Abstract
The interaction between the yeast G protein coupled receptor (GPCR), Ste2p, and its α-factor tridecapeptide ligand was subjected to double-mutant cycle scanning analysis by which the pairwise interaction energy of each ligand residue with two receptor residues, N205 and Y266, was determined. The mutations N205A and Y266A were previously shown to result in deficient signaling but cause only a 2.5-fold and 6-fold decrease, respectively, in the affinity for α-factor. The analysis shows that residues at the amine terminus of α-factor interact strongly with N205 and Y266 whereas residues in the center and at the carboxyl terminus of the peptide interact only weakly if at all with these receptor residues. Multiple-mutant thermodynamic cycle analysis was used to assess whether the energies of selected pairwise interactions between residues of the α-factor peptide changed upon binding to Ste2p. Strong positive cooperativity between residues 1 through 4 of α-factor was observed during receptor binding. In contrast, no thermodynamic evidence was found for an interaction between a residue near the carboxyl terminus of α-factor (position 11) and one at the N-terminus (position 3). The study shows that multiple-mutant cycle analyses of the binding of an alanine-scanned peptide to wild-type and mutant GPCRs can provide detailed information on contributions of inter- and intra-molecular interactions to the binding energy and potentially prove useful in developing 3D models of ligand docked to its receptor.
Determination of contacts between peptide ligands and their cognate G protein-coupled receptors (GPCRs)2 and the respective energetics of these interactions is essential for understanding how binding is transduced to intracellular signaling. Many studies have employed structure-activity relationships involving ligand analogs and mutagenesis of receptors to discern receptor-ligand interactions (reviewed in refs.1,2). Complementary investigations have used photocrosslinking to provide biochemical evidence for contacts (3, 4). In the 1980s, Fersht and coworkers and Horovitz introduced double-mutant cycle analysis to provide thermodynamic evidence for the interaction between groups within one protein or in ligand-protein complexes (5-7). The method has been applied to a number of receptor systems including the ligand-gated ion channel nicotinic (8, 9) and the GPCR muscarinic (10) acetylcholine receptors. The concept of this method is illustrated in a cycle for a ligand binding to its GPCR (Figure 1A). If the effect on the binding free energy (or the free energy of some other process) of the double mutation is not equal to the sum of effects of the single mutations then the two residues are coupled. Non-zero pairwise coupling energies calculated from such cycles reflect interactions that can be either direct or indirect. Coupling energies found for two directly interacting residues can be converted to a distance constraint and in this respect are similar in nature to nuclear Overhauser connectivities (11). Thus, detailed knowledge of coupling energies for a ligand and a receptor can be used to dock the ligand into a receptor whose structure is available (12-14).
Figure 1.
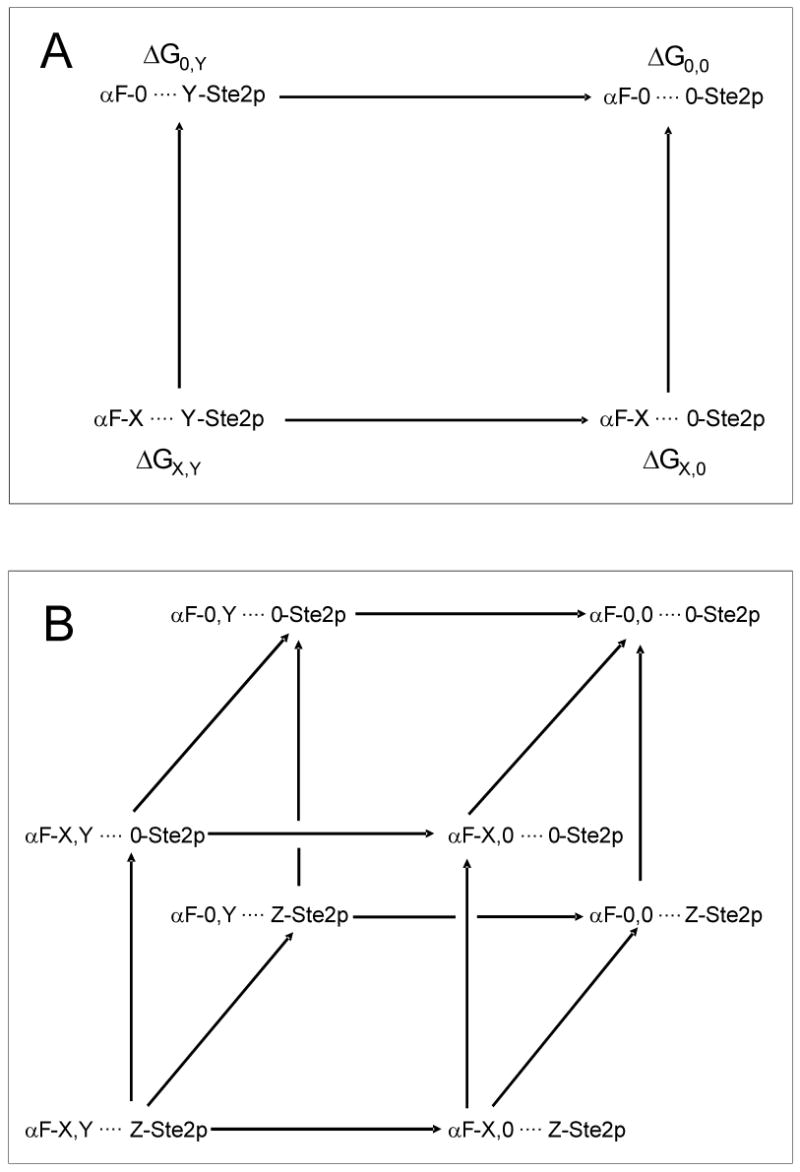
Thermodynamic mutant cycles for alanine scanned α-factor analogs interacting with Ste2p. A) Double-mutant cycle to study energetics of the interaction between a given residue X in α-factor with residue Y in the Ste2p receptor. B) Triple-mutant cycle to study the coupling between two α-factor residues (X, Y) in the presence of a receptor residue (Z).
