Summary
Sirtuin enzymes comprise a unique class of NAD+-dependent protein deacetylases. Although structures of a number of sirtuin complexes have been determined, structural resolution of intermediate chemical steps are needed to understand the deacetylation mechanism. We report crystal structures of the bacterial sirtuin, in complex with an S-alkylamidate intermediate, analogous to the naturally occurring O-alkylamidate intermediate, and a Sir2Tm ternary complex containing a dissociated NAD+ analogue and acetylated peptide. The structures and biochemical studies reveal critical roles for the invariant active site histidine in positioning the reaction intermediate, and for a conserved phenylalanine residue in shielding reaction intermediates from base exchange with nicotinamide. The new structural and biochemical studies provide key mechanistic insight into intermediate steps of the Sir2 deacetylation reaction.
Keywords: Sir2, Sirtuin, O-alkylamidate, S-alkylamidate, DADMe-NAD+
I. Introduction
Sir2 enzymes comprise an ancient and well-conserved family of NAD+ dependent deacetylases found in all domains of life (Frye, 2000). These remarkable enzymes, also known as sirtuins, utilize NAD+ to catalyze the removal of acetyl groups from acetyl-lysine residues on protein substrates including histones (Imai et al., 2000), Foxo transcription factors (Daitoku et al., 2004, Motta et al., 2004), HIV TAT (Pagans et al., 2005), acetyl-CoA synthetases (Starai et al., 2002), and P300 (Bouras et al., 2005). Numerous biological processes including aging (Kaeberlein et al., 1999), HIV transcription (Pagans et al., 2005), gene silencing (Johnson et al., 1990), chromatin structure (Fritze et al., 1997), fat metabolism (Picard et al., 2004), and neurodegeneration (Araki et al., 2004) are regulated by sirtuins.
The sirtuin enzymatic core is composed of a Rossmann fold domain and a smaller domain comprising a helical bundle and a zinc-binding motif (Avalos et al., 2002; Chang et al., 2002; Finnin et al., 2001; Min et al., 2001; Zhao et al., 2003). NAD+ and the acetyl lysine bind in the active site cleft, which is located between the two domains (Hoff et al., 2006). In the absence of peptide, the nicotinamide and N-ribose of NAD+ can adopt multiple conformations in the Sir2 active site (Avalos et al., 2004; Zhao et al., 2003). Upon binding acetyl lysine, NAD+ adopts a strained conformation that buries the nicotinamide ring deep within the active site (Hoff et al., 2006). In this state, the acetyl lysine is oriented to react with NAD+. The strained NAD+ conformation, which was trapped in the crystal structure of a Michaelis complex, suggests that Sir2 enzymes destabilize NAD+. Subsequent intermediate steps of the Sir2 deacetylation reaction, including the formation of the peptidyl-imidate intermediate, have not been structurally characterized.
In the deacetylation reaction, Sir2 enzymes consume one molecule of NAD+ and an acetylated lysine substrate, generating the deacetylated lysine, nicotinamide, and the metabolite, 2’-O-acetyl-ADP-ribose (Jackson et al., 2003; Sauve et al., 2001). In the first step of the Sir2 deacetylation reaction, the glycosidic bond between nicotinamide and the N-ribose is broken, and ADP ribose is transferred to the acetyl amide oxygen to generate a peptidyl-imidate species, also termed the O-alkylamidate intermediate (Sauve et al., 2001; Smith and Denu, 2006) (Figure 1A). The reaction mechanism for sirtuins could follow that of other ADP ribosyl-N-ribosyltransferases. These reactions are characterized by loss of the nicotinamide group prior to the addition of the nucleophile from the alpha-face of the ribosyl group. The reactive riboxacarbenium is then activated to react with unreactive nucleophiles, like the carbonyl oxygen of acetyl-lysine. These mechanisms involve migration of the anomeric carbon of the ribosyl group to accomplish nucleophilic displacement by electrophile migration, while the position of the leaving group and the attacking nucleophile remain stationary (Schramm and Shi, 2001). This mechanism is analogous to that utilized by the NAD+ hydrolase/ADP-ribosyl cyclase, CD38 and other N-ribosyltransferases (Sauve et al., 1998; Sauve and Schramm, 2004). An alternative mechanism has been suggested where the initial step proceeds through an SN2 nucleophilic attack of the acetyl amide oxygen on NAD+ with involvement of the amide oxygen, suggesting a transition state with significant SN2 character (Smith and Denu, 2007b).
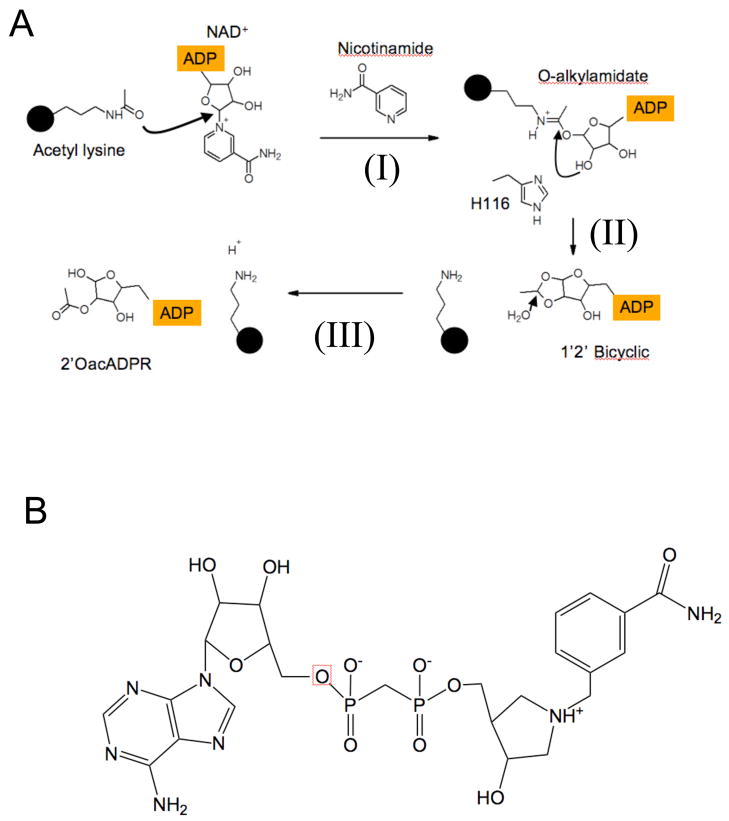
Figure 1. Sirtuin deacetylation reaction mechanism.
(A) In the first step of the Sir2 Deacetylation reaction (I), the ADP ribose moiety of NAD+ is transferred to acetyl lysine generating the O-Alkylamidate intermediate. In this step (I), the Nicotinamide-N-ribose bond is broken to generate free nicotinamide. Next, the N-ribose 2’OH group attacks the O-alkylamidate intermediate generating a 1’ 2’ bicyclic species (II). Subsequent hydrolysis of the 1’ 2’ bicyclic species yields deacetylated lysine and 2’ O-acetyl ADP ribose (III). The structure of the DADMe-NAD+ analogue, which represents a dissociated NAD+ species, is depicted in panel (B).
Following formation by Sir2 of the O-alkylamidate intermediate, a catalytic histidine activates the 2’ alcohol group of the N-ribose, which then attacks the intermediate, forming a 1,2 bicyclic species and the deacetylated lysine (Sauve et al., 2006). Subsequent hydrolysis of the bicyclic species yields the 2’ O-acetyl ADP ribose product (Jackson and Denu, 2002; Sauve et al., 2001) (Figure 1). The Sir2 deacetylation reaction is potently inhibited through a base-exchange reaction with the reaction product, nicotinamide. In this process, nicotinamide attacks the peptidyl-imidate intermediate to regenerate NAD+ and reform acetylated lysine (Jackson et al., 2003; Sauve and Schramm, 2003). Nicotinamide analogues such as isonicotinamide, that are chemically inert in the exchange reaction, activate sirtuins in vivo and in vitro by preventing nicotinamide rebinding (Howitz et al., 2003; Sauve et al., 2005). Other sirtuin activators function by different mechanisms (Howitz et al., 2003). To fully understand sirtuin deacetylation activity, a structure-based elucidation of the Sir2 deacetylation reaction is desirable.
