Abstract
microRNAs (miRNAs) are a class of small, non-coding, RNAs that regulate the stability or translation of mRNA transcripts. Although recent work has implicated miRNAs in development and in disease, the expression and function of miRNAs in the adult mammalian nervous system has not been extensively characterized. Here, we examine the role of two brain-specific miRNAs, miR-219 and miR-132, in modulating the circadian clock located in the suprachiasmatic nucleus. miR-219 is a target of the CLOCK/BMAL1 complex, exhibits robust circadian rhythms of expression and the in vivo knockdown of miR-219 lengthens the circadian period. miR-132 is induced by photic entrainment cues via a MAPK/CREB-dependent mechanism, modulates clock gene expression, and attenuates the entraining effects of light. Collectively, these data reveal miRNAs as clock- and light-regulated genes and provide a mechanistic examination of their roles as effectors of pacemaker activity and entrainment.
INTRODUCTION
An internal timekeeping mechanism—the ‘circadian’ clock—has arisen in nearly all organisms that allows them to adapt their physiological and behavioral process to a cyclic 24-hr world (Reppert et al., 2001). In mammals the suprachiasmatic nuclei (SCN) of the hypothalamus function as the master circadian clock (Ralph et al., 1990). As such, the SCN is able to drive endogenous rhythms, and can be entrained or reset by light and other external cues.
At the molecular level, the SCN clock is derived from interlocking positive and negative transcriptional feedback loops which drive rhythmic expression of critical clock components. In the positive limb, heterodimers of the transcription factors, CLOCK and BMAL1, induce transcription of the period (Per1, Per2, Per3) and cryptochrome (Cry1, Cry2) genes at the E-box elements within their promoters (Gekakis et al., 1998; Bunger et al., 2000). Negative feedback is achieved by the action of PER-CRY complexes, which inhibit CLOCK/BMAL1-dependent transcription (Shearman et al., 2000). An ancillary transcriptional loop consisting of the nuclear orphan receptor Rev-Erbα feedback inhibits transcription of the bmal1 gene by binding to an ROR element in the gene promoter (Preitner et al., 2002). In addition to transcription, post-translational regulation plays a critical role in clock timing processes (Lowrey et al., 2000). Importantly, key aspects of these feedback loops (period, amplitude and phase) can be modulated by changes in clock- and light-controlled genes (ccg). For example, with respect to phase, light-induced expression of the core clock genes per1 and per2 has been implicated in clock entrainment (Albrecht et al., 2001); with respect to clock gene amplitude, the clock-controlled, basic helix-loop-helix transcription factors Dec1 and Dec2, inhibit CLOCK/BMAL1-mediated transcription of the per1 gene via competitive binding for the E-box elements and direct protein-protein interactions with BMAL1 (Honma et al., 2002). In addition, clock- and light-controlled genes may indirectly affect timing through the physiological effects (eg., membrane excitability, transmitter release, etc.) of their protein products (Kraves et al., 2006; Cheng et al., 2002). Another possible route by which clock timing and entrainment could be affected is at the post-transcriptional level. Thus, microRNAs (miRNAs) represent a potentially novel avenue by which biological timing processes may be regulated.
miRNAs were first characterized in Canorhabditis elegans and implicated in cell fate specification (Lee et al., 1993; Wightman et al., 1993), but many hundreds have been identified since then across phyla. miRNAs are small, evolutionarily conserved molecules that act as potent silencers of gene expression via translational repression or mRNA degradation. miRNAs can be transcribed from discrete genes as larger precursors, and processed in two sequential steps to form the 19-24-nt mature miRNA (He et al., 2004). One strand of the mature miRNA is incorporated into a RNA-induced silencing complex (RISC), which is then targeted to mRNAs containing recognition elements for that particular miRNA (He et al., 2004). It is conceivable that transcriptional events involved in clock timing, be they rhythmic or acutely activated by entraining stimuli, would induce expression of a subset of miRNAs, which in turn would regulate the expression of proteins that underlie key aspects of the timing process.
Here we examined the potential involvement of microRNAs in the SCN clock. With a combination of interrogation techniques we have identified and functionally characterized two microRNAs (miR-219, miR-132) within the context of the SCN clock timing process.
RESULTS
Identification of miR-219-1 and miR-132 as CLOCK- and CREB-Regulated Targets
As a starting point to begin to examine miRNA expression in the SCN, we utilized data from a genome-wide screening technique to identify CREB-regulated miRNAs (Impey et al., 2004). CREB was the initial focus because photic stimulation has been shown to elicit robust CRE-dependent gene expression in the SCN (Obrietan et al., 1999). For these studies we initially examined miR-132, a CREB-regulated miRNA that is induced by neurotrophins (Vo et al., 2005). First, we validated the CREB SACO results using a combination of chromatin immunoprecipitation (ChIP) and real-time PCR (Fig. 1A). The miR-132 enhancer region was selectively immunoprecipitated by a CREB antibody but not IgG (Fig. 1A). The abundance of a bona fide CREB target, c-Fos, was analyzed as a positive control (Fig. 1A). The CREB ChIP did not enrich for a locus near the 18S ribosomal RNA repeat or other negative control regions (Fig. 1A and data not shown). Given the role of CREB as a light-inducible transcriptional factor within the SCN, our data raised the possibility that miR-132 expression might be activated by light.
Figure 1. microRNA miR-132 and miR-219-1 are differentially regulated by CREB and CLOCK.
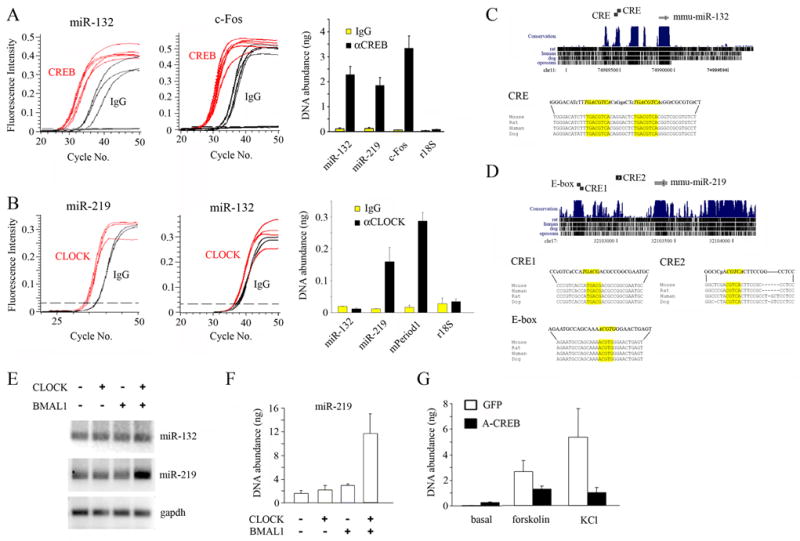
(A–B) Chromatin immunoprecipitation (ChIP) assay of PC12 cells using (A) an anti-CREB or (B) an anti-CLOCK antibody. RNA abundance was determined by real-time quantitative PCR. A nonspecific IgG was used as a negative control. In (A), the levels of immunoprecipitated (IP) DNA encompassing the promoter regions of miR-132, miR-219-1, c-Fos, and an r18S control were quantitated (right), and data for miR-132 (left) and c-Fos (middle) are presented as fluorescence intensity vs. number of PCR cycles. In (B), the levels of IP promoter DNA of miR-132, miR-219-1, mPeriod1 and an r18S control were determined (right), and data for miR-219-1 (left) and miR-132 (middle) are presented as fluorescence intensity vs. number of PCR cycles. (C) Position of murine (mmu-)miR-132 gene on chromosome 11 (top). Alignment scores are indicated by black bars in the conservation track of mouse, rat, human, dog and opossum (top). Two consensus CREs (italicized: yellow boxes) are located in the 5′ region of the miR-132 locus, and base mismatches from mouse in rat, human and dog are represented by lower case letters. Sequence alignments of mouse, rat, human and dog (bottom). (D) Position of murine (mmu-)miR-219-1 gene on chromosome 17 (top). Alignment scores are indicated in the conservation track (top). Two putative CREs (CRE1 and CRE2; italicized, yellow boxes) and an E-box motif (italicized, yellow box) are located in the 5′ region of the miR-219-1 locus (bottom). Mismatches from mouse are denoted by lower case letters. (E) Levels of pre-miR-219-1, but not of pre-miR-132 or gapdh, were elevated in PC12 cells co-transfected with expression constructs for CLOCK and BMAL1, as determined by semi-quantitative reverse transcription (RT)-PCR. (F) Quantitative analysis of CLOCK/BMAL1-dependent induction of pre-miR-219 in transfected PC12 cells as determined by real-time PCR. Data are presented as mean ± SEM nanograms of DNA. (G) Dominant-negative CREB (A-CREB) suppresses miR-132 induction. Primary cortical neurons were transfected with the expression constructs for A-CREB or GFP (negative control), were stimulated with forskolin (10 μM) or KCl (20 mM), and the abundance of pre-miR-132 transcript was determined by real-time PCR. Data are presented as mean ± SEM nanograms of cDNA.
