Abstract
BRD4, which is a member of the BET (bromodomains and extraterminal) protein family, interacts preferentially with acetylated chromatin and possesses multiple cellular functions in meiosis, embryonic development, the cell cycle, and transcription. BRD4 and its family members contain two bromodomains known to bind acetylated lysine, and a conserved ET domain whose function is unclear. Here we show the solution structure of the ET domain of mouse BRD4, which provides the first three-dimensional structure of an ET domain in the BET family. We determined the NMR structure of BRD4-ET with a root-mean-square deviation of 0.41 Å for the backbone atoms in the structured region of residues 608–676 on the basis of 1793 upper distance limits derived from NOE intensities measured in three-dimensional NOESY spectra. The structure of the BRD4-ET domain comprises three α-helices and a characteristic loop region of an irregular but well-defined structure. A DALI search revealed no close structural homologs in the current Protein Data Bank. The BRD4-ET structure has an acidic patch that forms a continuous ridge with a hydrophobic cleft, which may interact with other proteins and/or DNA.
Keywords: BET protein family, bromodomain, chromatin, ET domain, histone, NMR
The bromodomain-containing protein 4 (BRD4) is a member of the BET (bromodomains and extraterminal) protein family, a class of transcriptional regulators whose members are present in animals, plants, and fungi (Florence and Faller 2001; Wu and Chiang 2007). The BET proteins typically have two tandem N-terminal bromodomains followed by an ET domain. As predicted by the presence of bromodomains, these proteins have been found to be associated with acetylated chromatin. BET proteins are involved in diverse cellular phenomena such as meiosis, cell-cycle control, and homeosis and have been suggested to modulate chromatin structure and affect transcription via a sequence-independent mechanism (Wu and Chiang 2007).
For the bromodomains, including BET-type bromodomains, abundant structural information is available, and their function as acetylated lysine-binding domains is well characterized (Mujtaba et al. 2007; Nakamura et al. 2007; Wu and Chiang 2007). In addition, the crystal structure of papillomavirus DNA tethering complex E2 with the C-terminal fragment 1343–1362 of human BRD4 has recently been determined (Abbate et al. 2006). In contrast, so far no structural information is available for the ET domain of any of the BET proteins, presumably because attempts to crystallize the ET domain have been unsuccessful. The function of the ET domain is assumed to be protein binding. For example, the C-terminal region of yeast Bdf1 containing an ET domain is responsible for the interaction with the Taf67 (TAF7) subunit of TFIID (Matangkasombut et al. 2000). The C-terminal region of BRD4 containing the ET domain also interacts with Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen 1 (LANA-1) (Ottinger et al. 2006; You et al. 2006). However, the molecular mechanism for these interactions remained elusive because of the lack of structural information for the ET domain.
Here, we report the solution structure of the ET domain (residues 601–683) of mouse BRD4 (BRD4-ET). Based on structural features of the BRD4-ET domain, we discuss possible roles of the ET domain.
Results and Discussion
Structure determination
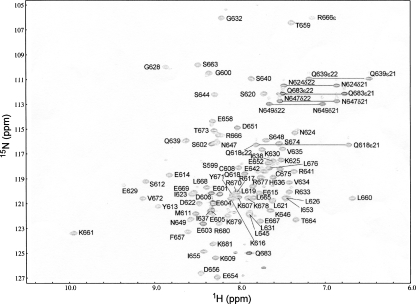
The protein construct used for the structure determination comprised 90 amino acid residues, including seven nonnative residues at its N terminus that are related to the expression system used (see Materials and Methods). The C-terminal 83 residues constitute the BRD4-ET domain, corresponding to residues 601–683 of the full-length mouse BRD4 protein. The line widths of the signals in the NMR spectra were indicative of a monomeric protein (Fig. 1). The resonance assignments of the backbone amide protons and the nonlabile protons are complete for the structured region of the protein, residues 608–676 (Fig. 2A). Among the labile side-chain protons, the amide groups of all asparagine and glutamine residues and the ε-proton resonance of Arg666 were assigned using intraresidual NOEs.
Figure 1.
Two-dimensional [15N,1H]-HSQC spectrum of BRD4-ET. Cross peaks are labeled with the one letter code and the sequence number of the corresponding amino acid.
