used to Generate the Phylogenetic Tree in Supplemental Figure 2 online.
Supplementary Material
Acknowledgments
We thank A. Untergasser and H. Kieft for technical assistance with cloning, transformations, and cell culturing. This work was supported in part by the Centre National de la Recherche Scientifique, partenariat Hubert Curien Van Gogh and partenariat Hubert Curien Picasso collaboration grants by Egide to A.-C.S. The Inter-Institute confocal microscopy platform was cofinanced by the Centre National de la Recherche Scientifique, the Université Louis Pasteur, the Région Alsace, the Association de la Recherche sur le Cancer, and the Ligue Nationale contre le Cancer. This work was also supported in part by a VENI grant and a Van Gogh collaboration grant from the Netherlands Organization for Scientific Research to J.W.V. The work of N.J. and L.H.P. was supported by the Ministère de l'Enseignement Supérieur et de la Recherche. L.H.P. was financed by the COMBIO European Commission (STREP Program Contract STREP 503568). Work in the Vernos laboratory was supported by Spanish grants (Grants HF-2006-0067, BFU2006-04694, and CSD2006-00023) and by European Union Grant MRTN/CT2004 512348.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Jan W. Vos (janw.vos@wur.nl), Isabelle Vernos (isabelle.vernos@crg.es), and Anne-Catherine Schmit (anne-catherine.schmit@ibmp-ulp.u-strasbg.fr).
Online version contains web-only data.
References
- Bayliss, R., Sardon, T., Ebert, J., Lindner, D., Vernos, I., and Conti, E. (2004). Determinants for Aurora-A activation and Aurora-B discrimination by TPX2. Cell Cycle 3 404–407. [PubMed] [Google Scholar]
- Bayliss, R., Sardon, T., Vernos, I., and Conti, E. (2003). Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell 12 851–862. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis. C. R. Acad. Sci. III. Life Sci. 316 1194–1199. [Google Scholar]
- Bogerd, H.P., Fridell, R.A., Benson, R.E., Hua, J., and Cullen, B.R. (1996). Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 16 4207–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittle, A.L., and Ohkura, H. (2005). Centrosome maturation: Aurora lights the way to the poles. Curr. Biol. 15 R880–R882. [DOI] [PubMed] [Google Scholar]
- Brunet, S., Sardon, T., Zimmerman, T., Wittmann, T., Pepperkok, R., Karsenti, E., and Vernos, I. (2004). Characterization of the TPX2 domains involved in microtubule nucleation and spindle assembly in Xenopus egg extracts. Mol. Biol. Cell 15 5318–5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaday, J., Stoppin-Mellet, V., Mutterer, J., Lambert, A.M., and Schmit, A.C. (2000). Higher plant cells: Gamma-tubulin and microtubule nucleation in the absence of centrosomes. Microsc. Res. Tech. 49 487–495. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas, R.E., Gruss, O.J., Mattaj, I.W., and Karsenti, E. (2001). Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat. Cell Biol. 3 228–234. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas, R.E., and Karsenti, E. (2003). Long-range communication between chromatin and microtubules in Xenopus egg extracts. Curr. Biol. 13 1728–1733. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., Freed, D.D., and Leinicke, A.J. (1991). Bipartite signal sequence mediates nuclear translocation of the plant potyviral Nla protein. Plant Cell 3 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron, M., Bunt, G., Bastiaens, P., and Karsenti, E. (2005). Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science 309 1373–1376. [DOI] [PubMed] [Google Scholar]
- Clarke, P.R., and Zhang, C. (2008). Spatial and temporal coordination of mitosis by Ran GTPase. Nat. Rev. Mol. Cell Biol. 9 464–477. [DOI] [PubMed] [Google Scholar]
- DeLano, W.L. (2002). Unraveling hot spots in binding interfaces: Progress and challenges. Curr. Opin. Struct. Biol. 12 14–20. [DOI] [PubMed] [Google Scholar]
- Demidov, D., Van Damme, D., Geelen, D., Blattner, F.R., and Houben, A. (2005). Identification and dynamics of two classes of Aurora-like kinases in Arabidopsis and other plants. Plant Cell 17 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducat, D., and Zheng, Y. (2004). Aurora kinases in spindle assembly and chromosome segregation. Exp. Cell Res. 301 60–67. [DOI] [PubMed] [Google Scholar]
- Eckerdt, F., Eyers, P.A., Lewellyn, A.L., Prigent, C., and Maller, J.L. (2008). Spindle pole regulation by a discrete Eg5-interacting domain in TPX2. Curr. Biol. 18 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt, M., Stoppin-Mellet, V., Campagne, S., Canaday, J., Mutterer, J., Fabian, T., Sauter, M., Muller, T., Peter, C., Lambert, A.M., and Schmit, A.C. (2002). The plant Spc98p homologue colocalizes with gamma-tubulin at microtubule nucleation sites and is required for microtubule nucleation. J. Cell Sci. 115 2423–2431. [DOI] [PubMed] [Google Scholar]