Sir2 enzymes must overcome several challenges to successfully deacetylate a substrate. These include transferring ADP ribose to an amide oxygen, protecting the peptidyl-imidate intermediate from hydrolysis (Smith and Denu 2006), deprotonating the N-ribose 2’ alcohol to form the bicyclic species (Figure 1), and protecting the peptidyl-imidate intermediate from base- exchange with nicotinamide (Jackson et al., 2003; Sauve and Schramm, 2003). Hydrolysis of the O-alkylamidate intermediate to ADP ribose is observed for a HST2 H135A mutant enzyme, demonstrating that the O-alkylamidate intermediate is susceptible to hydrolysis (Smith and Denu 2006). It is unclear how Sir2 enzymes shield the peptidyl-imidate intermediate from these off-pathway reactions. The final challenge for Sir2 enzymes is to use the N-ribose 2’ secondary alcohol to attack the peptidyl-imidate and form the 1,2 bicyclic intermediate (Figure 1A), as typical secondary alcohols require activation to become good nucleophiles. It has been proposed that a conserved active site histidine activates the 2’ alcohol either directly or through a shuttling mechanism with the 3’ alcohol, but mutation of this conserved histidine to alanine resulted in only a modest decrease in deacetylation activity of the Thermotoga maritima Sir2 enzyme, and a ten-fold decrease in kcat for HST2. (Hoff et al., 2006; Smith and Denu, 2006). Structural information on reaction intermediates is needed to provide insight into the formation of the bicyclic species, nicotinamide inhibition and other intermediate steps of the Sir2 deacetylation reaction.
In order to resolve key questions regarding the intermediate steps in the Sir2 deacetylation reaction, we have determined the structure of a bacterial sirtuin, Sir2Tm, bound to a trapped S-alkylamidate intermediate, which is a mimic of the natural peptidyl-imidate complex. This structure is the first direct visualization of a Sir2-ribosylated peptide intermediate complex and, in combination with biochemical data, helps to dissect key intermediate steps in the Sir2 deacetylation reaction. To explore the N-ribose nicotinamide glycosidic bond cleavage, we solved the structure of a Sir2-acetyl peptide-DADme-NAD+, an analogue of a dissociated NAD+, ternary complex. The structures and biochemical data presented here help expand our mechanistic understanding of Sir2 mediated deacetylation and serve as a foundation to begin to understand other Sir2 enzymatic activities.
III. Results
Structure of A Sir2-acetylated peptide –DADMe-NAD+ analogue ternary complex
The transfer of ADP ribose from NAD+ to the acetyl lysine and removal of the nicotinamide moiety is a poorly understood step of the Sir2 deacetylation reaction. This glycosidic bond cleavage and ADP ribosyl transfer reaction could proceed through a dissociative mechanism that has SN1 characteristics (Hoff et al., 2006). Because NAD+ can adopt numerous conformations in the Sir2 active site (Avalos et al., 2004; Zhao et al., 2003), we determined how an analogue of nicotinamide- dissociated NAD, DADMe-NAD+(Figure 1B), binds to the Sir2 active site. DADMe-NAD+ is an analogue of an elongated NAD+ designed to mimic dissociative transition states. In the ADP-ribosylating toxins from organisms causing cholera, pertussis and diptheria, the inhibition gave Km/Ki rations of 200, 0.48, and 0.3 respectively (Zhou et al., 2004). With km and Ki value of 125 and 360 uM respectively, the interaction of DADMe-NAD+ with this sirtuin is similar to that for pertussis and diptheria toxins (S1). To explore the nature of this transfer step, we solved the structure of Sir2Tm bound together with acetylated peptide and the NAD+ analogue, DADMe-NAD+ (Figure 1B and 2). The DADMe-NAD+ compound mimics dissociation of nicotinamide from ADP ribose by placing a methylene group between the N-ribose and nicotinamide moiety. The nitrogen in the nicotinamide ring is replaced by a carbon, giving the ring a neutral charge that would be expected following migration of the positive charge onto the ribosyl group. The C1’ carbon of the N-ribose is replaced by a nitrogen to mimic the cationic character of an oxycarbenium-like species. The pKa of this nitrogen is 8.4, providing a mixture of neutral and cationic forms under the crystallization conditions (Zhou et al., 2004).
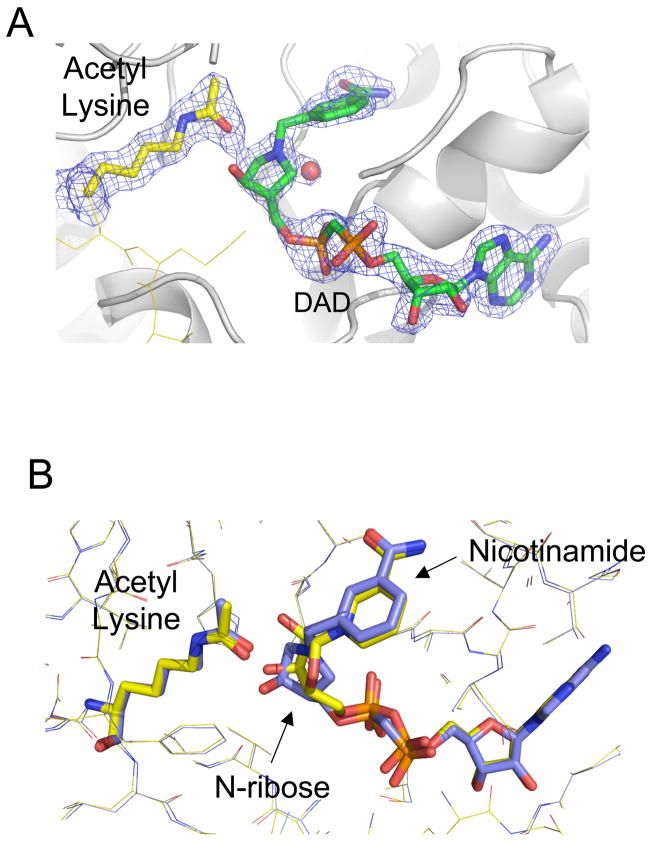
Figure 2. Structure of a Sir2TM-acetylated peptide – DADMe-NAD+ analogue ternary complex.
(A) The 2Fo-Fc (1σ) electron density maps defines the position of the acetyl group and DADMe-NAD+ analogue. (B) Overlaying the Sir2-acetyl peptide DADMe-NAD+ structure (colored in blue) with the Sir2 Michaelis complex (colored in yellow) reveals that the nicotinamide and acetyl lysine moieties remain fixed, while the N-ribose moiety of the DADMe-NAD+ molecule is closer to acetyl lysine.
The overall structure of the Sir2Tm –acetylated peptide – DADMe-NAD+ complex is similar to that of the Michaelis complex of the same sirtuin (Hoff et al., 2006) (Figure 2B). There is little rearrangement of the Sir2Tm active site. The electron density for the DADMe-NAD+ molecule and acetylated lysine residue is well defined and allows for the positioning of these moieties (Figure 2A). The DADMe-NAD+ analogue binds in the substrate binding cleft in a similar conformation to that of NAD+ in the Michaelis complex, differing from the NAD+ conformation in other Sir2-NAD+ complexes (Avalos et al., 2004; Zhao et al., 2003) (Figure 2B). The DAD ribose N1’, analogous to the C1’ of the NAD+ N-ribose, is positioned proximal to the acetylated lysine amide oxygen (Figure 2). The DADMe-NAD+ benzamide ring bound to the C pocket superimposes with the nicotinamide ring in the Sir2 Michaelis complex and is bound by identical Sir2 protein side chains and water contacts (Figure 2B). The ribose of DADMe-NAD+ adopts a similar orientation to the Michaelis complex N-ribose, except that it is closer to the acetyl amide oxygen (Figure 4). The migration of N1’ from DADMe-NAD+ to a position closer to the acetyl amide oxygen while the nicotinamide analogue remains in the same position is expected from a dissociative mechanism, where the ribosyl ribocation migrates from the nicotinamide, gains chemical reactivity at the transition state and completes the reaction coordinate by migrating to react with the fixed carbonyl oxygen. DADMe-NAD+ is larger than NAD+ by the insertion of the methylene bridge (Figure 1). In a SN2 mechanism, this increased atomic size in the NAD+ reaction coordinate would be expected to induce a steric clash to the acetyl-lysine, causing some displacement of the acetyl group. Instead, the structure shows alignment consistent with distances in the catalytic site for a mechanism of ribosyl migration.