To discover miRNAs that are regulated by the E-box-dependent core timing mechanism, we screened miRNAs using a CLOCK ChIP. Interestingly, the enhancer region of miR-219-1 was significantly enriched in the CLOCK IP fraction (Fig. 1B), whereas that of miR-132 was not (Fig. 1B). The mperiod1 gene, a bona fide target of CLOCK/BMAL1-mediated transcription, was used as the positive control, and 18S rRNA as the negative control (Fig. 1B). Notably, the miR-219-1 enhancer region was also enriched in the CREB antiserum IP fraction (Fig. 1A).
Sequence alignment of pre-miR-132 across multiple species, including mouse, human, dog, zebrafish and puffer fish indicates that the mature miR-132 sequence is highly conserved throughout evolution (Fig. 1C and not shown). With respect to the miR-219-1 gene, vertebrates possess multiple copies (Fig. 1D) and sequence alignment of pre-miR-219 from varied species shows a high degree of conservation (Fig. 1D). Zebrafish in situ hybridization has shown that miR-219 is enriched in the brain (Wienholds et al., 2005). As predicted by the ChIP assays, an E-box motif (non-canonical) is found in the promoter region of the miR-219-1 genes and two consensus CRE motifs (Fig. 1D, CRE1 and CRE2) were also identified. These response elements are found in mouse, rat, human and dog.
To determine whether the CLOCK/BMAL1 heterodimer regulates miR-219-1 expression, CLOCK and BMAL1 were expressed in PC12 cells and miR-219-1 expression was examined by semi-quantitative reverse transcription (RT)-PCR and real-time (RT) PCR approaches. Overexpression of CLOCK and BMAL1 together resulted in a significant increase in pre-miR-219-1 transcript levels as determined by semi-quantitative RT-PCR (Fig. 1E) and real-time PCR (Fig. 1F), whereas expression of CLOCK or BMAL1 singly had limited effect (Fig. 1E and 1F). Both pre-miR-132 and gapdh levels were largely unaffected by CLOCK/BMAL1 coexpression (Fig. 1E).
Next, we examined the role of CREB as a regulator of miR-132 transcript. In primary cortical neurons, both forskolin (an adenylate cyclase agonist) and KCl depolarization induced expression of pre-miR-132 as assessed by RT-PCR (Fig. 1G). Expression of A-CREB, which blocks CREB-mediated transcription, suppressed forskolin- and KCl-induced pre-miR-132 expression (Fig. 1G).
Circadian and Light-Inducible Expression of miR-219 and miR-132
We then turned to miR-132 and miR-219 expression in the murine brain. Using RT-PCR, we found that pre-miR-219-1 exhibited a robust circadian rhythm of expression in the SCN, with peak levels in the mid-subjective day (Fig. 2A). Levels of pre-miR-132 within the SCN were also rhythmic, albeit with a much dampened amplitude, and peaked in the subjective day (Fig. 2A). Rhythmic expression of pre-miR-132 and –219-1 was not observed in the cortex (Fig. 2A). Moreover, two other miRNAs, miR-16-1 and miR-128a, exhibited no significant change in expression over a circadian cycle (Supplementary Figure S1), indicating that the oscillations of miR-219 and -132 expression are unique to these miRNAs and do not merely reflect an upstream oscillation in the miRNA processing machinery. To test whether RT-PCR-based precursor quantitation reflected the abundance of mature microRNAs (21- or 22-mers), we monitored miR-219 and -132 levels using ribonucleoprotection assays (RPA). miR-219 levels in the SCN were significantly higher during the subjective day than during the subjective night, with the peak level observed at CT 6 (Fig. 2B). Likewise, miR-132 exhibited a modest, but significant rhythm, with the peak level occurring during the subjective day (Fig. 2B).
Figure 2. Circadian expression of miR-132 and miR-219 in the SCN.

(A) Abundance of pre-miR-132 and pre-miR-219-1 transcripts in pooled RNA samples of SCN and piriform cortex (CTX) tissue as a function of circadian time (CT 2, 6, 11, 15, 19, 22) was determined by semi-quantitative RT-PCR. Expression of gapdh served as the loading control. (B) Ribonuclease protection assays (RPA) for miR-219 (top) and miR-132 (bottom). Densitometric analysis of miR-219 (top) and miR-132 (bottom) is shown to the left. miRNA levels were normalized to 5S rRNA and expressed as fold expression. Mean values were determined from three independent experiments. (C) Quantitative real-time PCR analysis of pre-miR-219-1 (left) and pre-miR-132 (right) in the SCN of wild-type and mcry1/cry2 double mutant mice at CT 6 and CT 19. Data were normalized to gapdh levels and presented as fold expression.
To examine whether the miR-219 and miR-132 rhythms were driven by the SCN clock, microRNA expression levels were examined in crypotochrome (mCRY) 1/2-deficient mice (Masuki et al., 2005). As a component of the core clock timing mechanism, genetic deletion of both mCry 1 and mCry2 results in arrhythmic behavior and a loss of core clock gene expression (van der Horst et al., 1999; Okamura et al., 1999). For these experiments, wild-type and mCry1/mCry2 double mutant mice were dark-adapted for 2 days, and SCN tissue was collected during the mid subjective day (CT 6), and subjective night (CT 19). Quantitative RT PCR revealed that neither pre-miR-219-1 nor pre-miR-132 exhibited significant time-of-day variations in expression (Fig. 2C), indicating that rhythmic expression of the two microRNAs is dependent on the molecular clock.
We then addressed the light-inducible expression of both microRNAs. A light pulse (15 min, 100 lux) in the early (CT 15) and late (CT 22) subjective night significantly elevated pre-miR-132 expression within the SCN relative to dark control (Fig. 3A). This effect was phase-restricted and tissue-specific: a light pulse in the mid subjective day (CT 6) did not induce pre-miR-132 expression in the SCN, or in the cortex irrespective of circadian phase (Fig. 3A). In comparison, pre-miR-219-1 levels within the SCN were not affected by a light pulse delivered at any phase of the circadian cycle (Fig. 3A). RPA revealed that these changes in precursor transcript expression were reflected in the levels of the mature microRNA. Namely, miR-132 levels in the SCN were elevated following a light pulse delivered in the early and late subjective night, whereas miR-219 levels were not light-inducible (Fig. 3B).
Figure 3. Light-inducible expression of miR-132 in the SCN.

(A) Mice received a single light pulse (LF; 15 min, 100 lux) at CT 6, 15, or 22, and were sacrificed 1 hr later. Abundance of pre-miR-132 and pre-miR-219-1 transcripts in pooled RNA samples of SCN and piriform cortex (CTX) tissue was determined by RT-PCR. Mice which had not received a light pulse (dark) but were sacrificed at the same circadian time served as negative controls. Expression of gapdh served as the loading control. (B) RPA analysis of mature miR-132 and miR-219 in the SCN 1 hr after light (+) treatment at CT 6, 15, or 22. Mice which had not received a light pulse (−) but were sacrificed at the same circadian time served as negative controls. Expression of 5S rRNA served as the loading control. Radiolabeled RNA size markers (M; in nucleotides) are shown to the right. P denotes undigested probe. (C) Mice were infused with the MEK inhibitor U0126 (10 mM, 3 μl) or vehicle 30 min prior to a single light flash (LF: 15 min, 100 lux). Mice were sacrificed 1 hr later and the abundance of pre-miR-132 and gapdh transcripts in the SCN were determined by RT-PCR. (D) Animals were infused with U0126 (10 mM), KN-62 (10 mM), or KT-5823 (1 mM), and the effects on basal (dark) and light- (LF: 15 min, 100 lux) induced pre-miR-132 expression were analyzed via real-time quantitative PCR. Data were normalized to gapdh levels and presented as fold expression. Mean values were determined from three independent experiments.
