Abstract
We detail some of the pitfalls encountered when following live cultured somatic cells by light microscopy during mitosis. Principle difficulties in this methodology arise from the necessity to compromise between maintaining the health of the cell while achieving the appropriate temporal and spatial resolutions required for the study. Although the quality of the data collected from fixed cells is restricted only by the quality of the imaging system and the optical properties of the specimen, the major limiting factor when viewing live cells is radiation damage induced during illumination. We discuss practical considerations for minimizing this damage, and for maintaining the general health of the cell, while it is being followed by multi-mode or multi-dimensional light microscopy.
1. Introduction
Cell division, or mitosis (meiosis in germ cells), consists of a series of dynamic events that involve the coordinated interactions of many cellular components. During mitosis, the replicated DNA condenses into chromosomes, which then become attached to a complex structure known as the ‘mitotic spindle.’ The spindle acts as a scaffold for producing and directing the forces responsible for equal distribution of the chromosomes to daughter cells (karyokinesis), and it also defines the plane through which the cytoplasm will then be divided (cytokinesis). Mitosis was first described by Flemming [1], who reconstructed the general course of cell division from analysis of fixed cells. However, it took decades of technological developments in light microscopy (LM) before the details of cell division could be visualized in living cells.
Experimental studies of mitosis began in earnest when new imaging modes were introduced in the 1950s that allowed contrast to be generated between various components in living specimens. These modes included phase-contrast, polarization, and differential interference contrast light microscopy (reviewed in [2]). By the mid-1970s live-cell studies had established, with high temporal and spatial resolutions, how the major components of the spindle, including the chromosomes, centrosomes, and to some extent the microtubules, behave relative to one another during the various stages of division. With the development of video-enhanced LM (video-LM) in the early 1980s ([3,4] reviewed in [5,6]) live cell imaging technology became even more powerful. As a result the description of events that occur during mitosis have become even more accurate, and the corresponding molecular model(s) more meaningful (e.g. [7–9]).
During the past 20 years, the utility of video-LM has been greatly augmented by concurrent advances in protein labeling [10,11] and fluorescent imaging technologies, including the development of confocal and wide-field deconvolution systems [12–14]. These fluorescent approaches can also be combined with conventional transmitted imaging modes, like phase-contrast or differential interference contrast (DIC), to form 4-D “multimode” systems [15]. This, in turn, allows the dynamic behavior of one or more molecules to be correlated with the changing distribution of the chromosomes, centrosomes, and/or kinetochores [16–22].
The enhanced power of these new imaging techniques does, however, comes with a price. In order for the maximum signal/noise ratio and resolution to be achieved, cells must be illuminated with very high light intensities. Indeed, when focused at full power through a 100× 1.4 NA (numerical aperture) lens, the 100 W mercury lamps traditionally used for fluorescence imaging kill cells in just a few seconds. As a result, the intensity of light impacting the specimen needs to be highly attenuated, and it is important to remember from the start that a good biological LM workstation necessarily represents a compromise, in which some image quality is sacrificed to maintain cell viability.
In addition to protecting the specimen from light-induced damage, several other variables must be considered when conducting live-cell LM studies on mitosis, especially when imaging for >1–2 h. The specimen needs to be maintained in an environment that promotes its health, while it is under observation. In practice this means that it must be properly housed, fed, and kept at a comfortable temperature.
In this chapter we outline the problems encountered when following live cells in culture as they progress through the cell cycle, and we offer some practical solutions to these problems. Our emphasis, on the use of video-LM and fluorescence LM, reflects our own particular research interests, on how centrosomes and kinetochores function and interact to form the spindle in vertebrate somatic cells.
2. Basic considerations: keeping cells happy while they are under observation
There are critical periods during which cells are extremely sensitive to insult, and the transition from G2 into mitosis is one of these (reviewed in [23,24]). In the early literature the term ‘antephase’ was used to define that period in G2, just before the first visible signs of chromosome condensation (i.e., prophase), during which the cell can be arrested by a variety of stresses (e.g., [25,26]). These insults include, but are not limited to drugs that disrupt microtubules [27], hypothermia [28], oxidative stress [29], osmotic shock and X-rays [30–32]. Although some of these treatments delay the G2/M transition by triggering the ATM-kinase mediated DNA damage checkpoint, some work through other pathways (reviewed in [24,33]). Germane to this chapter is the well known but seldom discussed (or researched) fact that the visible wavelengths used to excite fluorescent probes in living cells, and even the monochromatic (e.g., 546 nm green) light used by most for conventional DIC or phase-contrast imaging, also produce damage in cells (e.g. [18,23] Fig. 1). This is a critical and too often overlooked issue that plagues live-cell LM studies on the cell and mitotic cycle.
Fig. 1.
Excessive illumination during imaging of live cells arrests the division cycle. In this example a mid-prophase PtK1 cell, in which chromosome condensation was already well advanced, decondenses its chromosomes and arrests in late G2 in response to multi-mode imaging. Top part of each frame presents DIC and bottom shows GFP-γ-tubulin epi-fluorescence. This visible change in nuclear morphology provides a convenient visible assay for over-illumination. From [18].
2.1. Circumventing radiation damage
Some cells, especially those from embryos (in, e.g., Drosophila, Caenorhabditis elegans), are relatively resistant to visible light, probably because they lack pathways to arrest the division cycle in response to DNA damage (e.g., [34]). By contrast, these checkpoints are normally present in cultured somatic cells, and can easily be triggered by excess illumination. As result, cells arrest in G2 before they become committed to the mitotic process (Fig. 1; [24]). Importantly, once the commitment to mitosis is made, the cell becomes relatively refractory to radiation damage [35–37], although excessive illumination can still produce delays during mitosis and/or an aborted division [38].
The amount of illumination tolerated before arresting at the G2/M transition varies among cell types, and may be due, in part, to the particular cells’ degree of transformation and/or presence or absence of G2 checkpoints. Some cells, like PtK1, LLC-PK, Indian Muntjac, and primary cell cultures from humans and salamanders, are extremely sensitive to light during antephase—to the point where it becomes very difficult to follow the G2/M transition at reasonable framing rates even with low-light level systems (e.g., [16,36,37]; reviewed in [23]). Other cell types may be more resistant. A good litmus test for determining if the illumination intensity is too high is to determine whether a cell in early to mid-prophase enters prometaphase (i.e., undergoes nuclear envelope breakdown) under the given viewing conditions. If the chromosomes decondense during the observation period, and the cell fails to re-enter mitosis in a reasonable time (several hours), it has likely been over-illuminated. However, since even changing the culture media can also induce a transient decondensation of condensing chromosomes, it is important to wait several hours after the construction of the preparation, before conducting this test.
Another important but often overlooked consideration is the wavelength (color) of the light used for illumination. The 546-nm illumination used in live-cell studies has become a standard simply because it matched one of the major spectral lines of mercury arc lamps, thus allowing for efficient use of this light source. However, since the intensity of the light source is not a concern in modern live-cell imaging (all source types produce ample light), this justification is now moot. Similarly, most modern lenses are well-corrected chromatically and thus do not require the use of green light for high resolution live-cell imaging. As a result, the wavelength of light that is the least deleterious to the specimen should be used. Unfortunately, as emphasized by Brakenhoff et al. [39], “The main problem when one tries to make an evaluation of the relative damage-inducing potential of radiationf in a biological object is that few hard facts are available.”