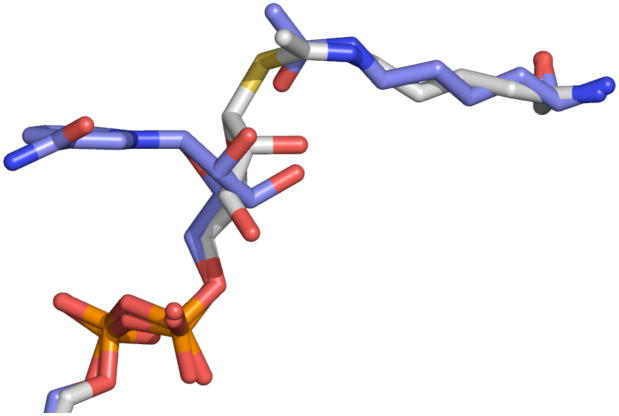
Figure 4. Comparison of the Sir2Tm-Michaelis and Sir2Tm-S-alkylamidate complexes.
The Sir2Tm-Michaelis structure (colored blue) was superimposed on to the Sir2Tm-S-alkylamidate structure (colored in gray). Only small movements of the NAD+ N-ribose and acetyl lysine in the Sir2 active site are necessary to form the S-alkylamidate, or the naturally occurring O-alkylamidate intermediate.
A S-alkylamidate reaction intermediate is trapped in Sir2 crystals
After the glycosidic bond linking nicotinamide to the N-ribose of NAD+ is cleaved, ADP ribose is transferred to the acetyl lysine, forming a proposed O-alkylamidate intermediate (Sauve et al., 2001). While this intermediate has not been directly observed, kinetic experiments (Smith and Denu 2006) and isotope labeling (Sauve, Celic et al. 2001) support its existence. Another study used a thioacetyl peptide to trap a species in the Sir2 deacetylation reaction, and used mass spectrometry supporting a trapped species with the molecular weight corresponding to an S-alkylamidate or 1’2’ bicyclic species (Smith and Denu, 2007a). An average carbon-sulfur bond is 1.8 Å, while a typical carbon-oxygen bond is around 1.4 Å. In the S-alkylamidate compound, the observed carbon sulfur bond distance is 1.79 Å. The elongated carbon-sulfur bond length in the S-alkylamidate could result in reduced turnover, as previously proposed (Smith and Denu 2007). Sir2Tm- acetyl peptide cocrystals are catalytically active when soaked with NAD+, but product turnover is not observed for hours (Hoff et al., 2006). The slow kinetics of the Sir2Tm crystals coupled with the reported eighty-fold decrease in turnover rate for the thioacetyl peptide suggested that it should be possible to trap a S-alkylamidate intermediate in Sir2Tm crystals (Hoff et al., 2006; Smith and Denu, 2007a)). To provide direct evidence for the existence of the O-alkylamidate intermediate, as well as mechanistic insights into the intermediate chemical steps of the Sir2 deacetylation reaction, we trapped an S-alkylamidate intermediate in the crystal and determined its structure at 2.5Å resolution (Figure 3).
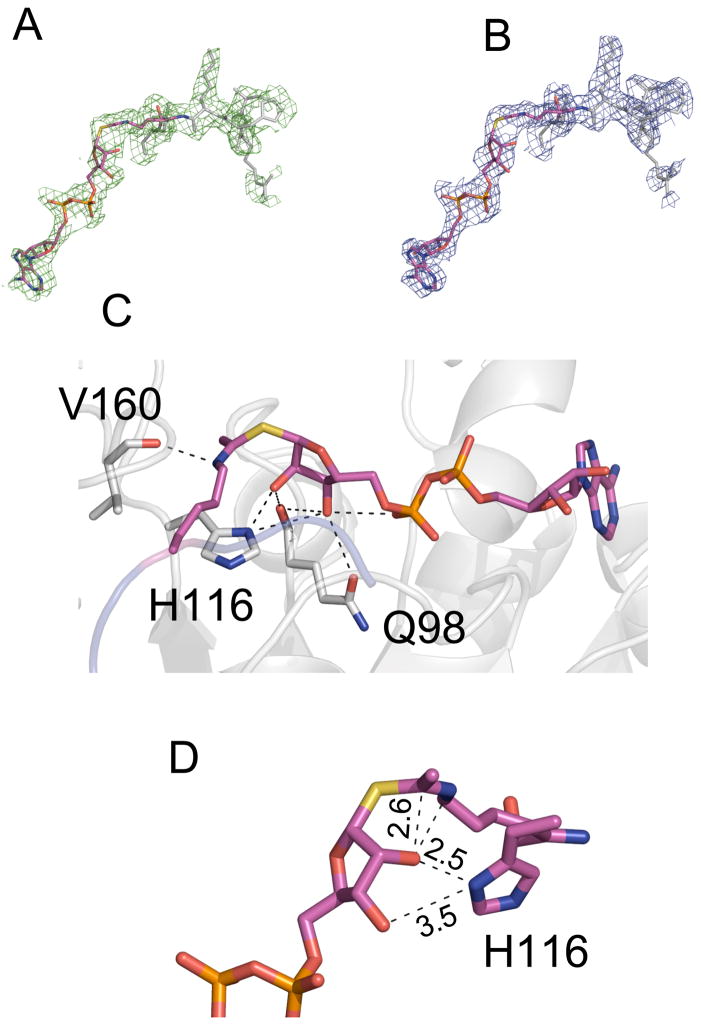
Figure 3. Structure of a Sir2-S-Alkylamidate intermediate complex.
The Fo-Fc omit electron density map (2.5σ) and 2Fo-Fc (1σ) electron density map of the solved complex defining the S-alkylamidate intermediate are displayed in panels (A) and (B) respectively. A hydrogen bond network between Sir2 and the S-alkylamidate (colored in pink) intermediate stabilize the observed conformation of the S-alkylamidate intermediate (C). Distances measured in angstroms between atoms of the S-alkylamidate and Sir2Tm H116 are displayed in panel (D).
A comparison of the Sir2-Michaelis and Sir2-S-alkylamidate complex structures indicates that only small movements are necessary to convert NAD+ and acetyl lysine to the O-alkylamidate intermediate in the Sir2 deacetylation reaction (Figure 4). The phosphate groups undergo little movement, and are in similar positions in both the Sir2-Michaelis and Sir2-S-alkylamidate intermediate complexes. The acetyl lysine amide oxygen moves 0.8 Å, accounting for a rotation of the acetyl group by 30°, while the N-ribose plane tilts 35°, resulting in the N-ribose 2’OH group moving 1.8 Å. The small displacements of the N-ribose and acetyl groups between the Sir2 Michaelis and S-alkylamidate complexes orients the 2’ OH group 2.6 Å from the acetyl group in the S-alkylamidate structure (Figure 3D). In this conformation, the S-alkylamidate 2’OH is oriented to form the 1’ 2’ bicyclic species, which is the next proposed intermediate in the Sir2 deacetylation reaction (Figure 1). As predicted, the relative stability of the S-alkylamidate intermediate could be do to inefficient attack of the 2’OH on the S-alkylamidate due to the increased C-S bond distance of 1.79 Å (Smith and Denu 2007).