The ERK/MAPK pathway is a key regulator of light-induced clock entrainment and activates CREB/CRE-dependent transcription (Obrietan et al., 1999; Butcher et al., 2002). Given our data implicating CREB in inducible miR-132 expression, we wanted to assess the involvement of the ERK/MAPK pathway in light-induced expression of miR-132. To this end, mice received a ventricular infusion of the MEK1/2 inhibitor, U0126, 30 min prior to a brief light pulse (15 min, 100 lux) at CT 15. Pretreatment of U0126 blocked the light-induced increase in pre-miR-132 levels in the SCN (Fig. 3C and 3D). Interestingly, infusion of U0126 in the subjective day (CT 5) likewise reduced basal miR-132 levels 1 hr later (Fig. 3D). In addition, we examined the role of two other signaling pathways which have been implicated in clock entrainment, the calcium/calmodulin kinase (CaMK) and cGMP-dependent protein kinase (PKG) pathways. Pretreatment with the PKG-selective inhibitor, KT-5823, had no effect on the light-induced increase in pre-miR-132 levels in the SCN relative to vehicle-infused controls (Fig 3D). In contrast, the CaMK inhibitor, KN-62, partially attenuated light-inducible expression of pre-miR-132 in the SCN (Fig 3D). Together, our data reveal that miR-219-1 is a clock-controlled gene, and that the robust light-induced activation of CREB-regulated gene miR-132 requires the ERK/MAPK cascade.
miR-219 Regulates Circadian Period Length and miR-132 Modulates Light-Induced Clock Resetting
Next, we addressed the functional roles of miR-219 and miR-132 in the SCN, using behavioral rhythms (ie., wheel-running activity) as a read-out. To this end, cholesterol-modified oligoribonucleotides complementary to microRNAs (antagomirs) were used to repress miRNA levels. Previous studies have shown that intravenous antagomir administration potently reduces microRNA expression in all tissues, except the brain (Krutzfeldt et al., 2005), suggesting that antagomirs cannot pass the blood brain barrier. To circumvent this issue, we utilized an intraventricular infusion approach that directly delivered antagomirs to the cerebral spinal fluid (CSF). From the CSF, we predicted that the antagomir would penetrate periventricular brain regions, including the SCN. Mice were infused with the miR-219 antagomir at CT 2 after 8–10 days of free-running in constant darkness (DD). CT 2 was selected as the time of infusion, because it precedes peak expression of miR-219. Based on RPA analysis, infusion of the miR-219 antagomir strongly suppressed expression of endogenous miR-219 in the SCN for up to 2 weeks (Fig. 4H and Supplementary Figure S2A). Vehicle infusion at CT 2 resulted in a modest period shortening post-treatment (−0.18 ± 0.03 hr: Supplementary Fig S2B and Fig. 4G). Period (tau) shortening likely resulted from experimental handling and infusion. Infusion of a negative control antagomir, a scrambled sequence of miR-219 antagomir which is not complementary to any known murine microRNA, likewise produced a period shortening; however, the effect was comparable to that of vehicle infusion (−0.17 ± 0.03 hr, p > 0.05 vs. vehicle: Fig. 4A and 4G). Hence, the scrambled antagomir per se had no effect on period length. Moreover, the change in period length as a result of infusion of miR-132 antagomir was not statistically different from infusion of drug vehicle (−0.12 ± 0.01 hr, p > 0.05 vs. vehicle: Fig. 4B and 4G). In contrast, mice infused with the miR-219 antagomir displayed a period change of a similar magnitude but in the opposite direction: the lengthening of period was statistically different from the period change produced by vehicle or scrambled antagomir (+0.15 ± 0.05 hr, p < 0.01 vs. vehicle and control antagomir: Fig. 4C and 4G). In some respects, this increase in period understates the impact of miR-219 inhibition in vivo: it counters the period-shortening effects of experimental handling and elicits a further increase. The tau of individual animals is plotted before and after infusion (Fig. 4D: scrambled, Fig 4E: 132 antagomir, Fig. 4F: 219 antagomir). Of note is the observation that every animal infused with miR-219 antagomir exhibited tau lengthening, and in 2 of the mice, tau lengthened by > 0.3 hrs. These results implicate miR-219 as a regulator of circadian period length.
Figure 4. miR-219 regulates circadian period length.

(A–C) Representative actograms of wheel-running activity of C57Bl/6J mice which had received an infusion of (A) a negative control antagomir (scrambled), (B) miR-132 antagomir (40 μM, 3 μl) or (C) miR-219 antagomir at CT 2. Periods of darkness are shaded in gray. Activity onsets are indicated by blue and red lines. Red asterisk denotes infusion. The x-axis (top) indicates the Zeitgeber (ZT) time over a 24-hr cycle. The y-axis (left) indicates the nth day of the experiment. (D–F) Graphical representation of the period length (tau) of individual animals that were infused with (D) the negative control antagomir (scrambled), (E) miR-132 antagomir, or (F) miR-219 antagomir before (PRE) and after (POST) the infusion. (G) Quantitation of the effects of miR-219 antagomir on period length under free-running conditions. Values are presented as the mean difference ± SEM between post- and pre-infusion tau values (in hr). n=8–10 per group. * p<0.01 (two-tailed Student’s t-test). (H) Knockdown of miR-219 expression. miR-219 antagomir (40 μM, 3 μl) or vehicle were infused into the lateral ventricle at CT 2, and SCN tissue was subsequently harvested at CT 10. Abundance of mature miR-219 was determined by RPA. Abundance of 5S rRNA was used as the loading control.
To examine the potential role of miR-132 in photic entrainment of the clock, we tested the phase-delaying effects of early night light following miR-132 knockdown. Initially, mice infused with antagomir vehicle 1 hour prior to a brief light pulse (15 min, 20 lux) at CT 15 exhibited a mean phase delay of −55 ± 9.7 min (Supplementary Fig. S2C and Fig. 5D). Control antagomir (scrambled) infusion had no effect on light-induced phase delays (−65 ± 8.4 min, p > 0.05 vs. vehicle: Fig. 5A and 5D). Similarly, mice infused with the miR-219 antagomir showed phase shifts that were comparable to those of vehicle-infused controls (−45 ± 9.8 min, p > 0.05 vs. vehicle: Fig. 5C and 5D). In contrast, light-induced phase shifting was greatly potentiated (by nearly 2-fold) in mice which received an infusion of the miR-132 antagomir (−116 ± 11.5 min, p < 0.01 vs. vehicle and control antagomir: Fig. 5B and 5D). RPA analyses revealed that pre-infusion of the miR-132 antagomir abolished light-induced expression of miR-132 in the SCN (Fig. 5E). Thus, our data indicate that light-induced miR-132 functions as a negative regulator of photic clock resetting.
Figure 5. miR-132 negatively modulates light-induced clock resetting.

(A–C) Representative actograms of wheel-running activity of C57Bl/6J mice which received an infusion of (A) a negative control antagomir (scrambled), (B) miR-132 antagomir (40 μM, 3 μl), or (C) miR-219 antagomir at CT 14. All subjects received a single light pulse (15 min, 20 lux) at CT 15. Periods of darkness are shaded in gray. Activity onsets are indicated by blue lines. Red asterisk denotes light pulse. The x-axis (top) indicates the Zeitgeber (ZT) time over a 24-hr cycle. The y-axis (left) indicates the nth day of the experiment. (D) Quantitation of the effects of miR-132 antagomir on CT 15 light-induced phase shifts. Values are presented as mean ± SEM phase shift (in min). n=8–12 per group. * p<0.01 (two-tailed Student’s t-test). (E) Knockdown of miR-132 expression. miR-132 antagomir (40 μM, 3 μl) was infused into the lateral ventricle at CT 14, and a brief light pulse (100 lux, 15 min) was administered at CT 15. SCN tissue was subsequently harvested at CT 17. Control subjects were infused with vehicle and/or did not receive a light treatment. Abundance of mature miR-132 was determined by RPA. Abundance of 5S rRNA was used as the loading control.