We (R.W. Cole and C.L. Rieder, unpublished) have used the illumination-induced reversion of chromosome condensation in mid-prophase cells as an assay for determining how well PtK1 cells tolerate various wavelengths of light. The conclusion from these studies is that, during prophase, cells have little tolerance for UV and infrared (IR) illumination, and are the least sensitive to red, followed by green and blue, in the visible spectrum. This is consistent with the data of Manders and co-workers [37] who, in a confocal LM study on chromosome condensation during G2, found that vertebrate cells (e.g., Indian muntjac, human fibroblasts, and HeLa cells) tolerate red light (647 nm) better than green (568 nm) or blue (488 nm). Thus, from the biological standpoint it is reasonable to use red (600–650 nm) light for live-cell observations. Unfortunately, there are several factors that can force a compromise on this point. First, because resolution depends on the wavelength of light used, red light is the worst possible choice for achieving high resolution, due to its long wavelength. Nevertheless, since the resolution limit of the microscope is rarely an issue in live-cell imaging, this point will be germane only to certain special cases. This is because, when filming a dividing cell, the resolution is limited by the internal movements in the cell, temperature drifts, and imperfections in the optics and illumination systems. Also, in multi-mode instruments, where trans-illumination (DIC or phase contrast) is combined with fluorescence, it often makes the most sense to match the wavelength of the trans-illumination to the wavelength dictated by the fluorophore. This eliminates the need for multiple band-pass dichroic mirrors which are always less-efficient than simple single-pass ones. Finally, many of the modern charge-coupled device (CCD) cameras are optimized for green fluorescence protein (GFP)-imaging and their sensitivity suffers significantly in the red portion of the spectrum. This limitation will likely be overcome in the near future, as cameras are developed that are more homogeneously sensitive across the visible spectrum, but for now it remains an important practical consideration.
In addition to choosing the correct wavelength, care should be taken to prevent contamination of the illuminating light with even trace amounts of UV and IR. While modern band-pass filters are generally quite good in the middle of the visible spectrum, they still tend to pass some irradiation at the extremely low and high wavelengths. Therefore, it is helpful to install inexpensive glass filters, such as GG400 (anti-UV) and KG5 (anti-IR), in the light path. The use of these additional filters is particularly important when Mercury arc, or to lesser extent, Xenon arc, lamps are used, since these produce very high amounts of UV.
While choosing the appropriate illumination wavelength can reduce photo-damage, it must be emphasized that high-intensity light of any color is inherently deleterious to live cells. Whether or not the wavelength is optimized, the light must be shuttered at all times except during actual image acquisition. Shuttering of the light used in every illumination path (trans-illumination, fluorescence excitation, etc.) is, without a doubt, the single most important factor in live-cell imaging. To achieve seamless coordination between exposure of the cells to light and image acquisition, all shutters must be electronically controlled. As a result, the question of how many shutters a certain type of software supports must be considered, when a live-cell imaging system is being constructed. Another important consideration is how fast the shutters used in a particular system operate. Here it is noteworthy that some commercial instruments perform shuttering via a blank position on a filter wheel, instead of by true fast (7–8 μs) shutters, which can add an extra 100–200 ms of light exposure to every frame recorded. While the absolute duration of the exposure does not appear too dramatic at face value, under average recording conditions the extra 100-ms acquisition time inherent in filter-wheel shuttering effectively doubles a cell’s exposure to light, which makes the system 100% less efficient than if a true μs-speed shutter were used.
As a rule, manufacturers design microscopes as stand-alone instruments that are completely self-contained and universally suited for all potential applications. However, for a particular application, many of the features embedded in the design are useless or even degradative. For example, a problem often encountered during work with fast shutters and/or filter wheels is that these devices produce significant vibrations. When attached directly to the body of the microscope, they induce vibrations that can last for hundreds of milliseconds. This, in turn, decreases the resolution of the recorded images. This problem is common in modern research-class microscopes, which contain many motorized parts and components. To restrict these vibrations manufacturers often limit the speeds of the attached devices, or introduce delays between, e.g., changes in the filter wheel position and the actual acquisition of the image. Needless to say, these delays at best decrease the performance of the system, and at worst expose the cell to unnecessary light.
From experience, we find that mounting the lamp-housing, filter wheel, and shutter assembly on a separate stand, external to the microscope, eliminates vibrations and allows for a no-delay synchronization between the operation of the filter wheel and image acquisition. This approach is also cost effective because it eliminates the need to purchase the mounting kits and flanges that are normally required to couple the filter wheel, burner, etc., to a particular model of microscope. Instead of purchasing these accessories (often ~$300–500), we simply buy steel rods (~$20–30 from companies like ThorLabs, Oriel Instruments, etc.), with which we mount all vibrating devices directly onto a vibration-isolating table (Fig. 2). The biggest problem with this design is in convincing the salesman that we do not need the “essential” parts that he wants to sell us. Obviously, a prerequisite for our solution is that a vibration-isolating table, with threaded mounting holes, be available to mount the microscope on. We consider the table to be essential for true high-resolution imaging.
Fig. 2.
Vibrations generated by shutters and filter wheels can be eliminated by mounting such devices external to the microscope chassis. In this example, the shutter/epi-light source assembly and filter wheel (bottom), as well as shutter/trans-illumination source assembly (top), are all mounted onto a vibration-isolation table using stainless steel rods. The whole assembly is disconnected from the microscope body, which in turn rests on the same vibration-isolation table.
By eliminating the delays normally required for vibration dampening, and by fast shuttering, we have been able to double the practical image acquisition efficiency of our microscopes. While a 100-ms exposure actually requires about a 250-ms light exposure using the original turn-key system, through our customization we have reduced this to ~112 ms. As a result, we can now record twice as many images of a specimen—a significant gain in research on mitosis. However, even this improvement may not always be sufficient to eliminate photodamage, particularly in studies in which high temporal resolution or significant number of individual Z-planes are required.
In some instances, configuring the imaging system for minimal photo damage can be complemented by a “biological” approach, which entails overriding the checkpoint pathways that delay cell-cycle progression in response to the damage. For example, the DNA damage checkpoint pathways can be blocked by treating cells 5–10 h prior to observations with caffeine (~4 mM), wortmanin (~5 μM), or UNC-01 (~5 μM). The first two drugs inhibit the ATM kinase, positioned near the beginning of signal transduction cascade [40,41] without apparent effects on progression through the cell cycle [23]. Alternatively, UNC-01 inhibits the Chk1 kinase that is downstream from ATM, and is required for maintaining the checkpoint [42]. We have found that early-prophase cells, present in PtK1 cultures that have been treated with 5 μM caffeine for 12 h, proceed into mitosis when the treatment is followed by doses of illumination that would be sufficient to induce a reversion in the absence of caffeine [23].
On the other hand, an illumination-mediated arrest of the cell cycle is not necessarily caused by DNA damage. Additional pathways exist in cells, involving, e.g., the p38 kinase, that can block cell cycle progression in response to other forms of stresses (reviewed in [24,33,43]).
Finally, because of the sensitivity of the G2/M transition to illumination, it may sometimes be prudent to simply wait until the cell has become committed to the mitotic process before initiating observations. This is especially true for high-resolution studies, in which the light intensity and framing rate must necessarily be high. As a rule, for most cell types, the commitment to mitosis occurs as the nucleoli begin to fade, several minutes prior to nuclear envelope breakdown.