The observed S-alkylamidate conformation in the Sir2-S-alkylamidate complex structure is stabilized by an extended hydrogen bond network consisting of backbone and side chain contacts (Figure 3C). Key conserved side chains in this network include His116, Gln98, and Asn99, which aid in positioning the N- ribose moiety of the intermediate in a conformation that is poised to form the bicyclic intermediate. The conserved active site histidine, His116, is 2.5Å and ribose 2’OH and 3.3 Å from the 3’OH group (Figure 3C). By comparison, this histidine is 3.3 Å from both the 2’OH and 3’OH groups in the unreacted Michaelis complex structure (Hoff et al., 2006). We previously suggested that His116 could deprotonate the 2’ alcohol group though a shuttling mechanism where H116 deprotonates the 3’OH, which in turn deprotonates the 2’ alcohol, thus promoting its reactivity towards forming the 1’2’bicyclic species (Hoff et al., 2006). However, the Sir2Tm S-alkylamidate complex structure demonstrates that if H116 is indeed a general base, it can directly deprotonate the N-ribose 2’ alcohol because His116 is in contact with the N-ribose 2’ alcohol (Figure 3D). Based on the Sir2 deacetylation reaction mechanism, the O-alkylamidate intermediate ε-nitrogen would carry a positive charge, which is stabilized by a hydrogen bond to the backbone carbonyl of Val160 (Figure 3C).
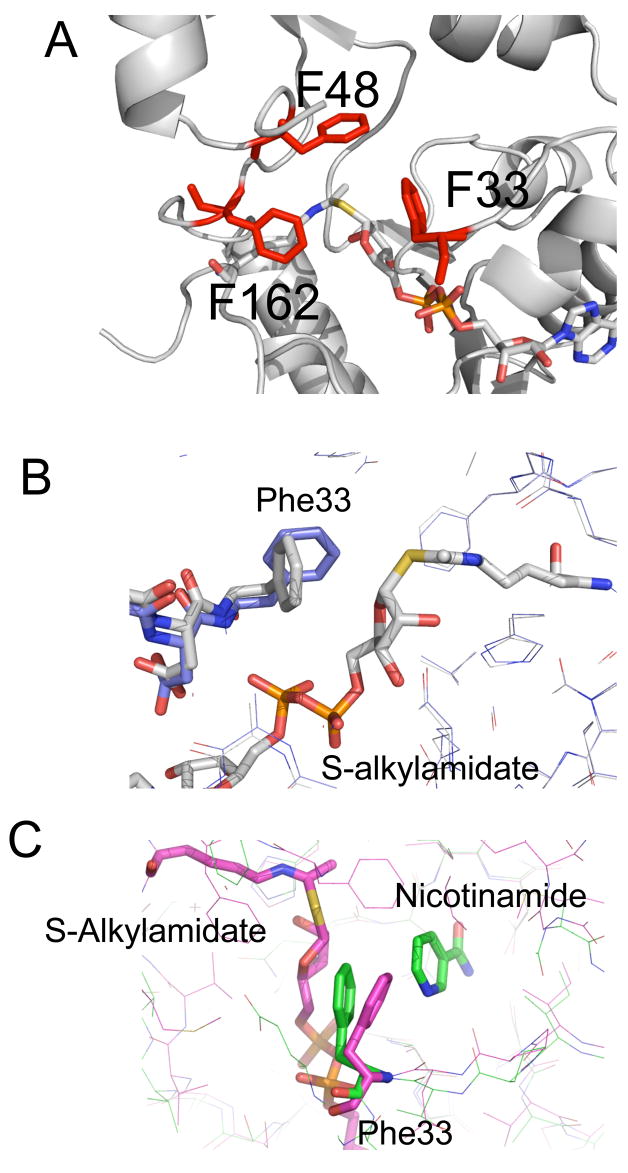
The O-alkylamidate is a sufficiently long-lived intermediate that Sir2 enzymes must shield it from hydrolysis by solvent for a lifetime sufficient to permit base exchange with nicotinamide. Failure to prevent hydrolysis would yield an ADP ribose hydrolysis product as observed for the HST2 H135A mutant enzyme (Smith and Denu 2006). The base exchange reaction - which regenerates the starting reactants, acetyl lysine and NAD+ and is exploited by the cell to regulate sirtuin activity (Sauve and Schramm, 2003) – must be controlled so that the deacetylation reaction is not largely aborted. The O-alkylamidate intermediate is susceptible to methanolysis, which yields β-1’-O-methyl ADP-ribose (Smith and Denu, 2006). Aborting the Sir2 deacetylation reaction could be biologically important in cases where deacetylation is not needed by the cell (Sauve and Schramm 2003). To protect the O-alkylamidate intermediate from these reactions, the Sir2 active site contains a cluster of conserved phenylalanines that shield the intermediate from attack by water. These residues, including Phe33, Phe48 and Phe162, stack against the S-alkylamidate intermediate, physically shielding it from solvent (Figure 5A). Phe48 and Phe162 in the Sir2Tm-S-alkylamidate structure stack against the acetyl lysine. In this conformation, Phe48 and Phe162 are also positioned to protect the O-alkylamidate intermediate. In the Michaelis complex, Phe33 stacks against the nicotinamide moiety (Figure 5B). This Phe33 conformation would not, however, protect the O-alkylamidate intermediate from base exchange or solvent hydrolysis. Instead, Phe33 moves following cleavage of the nicotinamide-ribose glycosidic bond to stack against the O-alkylamidate intermediate, in an orientation that blocks base exchange with nicotinamide (Figures 5B and C). The conformation of Phe33 in the Sir2 S-alkylamidate structure is similar to that observed in the structure of Sir2Tm bound to the products, O-acetyl-ADP ribose and nicotinamide (Avalos et al., 2005) (Figure 5C). In this state, the conformation of Phe 33 in the S-alkylamidate intermediate complex structure would sterically clash with the nicotinamide moiety in both the Sir2 products and Michaelis complex structures, suggesting that Phe 33 may be involved in expelling nicotinamide from the Sir2 active site as previously suggested (Avalos et al., 2005). In the presence of excess nicotinamide, a robust nicotinamide exchange reaction occurs, making the reverse reaction that reforms enzyme-bound N-acetyl peptide and NAD+ more probable than formation of 2’-O-Ac-ADPR, nicotinamide and deacetylated peptide (Jackson et al., 2003; Sauve and Schramm, 2003). There is no electron density corresponding to nicotinamide in the Sir2-S-alkylamidate electron density maps. Based on previous structures of Sir2 bound to nicotinamide, free nicotinamide binds in a cleft called the C-pocket (Avalos et al., 2005). In the S-alkylamidate structure, the dimensions of this C pocket are constricted, and would not accommodate nicotinamide, suggesting that nicotinamide was expelled from this site.
Figure 5. A conserved cluster of phenylalanines protect the peptidyl –imidate intermediate from hydrolysis and base exchange with nicotinamide.
A conserved patch of bulky aromatic residues, colored in red, including Phe33, 48, and 162 shield the S-alkylamidate intermediate from solvent. (B) Sir2Tm Phe33 reorients from its Michaelis complex position (colored in blue) to stack against the S-alkylamidate intermediate (colored in gray). (C) The conformation of Phe 33 in the Sir2Tm-S-alkylamidate intermediate (colored in pink) is similar to its position in the Sir2 products structure (colored in green). In this conformation, Phe33 would protect the S-alkylamidate from base-exchange with nicotinamide.