As a test of both miRNA functionality and antagomir efficacy in the SCN, we analyzed the expression of a bona fide miR-132 target, p250GAP. Expression of p250GAP has been shown previously to be decreased by miR-132 under in vitro conditions (Vo et al., 2005). Consistent with a light-induced upregulation of miR-132, we observed a suppression of p250GAP protein levels in the SCN 12 hrs after a light pulse at CT 15 (100 lux, 15 min: Supplementary Fig. S2D and S2E). Importantly, infusion of the miR-132 antagomir blocked light-dependent down-regulation of p250GAP (Supplementary Fig. S2D and S2E).
miR-219 and miR-132 Alter Cellular Excitability and Regulate Protein Expression of SCOP and RFX4, Respectively
As a starting point to address the mechanisms underlying their effects on clock timing, we compiled a list of potential targets of miR-132 and miR-219 (Tables 1 and 2) using three computational target prediction algorithms: TargetScan (www.targetscan.org), miRanda (www.microrna.org), and miRBase (www.microrna.sanger.ac.uk). Notably, several ion channels contained a miR-132 or miR-219 recognition sequence within the 3′-UTRs of their transcripts. Together with the recent work indicating that the clock is influenced by changes in cellular excitability (Nitabach et al., 2005), we speculated that miR-219 and/or miR-132 may influence the clock via alterations in cellular excitability. To address this question, cultured cortical neurons were transfected with expression vectors for miR-219 or miR-132, and the effects on depolarization- and glutamate receptor-evoked Ca2+ responses were examined. Relative to untransfected neurons, miR-132 significantly potentiated the responsiveness to K+ (20 mM), glutamate (20 μM) and NMDA (20 μM) administration (Fig. 6A and 6B). Interestingly, miR-219 triggered a modest, but significant attenuation of evoked responsiveness (Fig. 6B). In contrast, neither miR-1 nor empty vector control significantly affected Ca2+ responsiveness relative to untransfected neurons (Fig. 6B). Collectively, these data reveal that both light-responsive miR-132 and clock-regulated miR-219 influence cellular excitability.
Table 1.
Predicted miR-132 targets
| accession # | gene symbol | description | database§ | rhythmic (R)‡/light-induced (L)†/cell excitability (E) |
|---|---|---|---|---|
| NM_007767 | Pcdha6 | protocadherin alpha 6 | T | R (P) |
| NM_008258 | Hn1 | hematological and neurological expressed sequence 1 | M, T | R (P) |
| NM_010474 | Hs3st1 | heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | T | R (P) |
| NM_008770 | Cldn11 | claudin 11 | T | R (P) |
| NM_025356 | Ube2d3 | ubiquitin-conjugating enzyme E2D3 | T | R (P) |
| NM_016916 | Blcap | bladder cancer associated protein homolog (human) | M | R (P, U) |
| NM_016748 | Ctps | cytidine 5′-triphosphate synthase | S | R (P) |
| NM_009444 | Tgoln2 | trans-golgi network protein 2 | T | R (P) |
| NM_007604 | Capza2 | Capping protein alpha 2 | S | R (P) |
| NM_007508 | Atp6vla | ATPase, H+ transporting, lysosomal (vacular proton pump), alpha 70 kDa, isoform 1 | T | R (P) |
| NM_009481 | Usp9x | ubiquitin specific peptidase 9, X chromosome | M, T | R (P) |
| NM_015791 | Fbxo8 | f-box only protein 8 | T | R(P) |
| NM_011361 | Sgk | serum/glucocorticoid regulated kinase | M, S | R (P, U) |
| NM_011334 | Clcn4 | chloride channel 4 | T | R (P) |
| NM_008211 | H3f3b | H3 histone, family 3B | T | R (P) |
| NM_021510 | Hnrph1 | heterogeneous nuclear ribonucleoprotein H1 | T | R (P) |
| NM_011908 | Ubl3 | ubiquitin-like 3 | T | R (P) |
| NM_016690 | Hnrpdl | heterogeneous nuclear ribonucleoprotein D-like | T | R (P) |
| NM_015781 | Nap1l1 | nucleosonie assembly protein 1-like 1 | T | R (P) |
| NM_019912 | Ube2d2 | ubiquitin-conjugating enzyme E2D2 | T | R (P) |
| NM_007761 | Crcp | calcitonin gene-related peptide-receptor component protein | S | R (P) |
| NM_025570 | Mrpl20 | mitochondrial ribosomal protein L20 | T | R (P) |
| NM_009215 | Sst | somatostatin | S | R (P) |
| NM_008186 | Gtf2h1 | general transcription factor IIH, polypeptide 1 (62 kD subunit) | S | R (P) |
| NM_013795 | Atp51 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit g | M | R (P) |
| NM_008556 | Pea15 | phosphoprotein enriched in astrocytes 15 | T | R (P) |
| NM_027324 | Sfxn1 | sideroflexin 1 | T | R (P) |
| NM_008211 | H3f3b | H3 histone, family 3B | T | R (U) |
| U12142 | NR1D2 | Rev-ErbA beta | T | R (U) |
| NM_010518 | Igfbp5 | insulin-like growth factor binding protein 5 | T | R (U) |
| NM_017373 | E4BP4(Nfil3) | E4BP4 (Nuclear factor, interleukin 3, regulated) | S | R (U) |
| NM_017379 | Tuba8 | Tubtilin alpha 8 | T | R (U) |
| NM_080554 | Psmd5 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 | T | R (U) |
| NM_008863 | Pkibeta | Protein kinase inhibitor beta, cAMP dependent, testis specific | T | R (U) |
| NM_021704 | cxcl12 | stromal cell derived factor 1 | T | R (U) |
| NM_009758 | Bmpr1a | Bone morphogenetic protein receptor, type 1A | T | R (U) |
| L16992 | Bckdhb | Branched chain ketoacid dehydrogenase E1, beta polypeptide | S, T | R (U) |
| BC006865 | Pdia6 | protein disulfide isomerase associated 6 | S | R (U) |
| NM_007570 | btg2 | B-cell translocation gene 2, anti-proliferative | M, T | L |
| NM_001045913 | rrad | ras-related associated with diabetes | S | L |
| NM_013613 | nr4a2 | nuclear receptor subfamily 4, group A, member 2 | M, T | L |
| AB086957 | rfx4 | brain-specific regulatory factor X4 | M | L* |
| NM_008036 | FosB | FBJ osteosarcoma oncogene B | M | L |
| NM_008425 | KCNJ2 | Inward rectifier potassium channel subfamily j, member 2 | M | E |
| NM_010603 | KCNJ12 | ATP sensitive Inward rectifier potassium channel subfamily j, member 12 | M, T | E |
| NM_008432 | KCNK5 | potassium channel, subfamily K, member 5 | S | E |
| NM_080465 | KCNN2 | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 | S | E |
| NM_009899 | CLCA1 | chloride channel calcium activated 1 | S | E |
M = miRanda, S = miRBase (Sanger), T = TargetScan
reference for the microarray analysis of rhythmic genes in the SCN: P = Panda et al., 2002; U = Ueda et al., 2002
microarray analysis of light-inducible genes in the SCN: Araki et al., 2006
Araki et al., 2004
Table 2.
Predicted miR-219 targets
| accession # | gene symbol | description | database§ | rhythmic (R)‡/light-induced (L)†/cell excitability (E) |
|---|---|---|---|---|
| M96163 | Snk | serum-inducible kinase | M | R (U), L |
| NM_013842 | Xbp1 | X-box binding protein 1 | S | R (U) |
| NM_007454 | Ap1b1 | Adaptor protein complex AP-1, beta 1 subunit | M, S | R (U) |
| AK014338 | Armet | Arginine-rich, mutated in early stage tumors | S | R (U) |
| BC009654 | P4ha1 | Procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha 1 polypeptide | S | R (U) |
| NM_009026 | Dexras1 (Ags1,Rasd1) | RAS, dexamethasone-induced 1 | S | R (P, U) |
| NM_008832 | Phka1 | Phosphorylase kinase alpha 1 | T | R (U) |
| NM_007489 | Bmal1(Arnt1) | Brain and muscle Ah receptor unclear translocator-like protein 1 (Aryl hydrocarbon receptor unclear translocator-like) | S | R (P, U) |
| NM_013559 | Hsp110 | Heat shock protein, 105 kDa | S | R (U) |
| NM_009758 | Bmpr1a | Bone morphogenetic protein receptor, type 1A | T | R (U) |
| L16992 | Bckdhb | Branched chain ketoacid dehydrogenase E1, beta polypeptide | T | R (U) |
| BC006865 | pdia6 | protein disulfide isomerase associated 6 | S | R (U) |
| NM_011738 | Ywhah | tyrosine 3-monooxygenase/tryptophase 5-monooxygenase activation protein, eta polypetide | T | R (P) |
| NM_009863 | Cdc7 | Cell division cycle 71 homolog | S | R (P) |
| NM_020593 | Fbxo3 | f-box only protein 3 | M, T | R (P) |
| NM_021516 | MARK3 | MAP/microtubule affinity-regulating kinase 3 | S | R (P) |
| NM_012055 | Asns | asparagine synthetase | T | R (P) |
| NM_013699 | Ubp1 | upstream binding protein 1 | T | R (P) |
| NM_026139 | Armcx2 | armadillo repeat containing, X-linked 2 | S | R (P) |
| NM_031375 | Ngrn | neugrin | S, T | R (P) |
| NM_007830 | Dbi | diazepam binding inhibitor | T | R (P) |
| BC010270 | Bsg | basigin | S | R (P) |
| NM_011406 | Slc8a1 | solute carrier family 8 (sodium/calcium exchanger), member 1 | T | R (P) |
| NM_023464 | Ssna1 | Sjogren’s syndrome nuclear autoantigen 1 | S | R (P) |
| NM_027324 | Sfxn1 | sideroflexin 1 | T | R (P) |
| NM_010637 | Klf4 | kruppel-like factor 4 | S | L |
| NM_018781 | egr3 | early growth response 3 | M, T | L |
| BC059254 | phlpp (scop) | PH domain and leucine rich repeat protein phosphatase | T | R* |
| NM_001031811 | KCNH8 | potassium voltage-gated channel, subfamily H, member 8 | M, T | E |
| NM_021544 | SCN5A | sodium channel, voltage-gated, type V. alpha | T | E |
| NM_009899 | CLCA1 | chloride channel calcium activated 1 | S | E |
| NM_011695 | VDAC2 | voltage-dependent anion channel 2 | S | E |
M = miRanda, S = miRBase (Sanger), T= TargetScan
reference for the microarray analysis of rhythmic genes in the SCN: P = Panda et al., 2002;U = Ueda et al., 2002
microarray analysis of light-inducible genes in the SCN: Araki et al., 2006
Figure 6. miRNA-specific regulation of cell excitability and target protein abundance.