2.2. The growth or viewing chamber
Almost every laboratory that follows living cells with the LM has their own custom-designed viewing chamber, some of which are commercially available (e.g., search the Web via Google for “cell culture chambers” or “viewing chambers, cells”). In general, these chambers are designed to maintain specimen viability while at the same time providing optical properties optimal for LM. The type of chamber used will depend on several factors, including the temporal and spatial resolutions required for the study, as well as its duration. High-resolution video-enhanced (DIC or phase-contrast) LM requires that the NA of the condenser be matched to that of the objective lens. This means that, to achieve full resolution with a 60× 1.4 NA lens, a 1.4 NA oil-immersion condenser must be used. Unfortunately, the maximum working distance for this type of condenser is limited to less than 1.5 mm, which obviously limits the depth of the observation chamber.
In general most chamber designs for viewing cells on a microscope stage are based on the sandwiching of two cover-slips, separated from one another by a spacer, between two metal or plastic plates (reviewed in [5,44]). Because the thickness of the chambers used for high-resolution LM is usually limited to approximately 1 mm, they hold very small volumes of fluid, often only 50–250 μl. Depending on the type of cell this may not be a problem even for long-term studies. For example, cultures of amphibian tissues, which grow very slowly and at room temperature, remain healthy in sealed perfusion chambers that hold only 250 μl for up to 2 days without a change of medium [44]. By contrast, in the same chambers, mammalian cells, which need to be maintained at 35–37 °C for maximum growth, must be perfused with fresh medium every 20–40 min [45]. The requirement in some high-resolution studies to frequently replenish the media has led to the design of various chambers in which a continuous flow of fluid is maintained across the cell surface [46,47].
On the other hand, if the goal of the study is to collect high-resolution epi-fluorescent data, without the need for corresponding high-resolution transmitted LM information, thicker chambers can be used, because a condenser is either not required, or can be of a low-NA, long-working distance type. For such studies, the easiest way to maintain cells on the microscope long-term is to use tissue-culture dishes with 170-μm glass bottoms (available from several manufacturers, including World Precision Instruments). These dishes allow for easy changes of growth medium and also proper gas exchange. They can support normal growth of most mammalian cell types indefinitely.
For routine long-term (1–4 days) studies requiring high-resolution fluorescence, but lower-resolution transmitted LM, we use a modified version of the Rose chamber (Fig. 3; detailed in [48]). This closed chamber holds ~1 ml of medium which, when the medium is changed every 48 h, is sufficient to promote the exponential growth of vertebrate cells at 37 °C until the culture becomes confluent. Because the chamber is closed, the pH cannot be controlled by the usual NaHCO2/CO2-buffering system, so a CO2-independent medium should be used instead, such as Leibovitz’s L-15. Alternatively, conventional (e.g., MEM) media can be used after its buffering capacity is enhanced with Hepes (~10–20 mM). It is noteworthy that the phenol red dye, usually used as a pH indicator in the media, is highly light-absorbent in the visible spectrum and can easily photo-sensitize cells, making them more susceptible to photodamage during imaging. For this reason, we usually conduct fluorescence imaging in phenol red-free media, which is commercially available from numerous companies (e.g., Gibco/Invitrogen, Carlsbad, CA, USA).
Fig. 3.
We maintain cells for long-term microscopic observations in modified Rose chambers. A side view of the fully assembled chamber is seen in the middle, just above the 3″ (8 cm) mark on the ruler. The chamber is constructed from two 25 mm2 coverslips (top left and right corners), a silicon spacer (middle, above assembled chamber), a metal planchet milled to accept a condenser lens (left side, middle), and another planchet milled for objectives (right side, middle and bottom). The whole assembly is held together by four screws (bottom left). The chamber can be filled and drained using two 25G needles and a syringe. It can also be constructed using different objective planchets depending on the viewing conditions. For high-resolution oil-immersion work the top planchet, which is more extensively milled and thinner, is used (see also [48]).
During an experiment, it often becomes necessary to temporarily remove the culture from the microscope stage, e.g., to feed it, or in some cases to switch it to another microscope. This introduces the problem of relocating the same cell and/or field of view that was previously under observation. As a rule, relocation is done by first making fiducial marks on the coverslip. We usually draw a circle around the cell with a diamond-tipped objective scribe (approx. $1500 from Carl Zeiss, Cat. No. 462960), which is mounted on the microscope nosepiece adjacent to the objective used for the study. This scribe is designed to scratch a circle of a variable but definable diameter (20 μ–3 mm) onto the surface of the coverslip above the cell, so that the center of the circle contains the field of view [49]. Other methods for re-locating cells employ “marker” coverslips that can be constructed, e.g., from EM finder grids (www.borisylab.nwu.edu/pages/protocols/electmicrosc-text.html), or purchased from ProSciTech (Thuringowa Central, Australia; Product No. G-490-G491; www.proscitech.com.au/get_frames.htm?20.htm) or Bellco Glass (Vineland, NJ, USA; Stock No. 1916-92525; www.bellcoglass.com/us/1916-92525.htm).
2.3. Temperature control
Not surprisingly, the rate at which cells proceed through the cell cycle and mitosis is extremely sensitive to temperature [28]. For example, a temperature drop from 37 to 33 °C doubles the generation time of HeLa cells [50], while shifting mouse leukemia cells from 37 to 28 °C prolongs the cell cycle 7-fold [51]. While mitosis in most mammals (when defined from nuclear envelope breakdown to anaphase onset) takes ~25–30 min at 37 °C, it requires almost an hour at 33–34 °C. Surprisingly, at 33–34 °C, nocodazole or colcemid arrests PtK1 and other vertebrate somatic cells in a mitosis for ~8 h, before they leak through the block and enter G1 [52,53]. However, at 37 °C, the block only lasts for 3–4 h (S. La Terra and C.L. Rieder, unpublished). Clearly, any imaging study with a goal that requires collection of data on the duration of a mitotic event or the cell-cycle progression must carefully consider temperature control.
By far the simplest and best way to maintain a specimen on a microscope stage at a specific temperature is to keep the whole assembly in a room that can be adjusted to the desired temperature. For our studies on mammalian cells, we keep two phase-contrast microscopes in a 37 °C warm room, and we have experienced no problems with the computers, video cameras or the electronics. On the other hand, it may be impractical to dedicate a whole thermo-stable room to microscopes. As an alternative, enclosures can be built (e.g., from Plexiglas) that entirely surround the microscope but not the peripherals. The space inside the enclosure can then be maintained at a specific temperature with a blower or hair dryer, mounted well away from the microscope stage, that is controlled by an electronic feedback circuit (Fig. 4; see also [44]). We maintain the temperature in this manner for several microscopes, and they give similar results to the microscopes housed in the warm room.
Fig. 4.
Enclosing the microscope and some of its peripherals in a plexiglass box allows for precise control of the specimen temperature during live-cell imaging. The temperature inside of this box is maintained by a heat blower (bottom right hand corner of image), positioned well away from the specimen stage, which cycles on and off in response to a thermistor positioned near the specimen. Note that the oculars protrude from the box (but are sealed by cotton), and that focusing can be done externally. See text for details.