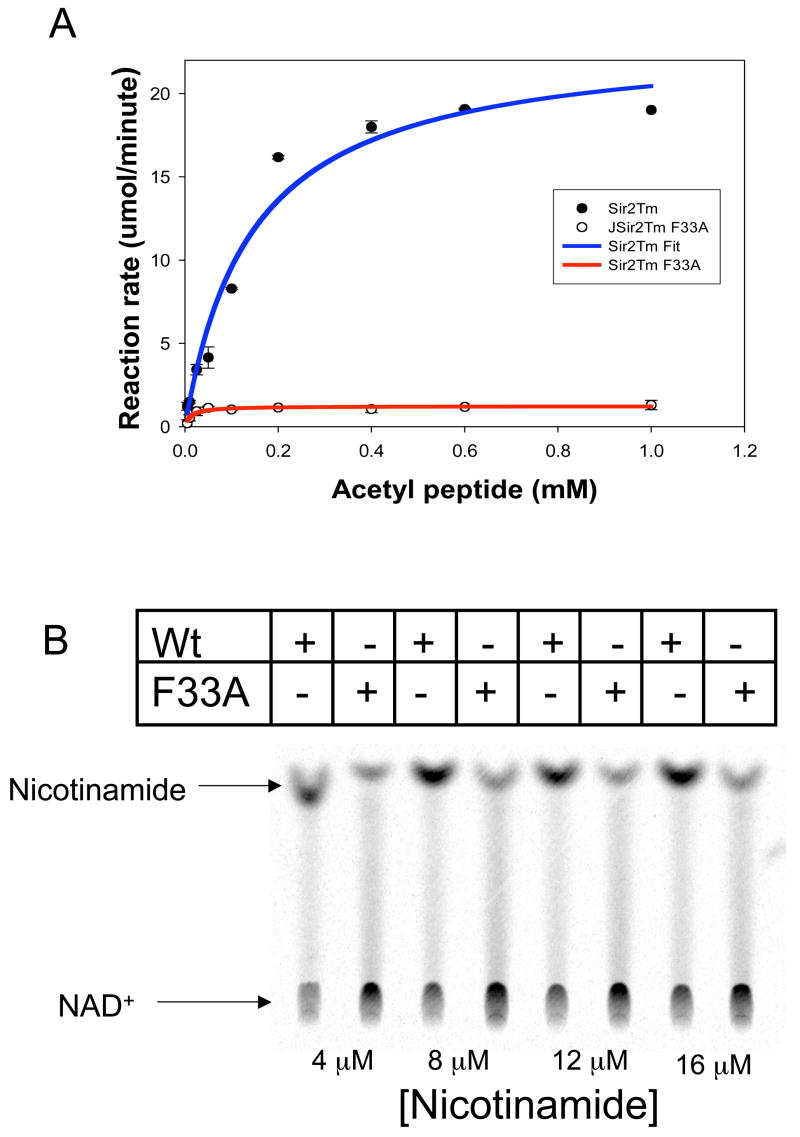
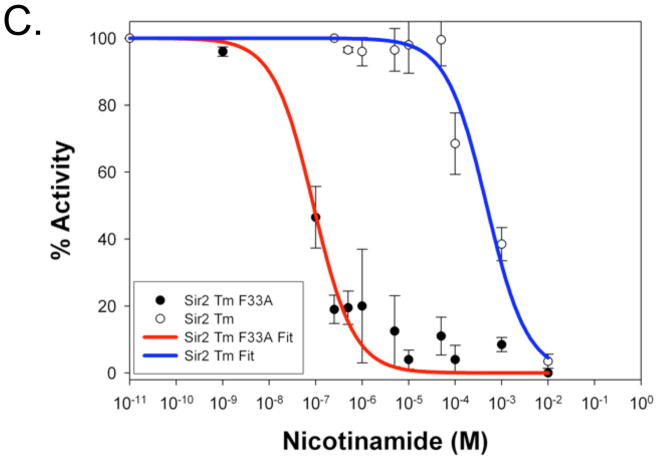
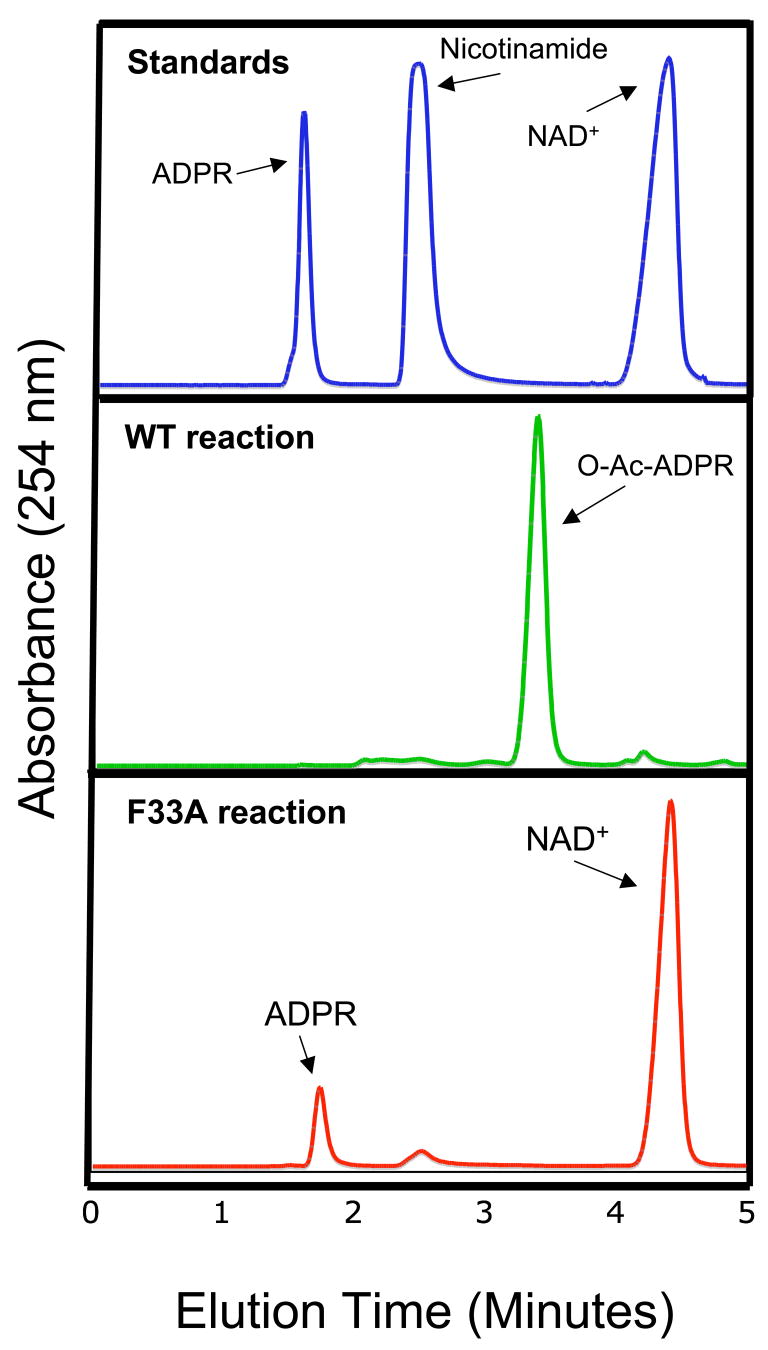
To verify the proposed role of Phe33 in protecting the O-alkylamidate intermediate from hydrolysis and the base exchange reaction with nicotinamide, which regenerates acetyl lysine and NAD+, we produced Sir2Tm F33A mutant protein and compared its enzymatic properties to wild type Sir2Tm. The other phenylalanine residues, F162 and F48, are located in positions critical for acetyl lysine binding or structural integrity of Sir2Tm, so these residues were not mutated. A standard NAD+ consumption assay, in which the NAD+ concentration was fixed at 1 mM and acetyl peptide was varied revealed that the kcat for NAD+ consumption of the Sir2 F33A mutant enzyme, 0.3 sec−1, is significantly lower than that of the wild type protein, 5.9 sec−1 (Figure 6A). To determine if the Sir2Tm F33A protein is more susceptible to nicotinamide inhibition, assays were performed with [14C]nicotinamide at various nicotinamide concentrations (Figure 6B), and the reaction products were run on silica TLC plates to separate unconsumed nicotinamide and NAD+. As can be seen in Figure 6B, there is significantly more base exchange with the Sir2F33A mutant, implicating Phe33 in preventing base exchange (Figure 6B). Consistent with this observation, the F33A mutation makes Sir2Tm more sensitive to nicotinamide inhibition, with an IC50 value for the Sir2Tm F33A enzyme of 0.1 μM, which is at least three orders of magnitude lower than that of the wild type enzyme, 480 μM (Figure 6C) Sir2Tm wild type and Sir2TmF33A reactions were analyzed by reverse phase HPLC (Figure 7). The major nucleotide product observed for the wild type enzyme is O-acetyl-ADP-ribose, while the F33A mutant enzyme reactions contained NAD+ and ADP-ribose. These results are consistent with the role of F33A in shielding the O-alkylamidate intermediate from solvent hydrolysis and base exchange with nicotinamide.
Figure 6. Enzymatic characterization of the Sir2Tm F33A enzyme.