(A) Cultured cortical neurons were transfected with expression constructs for pre-miR-132, pre-miR-219 (not shown) or pre-miR-1 (not shown), along with a DsRed.T3-NLS-PEST expression plasmid as a transfection marker (transfection ratio 3:1 of pre-miR construct: DsRed). Negative controls were cultures transfected with empty vector (pcDNA3.1), and adjacent untransfected neurons (DsRed-negative); internal Ca2+ levels were monitored using time-lapse digital microscopy of Fluo-4. Representative data from Fluo-4 loaded (green) untransfected and transfected (red) neurons stimulated with K+ (20 mM), glutamate (20 μM) and NMDA (20 μM). (B) Peak ± SEM Ca2+ responses of untransfected and transfected neurons. Data for each experiment were normalized to the peak response of untransfected neurons, which was set equal to a value of 1. * p<0.01. (C) HEK 293 cells were transfected with expression constructs for SCOP or FLAG-RFX4, along with pre-miR-132, pre-miR-219 or pre-miR-1 and DsRed.T3-NLS-PEST (transfection ratio 2:6:1 of target: pre-miR construct: DsRed). Forty-eight hours after transfection, cells were fixed and immunolabelled for SCOP or FLAG-RFX4. Representative panels show target expression when transfected with each pre-miRNA. Note the reduced expression of SCOP when cotransfected with pre-miR-219, and the reduced expression of FLAG-RFX4 when cotransfected with pre-miR-132. (D) Mean immunolabeling intensity for SCOP and FLAG-RFX4 under the three cotransfection conditions. Y-axis denotes fluorescent intensity units (0-4096 scale). Values are presented as mean ± SEM. n=200–250 cells per group. *p<0.05 (two-tailed Student’s t-test).
For a number of putative miR-132 or -219 targets (Tables 1 and 2), the corresponding transcripts have been shown to be rhythmically expressed or light-inducible in the SCN (Shimizu et al., 1999; Panda et al., 2002; Ueda et al., 2002; Araki et al., 2006). Interestingly, a putative miR-132 target is regulatory factor X4 (RFX4), a member of the winged subfamily of helix-turn-helix transcription factors that is highly expressed in the SCN; moreover, RFX4 transcript levels are rapidly induced in the SCN following nocturnal light exposure (Araki et al., 2004). SCOP, a potential target of miR-219, is a member of the leucine-rich repeat (LRR)-containing protein family and its expression in the SCN is rhythmic, peaking in the subjective night (Shimizu et al., 1999). Given the light-inducible nature of both miR-132 and RFX4, and the complementary circadian expression profiles of miR-219 and SCOP, we hypothesized that RFX4 and SCOP may be physiologically relevant targets of miR-132 and miR-219, respectively.
We used an in vitro overexpression based approach as a starting point for microRNA target validation. N-terminally tagged FLAG-RFX4 and SCOP expression constructs, containing the entire 3′-untranslated region (3′-UTR) of the respective transcript and, therefore, the microRNA target recognition sequence, were transfected into HEK293 cells. Cells were co-transfected with expression vectors for miR-132, miR-219 or miR-1. Expression of miR-132 significantly reduced RFX4 protein abundance relative to miR-1 or miR-219 (Fig. 6C and 6D). Likewise, cultures co-transfected with miR-219 showed significantly lower levels of SCOP protein compared with those expressing miR-1 or miR-132 (Fig. 6C and 6D). Thus, our data indicate the RFX4 and SCOP are in vitro targets of miR-132 and miR-219, respectively, suggesting the possibility that they may also be in vivo targets of these two microRNAs within the SCN.
miR-219 and miR-132 Influence the Core Clock
To move from cell physiology to potential involvement of miR-132 or miR-219 in core clock timing mechanisms, we used an in vitro reporter system to examine their effects on CLOCK/BMAL1-dependent Per1 transactivation. In HEK293 cells, overexpression of CLOCK and BMAL1 together resulted in a 2.3-fold increase in Per1-driven luciferase reporter activity (Fig. 7A). Co-expression of miR-219 or miR-132 augmented Per1-luciferase activity by 40% and 37%, respectively, relative to CLOCK/BMAL1-expressing cells (Fig. 7A). This effect was specific to miR-219 and miR-132; overexpression of miR-1 did not alter CLOCK/BMAL1-dependent transcription (Fig. 7A). Interestingly, expression of miR-219 or miR-132 alone (in the absence of heterologous CLOCK and BMAL1) produced a small, but significant, increase in Per1-luciferase activity. These combined effects of CLOCK/BMAL1 and miR-219 or miR-132 were greater than the additive effects of CLOCK/BMAL1 and the miRNAs when administered alone, suggesting that miR-219 and miR-132 are positive modulators of CLOCK/BMAL1-dependent Per1 transcription.
Figure 7. miR-132 modulates Per1 gene transcription and Per2 protein stability.
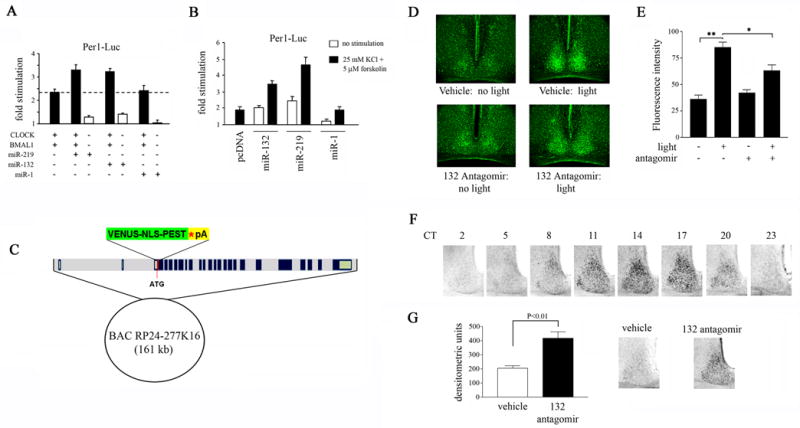
(A) Effect of miR-132 and miR-219 on CLOCK/BMAL1-mediated Per1 transactivation. HEK293T cells were transfected with a Per1-luciferase reporter construct, in combination with the following: CLOCK and BMAL1 expression plasmids and constructs encoding pre-miR-132, pre-miR-219 or pre-miR-1. Cells were lysed 48 hrs post-transfection and assayed for luciferase activity. Y-axis denotes fold difference in luciferase activity relative to control samples transfected with the Per1-luciferase construct alone. Experiments were performed four times, and representative data were averaged from quadruplicate determinations. (B) Effect of miR-132 and miR-219 expression on Per1 transactivation in primary neurons under basal and stimulated conditions. Primary rat embryonic neurons were transfected with a Per1-luciferase reporter construct in combination with the expression vector for pre-miR-132, pre-miR-219 or pre-miR-1. Forty-eight hrs post-transfection, neurons were stimulated with 25 mM KCl and 5 μM forskolin and cell lysates were prepared 6 hrs later. Y-axis denotes fold difference in luciferase activity relative to control samples transfected with the Per1-luciferase construct alone. Experiments were performed four times, and representative data were averaged from quadruplicate determinations. (C) Generation of Per1-Venus BAC transgenic mice. The Venus-NLS-PEST-polyA cassette was inserted into the per1 locus on a 161-kb bacterial artificial chromosome (BAC) by homologous recombination. The BAC clone contains the full intergenic sequences of mPer1 and its flanking genes, including the 5′ promoter region. (D) The effect of miR-132 knockdown on light-induced Venus expression in the SCN of Per1-Venus BAC transgenic mice. miR-132 antagomir (40 μM, 3 μl) or drug vehicle (saline), were infused into the lateral ventricles of Per1-Venus BAC mice at CT 14, and a brief light pulse was administered at CT 15. Tissue was harvested at CT 19 for analysis of Venus expression by immunofluorescence. Dark control animals were not exposed to light but were sacrificed at the same circadian time. (E) Quantitation of light-induced Venus expression as a function of miR-132 abundance. Values are presented as mean ± SEM. n=3–8 mice per group. *p<0.05 (two-tailed Student’s t-test). (F) Immunohistochemical analysis of PER2 protein abundance in the SCN as a function of circadian time. (G) In a separate experiment, mice were infused with miR-132 antagomir (40 μM, 3 μl) or vehicle at CT 14 and exposed to light (100 lux, 15 min) at CT 15 (right). SCN tissue was harvested 29 hr later and immunostained for PER2. Quantitation of PER2 immunoreactivity in the SCN of miR-132 antagomir- or vehicle-infused mice (left). Values are presented as mean ± SEM. n=8–10 per group. *p<0.05 (two-tailed Student’s t-test).