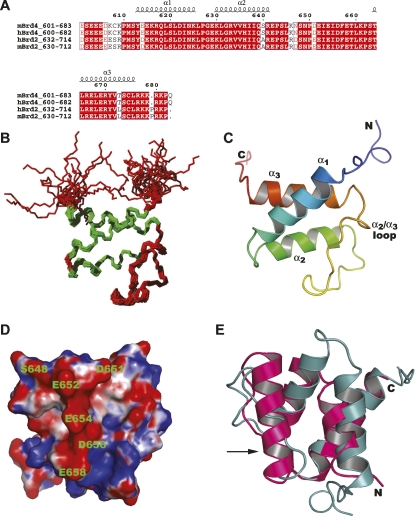
Figure 2.
Solution structure of mouse BRD4-ET. (A) Sequence alignment of BRD4-ET (residues 601–683) with its homologs. The colors are chosen according to the similarity (red box and white character for conserved residues; red character for similarity in a group; blue frame for similarity across groups). (B) Superposition of the backbone atoms of the 20 conformers that represent the solution structure of BRD4-ET for best fit of the backbone atoms N, Cα, C′ of residues 608–676. The α-helical segments are indicated in green. (C) Ribbon diagram of the BRD4-ET domain, viewed as in B, and colored from blue (N terminus) to red (C terminus). (D) Electrostatic surface potential of the BRD4-ET domain showing the acidic region. The surface is colored red and blue for potential values below −5k B T and above +5k B T, respectively, where k B is the Boltzmann constant and T the room temperature. (E) Superposition of the BRD4-ET domain (cyan) with the N-terminal SAP domain of SUMO ligase PIAS1 (pink) (PDB code 1V66). An extra helix (indicated by the arrow) in the SAP domain is replaced by the long α2/α3 loop in the BRD4-ET domain structure.
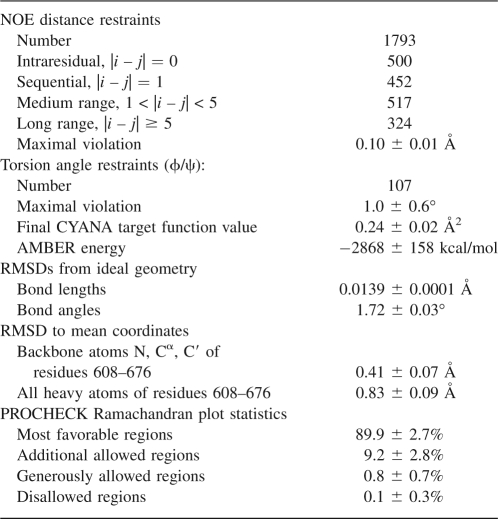
Of the 2944 cross peaks that were identified in the three-dimensional NOESY spectra, 99.6% could be assigned by the program CYANA (Güntert 2003). The fact that virtually all NOEs can readily be assigned and are consistent with a monomeric structure makes it unlikely that under the conditions of the NMR experiments the protein could form a stable multimeric structure. Also, surface plasmon resonance measurements (see below) showed that the BRD4-ET domain has no self-association activity, which further supports the existence of BRD4-ET as a monomer in solution. Almost 20 NOE distance restraints per residue, including 324 long-range distance restraints between protons five or more residues apart in the sequence, were used in the final structure calculations with CYANA and in the subsequent restrained energy refinement with the program OPALp (Koradi et al. 2000). The 20 energy-minimized conformers that represent the solution structure of the BRD4-ET domain (Fig. 2B) showed excellent agreement with the experimental data and are well defined with RMSD values to the mean coordinates of 0.41 Å for the backbone and 0.83 Å for all heavy atoms in the structured region of residues 608–676 (Table 1). More than 99% of the (φ,ψ) backbone torsion angle pairs were found in the most favored or additionally allowed regions of the Ramachandran plot. The solution structure of BRD4-ET and the conformational restraints were deposited in the Protein Data Bank (PDB) with accession code 2JNS. The chemical shifts were deposited in the Biological Magnetic Resonance Data Bank (BMRB; www.bmrb.wisc.edu) with accession number 15125.
Table 1.