Sir2Tm F33A (red curve) NAD+ consumption was measured and compared to wild type Sir2Tm enzyme (blue curve) (A). The Kcat of the Mutant enzyme, 0.3 sec−1, was 30 fold lower than wild type enzyme, 5.9 sec−1. To determine if Sir2Tm F33A enzyme was more sensitive to nicotinamide inhibition, base exchange assays with [14C] nicotinamide were preformed (B). Reaction products were separated on a silica TLC plate. As observed, the Sir2Tm F33A protein has higher base exchange activity. (C) Nicotinamide inhibition assays were preformed, and Sir2Tm F33A,IC50 of 0.1 μM, is at least three orders of magnitude more sensitive to nicotinamide inhibition than the wild type enzyme, IC50 of 480 μM.
Figure 7. HPLC analysis of Sir2Tm reactions.
The Sir2Tm (green) and Sir2Tm F33A reactions (red) were analyzed by reverse phase HPLC. Standards were run to identify reaction components (Blue trace)
Discussion
The Sir2Tm structures and biochemical studies presented here, taken together with previous work, provide important new insights into the intermediate steps in the Sir2 deacetylation mechanism. A previous study describing the wild-type Michaelis complex containing Sir2Tm bound to both acetylated peptide and NAD+ (Hoff et al., 2006) revealed that the acetyl oxygen of acetyl-lysine was poised to attack the C1’ of NAD+. In the Michaelis complex, the acetyl lysine inserts into the active site through a hydrophobic tunnel proximal to the N-ribose of NAD+, which promotes binding of the nicotinamide moiety of NAD+ in the hydrophobic C pocket. It was proposed (Hoff et al., 2006) that the observed configuration of co-substrates promoted cleavage of the first step in the deacetylation reaction, namely the cleavage of the glycosidic bond to release nicotinamide, in two ways: by burying the charged nicotinamide ring in the hydrophobic C pocket, and by rotating the carboxamide out of plane. In addition to these destabilizing interactions, the adjacent N-ribose is relatively flattened, as compared with the more highly puckered sugar seen in NAD+ that is not constrained to insert into the C pocket (Avalos et al., 2004). The net effect would be to promote the dissociation of nicotinamide and formation of a transition state with oxacarbenium-like character, which could then be attacked by the acetyl oxygen.
The structure of a Sir2Tm ternary complex containing an acetylated peptide and a mimic of a nicotinamide dissociated NAD+ species, DADMe-NAD+ (Figure 2), shows how the active site accommodates this dissociated NAD+ species. The DADMe-NAD+ molecule mimics a dissociated NAD+ species, where positive charge develops on the N-ribose ring, and the nicotinamide-N-ribose bond is elongated (Figure 1B). As expected for a dissociative-like transition state, the Sir2Tm-DADMe-NAD+-acetyl peptide shows that the acetyl lysine and nicotinamide remain fixed relative to their Michaelis complex positions, while the reactive riboxacarbenium migrates towards and is trapped by the acetyl lysine forming the O-alkylamidate intermediate.
The Sir2Tm –S-alkylamidate structure gives the first structural evidence for the existence of the unusual O-alkylamidate chemical species and provides mechanistic insight into the intermediate steps in the Sir2 reaction (Figure 3). The active site histidine, His116 in Sir2Tm, has been proposed to act as a general base by abstracting a hydrogen from the N-ribose 2’ OH to form the bicyclic intermediate (Hoff et al., 2006; Smith and Denu, 2006). Kinetic studies of Sir2 enzymes, including Sir2Tm and HST2, demonstrate that mutating the catalytic histidine, His116, to alanine modestly reduces the enzymatic activity (Smith and Denu, 2006). The Sir2-S-alkylamidate structure reveals why this histidine residue is dispensable for Sir2 deacetylation. In the S-alkylamidate structure, His116 positions the N-ribose of the intermediate (Figure 3C). Besides His116, other Sir2 side chain and backbone atoms contact the S-alkylamidate intermediate, including the universally conserved residue Gln98 (Figure 3C). Mutating His116 to alanine would result in the loss of two hydrogen bonds to the N-ribose, but because there are 4 other hydrogen bonds, this deletion would not be expected to significantly alter the conformation of the S-alkylamidate intermediate. This suggests that mutation of H116 to alanine only slightly lowers Sir2 activity because other Sir2 backbone and side chain hydrogen bonds are in place to position the peptidyl-imidate intermediate for subsequent chemical steps (Figures 3C and D). Subsequent formation of the bicylic species could then occur (Figure 3D). Once the O-alkylamidate intermediate adopts the proper conformation, the reactivity of this species could then drive the formation of the 1’ 2’ bicyclic intermediate.
Sir2 enzymes must protect the peptidyl-imidate intermediate from hydrolysis, which would abort the deacetylation reaction. Failure to shield the peptidyl-imidate intermediate would produce ADP ribose hydrolysis products and not the proper product, 2’-O-acetyl ADP ribose (Smith and Denu 2006). Two atoms in the peptidyl-imidate intermediate that are sensitive to hydrolysis are the C1’ of the N-ribose moiety and the carbon from the acetyl group (Figure 5A). Our structure reveals that, while a small portion of the intermediate is exposed to solvent, including the ribose ring oxygen, S-alkylamidate sulfur, and methyl group from the acetyl moiety, no atoms susceptible to hydrolysis are solvent exposed (Figure 5A). Conserved aromatic residues including Phe33, Phe48, and Phe162 protect the O-alkylamidate intermediate from hydrolysis. ADP-ribose is a reaction product of the Sir2F33A enzyme (Figure 7), demonstrating that this conserved residue protects the O-alkylamidate from solvent hydrolysis.
Sir2 enzymes permit the O-alkylamidate intermediate to react with nicotinamide in a base-exchange reaction, but this intermediate must also be converted to the 1’,2’-bicyclic intermediate to complete deacetylation (Figure 1). When nicotinamide reacts with the O-alkylamidate intermediate, it becomes a potent inhibitor of sirtuins in vitro and in vivo (Bitterman et al., 2002)Yeast treated with nicotinamide show a decreased level of transcriptional silencing, similar to that observed when Sir2 is deleted (Bitterman et al., 2002). It was previously demonstrated that free nicotinamide binds in a region called the C pocket, which is also were the nicotinamide moiety of NAD+ binds (Avalos et al., 2005). Free nicotinamide bound in the C pocket can then undergo base-exchange with the O-alkylamidate intermediate to regenerate NAD+ (Avalos et al., 2004; Avalos et al., 2005; Hoff et al., 2006). To prevent base exchange, Phe33 stacks against the ribose moiety of the S-alkylamidate intermediate, physically blocking the C1’ from attack by nicotinamide (Figure 5C). Mutation of this conserved phenylalanine in yeast dramatically lowers Sir2 mediated gene silencing and in vivo activity (Armstrong et al., 2002).
Previous structural studies had indicated that Phe33 can adopt multiple conformations and could serves as a “gate keeper” that regulates base exchange (Avalos et al., 2005). The active site from the Sir2 Michaelis complex (Hoff et al., 2006) and the Sir2 S-alkylamidate complex described here are highly similar except for the conformation of Phe33 and the diameter of the nicotinamide exit tunnel (Figure 6). The conformation of F33 in the Sir2- S-alkylamidate is consistent with our earlier proposal that this side chain could stabilize an oxacarbenium intermediate through pi-cation interactions (Hoff et al., 2006). Phe33 adopts a conformation in the Sir2-S-alkylamidate structure that constricts the putative nicotinamide exit tunnel observed in previous Sir2Tm structures (Figure 5). This conserved phenylalanine adopts multiple conformations in different structures, and in the Michaelis conformation, would allow for diffusion of nicotinamide out of the active site upon formation of the O-alkylamidate intermediate (Figure 5B). The conformation of Phe33 in the S-alkylamidate complex would clash with the nicotinamide moiety of NAD+ in the Sir2 Michaelis complex conformation, suggesting Sir2 enzymes could use Phe33 to expel nicotinamide from the active site through an exit tunnel (Figures 5B and C). As an added safeguard, the conformation of Phe33 further constricts the exit tunnel after expelling nicotinamide, thereby blocking its reentry into the active site for as long as the Phe33 side chain remains in the observed position. The importance of Phe33 is further highlighted by comparing the enzymatic properties of Sir2Tm F33A to the wild type enzyme. Sir2Tm F33A is three orders of magnitude more sensitive to nicotinamide inhibition, has a lower Kcat for NAD+ consumption, and greater amount of nicotinamide base exchange activity (Figure 6). Further, the Sir2Tm F33A mutant protein had ADP ribose as a reaction product, which was not observed for the Sir2Tm wild type enzyme (Figure 7). These biochemical results, in conjunction with the Sir2Tm-S-alkylamidate intermediate structure, emphasize that Phe33 is a “gate keeper” for regulating Sir2 mediated base exchange and solvent hydrolysis. This residue is found in virtually all sirtuins, pointing to the importance of this residue in regulating Sir2 chemistry. The structures and biochemical data presented here reveal insight into intermediate steps of the sir2 deacetylation, and help motivate future studies to further characterize Sir2 reaction intermediates.