In addition, we examined the effects of miR-219 or miR-132 on basal and inducible Per1 transcription in primary neurons. Under basal conditions, overexpression of miR-219 or miR-132 resulted in a 2.4- and 2-fold enhancement, respectively, of Per1-driven luciferase activity (Fig. 7B). A similar level of reporter expression was attained in empty vector-transfected controls following co-stimulation with forskolin (5 μM) and KCl (25 mM: 1.9-fold increase; Fig. 7B). Interestingly, the effects of forskolin/KCl were augmented by miR-219 or miR-132. For example, stimulation with forskolin/KCl in the presence of miR-219 triggered an ~ 4.5-fold increase in per1-luciferase expression (Fig. 7B). Importantly, these effects were miRNA-specific; miR-1 overexpression did not significantly alter per1-luciferase expression or augment the effects of forskolin/KCl (Fig. 7B). Together these data provide a route by which miR-219 and miR-132 can modulate the clock timing process.
Thus far, our data indicate that light-induced miR-132 expression acts to feedback inhibit the magnitude of photic resetting in vivo, and that ectopic expression of miR-132 stimulates Per1 gene transcription in vitro. To directly assess the role of miR-132 in per1 gene expression in vivo, we generated a strain of reporter mice that expresses the fluorescent protein Venus under the control of the per1 gene promoter. The enhanced YFP derivative Venus was selected as a reporter of per1 transcription because of its superior fluorescent intensity compared with GFP or YFP and its enhanced maturation rate compared with YFP (Nagai et al., 2002). To further increase fluorescent density and to ensure rapid protein turnover, a nuclear localization sequence (NLS) and a PEST destabilization domain were cloned in-frame to the coding sequence of Venus (Fig. 7C). Three of the transgene lines exhibited robust light-induced expression, as well as a circadian expression profile that mirrors that of mPer1 (data not shown).
To determine the effect of miR-132 on light-induced per1 transactivation, Per1-Venus transgenic mice were infused with the miR-132 antagomir prior to a brief light pulse (100 lux, 15 min) at CT 15. Four hours following light stimulation, Venus fluorescence was strongly upregulated in the SCN of vehicle-infused mice relative to dark controls (Fig. 7D and 7E). In contrast, pre-treatment with miR-132 antagomir significantly attenuated light-induced Venus fluorescence by ~ 50% relative to vehicle controls (Fig. 7D and 7E). Infusion of miR-132 antagomir had no effect on basal expression of Venus at CT 19 (Fig. 7D and 7E). These data indicate that miR-132 positively modulates light-induced per1 transactivation in vivo.
The transcriptional events underlying photic resetting are unclear. However, recent evidence suggests that light-induced PER1 influences the magnitude of the phase shift by triggering PER2 protein degradation and/or enhancing PER2 protein synthesis (Masubuchi et al., 2005). Not only do mPer1-deficient mice exhibit larger phase delays of behaviour rhythms following a 12-hr extension of the light cycle, but light-induced mPER2 protein expression is greater in the SCN of mPer1−/− mice compared with wild-type controls during both the interval of prolonged light exposure, as well as throughout much of the subsequent circadian cycle (Masubuchi et al., 2005). In addition to this, there are pronounced phase differences (~ 4 hr) in mPER2 expression between wild-type and mPer1−/− mice in the second cycle. These observations, coupled with our data showing that miR-132 regulates period1 expression and clock entrainment, led us to examine the potential effects of miR-132 on PER2 accumulation. As a starting point, we mapped PER2 protein expression in the SCN as a function of circadian time. As previously reported, PER2 protein levels reach a peak in the early subjective night (CT 14 and CT 17), with a distribution pattern which demarcates the dorsomedial SCN (Fig. 7F). PER2 protein abundance was low in the late subjective night (CT 23) and early to mid subjective day (CT 2 and CT 5: Fig. 7F). We then infused mice with either miR-132 antagomir or drug vehicle prior to a brief light pulse at CT 15. PER2 protein levels were assessed 29 hours later (in the subsequent circadian cycle). PER2 abundance in the SCN of vehicle-infused mice was low, in contrast with the much greater PER2 staining in the dorsomedial aspect of the SCN of miR-132-antagomir infused mice (Fig. 7G). Quantitative analyses indicated that pretreatment with miR-132 antagomir prior to a CT 15 light pulse resulted in a 2-fold enhancement of PER2 levels 29 hrs later relative to vehicle controls (Fig. 7G). Our data are consistent with the observations of Masubuchi et al. (2005) indicating elevated PER2 protein levels in the SCN of mPer1−/− mice in the second circadian cycle following prolonged light exposure. It remains to be determined whether the increase in PER2 immunoreactivity in our experiment is a result of altered phasing of PER2 (a reflection of the difference in light-induced behavioural phase-shifting between vehicle and antagomir-infused subjects), alterations in PER2 stability/degradation or protein synthesis, or a combination of both.
DISCUSSION
The data presented here reveal miRNAs as key regulators of the circadian timing process. miR-219-1 is a clock-controlled gene that plays a role in regulating the length of the circadian day, whereas miR-132 is light-inducible and modulates the phase shifting capacity of light. The effects of miR-132 culminate at the level of mPer1 transcription and PER2 protein expression, thus identifying a potential route by which these miRNAs influence the transcriptional rhythm which lies at the core of the timing process. Our data also reveal that both miR-219 and miR-132 affect cellular excitability, which, in turn, might regulate both clock periodicity and clock entrainment. Collectively, our observations reveal a new and previously unexplored layer of modulation to the circadian clock: inducible translation control via microRNAs.
Biological oscillators such as the circadian clock (and the cell cycle clock) are modeled as interacting positive and negative feedback loops that drive the rhythmic expression of a few molecular determinants. In the case of circadian clocks, the molecules in question are CLOCK and BMAL1, which together activate transcription of Per and Cry genes. PER and CRY proteins subsequently work together to inhibit their own transcription. The established view is that rhythmic gene transcription is the underlying molecular basis for circadian rhythm generation. This model has been enriched by a series of findings (described below) indicating that a complex, integrated, set of physiological processes act in coordination with the transcription-based oscillator to drive highly precise SCN pacemaker activity.
In this context, microRNAs represent a route by which the circadian clock may be regulated. These small noncoding RNAs, which are impressively numerous and phylogenetically extensive (there are currently 364 mouse miRNA genes listed in the MicroRNA Registry [http://www.microrna.sanger.ac.uk/sequences/index.shtml]), have been implicated in mRNA turnover of miRNA targets (Lim et al., 2005), suppression of their protein translation (Lee et al., 1993; Wightman et al., 1993), and, possibly, methylation of target genes (Bao et al., 2004). A role of microRNAs in circadian timing processes has not been demonstrated until now (this report).