We have been studying the biology of a yeast mating factor GPCR, Ste2p, and its contacts with α-factor [Trp1-His-Trp-Leu-Gln-Leu-Lys-Pro-Gly-Gln-Pro-Met-Tyr13], the tridecapeptide ligand of this GPCR, by employing α-factor analogs, site-specific mutagenesis and photocrosslinking analysis (15-18). These studies together with extensive molecular biology investigations (19-23) have indicated likely interactions between Y266 and residues near the amine terminus of α-factor. Very recently, biochemical evidence based on disulfide crosslinking indicated that N205 and Y266 might be close in an activated state of Ste2p as represented by a constitutively active mutant of this GPCR (24).
In order to probe the thermodynamics of the interactions that occur between α-factor and Ste2p, we have now conducted a double-mutant cycle analysis using binding data previously obtained for the interaction of a series of alanine-scanned α-factor analogs with the N205A and Y266A receptor mutants (16, 24). Noteworthy is the fact that both of these receptor mutants are deficient in signaling but bind α-factor with nM affinity. We also report new studies on the interactions between wild-type and mutant Ste2p with α-factor analogs in which two native residues were replaced by alanine. These double alanine mutants were used to construct triple-mutant thermodynamic cycles (7) to ascertain whether cooperative interactions between residues within α-factor occur during binding to its GPCR. The results show the value of using an alanine-scanned series of a medium sized peptide ligand in multiple-mutant cycle analysis of its interaction with a GPCR and provide thermodynamic values for the interaction free energies of specific residues in α-factor and Ste2p.
Experimental Procedures
Peptide Synthesis
All peptides were synthesized using an Applied Biosystems Model 433A automated peptide synthesizer. Fmoc/OtBu protection and HBTU coupling were employed in FastMoc mode recommended by the manufacturer. Peptides were assembled beginning with either Fmoc-Tyr(OtBu) Wang Resin or Fmoc-Ala-Wang resin. After assembly, peptides were cleaved and deprotected using trifluoractetic acid/ water with ethanedithiol as a scavenger and purified by reversed phase HPLC. All methods have been previously described (25). Crude yields were 79-94% based on the original resin loading and the crude peptide was >90% homogeneous. After purification by HPLC the double alanine analogs of α-factor were characterized by electron spray mass spectrometry and found to have the calculated molecular weight to ± 1 Da. All peptides used in the binding studies were >99% homogeneous on HPLC in two systems. In this study all α-factor analogs contain norleucine in place of Met12. Norleucine is isosteric with methionine and we have demonstrated that Nle12 and Met12 analogs of α-factor have identical biological activity and identical receptor affinities (26). We use norleucine because it does not have a tendency to oxidize during synthesis and storage.
Receptor Binding Competition Studies
Binding assays were performed using [3H]α-factor synthesized as described previously (26). The competition binding assay was started by the addition of [3H]α-factor and various concentrations of non-labeled α-factor or α-factor analogs (140 μl) to a 560-μl cell suspension such that the final concentration of radioactive peptide was 6 × 10−9 M (20 Ci/mmol). After a 30-min incubation, triplicate samples of 200 μl were filtered and washed over glass fiber filter mats using the Standard Cell Harvester (Skatron Instruments, Sterling, VA) and placed in scintillation vials for counting. Each experiment was carried out at least three times with similar results. Data curves for competition binding assays were fitted from at least eight triplicate data points using Prism™ software (GraphPad) with nonlinear regression for a one site competition model. The Ki values for competition binding assays were calculated by using the equation of Cheng and Prusoff (27), where Ki = IC50/(1 + [ligand]/Kd).