Statistics for the NMR solution structure of BRD4-ET
Solution structure of BRD4-ET
The BRD4-ET domain is an α-helical protein with three α-helices, Tyr613–Lys625 (α1), Lys630–Arg641 (α2), and Thr664–Leu676 (α3) (Fig. 2C). The helices have most of the characteristic CO(i)–NH(i + 4) hydrogen bonds. In addition, several 310-helix type hydrogen bonds are present in the majority of the conformers at the end of helix α1, in helix α2, and for CO(642)–NH(645), CO(645)–NH(648), CO(649)–NH(652), and CO(656)–NH(660) in the loop region connecting helices α2 and α3 (α2/α3 loop). Helix α2 is arranged antiparallel with respect to the helices α1 and α3. The hydrophobic core of the protein includes the residues with less than 10% solvent accessible surface area, Met611, Leu619, Ile623, Leu626, Val634, Val635, Ile637, Asn639, Ile653, Phe657, Leu660, Thr664, Leu665, Leu668, and Val672. Almost all of these buried residues are located in the three helices α1, α2, and α3 or in the α2/α3 loop (see below).
The most characteristic feature of the BRD4-ET structure is the α2/α3 loop region of residues 642–663 between helices α2 and α3. The α2/α3 loop has no canonical regular secondary structure. Nevertheless, it is well defined by a large number of NOE restraints, contributes to the hydrophobic core of the protein, and contains 310-helix-like turns at residues 643–645 and 650–652. Three proline residues, including Pro643 and Pro650 at the N termini of the 310-helix-like turns, prevent the formation of longer α-helices. The 135 long-range distance restraints involved in the α2/α3 loop region correspond to an average of 6.1 long-range distance restraints per residue in the α2/α3 loop, compared with 4.7 long-range distance restraints per residue over the entire structured region of the protein, residues 608–676. The presence of many long-range NOE restraints indicated that the loop possesses a well-defined conformation and is not more flexible than the rest of the protein. The loop is stabilized mainly by hydrophobic interactions. The five hydrophobic residues Leu645, Ile653, Ile655, Phe657, and Leu660 in the α2/α3 loop point toward the interior of the protein and interact with buried hydrophobic residues located in the three helices (i.e., Leu619 and Ile623 of α1; Val634, Val635, and Ile638 of α2; and Leu665 of α3). Five out of the six negatively charged residues in the α2/α3 loop, Asp651, Glu652, Glu654, Asp656, and Glu658 are exposed on the protein surface where they form a negatively charged stretch. In contrast, the Glu642 side-chain carboxyl group forms an interior salt bridge with the side-chain amide group of Lys661. Overall, the long α2/α3 loop has a special and rather fixed conformation that is the most characteristic feature of the BRD4-ET structure. This unique conformation results in two adjacent pockets which may be utilized for protein–protein interaction. One pocket is formed by Glu642, Leu645, and Ser648 and bordered by the negatively charged residues Glu652, Glu654, Asp656, and Glu658. The other pocket is formed by Tyr613, Arg617, and Asn624 from α1 and Leu63 from α2, bordered by Asp651, Glu654, Asp656, and Glu658 (Fig. 2D).
A search for structural homologs using the program DALI (Holm and Sander 1993) revealed no proteins in the PDB with a high structural similarity to BRD4-ET. The best match with the structured region of BRD4-ET, residues 608–677, was found for the N-terminal SAP domain of the SUMO ligase PIAS1 (PDB accession code 1V66) (Okubo et al. 2004), however, only with a marginal Z-score of 4.2 and an RMSD of 3.0 Å for the superposition of 55 out of the 69 residues (Fig. 2E). There is no detectable sequence homology between the two proteins. The other proteins for which DALI reported a weak structural homology are mainly DNA-binding proteins (data not shown). The N-terminal SAP domain of PIAS1 binds to the tumor suppressor p53, which is a target protein for sumoylation by PIAS1, and it has a strong binding affinity toward A/T-rich DNA (Okubo et al. 2004), indicating that the PIAS1 SAP domain binds to its protein partner as well as to the DNA. The N-terminal SAP domain of PIAS1 has a helix in the region corresponding to the α2/α3 loop in BRD4-ET, in which a “nascent” helical loop and several 310-helix-type hydrogen bonds are interrupted by proline residues.