The use of SACO analysis, which permitted an unbiased and systematic analysis of transcription factor binding across an entire mammalian genome, revealed that a substantial number of CREB binding targets were noncoding (S.I., unpublished data). We analyzed one of these CREB targets, miR-132, for its potential involvement in the circadian clock, based on the rationale that the CREB/CRE transcriptional pathway plays a pivotal role in coupling light to changes in gene expression in the SCN that underlie entrainment (Dziema et al., 2003). Similar to other CREB-regulated light-responsive immediate early genes (eg., Fos, Jun, and the core clock gene mPer1), miR-132 gene expression in the SCN is tightly regulated by light, but its induction is phase-restricted to the subjective night. It should be noted that the observed differences in pre-miR-132 levels are also reflected in the abundance of mature miRNA, ruling out the possibility that light has no functional consequence on miR-132 effects. As with other CREB-regulated genes, infusion of the MEK1/2 inhibitor, U0126, blocked light-inducible miR-132 expression, indicating that it is mediated by a MAPK-dependent signaling pathway. Multiple lines of evidence indicate MAPK signaling as a principal effector of light-induced clock entrainment (Obrietan et al., 1998; Butcher et al., 2002). The characteristics determined in this study (ie., regulation by CREB and MAPK, rapid light inducibility, phase restriction) raise the possibility that miR-132 contributes to the clock entrainment mechanism.
Conceivably, miRNAs within the SCN may be regulated not only by light but also by the molecular clock itself. As a starting point, we sought to identify potential miRNAs, within the CREB SACO subset of miRNAs, that are direct targets of CLOCK/BMAL1-mediated transcription. Using chromatin immunoprecipitation, the miR-219-1 locus was enriched in the CLOCK-binding DNA fraction and identified by a PCR-based approach. The murine miR-219-1 locus is architecturally analogous to the mPer1 and mPer2 loci, in that its 5′ region consists of an E-box motif as well as CRE consensus sites. The presence of an E-box motif and the CLOCK ChIP data strongly suggested to us that miR-219-1 is a clock-regulated gene. Similar to other E-box regulated core clock (ie., per1, per2, cry1, cry2) and clock output (eg., arginine vasopressin [AVP]) genes, we observed robust induction of miR-219 expression in CLOCK/BMAL1-overexpressing cells in vitro. In vivo, miR-219 exhibits a circadian rhythm of expression in the SCN. Our observation that miR-219 levels peak in the early- to mid-subjective day lends further support for a CLOCK/BMAL1-dependent miR-219 transcriptional rhythm: this time window coincides with maximal mPer1 expression (mRNA levels peak by mid-day and decline substantially by CT 12) and precedes the mPer2 peak (mid- to late-subjective day) by several hours. Moreover, the trough in miR-219 expression occurs in the subjective night, a time when PER/CRY protein complexes have reached sufficient levels to inhibit CLOCK/BMAL1-mediated transcription. Interestingly, although miR-219-1 contains CRE consensus sequences in its promoter and was identified in the CREB SACO, it is not rapidly upregulated by light. In this respect, miR-219-1 is analogous to mPer2, which contains a CRE (in addition to an E-box), but its transcription is not elicited by CREB-activating stimuli, such as cAMP (Travnickova-Bendova et al., 2002).
To define a functional role for miR-132 and miR-219 in the clock, we employed an intraventricular infusion technique to deliver antagomirs to the SCN. With this approach we detected a striking suppression of both clock-regulated (mir-219) and light-induced (miR-132) expression. miR-219 knockdown resulted in an increase in the length (tau) of the circadian clock period. Thus, period length increased by ~ 9 min. As noted above, this mean value underestimates the effects of the knockdown. Along these lines, tau lengthening is counter to the effects of the infusion of non-target antisense, or drug vehicle, where period shortening was consistently observed. Likewise, some animals exhibited > 2X the mean tau lengthening. This animal-to-animal variability in the effects of the antagomir may result from variability in the efficacy of the infusion. In some respects, the relative effect of miR-219 antagomir on circadian period vs. miR-132 on light-induced phase-shifting is reminiscent of the relative difference between period and phase responses of the circadian system: light-induced phase shifts are on the order of up to several hours, whereas changes in period to a single light pulse are measured in minutes (Comas et al., 2006).
When placed in a broad context, the tau shift resulting from miR-219 knockdown suggests that rhythmic miR-219 plays a significant role in determining phase length. Along these lines, genetic deletion of a number of protein factors that have proven roles in core clock timing processes or input pathways to the clock, nevertheless, has a subtle impact on the circadian system. The absence of core clock proteins mPer1, Cry1, Cry2, and Clock singly does not result in arrhythmicity under constant conditions. Another example is Rev-Erbα, a critical regulator of Bmal transcription, that, in its absence, produces only modest changes in period length and phase-shifting properties but does not alter circadian rhythm generation (Preitner et al., 2002). In fact, there are only a handful of examples in which the absence of a single factor (eg., Bmal1 and mPer2) has a profound impact on the circadian timing system. Thus, these findings indicate that the clock is quite resistant to molecular perturbations; either the clock loses all capacity to keep time (arrhythmic), or the clock compensates with typically a modest phenotypic effect. Thus, placed in this context of these observations, we conclude that miR-219 has a role in regulating period length.
Knockdown of miR-132 by antagomir infusion enhanced the phase-resetting effects of light by nearly 2-fold. The difference in response may be attributed to 1) the strength of the inputs to the circadian pacemaker (eg., strength of the signaling pathways that are activated by light), 2) circadian pacemaker amplitude, which determines whether or not the same stimulus is more or less effective at phase-shifting the clock, or 3) a combination of both. While our study did not address the role of miR-132 in clock gene rhythms (point #2), our observation that light-induced mPer1 expression is affected by miR-132 (discussed later) lends support to a change in input strength (point #1). In addition, bearing in mind the fact that light induces expression of miR-132, one should further note that light suppresses clock responsiveness: this is observed as a low-amplitude phase response curve (PRC) under an LD cycle vs. a high-amplitude PRC in DD-adapted animals (Refinetti 2003). Even a single 1-hr light pulse in dark-adapted mice has been shown to reduce the magnitude of phase-shifting elicited by a second light pulse (Refinetti 2003). This raises the attractive possibility that the physiological significance of miR-132 induction by light is to act as a feedback inhibitor of photic responsiveness.
The target genes that mediate the effects of miR-219 on clock timing and miR-132 on clock entrainment are unknown. The challenge of identifying specific genes and determining how they relate to a complex physiological process, such as SCN timing, can be appreciated by examining the large number of predicted targets. To emphasize this point, the miRanda software predicts 114 and 265 targets for miR-219 and miR-132, respectively. Likewise, there is often little overlap between targets predicted by different algorithms (reviewed in Rajewsky, 2006), and only a limited number of predicted targets have been confirmed. In this light, our in vitro identification of SCOP and RFX4 as miR-219 and miR-132 targets, respectively, is a first step in this characterization. Thus, rather than exclusively focusing on direct miRNA targets, we turned our attention on three well-characterized (and experimentally conducive) physiological effectors of clock timing and entrainment: cellular excitability, clock gene expression and clock protein stability/abundance.
To assess the effects of microRNAs on cellular excitability, we measured neuronal Ca2+ responses to depolarizing agents. Our studies were conducted in cortical neurons in order to circumvent the issue of varied phasing of SCN cells in vitro (Honma et al., 1998). Our studies indicate that miR-132 overexpression enhances neuronal excitability, whereas miR-219 has a modest, but significant, suppressive effect. The literature supports a role of membrane excitability in diverse aspects of clock physiology (McMahon and Block, 1987; Meredith et al., 2006). In this context, the effect of miR-219 and miR-132 on neuronal excitability may, in part, contribute to the behavioral phenotypes elicited by their in vivo knockdown.
To address the effect of microRNAs on clock gene expression, we used complementary in vitro gain-of-function and in vivo loss-of-function approaches. Using a luciferase reporter system in vitro, we demonstrated that CLOCK/BMAL1- and depolarization-dependent mPer1 transcription is augmented by miR-219 and miR-132. Conversely, in the case of miR-132, antagomir-mediated knockdown resulted in an attenuation of light-induced mPer1 expression. Whether or not the effects of miR-132 on mPer1 expression are causal to the potentiation in photic resetting in antagomir-infused animals, or is merely coincident with the behavioral phenotype, remains to be determined. However, it is interesting to note that mPer1−/− mice exhibit exaggerated phase delays in response to prolonged light exposure (Masubuchi et al 2005), and in general phenocopies the behavior of miR-132-antagomir-infused mice.
CONCLUSION
A series of second-generation studies will be required to identify the specific target transcripts and/or genetic networks through which miR-219 and miR-132 influence the timing process. Collectively, these observations reveal a new class of clock-regulated genes, and as such, provide a new dimension in our understanding of circadian clock regulation: inducible translation control via microRNAs.
EXPERIMENTAL PROCEDURES
More detailed description of Experimental Procedures is available in theSupplemental Data.