Mutant Cycle Analysis
The free energy of coupling, Δ2Gint, between two residues, X and Y, is given by:
| (1) |
where 0 stands for the residue that was mutated (in this study all the mutations were to alanine). Residues X and Y can be in the same molecule (peptide or receptor) in which case an intra-molecular coupling energy is determined or in different molecules (e.g. X in the peptide and Y in the receptor) in which case an inter-molecular coupling energy is determined. In the latter case, ΔGX,Y and ΔG0,Y are the respective binding energies of the wild-type receptor to the wild-type and mutant peptides and ΔGX,0 and ΔG0,0 are the respective binding energies of the mutant receptor to the wild-type and mutant peptides. Higher-order coupling energies, ΔnGint, between n residues can be determined by constructing the appropriate n-dimensional mutant cycles. Here, three-dimensional mutant constructs were created that involve two peptide residues, X and Y, and one receptor residue Z. The third-order coupling energy, Δ3Gint, calculated from such a construct is given by:
| (2) |
The first four terms in Eq. 2 correspond to a double-mutant cycle for X and Y in the presence of the wild-type receptor whereas the last four terms in Eq. 2 correspond to a double-mutant cycle for X and Y when the receptor residue Z has been mutated. Hence, Δ3Gint is a measure of the effect of mutating Z on the pairwise interaction between X and Y. Owing to symmetry in the triple-mutant cube, Δ3Gint is also a measure of the effect of mutating X or Y on the pairwise interaction energy between the remaining two residues. The errors in ΔnGint are obtained by calculating the square root of the sum of squares of the errors in the appropriate 2n free energies. The Δ2Gint energies calculated herein were based on two previously published data sets (16, 24) for binding of alanine scanned α-factor analogs to Ste2p, Ste2p(N205A) and Ste2p(Y266A). To conduct these calculations we averaged the values of the binding constants for the wild-type receptor and the alanine-scanned α-factor analogs obtained in the separate studies. The ΔnGint values for the cubic analysis are based on the binding constants obtained in the present study for the double-alanine analogs of α-factor and the average values for the single alanine-scanned analogs as discussed above. In analyzing the coupling between receptor residues and α-factor residues, we use the terms strong and weak coupling when Δ2Gint has a relatively small error and is less than −1 kcal/mol, or is from 0.2 to 0.4 kcal/mol, respectively, and the term no coupling when ΔnGint is of the same order of magnitude as its error.
Results
Interaction energies of α-factor residues with Y266
The coupling energies, Δ2Gint, between each peptide residue and position Y266 in the Ste2p receptor were calculated from previously published data (16, 24) using Eq. 1 (Figure 2). The calculations indicate strong coupling between the first four residues of α-factor and Y266 with Δ2Gint values between -1.40 (± 0.23) and -1.81 (± 0.17) kcal/mol. In contrast, Y266 is found to not interact, within experimental error, with peptide residues Gln5, Lys7, Pro8, Gly9, Gln10 and Pro11, and weakly, if at all, with residues Leu6 (-0.56 ± 0.15 kcal/mol), Nle12 (0.40 ± 0.18 kcal/mol), and Tyr13 (-0.36 ± 0.16 kcal/mol). Most significant is the striking difference between the interactions of Y266 with residues at the amine terminus of α-factor and with the remaining residues of this peptide.
Figure 2.
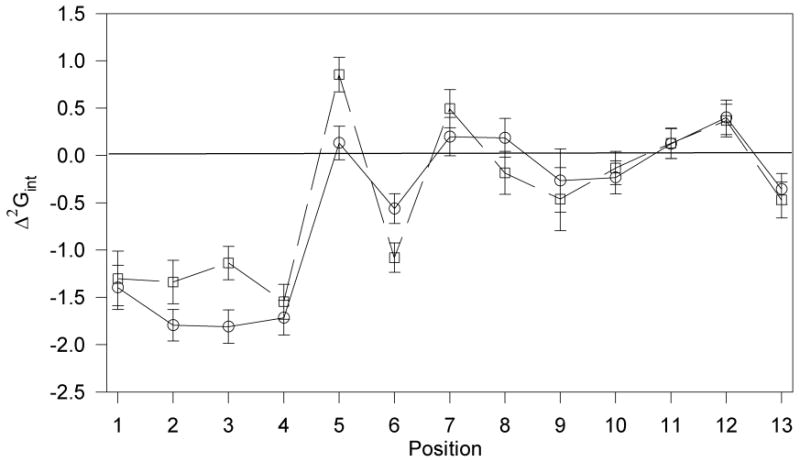
Free energies of pairwise interactions between residues in α-factor (Trp1-His-Trp-Leu-Gln-Leu-Lys-Pro-Gly-Gln-Pro-Nle-Tyr13) and Y266(○) or N205 (□). The free energies of interaction, (Δ2Gint), were calculated using Eq. 1. The data used in the calculation was published for Ste2p(Y266A) receptor (16) and for Ste2p(N205A)(24). As this latter data set did not contain a precise Ki value for [Ala13]α-factor, the Ki value for this peptide was redetermined using α-factor and [Ala1]α-factor as controls, and the data was normalized to the original data set.
Interaction energies of α-factor residues with N205
Using an identical approach, coupling energies were calculated between each peptide residue and N205 in Ste2p (Figure 2). The results indicate that coupling occurs between position 205 of the receptor and residues 1-4 of α-factor while residues Pro8, Gly9, Gln10 and Pro11 are not, within experimental error, coupled with this receptor residue, and residues Lys7, Nle12 and Tyr13 are only weakly coupled to it. This correlates very well with the interactions found between the pheromone and Y266 (Figure 2). The average interaction energy for residues 1-4 with N205 is -1.33 kcal/mol. This compares with an average interaction energy of -1.68 kcal/mol for these same residues with Y266. In contrast with Y266, with which residues 5 and 6 of α-factor are not or are relatively weakly coupled, respectively, the Δ2Gint energies of residues Gln5 (0.85 ± 0.23 kcal/mol) and Leu6 (-1.08 ± 0.21 kcal/mol) with N205 are relatively strong.