Finally, for high-resolution studies, we sometimes use a microscope at ambient (23 °C) temperature, but with the specimen temperature maintained with a (Rose) chamber heater (Fig. 5). Most modern stage heaters employ Peltier devices that can maintain specimens at any temperature, from below 0 to 100 °C, for many hours. They can either be built at little cost (see design in Rieder and Cole [36]) or purchased commercially (e.g., the 14417 heated-stage insert; WPI, Sarasota, FL; www.wpieurope.com/microscopy/HSI.html, or several different stages from 20-20 Technologies; Wilmington, NC; www.20-20tech.com). The major problem with this approach is that the objective lens, and to a lesser extent the condenser, both act as large radiant bodies to cool the chamber, especially when oil-immersion lenses are used. This problem can be alleviated by the use of special objective heaters, available from the same companies that offer stage heaters. However, some of the modern lenses, particularly the newest 60× 1.4NA, are incompatible with these devices due to large diameter of the lens barrel and the geometry of the lens tip. In practice, we find that, if we keep the room temperature at 23 °C and our block heater at higher temperature (41 °C), the temperature of the cells directly under an oil immersion lens remains relatively constant at ~35 °C. The particular temperature shift between the heater and the center of the coverslip can be determined by inserting a thermo-probe into the chamber, just under the objective. This approach to heating is, however, very sensitive to drafts generated by open doors and air-handling systems in the building. These fluctuations can be minimize placing a cardboard box (with aluminum foil glued over it to prevent dust accumulation) over the microscope, with one side left open. When the microscope is in use, this side can be sealed with aluminum foil with the eyepieces protruding (Fig. 6). Such an enclosure is also useful because it isolates the microscope from external light sources.
Fig. 5.
(A) Peltier-based heater to keep the viewing chamber at a desired temperature (also see [44]). This heater, in which our modified Rose chamber is mounted, can be firmly attached to the microscope stage by clamps (B).
Fig. 6.
An overview of a thermal-stable microscope that we use in our research. The microscope proper is mounted on a vibration-isolation table and is equipped with filter wheels, multiple shutters, and a Rose-chamber stage heater. All of these devices are covered by a cardboard box (covered with aluminum foil), to shield the entire assembly from airflow and light. Electronic controllers for all of these devices are placed outside of the microscope to prevent their overheating. The whole system is driven by a workstation (on the left) that runs image-acquisition software (in this case Isee, Isee Imaging, Raleigh, NC, USA).
There are several live-cell viewing chamber systems on the market that are designed to simultaneously maintain the temperatures of both the viewing chamber and the objective lens. One of the more popular is the Bioptechs Focht Chamber System 2 (FCS2; available through ASI, Eugene, OR; USA; www.asiimaging.com/index.html). This is a versatile, high-resolution closed perfusion system, compatible with all types of LM, that uses resistance heating to maintain the temperature of both the specimen chamber and the objective. Another is the DSC200 Dvorak-Stotler controlled environment culture chamber, which uses an ASI 400 air stream incubator to warm both the objective and the specimen chamber (available through Nevtek; Burnsville, VA, USA; www.nevtek.com/incubatr.htm). Although we have had no experience with the former system, we have used air stream incubators in the past. Whereas they do maintain the temperature of the specimen within a few degrees, the coverslip often “bounces” between the “on” and “off” cycles which produces constant and distracting changes in focus.
3. Choosing the appropriate imaging system and peripherals
While the use of high light intensities does not necessarily pose a risk when fixed cells are observed, it is imperative to avoid over-sampling when working with living material. As noted at the outset, live-cell microscopy always represents a compromise between achieving the best image quality and preserving the health of the cell. This means that the spatial and temporal resolutions should always be matched to the goals of the study. For example, if a moving object (e.g., a chromosome) is followed by time-lapse microscopy, there is no need to capture images more frequently than the duration it takes the object to systematically shift by 2–3 pixels in the image (for the ~2 μm/min that vertebrate chromosomes move, this is ~15 frames/min at 60× with a full-resolution camera; see below). Similarly, it is rarely beneficial to collect Z-series of living cells at steps smaller than 0.5–0.75 μm, even for 1.4 NA lenses (see below).
Regardless of the manufacturer, all major brands of microscopes are capable of live cell imaging; and while various brands may have certain advantages for specific applications, no one of them can be considered to be superior to the rest of the pack. When a microscopy system is being configured, it is important to realize that off-the-shelf research microscopes are usually over-designed for most applications. The incorporation of intermediate lenses, fancy multi-band dichroic mirrors, beam-splitters, and other similar devices decreases the transmission efficiency of the system, which in turn forces the researcher to use higher intensities of excitation light. As we have emphasized above, this is deleterious to the specimen. Therefore, it is important to configure the microscope to match the imaging needs of the study.
3.1. Objective lenses
If a microscope is to be optimized for an application, it is first necessary to understand how the brightness and resolution of an image are defined. The theory of image formation is beyond the scope of this review and can be found elsewhere (we strongly recommend comprehensive, and yet remarkably easy to understand, lectures by Dr. José-Angel Conchello, Washington University, St. Louis, MO; rayleigh.wustl.edu/~josec/tutorials/). Nevertheless, we need to introduce a couple of very basic equations that help to explain the fundamental characteristics of a microscope. The most important parameter describing the capability of an objective lens is its NA. The NA defines how much light from a single point-source can be gathered by the lens. Mathematically, the NA is expressed as NA = η sin α, where η is the refractive index of the medium, and α is the angle of maximally diffracted light rays that still contribute to image formation. Since sinα can not exceed a value of 1, it is obvious that the NA can not be greater than the value of the refractive index of the medium between the object and the lens.
The NA directly defines two important features of the lens. First, it determines the ultimate optical resolution of the system. According to the Rayleigh criterion (which is very conservative) resolution is equal to 0.61λ/NA (where λ = the wavelength of the rays of light). Second, the NA defines the intensity (brightness) of the image, which is proportional to the fourth power of the NA, and inversely proportional to the second power of the magnification. As a direct consequence, a given fluorescent object should appear almost three times brighter when imaged with a 60× 1.4 NA lens than when imaged with a 100× 1.4 NA lens. However, the intensity of the object is also significantly affected by variations in the transmission quality of the lens. Modern multi-color fluorescence and multi-mode microscopy requires that lenses are well corrected for chromatic aberration throughout the spectrum (apochromatic) and have a flat imaging plane (“plano” lenses). Unfortunately, apochromatic and plan-apochromatic lenses, as well as those lenses that are designed to capture a wider field of view, inevitably contain more optical elements and, thus absorb more light then do other less precise lenses (achromatic lenses and lenses with fluorite elements). This, in turn, means that a plan-apochromatic 60× 1.4 NA lens, which theoretically should be 2.8 times brighter than a 100× 1.4 NA lens, is in practice only about twice as bright. Nevertheless, the 60× 1.4 NA lens still provides the brightest image achievable at the limit of optical resolution.
3.2. CCD cameras
We cannot evaluate here the plethora of CCD cameras available on the market. This technology is developing very rapidly, and improved models will no doubt be available by the time that this volume appears. As for microscopes, all major brands provide cameras that produce very high quality images. However, there are several important features of a CCD camera that should be considered when a camera is being chosen for live-cell work.
It may not be apparent to an inexperienced microscopist why two different cameras that use the same CCD chip can differ in price by as much as 100%. Intuitively, it would seem that all cameras that use the “right” chip should be equally sensitive and capable of providing the same image quality. Actually, this perception is not too far from reality, if the camera is used for acquiring images from static fixed-cell preparations. However, for live-cell image acquisition, the situation is dramatically different.
A fundamental difference between fixed- and live-cell imaging is that, when working with the former, the researcher has ample latitude in defining the image acquisition time, electronic gain setting, read-out time, and other relevant imaging parameters. As a result, it is always possible to acquire an image that uses the full dynamic range of the camera and, thus, that has the best signal/noise ratio. Unfortunately, this is not the case when working with live cells, because the imaging parameters are dictated by the necessity to provide conditions that are safe for the cell (see first part of this chapter). In some situations this means that the intensity of the object of interest, in the recorded images, is only a few counts (grey levels) higher than the background intensity. Under this condition, the most important consideration becomes the “quietness” of the camera’s electronics and the precision of the charge read-out. Less precise electronics in the less expensive cameras lead to more electronic noise, which in turn leads to a less uniform background image. As a result, a low-intensity object will not be discriminated as readily from the background, producing an apparent loss of sensitivity. Similar effects can be observed when the same camera is used at two different readout parameters (Fig. 7). Therefore, you must pay more for a “quiet” camera, with more sophisticated electronics, than for a less quiet camera that utilizes the same CCD chip.