II. Experimental procedures
Protein expression, purification and crystallization
Thermotoga maritima Sir2 enzymes was expressed and purified as previously described (Smith, Avalos et al. 2002). A dissociative NAD+ analogue, DADMe-NAD+ was designed to contain a neutral analogue of nicotinamide and a cationic mimic of the ribocationic group for a dissociated nicotinamide-ribosyl bond. Synthesis also included a methylene bridge to provide stability in the pyrophosphate bridge linking the AMP and NMN analogue groups (Zhou et al., 2004). Geometry appropriate to a dissociated NAD+ state was provided by a methylene group linking the nicotinamide and ribosyl cation mimics (Figure 1).
To produce crystals of the Sir2Tm DAD NAD+-acetyl peptide complex, Sir2Tm at 16 mg/ml was mixed with 5 ul of 40 mM P53 acetylated p53 peptide, KKGQSTSRHK(Kac)LMFKTEG, to give a final solution containing 10 mg/ml Sir2Tm and 4 mM acetyl peptide. Crystals of the Sir2Tm-acetyl P53 peptide were grown in 100 mM CHES (pH 9.5) and 15 % PEG 3350 by vapor diffusion. The crystals formed in the space group P212121 with unit cell dimensions a = 45.8 Å, b = 59.7 Å , and c = 106 Å. Crystals were then soaked for 13 hours in cryoprotectant with DADMe-NAD+ ( 100 mM CHES pH 9.5, 5 mM DAD NAD, 20 % PEG 3350).
To produce crystals of the Sir2- S-alkylamidate intermediate complex, 25 ul of Sir2Tm (20 mg/ml) was incubated one hour on ice with 5 ul of an N-ε-thioacetyl-lysine peptide (22 mM) derived from the p53 sequence, KKGQSTSRHK(Ks-ac)LMFKTEG (Fatkins et al., 2006). 1 ul of Sir2-thiopeptide solution was then mixed with 1 uL of a crystallization solution consisting of 13–15% PEG3350 (w/v) and 0.1 M CHES (pH 9.5). The hanging drop vapor diffusion method was used to grow crystals of the Sir2-thiopeptide complex. After 2 days, crystals appeared of the Sir2-peptide complex. The crystals formed in the space group P212121 with unit cell dimensions a = 46.5 Å, b = 60.4 Å , and c = 107 Å. Crystals were allowed to grow for an additional week, and then soaked for 0.5 h – 13 h in a cryoprotective solution containing 20% PEG3350, 0.1 M ChES (pH 9.5), and 0.5 mM NAD+. Soaks with higher NAD+ concentrations resulted in significant crystal cracking. After soaking, crystals were flash frozen in liquid nitrogen.
Sir2-DADMe-NAD+-acetyl P53 peptide complex crystallographic data collection and processing
Electron diffraction data were recorded from frozen Sir2-DADMe-NAD+-acetyl P53 peptide crystals at beamline GM/CA-CAT24D at the Argonne National Laboratory equipped with a quantum CCD detector. Data were reduced with HKL2000 (Otwinowski and Minor, 1997) and CCP4 (CCP4, 1994). Initial phases were calculated using Sir2TM bound to P53 acetyl peptide (PDB id 2H4F). Model building was done with Coot (Emsley and Cowtan 2004) and refinement was performed with CNS (Brunger et al., 1998) and CCP4 (CCP4, 1994). Crystallographic statistics are summarized in Table 1.
Table 1.
Crystallographic statistics. Numbers in parentheses indicate statistics for the outermost shell.
| Crystal | Thio intermediate | DAD TS structure |
|---|---|---|
| Diffraction Data | ||
| Space group | P212121 | P212121 |
| Unit cell (Å) | 46.5, 60.4, 107 | 45.8, 59.7,106 |
| Resolution | 2.5 Å | 1.9 Å |
| Measured reflections | 40,888 | 60,754 |
| Unique reflections | 10,834 | 22,339 |
| Completeness (%) | 99.5 (96.0) | 99.6 (96.4) |
| Avg I/Sigma | 14.3 | 15.2 |
| Multiplicity | 2.1 | 3.2 |
| Rmerge | 0.11 (.45) | 0.10 (.37) |
| Refinement | ||
| Resolution range (Å) | 2.5 | 50–1.9 |
| Reflections | 10834 | 22339 |
| Working | 10287 | 21169 |
| Test (5%) | 547 | 1170 |
| Total atoms | 1982 | 2039 |
| Protein | 1845 | 1770 |
| peptide | 68 | 92 |
| S-alkylamidate | 40 | ------- |
| Water molecules | 87 | 145 |
| R (%) | 22.4% | 19.7% |
| Rfree (%) | 24.5% | 22.9% |
| rmsd | ||
| Bond lengths (Å) | 0.015 | .016 |
| Bond angles (°) | 1.6 | 1.4 |
Sir2-S-Alkylamidate complex crystallographic data collection and processing
Diffraction data were recorded from frozen Sir2-thiopeptide crystals at the BioCARS facility at Argonne National Laboratory on the 14-BM-C beamline equipped with a ADSC Quantum-315 detector. Data were processed and reduced with HKL2000 (Otwinowski and Minor, 1997) and CCP4 (CCP4, 1994). The structure was solved by molecular replacement with MOLREP implemented in CCP4 using the apo structure of Sir2Tm (PDB id 2H4F) as the search model. The initial molecular replacement solution was subjected to rigid body refinement, followed by conjugate gradient and simulated annealing refinement in CNS (Brunger et al., 1998).
The structure of Sir2Tm bound to a reactive intermediate was solved by molecular replacement using the Sir2 apoenzyme as the search model, and difference maps were inspected to determine if the S-alkylamidate intermediate was present. The intermediate was fit to the electron density and included in the remaining stages of refinement. Removal of the S-alkylamidate intermediate and refinement of the Sir2-peptide complex increase R and Rfree by 1.5 percentage points. Because the sulfur is electron rich, its position was confirmed by contouring the simulated annealing omit Fo-Fc map at 5 sigma. To investigate whether other intermediates were present in the crystal, the proposed 1,2 bicyclic intermediate (Figure 1), substrates, and Sir2 reaction products were also modeled but did not fit the electron density. With the occupancy set at 1, the average B factor for the S-alkylamidate intermediate is 77 Å2. Since the B factor and occupancy are coupled parameters, an elevated B factor could suggest a lower occupancy for the S-alkylamidate intermediate. Reducing the occupancy of the intermediate to 0.7 gave an average B factor for the intermediate of 33 Å2 with similar R and Rfree values, and virtually identical electron density. Because the intermediate was obtained by soaking crystals, it is likely that some of the Sir2 complexes in the crystal do not contain the S-alkylamidate intermediate, and this would result in a lower occupancy for the S-alklamidate intermediate.
The S-alkylamidate intermediate and flexible loop regions were built into difference electron density maps using Coot (Emsley and Cowtan 2004). There was clear density defining the S-alkylamidate covalent linkage, N-ribose moiety, and phosphate groups. The density defining the adenine ring was ambiguous in the Fo-Fc omit maps, but improves in the 2Fo-Fc electron density maps of the solved structure. Crystallographic statistics are summarized in Table 1. Figures were generated with Pymol (Delano, W.L.).