Effects of Y266 and N205 on intra-peptide pairwise interactions
In principle, it is possible by use of multiple mutant thermodynamic cycles to study how interaction energies between residues within a peptide ligand are affected by residues in the receptor protein. Given the above results indicating strong coupling energies between each of the four residues at the amine terminal of α-factor and residues N205 and Y266 of Ste2p, we synthesized a series of peptides containing double replacements involving the Trp1-His2-Trp3-Leu4 region of α-factor and examined the binding of these peptides to both mutant receptors. As a control, we synthesized [Ala3, Ala11]α-factor since Pro11 had a coupling energy near zero with both N205 and Y266 (Figure 2). Although none of these double-alanine α-factor analogs were found to be agonists, they competed with radioactive α-factor for the receptor (Figure 3), and it was possible to determine accurate Ki values (Table 1). These varied from a low value of 48.9 (±8) nM for [Ala1, Ala3]α-factor with Ste2p(Y266A) to >2 μM for [Ala3, Ala11]α-factor with Ste2p and Ste2p(N205A). Using these Ki data, we constructed triple-mutant cubes to determine higher-order coupling energies for Y266 or N205 and various pairs of residues of the tridecapeptide (Table 2). The coupling energy of residues Trp3 and Pro11 in the presence of Ste2p is -0.45 (± 0.14) kcal/mol. In contrast, the coupling energies between any pair of residues 1-4, in the presence of Ste2p, are between -0.82 and -1.88 kcal/mol. In the presence of the mutant receptors, the coupling between Trp3 and Pro11 remains unchanged [Ste2p(Y266A)] or is zero within experimental error [Ste2p(N205A)]. Most of the intra-peptide coupling energies for residues 1 to 4 of α-factor are close to zero in the presence of the mutant receptors with the notable exception of the coupling energies of Trp3 and Leu4 in the presence of Ste2p(Y266A) and Ste2p(N205A) which are 1.02 (±0.13) and 0.73 (±0.19) kcal/mol, respectively, and that of Trp1 and Trp3 in the presence of Ste2p(Y266A) which is -0.68 (±0.14) kcal/mol.
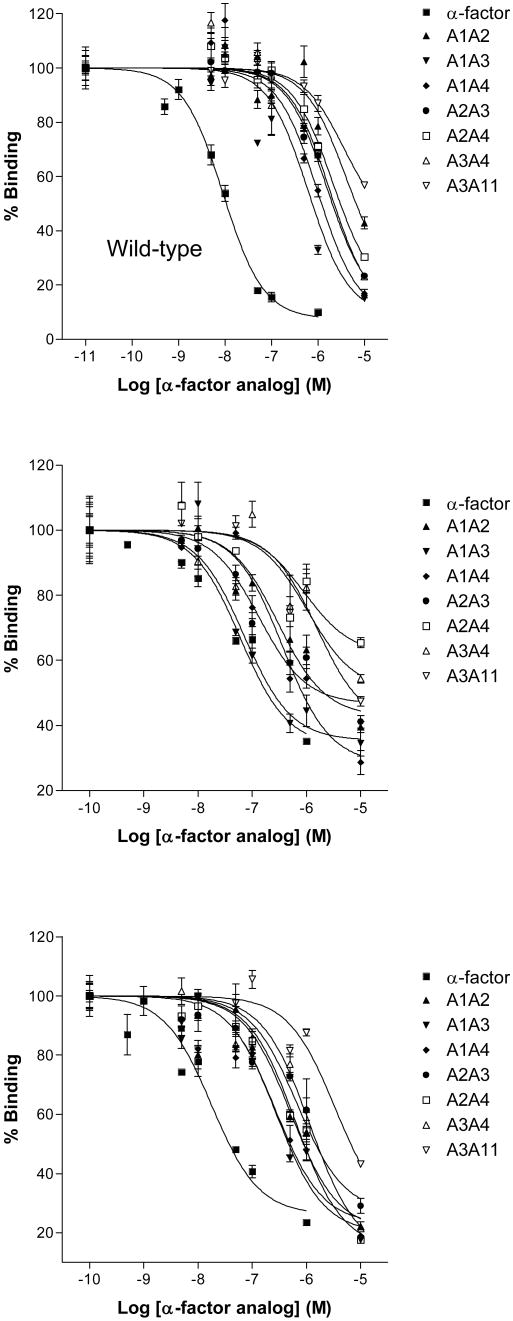
Figure 3.
Inhibition of binding of [3H-Pro11, Nle12]α-factor to Wild-type Ste2p [Panel A], Ste2p(Y266A) [Panel B], and Ste2p(N205A) [Panel C]. Binding competition was performed using S. cerevisiae LM102 strain transformed with a plasmid coding for Ste2p, Ste2p(Y266A), or Ste2p(N205A) as described in the Experimental Section. Competing peptides were ((-factor (■), [Ala1, Ala2]α-factor (▲), [Ala1, Ala3]α-factor (▼), [Ala1, Ala4]α-factor (◆), [Ala2, Ala3]α-factor (●), [Ala2, Ala4]α-factor (□), [Ala3, Ala4]α-factor (Δ), and [Ala3, Ala11]α-factor (∇).
Table 1.