Fig. 7.
The dependence of camera sensitivity on the readout speed. In this example, the images of a live PtK1 cell, expressing α-tubulin/GFP, were recorded on an Orca II camera operated in “fast” (10 MH; top row), and “precise” (1.25 MH; bottom row) modes. As evident from comparison of (A and A′), the background noise (no light to the camera) is dramatically lower when the slow readout speed is used. This difference does not affect camera performance when the exposure time can be adjusted to use the entire dynamic range (cf. B and B′). However, under identical conditions, but in a light-limiting situation, slow (precise) readout provides better image quality (cf. C and C′ ~10 times less light than in A and A′). The difference becomes even more dramatic under extremely low light conditions (cf. D and D′, ~5 times less light than in B and B′) when slow readout provides an acceptable image while the fast-readout mode does not.
Another important consideration for live-cell imaging is to properly match the objective lens with the detector (camera) resolution. Since a CCD detector is formed by an array of wells, the size of the well can limit the final resolution of the image. Theoretically, to achieve full resolution you should have 3 pixels for each Airy disk (Nyquist sampling criterion). This means that, for an optical resolution limit of 250 nm (1.4 NA lens), the pixel size in the image should be ~83 nm. In live-cell imaging, however, the Nyquist criterion can be reduced to ~2.0–2.2 pixels per Airy disk without noticeable degradation of the image. Reducing the number of pixels per Airy disk helps increase the brightness of the image, since larger CCD wells accumulate more charge. Thus, in practice the ideal pixel size for a 1.4 NA lens is ~100–120 nm. For translation of this number into the appropriate physical size of the CCD wells, the magnification factor of the lens must also be considered. For example, for a 60× 1.4 NA objective lens, the full image resolution will be reached at a well size of 6 μm (60 ×0.12 μm = 7.20 μm). However, for a 100× 1.4 NA lens the non-limiting CCD well size is 10 μm. These simple computations reveal that each CCD camera performs best when it is properly matched to an appropriate lens. For example, use of a 10-μm well CCD camera with a 100× lens will result in a 100-nm pixel size, which is well within the non resolution-limiting range. However, the same CCD camera will yield 167-nm pixel size when used with a 60× lens, and thus will somewhat limit the resolution of the recorded images (see [54] for example of under-sampling image degradation). Conversely, a 6-μm well CCD camera will be perfectly matched to a 60× lens, but will result in over-sampling when images are recorded with a 100× lens. Such over-sampling will severely decrease the brightness of the image, while not improving resolution.
The only “perfect” solution for matching the optics with the pixel resolution is to use different cameras for different lenses. However, some sort of compromise must be found for those of us who cannot afford to purchase multiple CCD cameras for each microscope. In our opinion, the most versatile CCD cameras are those with 6–7 μm wells and on-chip binning. The binning feature allows several neighboring pixels (4 for bin 2, 9 for bin 3, etc.) to be read as a single “super-pixel,” thus increasing the effective size of the well. We usually use the full resolution of the CCD camera when imaging with a 60× lens (100–115 nm pixels), and bin 2 for a 100× lens (120–140 nm pixels). This approach allows us to achieve practically full resolution with the brighter 60× lens. Additionally, since 100× 1.4 NA lenses are approximately half as bright as 60× 1.4 NA lenses (see above), and given that a binning factor of 2 increases sensitivity of the camera 4-fold, the images recorded with 100× at bin 2 are approximately twice as bright (and of almost the same resolution) as are the images recorded with 60× and no binning. Finally, when we need to significantly increase the sensitivity, we use a 60× 1.4 NA lens and bin 2 on the camera. Under these conditions the image suffers certain loss of resolution (pixel size 200–230 nm). However, the brightness is increased 4-fold over the combination of 60× 1.4 NA lens and no binning, and approximately 2-fold over the combination of 100× 1.4 NA lens and bin 2. Additionally, the same equipment allows us to record true full optical-resolution images by using no binning with the 100× 1.4 NA lens (60–70 nm pixels).
3.3. Improving image quality
One disadvantage of high-NA lenses is that their depth of focus is very shallow (~500 nm for 1.4 NA lens). This means that, at any given time, the lens is only at focus in a thin slice of the cell’s volume. The light coming from other levels in the cell, outside of the focal plane, does not contribute constructively to image formation. Worse, this out-of-focus light or “blur” contaminates the in-focus image and decreases its contrast and resolution.
A critical advance to the study of mitosis in living cells has been the development of fluorescence-based “multidimensional” imaging. It is now possible to record a series of individual focal plane images, by automatically re-focusing the objective lens, so that it “steps” along its Z-axis. The resulting (Z) series of images contains in-focus information on all intracellular structures and can be used to create a single (maximum-intensity) “projection” image that represents the entire cell volume in 2-D. The problem, however, is that this image also contains significant amount of blur, which often makes the structure of interest unrecognizable in the projected image. This blur can be reduced in two ways: one involves capturing the images via confocal LM systems, while the other uses mathematical computations to allow out-of-focus photons to be reassigned to their points of origin.
3.3.1. Use of confocal LM
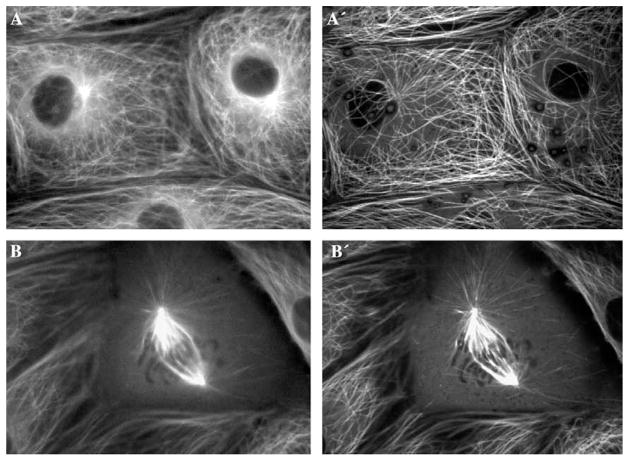
The most common method for reducing the contributions of out-of-focus light is to obtain fluorescent images using a confocal microscope. In confocal LM, the fluorophores within the specimen are excited only in a diffraction-limited spot, and the out-of-focus photons are then rejected by a small aperture in the image-conjugated plane [55]. As result, the image contains information only about those fluorophores that reside in one focal plane. Although confocal LM was the first practical method for obtaining 3-D fluorescent data sets of living cells, similar data sets can now also be obtained through the use of wide-field deconvolution techniques. One widely held but erroneous belief is that confocal microscopes, when compared to conventional wide-field systems (which rely on simultaneous excitation of all fluorophores in the field of view), achieve superior resolution. In fact, in both cases, the true optical resolution is limited by the same diffraction principles, and the apparent crispness of confocal images reflects an improvement in contrast rather than increase in the resolving power [56]. There are many advantages to confocal microscopy which have been detailed elsewhere [55]. The point that we wish to make here is that, for imaging the division process in somatic cells in tissue culture, wide-field microscopy often performs on a par with, or even better than, confocal microscopy (Fig. 8).
Fig. 8.