- Eudes, R., Le Tuan, K., Delettre, J., Mornon, J.P., and Callebaut, I. (2007). A generalized analysis of hydrophobic and loop clusters within globular protein sequences. BMC Struct. Biol. 7 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard, J.L., Nguyen, I., Bergdoll, M., Mutterer, M., Steinmetz, A., and Lambert, A.M. (2002). A novel pollen-specific α-tubulin in sunflower: Structure and characterization. Plant Mol. Biol. 49 611–620. [DOI] [PubMed] [Google Scholar]
- Francis, D. (2007). The plant cell cycle—15 years on. New Phytol. 174 261–278. [DOI] [PubMed] [Google Scholar]
- Geelen, D.N., and Inzé, D.G. (2001). A bright future for the Bright Yellow-2 cell culture. Plant Physiol. 127 1375–1379. [PMC free article] [PubMed] [Google Scholar]
- Goodin, M.M., Dietzgen, R.G., Schichnes, D., Ruzin, S., and Jackson, A.O. (2002). pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31 375–383. [DOI] [PubMed] [Google Scholar]
- Gruss, O.J., Carazo-Salas, R.E., Schatz, C.A., Guarguaglini, G., Kast, J., Wilm, M., Le Bot, N., Vernos, I., Karsenti, E., and Mattaj, I.W. (2001). Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104 83–93. [DOI] [PubMed] [Google Scholar]
- Gruss, O.J., and Vernos, I. (2004). The mechanism of spindle assembly: Functions of Ran and its target TPX2. J. Cell Biol. 166 949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss, O.J., Wittmann, M., Yokoyama, H., Pepperkok, R., Kufer, T., Sillje, H., Karsenti, E., Mattaj, I.W., and Vernos, I. (2002). Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 4 871–879. [DOI] [PubMed] [Google Scholar]
- Haas, M., Geldreich, A., Bureau, M., Dupuis, L., Leh, V., Vetter, G., Kobayashi, K., Hohn, T., Ryabova, L., Yot, P., and Keller, M. (2005). The open reading frame VI product of Cauliflower mosaic virus is a nucleoplasmic protein: Its N-terminus mediates its nuclear export and formation of electron-dense viroplasm. Plant Cell 17 927–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42 819–832. [DOI] [PubMed] [Google Scholar]
- Hetzer, M., Gruss, O.J., and Mattaj, I.W. (2002). The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat. Cell Biol. 4 E177–E184. [DOI] [PubMed] [Google Scholar]
- Hunold, R., Burrus, M., Bronner, R., Duret, J.P., and Hahne, G. (1995). Transient gene expression in sunflower (Helianthus annuus L.) following microprojectile bombardment. Plant J. 105 95–109. [Google Scholar]
- Hyman, A.A., and Karsenti, E. (1996). Morphogenetic properties of microtubules and mitotic spindle assembly. Cell 84 401–410. [DOI] [PubMed] [Google Scholar]
- Jeong, S.Y., Rose, A., Joseph, J., Dasso, M., and Meier, I. (2005). Plant-specific mitotic targeting of RanGAP requires a functional WPP domain. Plant J. 42 270–282. [DOI] [PubMed] [Google Scholar]
- Karsenti, E. (2005). TPX or not TPX? Mol. Cell 19 431–432. [DOI] [PubMed] [Google Scholar]
- Karsenti, E., and Vernos, I. (2001). The mitotic spindle: A self-made machine. Science 294 543–547. [DOI] [PubMed] [Google Scholar]
- Kawabe, A., Matsunaga, S., Nakagawa, K., Kurihara, D., Yoneda, A., Hasezawa, S., Uchiyama, S., and Fukui, K. (2005). Characterization of plant Aurora kinases during mitosis. Plant Mol. Biol. 58 1–13. [DOI] [PubMed] [Google Scholar]
- Kufer, T.A., Sillje, H.H., Korner, R., Gruss, O.J., Meraldi, P., and Nigg, E.A. (2002). Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 158 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai, K., Uchida, K., Miyamoto, T., Ushigusa, T., Shinohara, S., Yamaguchi, R., and Tateyama, S. (2003). Three cases of canine gastrointestinal stromal tumors with multiple differentiations and c-kit-expression. J. Vet. Med. Sci. 65 1119–1122. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Liu, B., Marc, J., Joshi, H.C., and Palevitz, B.A. (1993). A gamma-tubulin-related protein associated with the microtubule arrays of higher plants in a cell cycle-dependent manner. J. Cell Sci. 104 1217–1228. [DOI] [PubMed] [Google Scholar]
- Lloyd, C., and Chan, J. (2006). Not so divided: The common basis of plant and animal cell division. Nat. Rev. Mol. Cell Biol. 7 147–152. [DOI] [PubMed] [Google Scholar]
- Meier, I. (2007). Composition of the plant nuclear envelope: Theme and variations. J. Exp. Bot. 58 27–34. [DOI] [PubMed] [Google Scholar]
- Murray, A.W. (1991). Cell cycle extracts. Methods Cell Biol. 36 581–605. [PubMed] [Google Scholar]