Binding affinities of double-alanine substituted α-factor analogs to wild-type, Y266A and N205A α-factor receptors.
| Peptide | Binding (Ki = nM)a | ||
|---|---|---|---|
| WT | Y266A | N205A | |
| α-factor | 5.6±0.9 | 37.5±4.0 | 16.8±2 |
| [Ala1,Ala2]α-factor | 2910±200 | 331±18 | 537±29 |
| [Ala1,Ala3]α-factor | 411±35 | 48.9±8 | 255±13 |
| [Ala1,Ala4]α-factor | 387±28 | 223±15 | 361±20 |
| [Ala2,Ala3]α-factor | 816±45 | 117±10 | 736±38 |
| [Ala2,Ala4]α-factor | 950±52 | 498±34 | 526±27 |
| [Ala3,Ala4]α-factor | 710±32 | 999±76 | 1018±85 |
| [Ala3,Ala11]α-factor | 2341±115 | 789±67 | 2829±110 |
Ki values for α-factor and double Ala substituted α-factor analogues were determined in competition binding assays by displacement of [3H]α-factor. All values are the mean ± S.E. from three separate experiments.
Table 2.
Multiple-mutant cycle analysis of pairwise interactions in α-factor complexed with Ste2p.
| Pairwise Interactions in α-Factor | Δ2Ginta | Δ3Gintb | Δ2Ginta | Δ3Gintc | ||
|---|---|---|---|---|---|---|
| Wild typed | A266d | Wild typed | A205d | |||
| Trp1,His2 | -0.82±0.22 | 0.01±0.11 | -0.83±0.24 | -0.82±0.22 | 0.28±0.20 | -1.11±0.30 |
| Trp1,Trp3 | -1.55±0.23 | -0.68±0.14 | -0.87±0.27 | -1.55±0.23 | 0.06±0.19 | -1.61±0.30 |
| Trp1,Leu4 | -1.82±0.22 | -0.11±0.13 | -1.71±0.26 | -1.82±0.22 | 0.44±0.21 | -2.26±0.30 |
| His2,Trp3 | -1.74±0.15 | -0.35±0.11 | -1.38±0.19 | -1.74±0.15 | 0.14±0.18 | -1.88±0.24 |
| His2,Leu4 | -1.88±0.15 | 0.17±0.13 | -2.05±0.19 | -1.88±0.15 | 0.11±0.19 | -2.00±0.24 |
| Trp3,Leu4 | -1.63±0.15 | 1.02±0.13 | -2.67±0.20 | -1.63±0.15 | 0.73±0.19 | -2.36±0.24 |
| Trp3,Pro11 | -0.45±0.14 | -0.48±0.10 | 0.03±0.18 | -0.45±0.14 | 0.13±0.17 | -0.59±0.22 |
2nd order coupling energy determined for the indicated pairwise interactions between specified residues in the α-factor in the presence of either the wild-type or mutant receptor. The cycles are illustrated in Figure 1A.
3rd order coupling energy calculated for the triple mutant cycle (Figure 1B) involving the specified α-factor residues and residue 266 of Ste2p.
3rd order coupling energy calculated for the triple mutant cycle (Figure 1B) involving the specified α-factor residues and residue 205 of Ste2p.
Y266 and N205 represent wild-type Ste2p residues. A266 and A205 represent Ste2p mutants with Y266A and N205A replacements, respectively.
The overall Δ3Gint values of the triple-mutant cubes for the interaction of N205 in the receptor with pairs of residues in α-factor are <-1.1 kcal/mol except for the cube with replacements at positions 3 and 11 which was found to have a Δ3Gint of -0.59 (±0.22) kcal/mol (Table 2). Similarly, Δ3Gint for Y266 and positions 3 and 11 was found to be 0.03 (±0.18) kcal/mol whereas the other Δ3Gint values for Y266 and peptide residues at positions 1-4 were found to have moderate (-0.83 or -0.87 kcal/mol) to quite strong (≤-1.38 kcal/mol) Δ3Gint values (Table 2).
Discussion
The binding energy of a medium sized peptide with its protein receptor reflects a complex set of inter-residue contacts as well as other factors such as conformational entropy and solvent effects. Single mutations in one of the binding partners can influence many of these factors, and thus their effects on the binding energy are very difficult to interpret. For example, analysis of binding data indicates that replacement of Leu6 in α-factor by Ala causes an approximately 200-fold decrease in affinity whereas replacement of Trp1 by Ala causes a 20 to 30-fold decrease (16, 24) but finding a direct molecular explanation for these results is very tricky (even if the structure of the complex were known) since mutation of a single residue may impact multiple interactions. Double-mutant cycle analysis provides a way for isolating the energetics of specific pairwise interactions involved in complex formation. The double-mutant cycle calculations presented in this communication show that the interaction energies of the residues at positions 1 to 4 of α-factor with Y266 of Ste2p are similar (-1.40 to -1.81 kcal/mol) and stabilizing with respect to complex formation. Indeed, the first four residues of α-factor (Trp1-His2-Trp3-Leu4) form an aromatic/hydrophobic patch that would be expected to interact favorably with an aromatic group such as Y266. However, it is important to note that some of the interactions of the amine terminus residues with Y266 may not be direct as will be discussed further below. Interestingly, although less definitive, Leu6 also contributes favorably to the binding interaction of α-factor with Y266 whereas the polar residues Gln5 and Lys7 do not (Figure 2). Hence, it is tempting to speculate that the side-chains of Gln5 and Lys7 face away from the receptor and that this part of the pheromone adopts a β-strand like conformation in its bound state. This proposed orientation of Lys7 is in agreement with a fluorescence analysis that indicated that the side-chain of Lys7 most likely faces away from the transmembrane region and interacts with loop residues (28, 29).