A comparison between wide-field fluorescence (A and B) and spinning-disk confocal (A′ and B′) images of interphase (A and A′) and mitotic (B and B′) cells expressing α-tubulin/GFP. As is clear from the comparison, confocal imaging provides a much clearer picture of how microtubules are distributed in interphase cells (cf. A and A′). However, the improvement in image quality is not as dramatic when mitotic spindle is imaged (cf. B and B′). Note that the confocal images presented here were recorded at approximately 75% higher excitation light intensity, than were the wide-field fluorescence ones.
Early confocal microscopes suffered from several features that restricted their use for live-cell studies of cultured cells. Although technological advances have mitigated some of these concerns, others remain. As an example, because images in the confocal microscope are formed by scanning the field with a diffraction-limited spot, the time required to generate each image is inevitably longer than in wide-field LM. This problem was especially severe in the older, single flying-aperture confocal microscopes, although it has become much less of a problem in the newer systems, and practically is non-existing in the spinning-disk confocal type. However, a persistent limitation of confocal technology is its inefficient use of light: even in-focus light in a confocal system must pass through a small aperture, which inevitably makes its transmission efficiency lower than in wide-field systems. Although this loss can be minimized, it will never be eliminated. As a result, a much higher intensity of excitation light must be used for confocal LM, which increases the potential for photodamage to the specimen.
We prefer to study mitosis in cultures of live somatic cells by wide-field LM. Not only does this approach require less light, but image restoration techniques are now available that improve the quality of 3-D wide-field data sets to a level similar to, or even exceeding, that of confocal LM (see [57] for a detailed comparison). It is important to note, however, that confocal LM still has an important niche in the study of cell division in vivo, since it remains the approach of choice for obtaining 3-D data sets from thicker specimens, including, e.g., Drosophila and Caenorhabditis elegans embryos. In these systems the use of confocal microscopy is particularly reasonable since the embryos themselves tend to also be very tolerant to high light intensities.
3.3.2. Use of image restoration (deconvolution)
The alternative method for reducing the amount of blur in fluorescent imaging is to reassign each photon back its point of origin. This approach is known as “deconvolution,” and it can be universally used in combination with wide-field and/or confocal fluorescence imaging. Unlike confocal imaging, deconvolution can actually surpass the diffraction-limited resolution and can resolve structures as small as ~100 nm. It should be emphasized, however, that a significant difference exists between true iterative deconvolution and simple image filtering. Deblurring based on filtering, which is often marketed as “deconvolution,” is much less expensive computationally and monetarily. Although it is fast, filtering requires relatively little information about the optical properties of the imaging system. As a result, it seldom provides the same quality of image restoration as does true iterative deconvolution.
All iterative algorithms operate in 3-D space and require that the point spread function (PSF) of the optical system be determined or calculated. The three major deconvolution approaches are defined by the way in which the PSF is determined (we recommend the lectures, noted previously, by José-Angel Conchello for a comprehensive comparison of the different algorithms). The earliest LM deconvolution approach was that of Agard and Sedat [58], and is now marketed by Applied Precision Inc. (Issaquah, WA, USA; http://www.api.com/) as the DeltaVision system. It involves fully optimizing the microscope and recording the best achievable PSF, which is then used for all further computations. An alternative method is to determine the individual PSF for each sample, using fluorescence beads added at the time of specimen preparation. This technique was developed by Carrington and colleagues [59] and is now marketed by Scanalytics (Fairfax, VA, USA; http://www.scanalytics.com/index.shtml). Finally, there is the so-called “blind” deconvolution method developed by Holmes and co-workers [60], which employs the mathematical restoration of both the PSF and the object (based on maximal likelihood of the result). This system is available through several companies, including AutoQuant Imaging (Watervliet, NY, USA. http://www.aqi.com/).
Since none of the existing deconvolution algorithms reigns supreme, we recommend testing several systems on your own data sets before investing in one product. There are multiple factors that affect the performance of deconvolution, any one of which can skew the choice toward one of the software packages for your particular set of problems. The geometry of the object is important (some systems work best for continuous objects while others make a microtubule look like beads on a string), as is the signal/noise ratio in your data sets. Also, there are several practical considerations that need to be weighed when a deconvolution software vendor is chosen. The difficulty entailed in exchanging the information between the deconvolution software and your image-acquisition package(s) is an important factor and should be considered. In most cases, plain images can be easily imported/exported via the standard “import/export as TIFF” option. However, this results in the loss of all “meta-data”—those parameters of image acquisition (pixel size, exposure time, wavelength, imaging mode, etc.) that are often essential for deconvolution. Needless to say, it requires significant effort to manually preserve (re-input) these data every time an image is transferred between the deconvolution and image acquisition software. Another practical consideration is whether the nature of your work requires that the PSF be re-collected for different types of preparations, or whether it supports the assumption that the PSF of your system does not vary strongly among different experiments. If the microscope is well-optimized and stable, and if the quality of your preparation is consistently high, one PSF can often be used successfully for all data sets recorded with the same lens. The success of the DeltaVision microscopy workstations, which represent the most highly optimized commercial light microscopes to date, validates this approach. In this class of integrated microscopy systems, we also recommend considering an Intelligent Imaging Innovations workstation (Denver, CO, US; www.intelligent-imaging.com/home.php). Both of these systems perform remarkably well for many applications and can fulfill most imaging needs for ~$200K.
Those with restricted budgets can nevertheless deconvolve data-sets that have been recorded on less-than-perfect microscopes. Under these conditions, it often makes sense to collect an individual PSF for each preparation. The individual-PSF approach to deconvolution is by no means inferior to the Agard and Sedat approach, and the results can be equally impressive [61]. This approach does, however, double the amount of work needed for analysis of each preparation, because it requires that an individual PSF be collected for each experimental data set. Finally, image restoration can also be achieved by blind deconvolution methods. As noted above, the PSF in this approach is not determined experimentally, but is instead reconstructed concurrently with restoration of the real data set. The fact that blind deconvolution does not involve characterizing the PSF before an experiment makes it an attractive method, particularly for laboratories in which image restoration is used only occasionally. However, it requires several times more computing power than do the experimental-PSF methods, and hours may be needed to restore an average-size data set. As computers become faster and cheaper, and the mathematics behind blind deconvolution methods improve, this will likely become the method of choice for biological microscopy.
3.4. Hardware requirements for multi-dimensional imaging
Assembling a fully automated and optimized multidimensional imaging system is a complex task. Despite the hefty price tag of user-friendly commercial systems, including the DeltaVision system, they are often well worth the investment, especially for a multi-user core facility. Alternatively, it is possible to reduce costs by as much as 50% by assembling a system in-house, but this can only be done in laboratories that possesses suffcient expertise and experience in LM. Few investigators can simply buy the peripheral devices and then attach them to a microscope to instantly form a high-quality multi-mode or multi-dimensional fluorescence workstation. The major problem here is in integrating the various hardware and software components into a well-coordinated, efficient system. Below, we briefiy discuss some of the elements that need to be considered when either choosing a turnkey commercial instrument, or when assembling a system in-house.
One item critical to all multi-color fluorescence systems is a device for switching between different wavelengths of light. Currently, there are two practical ways to do this. The first involves filter wheels. Filter wheels are reliable, relatively inexpensive, and are supported (i.e., driven) by a large number of imaging programs. Their primary disadvantage is in speed, or lack of thereof. It takes ~100 ms to switch between positions on a standard filter wheel, and ~25 ms to switch for “fast” ones. Although this sounds rapid, at 2 wavelengths it adds at least 4 s to a Z-series of 20 sections. Another problem is that filter wheels create mechanical vibrations that can degrade the image quality. As noted previously, this problem can be largely eliminated by mounting the wheel independently of the microscope (Fig. 2).