Generation of mPeriod1-Venus BAC Transgenic Mouse Strain
A BAC construct encompassing the entire mperiod1 gene (Clone ID number RP24-277K16) was obtained from BACPAC Resources Center (Oakland, CA). The Venus-NLS-PEST fusion gene was inserted in-frame with, and immediately downstream of, the PER1 translational start site by bacterial homologous recombination.
Generation of CREB SACO Library
The CREB SACO library was generated as described previously (Impey et al., 2004).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation of CREB-DNA and CLOCK-DNA complexes were performed exactly as described (Impey et al., 2004).
Cannulation and Infusion
Third ventricle cannulations and drug infusions were performed as described previously (Butcher et al., 2002).
Behavioural analyses
Behavioral assessment was conducted as described previously (Cheng et al., 2006). Mice were maintained at the animal facility of the Ohio State University in accordance with institutional guidelines. All animal handling and experimental procedures were approved by the Animal Welfare Committee of the Ohio State University.
Cell Culture and Transfection
PC12 cells and HEK293T cells were cultured as described previously (Impey et al., 2004). Embryonic rat cortical neuron cultures were prepared as described (Cheng et al., 2006).
Real-Time and Reverse Transcription (RT)-PCR
Mice under entrained conditions were dark adapted for 2 days prior to treatment. To profile for circadian expression, mice were sacrificed at defined circadian times under dim red light, tissues of the SCN and piriform cortex were dissected and stored at −80°C until use. RT-PCR was performed using the Access RT-PCR System (Promega, Madison, WI) according to manufacturer’s instructions.
Ribonuclease Protection Assay (RPA)
Tissue collection and RNA extraction were performed exactly as described above. Antisense RPA probes were generated using the mirVana miRNA Probe Construction Kit (Ambion, Austin, TX). Samples were pooled and 500 nanograms of total RNA was used with the mirVana miRNA Detection Kit (Ambion) according to manufacturer’s instructions.
Tissue Collection and Immunohistochemistry
Tissue collection and immunodetection were performed as described previously (Cheng et al., 2006).
Fura-2 Calcium Digital Imaging
Time-lapse fluorescent digital microscopy was performed as described previously (Lee et al., 2005).
Supplementary Material
Acknowledgments
We thank C. Weitz, A. Miyawaki, A. Nagy, N. Copeland, and M. Abe for sharing of reagents and K. Hoyt, J. Enyeart, S. Kharzai, and X. Pu for technical assistance. We also thank T. Todo for providing access to mcry1/mcry2 double mutant mice. This work is supported by NIH Grants MH62335 and NS47176 to KO, MH64954 to SI and Center Core Grant 5P30NS045758. HYMC is supported by a post-doctoral fellowship from the Canadian Institute of Health Research (CIHR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC. MPer1 and mper2 are essential for normal resetting of the circadian clock. J Biol Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- Araki R, Nakahara M, Fukumura R, Takahashi H, Mori K, Umeda N, Sujino M, Inouye ST, Abe M. Identification of genes that express in response to light exposure and express rhythmically in a circadian manner in the mouse suprachiasmatic nucleus. Brain Res. 2006;1098:9–18. doi: 10.1016/j.brainres.2006.04.096. [DOI] [PubMed] [Google Scholar]
- Bao N, Lye KW, Barton MK. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell. 2004;7:653–662. doi: 10.1016/j.devcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher GQ, Dziema H, Collamore M, Burgoon PW, Obrietan K. The p42/44 mitogen-activated protein kinase pathway couples photic input to circadian clock entrainment. J Biol Chem. 2002;277:29519–29525. doi: 10.1074/jbc.M203301200. [DOI] [PubMed] [Google Scholar]
- Butcher GQ, Lee B, Cheng HY, Obrietan K. Light stimulates MSK1 activation in the suprachiasmatic nucleus via a PACAP-ERK/MAP kinase-dependent mechanism. J Neurosci. 2005;25:5305–5313. doi: 10.1523/JNEUROSCI.4361-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MY, Bullock CM, Li C, Lee AG, Bermak JC, Belluzzi J, Weaver DR, Leslie FM, Zhou QY. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature. 2002;417:405–410. doi: 10.1038/417405a. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Dziema H, Papp J, Mathur DP, Koletar M, Ralph MR, Penninger JM, Obrietan K. The molecular gatekeeper Dexras1 sculpts the photic responsiveness of the mammalian circadian clock. J Neurosci. 2006;26:12984–12995. doi: 10.1523/JNEUROSCI.4253-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas M, Beersma DG, Spoelstra K, Daan S. Phase and Period Responses of the Circadian System of Mice (Mus musculus) to Light Stimuli of Different Duration. J Biol Rhythms. 2006;21:362–372. doi: 10.1177/0748730406292446. [DOI] [PubMed] [Google Scholar]
- Dziema H, Oatis B, Butcher GQ, Yates R, Hoyt KR, Obrietan K. The ERK/MAP kinase pathway couples light to immediate-early gene expression in the suprachiasmatic nucleus. Eur J Neurosci. 2003;17:1617–1627. doi: 10.1046/j.1460-9568.2003.02592.x. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- Honma S, Shirakawa T, Katsuno Y, Namihira M, Honma K. Circadian periods of single suprachiasmatic neurons in rats. Neurosci Lett. 1998;250:157–160. doi: 10.1016/s0304-3940(98)00464-9. [DOI] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Kauppinen S, Vester B, Wengel J. Locked nucleic acid: high-affinity targeting of complementary RNA for RNomics. Handb Exp Pharmacol. 2006;173:405–422. doi: 10.1007/3-540-27262-3_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraves S, Weitz CJ. A role for cardiotrophin-like cytokine in the circadian control of mammalian locomotor activity. Nat Neurosci. 2006;9:212–219. doi: 10.1038/nn1633. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lee B, Butcher GQ, Hoyt KR, Impey S, Obrietan K. Activity-dependent neuroprotection and cAMP response element-binding protein (CREB): kinase coupling, stimulus intensity, and temporal regulation of CREB phosphorylation at serine 133. J Neurosci. 2005;25:1137–1148. doi: 10.1523/JNEUROSCI.4288-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masubuchi S, Kataoka N, Sassone-Corsi P, Okamura H. Mouse Period1 (mPER1) acts as a circadian adaptor to entrain the oscillator to environmental light/dark cycles by regulating mPER2 protein. J Neurosci. 2005;25:4719–4724. doi: 10.1523/JNEUROSCI.4761-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuki S, Todo T, Nakano Y, Okamura H, Nose H. Reduced alpha-adrenoceptor responsiveness and enhanced baroreflex sensitivity in Cry-deficient mice lacking a biological clock. J Physiol. 2005;566:213–224. doi: 10.1113/jphysiol.2005.086728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon DG, Block GD. The Bulla ocular circadian pacemaker. I. Pacemaker neuron membrane potential controls phase through a calcium-dependent mechanism. J Comp Physiol [A] 1987;161:335–346. doi: 10.1007/BF00603959. [DOI] [PubMed] [Google Scholar]
- Meredith AL, Wiler SW, Miller BH, Takahashi JS, Fodor AA, Ruby NF, Aldrich RW. BK calcium-activated potassium channels regulate circadian behavioral rhythms and pacemaker output. Nat Neurosci. 2006;9:1041–1049. doi: 10.1038/nn1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Sheeba V, Vera DA, Blau J, Holmes TC. Membrane electrical excitability is necessary for the free-running larval Drosophila circadian clock. J Neurobiol. 2005;62:1–13. doi: 10.1002/neu.20053. [DOI] [PubMed] [Google Scholar]
- Obrietan K, Impey S, Smith D, Athos J, Storm DR. Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. J Biol Chem. 1999;274:17748–17756. doi: 10.1074/jbc.274.25.17748. [DOI] [PubMed] [Google Scholar]
- Obrietan K, Impey S, Storm DR. Light and circadian rhythmicity regulate MAP kinase activation in the suprachiasmatic nuclei. Nat Neurosci. 1998;1:693–700. doi: 10.1038/3695. [DOI] [PubMed] [Google Scholar]
- Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, Muijtjens M, Hoeijmakers JH, van der Horst GT. Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science. 1999;286:2531–2534. doi: 10.1126/science.286.5449.2531. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Okada M, Takano A, Nagai K. SCOP, a novel gene product expressed in a circadian manner in rat suprachiasmatic nucleus. FEBS Lett. 1999;458:363–369. doi: 10.1016/s0014-5793(99)01190-4. [DOI] [PubMed] [Google Scholar]
- Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, Iino M, Shigeyoshi Y, Hashimoto S. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