The electrostatic surface potential analysis of the BRD4-ET domain revealed one acidic and two basic patches on the protein surface. The acidic patch forms a continuous ridge, largely contributed by the long α2/α3 loop, with a hydrophobic cleft/base produced by the α1 and α2 helices (Fig. 2D). This acidic stretch of the BRD4-ET domain (and its homologs in the BET family) may interact with other proteins and/or DNA. The two basic patches, which are mainly due to positively charged residues from the C-terminal region, 677–683, or the α2 and α3 helices, respectively, may be potential sites for DNA recognition.
The BRD4 and BRD2 proteins recognize acetylated N-terminal tails in the histone H4 through their two bromodomains and remain bound to condensed chromosomes throughout mitosis (Florence and Faller 2001; Dey et al. 2003). We have recently shown that BRD2 forms a homodimer in solution as well as in the crystal structure (Nakamura et al. 2007). The self-association of BRD2 occurs through the N-terminal bromodomain. The ET domain did not show self-association activity as the N-terminal bromodomain did (data not shown).
Binding analysis
Recent studies showed that the ET domain of BRD4, and its homologs in BRD2 and BRD3, directly interact with Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen 1 (LANA-1) (Platt et al. 1999; Viejo-Borbolla et al. 2005; Ottinger et al. 2006; You et al. 2006). The C-terminal region comprising the last 20 residues of LANA-1 (residues 1139–1162) is sufficient for interacting with the BRD4-ET domain (Ottinger et al. 2006). Furthermore, the basic (pI = 10.82) stretch of residues 1134–1143 (SSIVKFKKPL) was reported to be indispensable for the BRD4-ET interaction.
To elucidate whether the BRD4-ET domain, especially the two pockets formed by the unique α2/α3 loop structure, contributes to the BRD4 interaction with LANA-1, we performed binding experiments between the BRD4-ET domain and a C-terminal peptide (residues 1134–1163) of LANA-1 by surface plasmon resonance analysis. We prepared the wild-type BRD4-ET domain and five BRD4-ET domain variants with multiple mutations in the pocket-forming residues (see Materials and Methods). Although we examined this binding with high concentration of the protein (up to 1 mM), we could not detect an interaction between the BRD4-ET proteins and the LANA-1 peptide (data not shown). We also tested the ability of the BRD4-ET domain to interact with the BRD4-ET domain itself, two BRD4 bromodomains, or the N-terminal tail peptide of histone H4 (residues 1–15). In all these studies, we could not observe an interaction with the BRD4-ET domain (data not shown). The BRD-ET domain might require a longer C-terminal region of LANA-1 or other endogenous factors for association. Further structural and biochemical studies on ET complexes are required to understand the molecular mechanisms of the ET domain in BET family proteins.
Materials and Methods
Protein expression and purification
The histidine-tagged ET domain of mouse BRD4 (SwissPROT: BRD4_MOUSE) protein was expressed for 6 h at 30°C, using an E. coli cell-free protein synthesis system (Matsuda et al. 2007). The protein was adsorbed to a 5 mL column of HisTrap HP (GE Healthcare) and eluted with 20 mM Tris-HCl buffer (pH 8.0) containing 500 mM NaCl and 500 mM imidazole. The protein solution was incubated with TEV protease for 1 h at 30°C to remove the histidine tag. The protein solution was desalted and passed through a 1 mL column of HisTrap HP (GE Healthcare). The TEV protease-digested protein comprised 90 amino acid residues, including seven nonnative residues (GSSGSSG) at its N terminus. The C-terminal 83 residues constitute the BRD4-ET domain, corresponding to residues 601–683 of the full-length mouse BRD4 protein. For the structure determination, a single, uniformly 13C/15N-labeled sample was concentrated to 1.05 mM in 20 mM Tris-HCl buffer (pH 7.0), containing 100 mM NaCl, 1 mM dithiothreitol, and 0.02% NaN3 with the addition of D2O to 10% v/v.