Enzymatic assays
NAD+ consumption assays were preformed with the same reaction conditions as previously described (Avalos et al., 2004). Briefly, 50 ug/ml of Sir2Tm wild type of Sir2Tm F33A protein were incubated at 37°C for 15 minutes with 1 mM of NAD+, but the NAD+ concentration was quantified using a fluorescence based assay described in Putt et al., 2004 for Sir2Tm and Sir2tm F33A. Data were fit with the Michaelis Menton equation in SigmaPlot. Nicotinamide base exchange and inhibition reactions were preformed as previously described (Avalos et al., 2005). 50 ug/ml of Sir2Tm wild type of Sir2Tm F33A protein were incubated at 37°C for 15 minutes with 125 uM of NAD+, 500 um of acetyl P53 peptide, and various concentrations of [14C]nicotinamide. Base exchange reactions were resolved on a silica TLC plate with and 80 ethanol: 20 ammonium acetate buffer. Nicotinamide inhibition reactions were preformed with 50 ug/ml of Sir2Tm wild type or Sir2Tm F33A protein were incubated at 37°C for 15 minutes with 125 uM of NAD+, 500 uM of acetyl P53 peptide, and various concentrations of nicotinamide. Nicotinamide inhibition data were fit with an equation 1 to determine the IC50 value for nicotinamide in SigmaPlot :
Where Vo is the initial rate of uninhibited reaction and vI is the initial rate of reaction in the presence of inhibitor.
HPLC analysis of Sir2 reactions
Sir2Tm enzyme, 50 ug/ml, was incubated with 125 uM NAD+ and 125 um of acetyl p53 peptide at 37°C for various times (5 minutes – 6 hours). Reactions were resolved by reverse phase HPLC on a Waters Nova-PAK C18 3.9x150 mm column. Buffer A was 0.05% TFA and buffer B was 0.05 % TFA in acetonitrile. A 0–40% gradient of Buffer B was run with a flow rate of 1 ml/minute.
Supplementary Material
Acknowledgments
We thank Robert Henning, Michael Bolbat, and the staff at BioCARS beamline 14-BMC for assistance during data collection; R. Sanishvili for help at beamline GM/CA-CAT; Michael Love (Johns Hopkins University) for computational support; Kamau Fahie for technical advice for HPLC; and Trevor Huyton for assistance with crystallography. C.W. and V.L.S. are members of the scientific advisory board of Sirtris Pharmaceuticals.
This work was supported by NSF grant 0615815 to C.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armstrong CM, Kaeberlein M, Imai SI, Guarente L. Mutations in Saccharomyces cerevisiae gene SIR2 can have differential effects on in vivo silencing phenotypes and in vitro histone deacetylation activity. Mol Biol Cell. 2002;13:1427–1438. doi: 10.1091/mbc.01-10-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalos JL, Bever KM, Wolberger C. Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol Cell. 2005;17:855–868. doi: 10.1016/j.molcel.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Avalos JL, Boeke JD, Wolberger C. Structural basis for the mechanism and regulation of Sir2 enzymes. Mol Cell. 2004;13:639–648. doi: 10.1016/s1097-2765(04)00082-6. [DOI] [PubMed] [Google Scholar]
- Avalos JL, Celic I, Muhammad S, Cosgrove MS, Boeke JD, Wolberger C. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol Cell. 2002;10:523–535. doi: 10.1016/s1097-2765(02)00628-7. [DOI] [PubMed] [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Chang JH, Kim HC, Hwang KY, Lee JW, Jackson SP, Bell SD, Cho Y. Structural basis for the NAD-dependent deacetylase mechanism of Sir2. J Biol Chem. 2002;277:34489–34498. doi: 10.1074/jbc.M205460200. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fatkins DG, Monnot AD, Zheng W. Nepsilon-thioacetyl-lysine: a multi-facet functional probe for enzymatic protein lysine Nepsilon-deacetylation. Bioorg Med Chem Lett. 2006;16:3651–3656. doi: 10.1016/j.bmcl.2006.04.075. [DOI] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Pavletich NP. Structure of the histone deacetylase SIRT2. Nat Struct Biol. 2001;8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- Garcia-Salcedo JA, Gijon P, et al. A chromosomal SIR2 homologue with both histone NAD-dependent ADP-ribosyltransferase and deacetylase activities is involved in DNA repair in Trypanosoma brucei. Embo J. 2003;22(21):5851–62. doi: 10.1093/emboj/cdg553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity J, Gardner JG, et al. N-lysine propionylation controls the activity of propionyl-CoA synthetase. J Biol Chem. 2007 doi: 10.1074/jbc.M704409200. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126(5):941–54. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Hoff KG, Avalos JL, Sens K, Wolberger C. Insights into the sirtuin mechanism from ternary complexes containing NAD+ and acetylated peptide. Structure. 2006;14:1231–1240. doi: 10.1016/j.str.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Jackson MD, Denu JM. Structural identification of 2’- and 3’-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta-NAD+-dependent histone/protein deacetylases. J Biol Chem. 2002;277:18535–18544. doi: 10.1074/jbc.M200671200. [DOI] [PubMed] [Google Scholar]
- Jackson MD, Schmidt MT, Oppenheimer NJ, Denu JM. Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J Biol Chem. 2003;278:50985–50998. doi: 10.1074/jbc.M306552200. [DOI] [PubMed] [Google Scholar]
- Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Celic I, Avalos J, Deng H, Boeke JD, Schramm VL. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry. 2001;40:15456–15463. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Moir RD, Schramm VL, Willis IM. Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition. Mol Cell. 2005;17:595–601. doi: 10.1016/j.molcel.2004.12.032. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Munshi C, Lee HC, Schramm VL. The reaction mechanism for CD38. A single intermediate is responsible for cyclization, hydrolysis, and base-exchange chemistries. Biochemistry. 1998;37:13239–13249. doi: 10.1021/bi981248s. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Schramm VL. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry. 2003;42:9249–9256. doi: 10.1021/bi034959l. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Schramm VL. SIR2: the biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates. Curr Med Chem. 2004;11:807–826. doi: 10.2174/0929867043455675. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Wolberger C, Schramm VL, Boeke JD. The Biochemistry of Sirtuins. Annu Rev Biochem. 2006 doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- Schramm VL, Shi W. Atomic motion in enzymatic reaction coordinates. Curr Opin Struct Biol. 2001;11:657–665. doi: 10.1016/s0959-440x(01)00269-x. [DOI] [PubMed] [Google Scholar]
- Smith BC, Denu JM. Sir2 protein deacetylases: evidence for chemical intermediates and functions of a conserved histidine. Biochemistry. 2006;45:272–282. doi: 10.1021/bi052014t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BC, Denu JM. Mechanism-based inhibition of sir2 deacetylases by thioacetyl-lysine Peptide. Biochemistry. 2007a;46:14478–14486. doi: 10.1021/bi7013294. [DOI] [PubMed] [Google Scholar]
- Smith BC, Denu JM. Sir2 deacetylases exhibit nucleophilic participation of acetyl-lysine in NAD+ cleavage. J Am Chem Soc. 2007b;129:5802–5803. doi: 10.1021/ja070162w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Avalos J, Celic I, Muhammad S, Wolberger C, Boeke JD. SIR2 family of NAD(+)-dependent protein deacetylases. Methods Enzymol. 2002;353:282–300. doi: 10.1016/s0076-6879(02)53056-1. [DOI] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, et al. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99(7):735–45. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- Zhao K, Chai X, Marmorstein R. Structure of the yeast Hst2 protein deacetylase in ternary complex with 2’-O-acetyl ADP ribose and histone peptide. Structure. 2003;11:1403–1411. doi: 10.1016/j.str.2003.09.016. [DOI] [PubMed] [Google Scholar]
- Zhou GC, Parikh SL, Tyler PC, Evans GB, Furneaux RH, Zubkova OV, Benjes PA, Schramm VL. Inhibitors of ADP-ribosylating bacterial toxins based on oxacarbenium ion character at their transition states. J Am Chem Soc. 2004 doi: 10.1021/ja038159+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.