The alternatives to a mechanical filter wheel include monochromators (e.g., Till Photonics Polychrom IV; Gräfelfing, Germany; http://www.till-photonics.de/home_e.htm) and optical changers (e.g., the DG4; Sutter; Novato, CA USA; http://www.sutter.com/). These devices, which are more expensive than filter wheels, use galvanometer-mounted mirrors to provide very rapid (several microseconds) switching between different wavelengths. Importantly, their action can be synchronized with that of the camera, and the rest of the peripherals, via TTL pulses easily generated via the computer’s parallel port. With these devices it is possible to collect a full two-color Z-series through a mitotic cell (15 sections) in <2 s. Since they are mounted externally they do not affect the stability of the system.
Most high-quality automated multi-color microscopy systems use complex multi-band pass dichroic mirrors instead of a set of individual single pass mirrors. For true multi-colored imaging this approach is justified, as it prevents shifting between the images recorded at different wavelengths. However, the use of these complex mirrors is so common nowadays that many researchers continue to use them even while working with cells labeled with only one fluorophore. This is despite of the fact that they are less efficient than are the simpler individual-pass mirrors. We find that the exposure time for GFP-labeled cells can be reduced by as much as 200% on the same workstation when an Endow GFP filter cube is used instead of the standard (on DeltaVision systems) quadruple-pass filter set (both from Chroma Technology, Brattleboro, VT, USA; http://www.chroma.com/). The take home message here again is to optimize the system for the task at hand.
To collect a Z-series, the objective must be “stepped” through the specimen at precise intervals. One way to do this is to use a stepping motor (e.g., Ludl, or Prior) to drive the microscope’s focusing knob; which, depending on the microscopes design, then translates the entire nosepiece or the microscope stage. Alternatively, a piezoelectric device can be attached to the nosepiece so that the objective lens moves up and down. The advantages of a stepping motor system include its lower cost and virtually unlimited travel distance. Additionally, since these motors work through the microscope’s own transmission, they allow one to use all lenses in the revolving nosepiece and switch them rapidly. The disadvantage is that it is slower than piezoelectric devices and has greater hysteresis. Because of their precision and speed piezoelectric devices are, as a rule, the approach of choice for collecting fast Z-sequences. However, they have a limited travel distance (~100–200 μm) and are generally more expensive than a stepping motor system.
Other factors may warrant consideration in the decision on a method for collecting Z-series. Piezoelectric devices, for example, block at least two positions (and sometimes three) in the nosepiece, and the lens attached to the device is raised 2–3 cm from its original position. In practice this means that the stage on an inverted microscope must be also elevated by insertion of special spacers. Also, addition of a piezoelectric device changes the distance between the lens and additional optics located in the nosepiece (e.g., the Wollaston prism) which makes some microscopy methods (e.g., DIC microscopy) impossible without special adapter (although not currently commercially available, upon request we can provide information on this issue). Although these disadvantages are not trivial, there are some benefits as well to using piezoelectric devices. One of them is that they do not use the microscope’s focusing mechanism and, so that the microscope can be refocused manually during time-lapse data collection without interrupting the series. This can be important when collecting a Z-series of a dividing cell, because as the cell rounds it often shifts outside of the range covered by the Z-sequence. To compensate for this rounding on a microscope equipped with a standard stepping motor, it is necessary to interrupt the recording and adjust the Z-level.
4. Concluding remarks
When purchasing or assembling a LM workstation for live-cell imaging it is important to keep in mind that the system must necessarily represent a compromise between keeping the cell healthy, while achieving the best temporal and spatial resolution. Most standard off-the-shelf microscopes are designed to attain the highest possible image quality and not to protect the cell. However, with some forethought, a standard microscope can be modified into a true live-cell imaging system. In this chapter we have discussed the more essential considerations necessary for successfully following live cells, at high resolution, as they divide. Although there are alternatives to many of the solutions we offer, the ones described are used by us and have proven their utility to our work. They allow us, e.g., to follow GFP labeled dividing human cells from prophase through cytokinesis, by multi-mode 4-D microscopy, which requires recording as many as 6000 fluorescence frames. Importantly, this is done without inducing side-effects indicative of radiation damage, including, e.g., prophase reversion, metaphase arrest, or the significant prolongation of mitosis. Similarly, by paying attention to detail, we can follow vertebrate tissue culture cells using multi-mode 4-D microscopy for up to three consecutive cell cycles (~75–80 h), recording one Z-section series (15 slices) and a DIC or phase-contrast frame every 20–30 min. Through the use of the information described in this chapter to optimize an imaging system, similar data can be collected using standard, albeit carefully chosen, equipment and a CCD camera costing <$20K.
References
- 1.Flemming W. Archiv fur Mikroskopische Anatomie. 1879;18:302–436. [Google Scholar]
- 2.Rieder CL, Khodjakov A. Science. 2003;300:91–96. doi: 10.1126/science.1082177. [DOI] [PubMed] [Google Scholar]
- 3.Allen RD, Allen NS, Travis JL. Cell Motil Cytoskeleton. 1981;1:291–302. doi: 10.1002/cm.970010303. [DOI] [PubMed] [Google Scholar]
- 4.Inoue S. J Cell Biol. 1981;91:131s–147s. doi: 10.1083/jcb.91.3.131s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inoue S, Spring K. Video Microscopy. Plenum Press; New York: 1997. pp. 1–741. [Google Scholar]
- 6.Salmon ED, Tran PT. Methods Cell Biol. 1998;56:153–184. doi: 10.1016/s0091-679x(08)60426-4. [DOI] [PubMed] [Google Scholar]
- 7.Skibbens RV, Rieder CL, Salmon ED. J Cell Biol. 1993;122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khodjakov A, Cole RW, McEwen BF, Buttle KF, Rieder CL. J Cell Biol. 1997;136:229–240. doi: 10.1083/jcb.136.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitaker M. Bioessays. 2000;22:180–187. doi: 10.1002/(SICI)1521-1878(200002)22:2<180::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Campbell RE, Ting AY, Tsien RY. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 12.Thomas CF, White JG. Trends Biotechnol. 1998;16:175–182. doi: 10.1016/s0167-7799(97)01169-4. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y-L. In: Video Microscopy. Sluder G, Wolf DE, editors. Academic Press; New York: 1998. pp. 305–315. [Google Scholar]