NMR spectroscopy
By use of Bruker DRX 600 or Bruker AV 800 spectrometers [15N,1H]-HSQC (Fig. 1), [13C,1H]-HSQC, HNCO, HN(CA)CO, HN(CO)CA, HNCA, CBCA(CO)NH, HNCACB, HBHA(CO)NH, CC(CO)NH, H(CCCO)NH, HCCH-TOCSY, HCCH-COSY, 15N-resolved NOESY, and 13C-resolved NOESY (both with 80-ms mixing time), spectra were recorded at 25°C. The raw NMR data were processed using the program NMRPipe (Delaglio et al. 1995). The programs KUJIRA (Kobayashi et al. 2007) and NMRView (Johnson and Blevins 1994) were used for the spectrum analysis.
Structure calculation
Peak lists for the NOESY spectra were obtained by interactive peak picking with the program NMRView. Torsion angle restraints were obtained with the program TALOS (Cornilescu et al. 1999). The three-dimensional structure was determined by combined automated NOESY cross-peak assignment (Herrmann et al. 2002) and structure calculation with torsion angle dynamics (Güntert et al. 1997) implemented in the program CYANA (Güntert 2003). The standard CYANA protocol of seven iterative cycles of NOE assignment and structure calculation, followed by a final structure calculation was applied. Each cycle of the structure calculation started from 100 randomized conformers, and 10,000 torsion angle dynamics steps were performed per conformer. The 20 conformers with the lowest final CYANA target function values were subjected to restrained energy refinement in explicit solvent against the AMBER force field (Cornell et al. 1995) using the program OPALp (Luginbühl et al. 1996; Koradi et al. 2000). PROCHECK-NMR (Laskowski et al. 1996), MOLMOL (Koradi et al. 1996), and PyMOL version 0.99 (www.pymol.org) were used to validate and to visualize the final structures, respectively.
Surface plasmon resonance analysis
Surface plasmon resonance analysis was carried out on a Biacore 3000 instrument, using CM5 sensor chips (GE Healthcare). The polypeptide SSIVKFKKPLPLTQPGENQGPGDSPQEMT corresponding to the C-terminal residues 1134–1162 of LANA-1 (purchased from Toray Research Center) was immobilized on the sensor chip by the amine-coupling method. Purified wild-type or mutant BRD4-ET proteins (concentrations: 10 μM–1 mM) were each loaded for 1 min with a 20 μL/min flow rate onto the chip, and the flow was further continued for 1 min to detect the dissociation of the proteins from the immobilized peptides in the HBS-EP running buffer composed of 10 mM HEPES buffer (pH 7.4), 150 mM NaCl, 3 mM EDTA, and 0.005% Tween-20. Analogous binding assays were performed using a BRD4-ET domain-liganded chip and LANA-1 analytes. Six BRD4-ET proteins were studied: (a) wild-type, (b) D651A + E652A + E654A + D656A + E658A, (c) E642A + L645E + S648L, (d) R617 + N624A + L631E, (e) simultaneous mutations at the eight positions of b and c, and (f) simultaneous mutations at the eight positions b and d. The integrity of the proteins used for the binding assays was confirmed by SDS-PAGE analysis.
Acknowledgments
We thank Yasuko Tomo, Eiko Seki, Kazuharu Hanada, Yukiko Fujikura, and Satoru Watanabe for sample preparation and Dr. Hua Li for help with the programs KUJIRA and NMRView. This work was financed by the National Project on Protein Structural and Functional Analyses of the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT). P.G. is supported by a Grant-in-Aid for Scientific Research of the Japan Society for the Promotion of Science (JSPS) and by a Lichtenberg Professorship of the Volkswagen Foundation.
Footnotes
Reprint requests to: Peter Güntert, Institute of Biophysical Chemistry, J.W. Goethe University Frankfurt am Main, Max-von-Laue-Strasse 9, 60438 Frankfurt am Main, Germany; e-mail: guentert@em.uni-frankfurt.de; fax: 49-69-79829643; or Balasundaram Padmanabhan, Biology Division, Aptuit Laurus Private Limited, ICICI Knowledge Park, Turkapally Shameerpet, Hyderabad - 500 078, India; e-mail: paddy.b@AptuitLaurus.com; fax: 91-40-23480481.
Abbreviations: BET, bromodomain and extraterminal; BRD4, bromodomain-containing protein 4; ET, extraterminal; HSQC, heteronuclear single quantum coherence; LANA-1, latency-associated nuclear antigen-1; NOE, nuclear Overhauser effect; NOESY, NOE spectroscopy; PCR, polymerase chain reaction; RMSD, root-mean-square deviation; TEV, tobacco etch virus.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.037580.108.