Two complications in the application of the double-mutant cycle method to peptide-GPCR interactions are that mutations in either one or both of the binding partners may (i) cause non-local conformational changes that affect the structure and energetics of the complex and/or (ii) alter the conformational sampling by the receptor of its various signaling and non-signaling states (30). The observation in this study that the pattern of interaction energies of the peptide residues with N205 is similar to that with Y266 (Figure 2), suggests that neither of the above-mentioned complications are of concern here. This key observation also indicates that Y266 and N205 are close in space as previously inferred (24), and that the pairwise interactions we identify between α-factor residues and receptor residues are more likely to be direct. The similar alanine-scanning, double-mutant cycle data for the two mutant receptors also strengthens the conclusion that the contribution of residues 1-4 of the α-factor ligand to binding of Ste2p, through interactions with residues Y266 and N205 of the receptor is dominant, although single substitutions elsewhere in the ligand have effects on binding.
Having evidence for strong interactions between N-terminal residues of α-factor and positions N205/Y266 of Ste2p we used triple-mutant cycle analysis to study the cooperativity of various ligand residues in binding to this receptor. Double-mutant cycle analysis allowed us to conclude that Trp3 and Pro11 are weakly coupled during binding to wild-type Ste2p (Δ2Gint = -0.45 (± 0.14) kcal/mol). The opposite surface of the triple-mutant cube (Figure 1B) for these residues indicates that Trp3 and Pro11 are weakly coupled also during binding to the Y266A mutant (Δ2Gint = -0.48 (± 0.10) kcal/mol). The higher-order interaction energy, Δ3Gint, for the mutant cube tells us how a pairwise interaction (e.g. of a residue in α-factor with a residue in the receptor) depends on another residue. This analysis shows that Pro11 has no thermodynamic influence on the interaction of Trp3 with Y266 (Δ3Gint = 0.03 ± 0.18 kcal/mol). Similarly, there is little influence of N205 on the interaction of Trp3 with Pro11 with N205 (Δ3Gint = -0.59 ± 0.22 kcal/mol). Owing to symmetry in these cubes, this also means, for example, that the pairwise interactions between N205 or Y266 with Trp3 are not affected by Pro11.
The results for positions 3 and 11 in the peptide suggest that strong coupling found between other residues of α-factor are likely due to localized interactions rather than global changes associated with binding. Strikingly, we found that residues 1-4 show strong positive cooperativity during binding to Ste2p with Δ2Gint values of -0.82 to -1.88 kcal/mol. However, in the presence of the mutant receptors, the coupling energies (Δ2Gint) for 9 out of 12 such cycles involving residues 1-4 of the pheromone are found to be close to zero. Only in the case of the interaction of Trp1 with Trp3, in the presence of the Y266A receptor, and of the interaction of Trp3 with Leu4 in the presence of both mutant receptors, were the coupling energies found to be significantly above experimental error. Most notable, the sign of these latter interactions differs from all other cycles and indicates a negative cooperativity or a destabilizing contribution to binding. The triple-mutant cycle analysis also shows that the interactions of Trp3 with both Y266 and N205 are strongly dependent on residues 1, 2 and 4, with Δ3Gint values of -0.87 (±0.27) to -2.67 (±0.24) kcal/mol. Similar strong influences on the interactions of residues 1, 2 and 4 of the tridecapeptide with positions Y266 and N205 of the receptor are exhibited by the other N-terminal residues.
Two models have been proposed for binding of α-factor to Ste2p. Both of these are based on the observations that α-factor is bent around the Pro-Gly sequence during its binding to Ste2p (17) and that Trp1 of the ligand interacts with the receptor near Y266 (16). One model suggests that the carboxyl terminus of the peptide is interacting with residues near the extracellular side of transmembrane helix one (TM1) (31) whereas the second has the carboxyl terminus interacting with F204, which is located at the junction between extracellular loop 2 and TM5 (22). The pairwise interactions uncovered by the mutant cycle analysis of the Y266A and N205A receptors indicate that Tyr13 of α-factor does not interact strongly with N205 or Y266 (Figure 2). Recently, these receptor residues were shown to be close enough to form a disulfide bond in a constitutively active Ste2p where N205 and Y266 were replaced by cysteine residues (24). A mutant cycle analysis of residues 45-60 of Ste2p should further elucidate whether the Tyr13 of α-factor binds to the region of the receptor located at the N-terminus-TM1 interface.
In summary, the double-mutant cycle analysis presented here shows the strong energetic coupling between the first four N-terminal residues of α-factor and residues Y266 and N205 of Ste2p. It also shows that the strength of these inter-molecular interactions is greatly influenced by other residues in this part of the peptide but not necessarily elsewhere. The value of double-mutant cycle-scanning of a medium sized peptide hormone is clearly apparent as it can reveal periodicities that reflect intermolecular or intra-peptide interactions. In principle, the distance constraints that can be derived from mutant cycle analysis can be used to help dock the pheromone to the receptor as done previously for other systems (14, 32, 33). The structure of Ste2p to be used in such an analysis can be a model based on the bovine rhodopsin structure (34).