- Nagata, T., Nemoto, Y., and Hasezawa, S. (1992). Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. Int. Rev. Cytol. 132 1–30. [Google Scholar]
- O'Brien, L.L., and Wiese, C. (2006). TPX2 is required for post-mitotic nuclear assembly in cell-free Xenopus laevis egg extract. J. Cell Biol. 173 685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özlü, N., Srayko, M., Kinoshita, K., Habermann, B., O'Toole, E.T., Muller-Reichert, T., Schmalz, N., Desai, A., and Hyman, A.A. (2005). An essential function of the C. elegans ortholog of TPX2 is to localize activated Aurora A kinase to mitotic spindles. Dev. Cell 9 237–248. [DOI] [PubMed] [Google Scholar]
- Perrin, R.M., Wang, Y., Yuen, C.Y., Will, J., and Masson, P.H. (2007). WVD2 is a novel microtubule-associated protein in Arabidopsis thaliana. Plant J. 49 961–971. [DOI] [PubMed] [Google Scholar]
- Rajangam, A.S., et al. (September 19, 2008). MAP20, a microtubule-associated protein in the secondary cell walls of Populus tremula L. × tremuloides Michx is a target of the cellulose synthesis inhibitor, 2,6-dichlorobenzonitrile. Plant Physiol. http://dx.doi.org/10.1104/pp.108.121913. [DOI] [PMC free article] [PubMed]
- Sali, A., and Blundell, T.L. (1993). Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234 779–815. [DOI] [PubMed] [Google Scholar]
- Schatz, C.A., Santarella, R., Hoenger, A., Karsenti, E., Mattaj, I.W., Gruss, O.J., and Carazo-Salas, R.E. (2003). Importin alpha-regulated nucleation of microtubules by TPX2. EMBO J. 22 2060–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmit, A.C., Vantard, M., and Lambert, A.M. (1985). Microtubule and F-actin rearrangement during the initiation of mitosis in acentriolar higher plant cells. Cell Motility: Mechanism and Regulation, H. Ishikawa, S. Hatano, and H. Sato, eds (Tokyo: University of Tokyo Press), pp. 415–433.
- Stewart, S., and Fang, G. (2005). Anaphase-promoting complex/cyclosome controls the stability of TPX2 during mitotic exit. Mol. Cell. Biol. 25 10516–10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppin, V., Vantard, M., Schmit, A.C., and Lambert, A.M. (1994). Isolated plant nuclei nucleate microtubule assembly: The nuclear surface in higher plants has centrosome-like activity. Plant Cell 6 1099–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulu, U.S., Fagerstrom, C., Ferenz, N.P., and Wadsworth, P. (2006). Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr. Biol. 16 536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme, D., Bouget, F.Y., Van Poucke, K., Inze, D., and Geelen, D. (2004). Molecular dissection of plant cytokinesis and phragmoplast structure: A survey of GFP-tagged proteins. Plant J. 40 386–398. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn, R.A., Laurent, F., Roth, R., and De Wit, P.J. (2000). Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant Microbe Interact. 13 439–446. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33 949–956. [DOI] [PubMed] [Google Scholar]
- Vos, J.W., Dogterom, M., and Emons, A.M. (2004). Microtubules become more dynamic but not shorter during preprophase band formation: A possible “search-and-capture” mechanism for microtubule translocation. Cell Motil. Cytoskeleton 57 246–258. [DOI] [PubMed] [Google Scholar]
- Vos, J.W., Safadi, F., Reddy, A.S., and Hepler, P.K. (2000). The kinesin-like calmodulin binding protein is differentially involved in cell division. Plant Cell 12 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, J.W., Valster, A.H., and Hepler, P.K. (1999). Methods for studying cell division in higher plants. In Mitosis and Meiosis 61, C.L. Rieder, ed (San Diego, CA: Academic Press), pp. 413–437. [DOI] [PubMed]
- Walczak, C.E., Vernos, I., Mitchison, T.J., Karsenti, E., and Heald, R. (1998). A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 8 903–913. [DOI] [PubMed] [Google Scholar]
- Wittmann, T., Wilm, M., Karsenti, E., and Vernos, I. (2000). TPX2, a novel Xenopus MAP involved in spindle pole organization. J. Cell Biol. 149 1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen, C.Y., Pearlman, R.S., Silo-Suh, L., Hilson, P., Carroll, K.L., and Masson, P.H. (2003). WVD2 and WDL1 modulate helical organ growth and anisotropic cell expansion in Arabidopsis. Plant Physiol. 131 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Q., Leung, S., Corbett, A.H., and Meier, I. (2006). Identification and characterization of the Arabidopsis orthologs of nuclear transport factor 2, the nuclear import factor of ran. Plant Physiol. 140 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.