References
- Abbate E.A., Voitenleitner C., Botchan M.R. Structure of the papillomavirus DNA-tethering complex E2:Brd4 and a peptide that ablates HPV chromosomal association. Mol. Cell. 2006;24:877–889. doi: 10.1016/j.molcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- Cornilescu G., Delaglio F., Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: A multidimensional spectral processing system based on Unix pipes. J. Biol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dey A., Chitsaz F., Abbasi A., Misteli T., Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florence B., Faller D.V. You bet-cha: A novel family of transcriptional regulators. Front. Biosci. 2001;6:D1008–D1018. doi: 10.2741/florence. [DOI] [PubMed] [Google Scholar]
- Güntert P. Automated NMR protein structure calculation. Prog. Nucl. Magn. Reson. Spectrosc. 2003;43:105–125. [Google Scholar]
- Güntert P., Mumenthaler C., Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- Herrmann T., Güntert P., Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Johnson B.A., Blevins R.A. NMR view: A computer program for the visualization and analysis of NMR data. J. Biol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kobayashi N., Iwahara J., Koshiba S., Tomizawa T., Tochio N., Güntert P., Kigawa T., Yokoyama S. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J. Biol. NMR. 2007;39:31–52. doi: 10.1007/s10858-007-9175-5. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter M., Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter M., Güntert P. Point-centered domain decomposition for parallel molecular dynamics simulation. Comput. Phys. Commun. 2000;124:139–147. [Google Scholar]
- Laskowski R.A., Rullmann J.A.C., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Luginbühl P., Güntert P., Billeter M., Wüthrich K. The new program OPAL for molecular dynamics simulations and energy refinements of biological macromolecules. J. Biol. NMR. 1996;8:136–146. doi: 10.1007/BF00211160. [DOI] [PubMed] [Google Scholar]
- Matangkasombut O., Buratowski R.M., Swilling N.W., Buratowski S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes & Dev. 2000;14:951–962. [PMC free article] [PubMed] [Google Scholar]
- Matsuda T., Koshiba S., Tochio N., Seki E., Iwasaki N., Yabuki T., Inoue M., Yokoyama S., Kigawa T. Improving cell-free protein synthesis for stable-isotope labeling. J. Biol. NMR. 2007;37:225–229. doi: 10.1007/s10858-006-9127-5. [DOI] [PubMed] [Google Scholar]
- Mujtaba S., Zeng L., Zhou M.M. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Umehara T., Nakano K., Jang M.K., Shirouzu M., Morita S., Uda-Tochio H., Hamana H., Terada T., Adachi N., et al. Crystal structure of the human BRD2 bromodomain: Insights into dimerization and recognition of acetylated histone H4. J. Biol. Chem. 2007;282:4193–4201. doi: 10.1074/jbc.M605971200. [DOI] [PubMed] [Google Scholar]
- Okubo S., Hara F., Tsuchida Y., Shimotakahara S., Suzuki S., Hatanaka H., Yokoyama S., Tanaka H., Yasuda H., Shindo H. NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers. J. Biol. Chem. 2004;279:31455–31461. doi: 10.1074/jbc.M403561200. [DOI] [PubMed] [Google Scholar]
- Ottinger M., Christalla T., Nathan K., Brinkmann M.M., Viejo-Borbolla A., Schulz T.F. Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. J. Virol. 2006;80:10772–10786. doi: 10.1128/JVI.00804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt G.M., Simpson G.R., Mittnacht S., Schulz T.F. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 1999;73:9789–9795. doi: 10.1128/jvi.73.12.9789-9795.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viejo-Borbolla A., Ottinger M., Brüning E., Bürger A., König R., Kati E., Sheldon J.A., Schulz T.F. Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. J. Virol. 2005;79:13618–13629. doi: 10.1128/JVI.79.21.13618-13629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.Y., Chiang C.M. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- You J.X., Srinivasan V., Denis G.V., Harrington W.J., Ballestas M.E., Kaye K.M., Howley P.M. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J. Virol. 2006;80:8909–8919. doi: 10.1128/JVI.00502-06. [DOI] [PMC free article] [PubMed] [Google Scholar]