Acknowledgments
We thank Melinda Hauser, Sanjay Khare and Boris Arshava for technical assistance.
Abbreviations
- GPCR
G protein-coupled receptor
- G protein
heterotrimeric GTP-binding protein
- Ste2p
α-factor receptor encoded by the STE2 gene
- TM
transmembrane domain
- Nle
norleucine; standard one-letter abbreviations for amino acids are used.
Footnotes
This work was supported by research grants GM22086 and GM22087 from the National Institutes of Health.
References
- 1.Kristiansen K. Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G-protein-coupled receptors: molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacol Ther. 2004;103:21–80. doi: 10.1016/j.pharmthera.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Marshall GR. Peptide interactions with G-protein coupled receptors. Biopolymers. 2001;60:246–277. doi: 10.1002/1097-0282(2001)60:3<246::AID-BIP10044>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 3.Pham VI, Sexton PM. Photoaffinity scanning in the mapping of the peptide receptor interface of class II G protein-coupled receptors. J Pept Sci. 2004;10:179–203. doi: 10.1002/psc.541. [DOI] [PubMed] [Google Scholar]
- 4.Fillion D, Deraet M, Holleran BJ, Escher E. Stereospecific synthesis of a carbene-generating angiotensin II analogue for comparative photoaffinity labeling: improved incorporation and absence of methionine selectivity. J Med Chem. 2006;49:2200–2209. doi: 10.1021/jm050958a. [DOI] [PubMed] [Google Scholar]
- 5.Carter PJ, Winter G, Wilkinson AJ, Fersht AR. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus) Cell. 1984;38:835–840. doi: 10.1016/0092-8674(84)90278-2. [DOI] [PubMed] [Google Scholar]
- 6.Horovitz A. Non-additivity in protein-protein interactions. J Mol Biol. 1987;196:733–735. doi: 10.1016/0022-2836(87)90045-3. [DOI] [PubMed] [Google Scholar]
- 7.Horovitz A, Fersht AR. Strategy for analysing the co-operativity of intramolecular interactions in peptides and proteins. J Mol Biol. 1990;214:613–617. doi: 10.1016/0022-2836(90)90275-Q. [DOI] [PubMed] [Google Scholar]
- 8.Malany S, Osaka H, Sine SM, Taylor P. Orientation of alpha-neurotoxin at the subunit interfaces of the nicotinic acetylcholine receptor. Biochemistry. 2000;39:15388–15398. doi: 10.1021/bi001825o. [DOI] [PubMed] [Google Scholar]
- 9.Osaka H, Malany S, Molles BE, Sine SM, Taylor P. Pairwise electrostatic interactions between alpha-neurotoxins and gamma, delta, and epsilon subunits of the nicotinic acetylcholine receptor. J Biol Chem. 2000;275:5478–5484. doi: 10.1074/jbc.275.8.5478. [DOI] [PubMed] [Google Scholar]
- 10.Vogel WK, Peterson GL, Broderick DJ, Mosser VA, Schimerlik MI. Double mutant cycle analysis of aspartate 69, 97, and 103 to asparagine mutants in the m2 muscarinic acetylcholine receptor. Arch Biochem Biophys. 1999;361:283–294. doi: 10.1006/abbi.1998.0985. [DOI] [PubMed] [Google Scholar]
- 11.Horovitz A. Double-mutant cycles: a powerful tool for analyzing protein structure and function. Fold Des. 1996;1:R121–126. doi: 10.1016/S1359-0278(96)00056-9. [DOI] [PubMed] [Google Scholar]
- 12.Hidalgo P, MacKinnon R. Revealing the architecture of a K+ channel pore through mutant cycles with a peptide inhibitor. Science. 1995;268:307–310. doi: 10.1126/science.7716527. [DOI] [PubMed] [Google Scholar]
- 13.Gilquin B, Racape J, Wrisch A, Visan V, Lecoq A, Grissmer S, Menez A, Gasparini S. Structure of the BgK-Kv1.1 complex based on distance restraints identified by double mutant cycles. Molecular basis for convergent evolution of Kv1 channel blockers. J Biol Chem. 2002;277:37406–37413. doi: 10.1074/jbc.M206205200. [DOI] [PubMed] [Google Scholar]
- 14.Roisman LC, Piehler J, Trosset JY, Scheraga HA, Schreiber G. Structure of the interferon-receptor complex determined by distance constraints from double-mutant cycles and flexible docking. Proc Natl Acad Sci U S A. 2001;98:13231–13236. doi: 10.1073/pnas.221290398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henry LK, Khare S, Son C, Babu VV, Naider F, Becker JM. Identification of a contact region between the tridecapeptide alpha-factor mating pheromone of Saccharomyces cerevisiae and its G protein-coupled receptor by photoaffinity labeling. Biochemistry. 2002;41:6128–6139. doi: 10.1021/bi015863z. [DOI] [PubMed] [Google Scholar]
- 16.Lee BK, Lee YH, Hauser M, Son CD, Khare S, Naider F, Becker JM. Tyr266 in the sixth transmembrane domain of the yeast alpha-factor receptor plays key roles in receptor activation and ligand specificity. Biochemistry. 2002;41:13681–13689. doi: 10.1021/bi026100u. [DOI] [PubMed] [Google Scholar]