- 14.McNally JG, Karpova T, Cooper JA, Conchello JA. Methods. 1999;19:373–385. doi: 10.1006/meth.1999.0873. [DOI] [PubMed] [Google Scholar]
- 15.Salmon ED, Shaw SL, Waters JC, Waterman-Storer CM, Maddox PS, Yeh E, Bloom K. Methods Cell Biol. 1997;56:185–215. [PMC free article] [PubMed] [Google Scholar]
- 16.Haraguchi T, Kaneda T, Hiraoka Y. Genes Cell. 1997;2:369–380. doi: 10.1046/j.1365-2443.1997.1280326.x. [DOI] [PubMed] [Google Scholar]
- 17.Waterman-Storer CM, Desai A, Bulinski JC, Salmon E. Curr Biol. 1998;8:1227–1230. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- 18.Khodjakov A, Rieder CL. J Cell Biol. 1999;146:585–596. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piel M, Meyer P, Khodiakov AL, Rieder CL, Bornens M. J Cell Biol. 2000;149:317–329. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abal M, Piel M, Bouckson-Castaing V, Mogensen MM, Sibarita JB, Bornens M. J Cell Biol. 2002;159:731–737. doi: 10.1083/jcb.200207076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howell BJ, Hoffman DB, Fang G, Murray AW, Slaninova I. J Cell Biol. 2001;150:1233–1249. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tavormina PA, Come MG, Hudson JR, Mo YY, Beck WT, Gorbsky GJ. J Cell Biol. 2002;158:23–29. doi: 10.1083/jcb.200202053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rieder CL, Cole RW. Cold Spring Harbor Symp Quant Biol. 2000;65:369–376. doi: 10.1101/sqb.2000.65.369. [DOI] [PubMed] [Google Scholar]
- 24.Mikhailov A, Rieder CL. Curr Biol. 2002;12:R331–R333. doi: 10.1016/s0960-9822(02)00833-3. [DOI] [PubMed] [Google Scholar]
- 25.Bullough WS, Johnson M. Proc R Soc. 1951;B138:562–575. doi: 10.1098/rspb.1951.0041. [DOI] [PubMed] [Google Scholar]
- 26.Gelfant S. Exp Cell Res. 1958;15:423–425. doi: 10.1016/0014-4827(58)90046-6. [DOI] [PubMed] [Google Scholar]
- 27.Rieder CL, Cole RW. Curr Biol. 2000;10:1067–1070. doi: 10.1016/s0960-9822(00)00678-3. [DOI] [PubMed] [Google Scholar]
- 28.Rieder CL, Cole R. Cell Cycle. 2002;1:169–175. [PubMed] [Google Scholar]
- 29.Bijur GN, Briggs B, Hitchcock CL, Williams MV. Environ Mol Mutagen. 1999;33:144–152. doi: 10.1002/(sici)1098-2280(1999)33:2<144::aid-em6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 30.Strangeways TSP, Hopwood FL. Proc R Soc Lond (Ser B) 1926;100:283–293. [Google Scholar]
- 31.Puck TT, Steffen P. Biophys J. 1963;3:379–397. doi: 10.1016/s0006-3495(63)86828-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doida Y, Okada S. Radiat Res. 1969;38:513–529. [PubMed] [Google Scholar]
- 33.Pearce AK, Humphrey TC. Trends Cell Biol. 2001;11:426–433. doi: 10.1016/s0962-8924(01)02119-5. [DOI] [PubMed] [Google Scholar]
- 34.Glover DM. Trends Genet. 1991;7:125–132. doi: 10.1016/0168-9525(91)90457-2. [DOI] [PubMed] [Google Scholar]
- 35.Strangeways TSP, Obrietan K. Proc R Soc. 1923;B95:373–381. [Google Scholar]
- 36.Rieder CL, Cole RW. J Cell Biol. 1998;142:1013–1022. doi: 10.1083/jcb.142.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manders EMM, Kimura H, Cook PR. J Cell Biol. 1999;144:813–822. doi: 10.1083/jcb.144.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mikhailov A, Cole RW, Rieder CL. Curr Biol. 2002;12:1797–1806. doi: 10.1016/s0960-9822(02)01226-5. [DOI] [PubMed] [Google Scholar]
- 39.Brakenhoff GJ, Muller M, Ghauharali RI. J Microsc. 1996;183:140–144. doi: 10.1046/j.1365-2818.1996.870647.x. [DOI] [PubMed] [Google Scholar]
- 40.Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill ED, Abraham RT. Cancer Res. 1998;58:4375–4382. [PubMed] [Google Scholar]
- 41.Blasina A, Price BD, Turenne GA, McGowan CH. Curr Biol. 1999;9:1135–1138. doi: 10.1016/s0960-9822(99)80486-2. [DOI] [PubMed] [Google Scholar]
- 42.Graves PR, Yu L, Schwarz JK, Gales J, Sausville EA, O’Connor PM, Piwnica-Worms H. J Biochem. 2000;275:5600–5605. doi: 10.1074/jbc.275.8.5600. [DOI] [PubMed] [Google Scholar]
- 43.Bulavin DV, Amundson SA, Fornace AJ. Curr Opin Genet Dev. 2002;12:92–97. doi: 10.1016/s0959-437x(01)00270-2. [DOI] [PubMed] [Google Scholar]
- 44.Rieder CL, Cole RW. In: Video Microscopy. Sluder G, Wolf DE, editors. 1998. pp. 253–275. [Google Scholar]
- 45.Jensen CG, Jensen LC, Ault JG, Osorio G, Cole R, Rieder CL. In: Cellular and Molecular Effects of Mineral and Synthetic Dusts and Fibers. Davis JM, Jurand MC, editors. Springer; Berlin: 1994. pp. 63–78. [Google Scholar]
- 46.Sprague E, Steinbach BL, Nerem RM, Schwartz CJ. Circulation. 1987;76:648–656. doi: 10.1161/01.cir.76.3.648. [DOI] [PubMed] [Google Scholar]
- 47.Salih V, Greenwald SE, Chong CF, Coumbe A, Berry CL. Int J Exp Pathol. 1992;73:625–632. [PMC free article] [PubMed] [Google Scholar]
- 48.Rieder CL, Hard R. Int Rev Cytol. 1990;122:153–220. doi: 10.1016/s0074-7696(08)61208-5. [DOI] [PubMed] [Google Scholar]
- 49.Rieder CL, Cassels GO. Methods Cell Biol. 1999;61:297–315. doi: 10.1016/s0091-679x(08)61987-1. [DOI] [PubMed] [Google Scholar]
- 50.Rao PN, Engelberg J. Science. 1965;148:1092–1093. doi: 10.1126/science.148.3673.1092. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe I, Okada S. J Cell Biol. 1967;32:309–323. doi: 10.1083/jcb.32.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rieder CL, Palazzo RE. J Cell Sci. 1992;102:387–392. doi: 10.1242/jcs.102.3.387. [DOI] [PubMed] [Google Scholar]
- 53.Rieder CL, Schultz A, Cole R, Sluder G. J Cell Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams MC, Salmon WC, Gupton SL, Cohan CS, Wittman T, Prigozhina N, Waterman-Storer CM. Methods. 2003;29:29–41. doi: 10.1016/s1046-2023(02)00282-7. [DOI] [PubMed] [Google Scholar]
- 55.Pawley JB. Handbook of Biological Confocal Microscopy. Plenum Press; New York: 1995. pp. 1–632. [Google Scholar]
- 56.Oldenbourg R, Terada H, Tiberio R, Inoue S. J Microsc. 1993;172:31–39. doi: 10.1111/j.1365-2818.1993.tb03390.x. [DOI] [PubMed] [Google Scholar]
- 57.Swedlow J, Hu K, Andrews PD, Roos DS, Murray JM. Proc Natl Acad Sci USA. 2002;99:2014–2019. doi: 10.1073/pnas.022554999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Agard DA, Sedat JW. Nature. 1983;302:676–681. doi: 10.1038/302676a0. [DOI] [PubMed] [Google Scholar]
- 59.Carrington W, Lynch RM, Moore EDW, Isenberg G, Fogarty KE, Fay FS. Science. 1995;268:1483–1487. doi: 10.1126/science.7770772. [DOI] [PubMed] [Google Scholar]
- 60.Krishnamurthi V, Liu YH, Bhattacharyya S, Turner JN, Holmes TJ. Appl Opt. 1995;34:6633–6647. doi: 10.1364/AO.34.006633. [DOI] [PubMed] [Google Scholar]
- 61.Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, Doxsey SJ. J Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]