- 17.Naider F, Becker JM. The alpha-factor mating pheromone of Saccharomyces cerevisiae: a model for studying the interaction of peptide hormones and G protein-coupled receptors. Peptides. 2004;25:1441–1463. doi: 10.1016/j.peptides.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 18.Son CD, Sargsyan H, Naider F, Becker JM. Identification of ligand binding regions of the Saccharomyces cerevisiae alpha-factor pheromone receptor by photoaffinity cross-linking. Biochemistry. 2004;43:13193–13203. doi: 10.1021/bi0496889. [DOI] [PubMed] [Google Scholar]
- 19.Dube P, Konopka JB. Identification of a polar region in transmembrane domain 6 that regulates the function of the G protein-coupled alpha-factor receptor. Mol Cell Biol. 1998;18:7205–7215. doi: 10.1128/mcb.18.12.7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dube P, DeCostanzo A, Konopka JB. Interaction between transmembrane domains five and six of the alpha -factor receptor. J Biol Chem. 2000;275:26492–26499. doi: 10.1074/jbc.M002767200. [DOI] [PubMed] [Google Scholar]
- 21.Lin JC, Parrish W, Eilers M, Smith SO, Konopka JB. Aromatic residues at the extracellular ends of transmembrane domains 5 and 6 promote ligand activation of the G protein-coupled alpha-factor receptor. Biochemistry. 2003;42:293–301. doi: 10.1021/bi026766o. [DOI] [PubMed] [Google Scholar]
- 22.Lin JC, Duell K, Konopka JB. A microdomain formed by the extracellular ends of the transmembrane domains promotes activation of the G protein–coupled α–factor receptor. Mol Cell Biol. 2004;24:2041–2051. doi: 10.1128/MCB.24.5.2041-2051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin JC, Duell K, Saracino M, Konopka JB. Identification of residues that contribute to receptor activation through the analysis of compensatory mutations in the G protein-coupled alpha-factor receptor. Biochemistry. 2005;44:1278–1287. doi: 10.1021/bi048050u. [DOI] [PubMed] [Google Scholar]
- 24.Lee YH, Naider F, Becker JM. Interacting residues in an activated state of a G protein-coupled receptor. J Biol Chem. 2006;281:2263–2272. doi: 10.1074/jbc.M509987200. [DOI] [PubMed] [Google Scholar]
- 25.Abel MG, Lee BK, Naider F, Becker JM. Mutations affecting ligand specificity of the G protein–coupled receptor for the Saccharomyces cerevisiae tridecapeptide pheromone. Biochim Biophys Acta. 1998;1448:12–26. doi: 10.1016/s0167-4889(98)00109-8. [DOI] [PubMed] [Google Scholar]
- 26.Raths SK, Naider F, Becker JM. Peptide analogues compete with the binding of alpha-factor to its receptor in Saccharomyces cerevisiae. J Biol Chem. 1988;263:17333–17341. [PubMed] [Google Scholar]
- 27.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 28.Ding FX, Lee BK, Hauser M, Patri R, Arshava B, Becker JM, Naider F. Study of the binding environment of alpha-factor in its G protein-coupled receptor using fluorescence spectroscopy. J Pept Res. 2002;60:65–74. doi: 10.1034/j.1399-3011.2002.21004.x. [DOI] [PubMed] [Google Scholar]
- 29.Ding FX, Schreiber D, VerBerkmoes NC, Becker JM, Naider F. The chain length dependence of helix formation of the second transmembrane domain of a G protein-coupled receptor of Saccharomyces cerevisiae. J Biol Chem. 2002;277:14483–14492. doi: 10.1074/jbc.M111382200. [DOI] [PubMed] [Google Scholar]
- 30.Kenakin T. Ligand-selective receptor conformations revisited: the promise and the problem. Trends Pharmacol Sci. 2003;24:346–354. doi: 10.1016/S0165-6147(03)00167-6. [DOI] [PubMed] [Google Scholar]
- 31.Son CD, Sargsyan H, Hurst GB, Naider F, Becker JM. Analysis of ligand-receptor cross-linked fragments by mass spectrometry. J Pept Res. 2005;65:418–426. doi: 10.1111/j.1399-3011.2005.00248.x. [DOI] [PubMed] [Google Scholar]
- 32.Sohn J, Parks JM, Buhrman G, Brown P, Kristjansdottir K, Safi A, Edelsbrunner H, Yang W, Rudolph J. Experimental validation of the docking orientation of Cdc25 with its Cdk2-CycA protein substrate. Biochemistry. 2005;44:16563–16573. doi: 10.1021/bi0516879. [DOI] [PubMed] [Google Scholar]
- 33.Eriksson MA, Roux B. Modeling the structure of agitoxin in complex with the Shaker K+ channel: a computational approach based on experimental distance restraints extracted from thermodynamic mutant cycles. Biophys J. 2002;83:2595–2609. doi: 10.1016/S0006-3495(02)75270-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eilers M, Hornak V, Smith SO, Konopka JB. Comparison of class A and D G protein-coupled receptors: common features in structure and activation. Biochemistry. 2005;44:8959–8975. doi: 10.1021/bi047316u. [DOI] [PMC free article] [PubMed] [Google Scholar]