Abstract
Biologically important human proteins often require mammalian cell expression for structural studies, presenting technical and economical problems in the production/purification processes. We introduce a novel affinity peptide tagging system that uses a low affinity anti-peptide monoclonal antibody. Concatenation of the short recognition sequence enabled the successful engineering of an 18-residue affinity tag with ideal solution binding kinetics, providing a low-cost purification means when combined with nondenaturing elution by water-miscible organic solvents. Three-dimensional information provides a firm structural basis for the antibody–peptide interaction, opening opportunities for further improvements/modifications.
Keyword: affinity tag, purification, monoclonal antibody, X-ray crystallography, Fab fragment, scFv fragment, F-spondin, reelin
Epitope tagging, in which a reporter peptide sequence is affixed to a target, is a paramount and universal technology in modern day biomedical research by enabling molecular localization within the cell, identification of molecular interactions, and isolation of target proteins from crude mixtures (Waugh 2005; Chang 2006; O'Hare et al. 2007). Particularly important in the field of structural biology is the use of epitope tags as a means to purify recombinant proteins (Terpe 2003; Arnau et al. 2006). Although many options exist for epitope tagging of bacterially expressed proteins including His, GST, MBP tags, available technology for purifying proteins expressed in mammalian cell systems is limited. The most versatile and widely used tagging system is the His-tag that binds to immobilized metal, which often suffers from low purification yield due to the incomplete absorption to the column and high levels of contamination with other metal-binding proteins (Lichty et al. 2005). Use of a unique peptide (Strep II) and binding protein (Strep-Tactin) pair has been successfully applied to many structural studies, but the high cost and the proprietary rights associated with the products limit the applicability of this system. Immunoaffinity chromatography using anti-peptide monoclonal antibodies (mAbs) offers a very powerful means to purify recombinant proteins of low abundance because of their very high specificity and affinity. The Flag epitope in combination with the anti-Flag mAb M2 is probably the most frequently used and successful system currently available (Einhauer and Jungbauer 2001). The bound proteins can be eluted by FLAG peptide without any adverse effect on their structure. Because the antibody column will be loaded with antigen peptide after the elution, however, it must be regenerated by washing in a harsh condition before the next run, decreasing its lifespan. Other less popular anti-peptide mAb systems include mAbs 1D4 directed against C-terminal sequence of bovine rhodopsin (Shimada et al. 2002), 2E11 directed against peptide 38 from TCR (Boldicke et al. 2000), and Tab2 directed against N-terminal peptide from TGFα (Crusius et al. 2006). Despite some successful applications of these anti-tag antibodies, none have proven as robust and universal as FLAG/M2, nor have they become publicly available to replace expensive commercial products. It is also important to note that all of the available anti-tag mAbs lack structural information to allow further engineering to improve the affinity and/or the specificity.
In this study, we introduce a novel affinity tagging system that is suitable for use in protein purification for structural as well as functional studies. Unique features of the mAb (clone P20.1) enable the efficient capture of tagged proteins from the culture supernatants of transfected cells and gentle elution by water-miscible organic solvents. By using this tagging/purification system, diffraction quality crystals were immediately obtained for one target protein domain. It also enabled us to purify full-length, intact, and biologically active giant glycoprotein reelin from the culture supernatant to homogeneity for the first time. Three-dimensional information for antibody-tag interaction derived from the crystal structure of the P20.1 Fab fragment in complex with the epitope peptide, along with the availability of single-chain Fv fragment (scFv) of P20.1, may guide further improvement of the system by protein engineering.
Results and Discussion
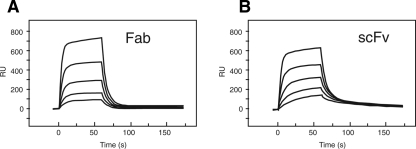
Monoclonal antibody P20.1 is a mouse IgG directed against the N-terminal segment of human protease activated receptor-4 (PAR-4), a G-protein coupled receptor implicated in platelet activation (Coughlin 2000). We have recently shown that it recognizes a linear stretch of six amino acids (GYPGQV), among which only three (underlined) were absolutely required for recognition (Sangawa et al. 2008). Biacore analysis using the Fab fragment of P20.1 showed that the monovalent affinity of P20.1 toward the epitope peptide was relatively low (Fig. 1A). Kinetic analysis of the data revealed that the association rate was in the range of typical protein–protein interaction (k on = 2.3 × 105 M−1s−1) while dissociation was significantly fast (k off = 0.13 s−1), causing the low overall affinity of ∼0.6 μM. Nucleotide sequences for the variable regions of P20.1 H and L chains were used to construct a recombinant scFv, which was shown to exhibit almost identical binding characteristics to the Fab (Fig. 1B). This indicates that the weak binding (i.e., fast k off) is an intrinsic feature of the P20.1 antigen-combining site rather than a consequence of structural damage caused by Fab preparation.
Figure 1.
Surface plasmon resonance kinetic analysis of P20.1 binding to peptide. C-terminally biotinylated fibronectin fragment bearing a 20-residue antigenic peptide (i.e., GGDDSTPSILPAPRGYPGQV, P4 sequence is underlined) at the N terminus was immobilized onto a straptavidin-coated sensorchip (Sangawa et al. 2008). Serially diluted Fab (A) or scFv fragment (B) of P20.1 (625, 313, 156, 78, and 39 nM) were then injected at a flow rate of 20 μL/min.
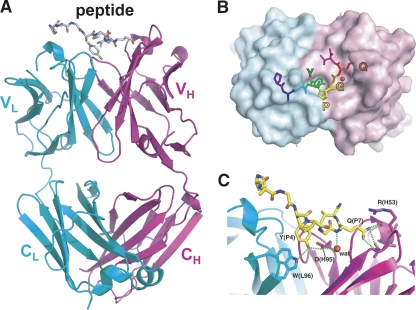
To investigate the chemical basis of the P20.1–peptide interaction, we determined the crystal structure of the P20.1 Fab in complex with an eight-residue antigenic peptide at 1.8 Å (Fig. 2A). There were two sets of Fab–peptide complexes within the asymmetric unit, of which the conformation of the interface residues was almost identical with minor differences at the periphery (see below). The structure revealed that the antigenic peptide binds to the CDR loops in a curved configuration (Fig. 2B). The critical requirement for the YXGQ sequence for the recognition by P20.1 determined by epitope mapping study (Sangawa et al. 2008) is nicely explained by the structure. In fact, only this four-residue segment is in direct contact with the antibody, burying merely 814 Å2 of total solvent accessible surface area upon the binding. Tyr P4 is deeply inserted into a hydrophobic crevice between the H and L chains, with the phenolic hydroxyl group forming a hydrogen bond with the carboxyl side chain of Glu H95 in CDR H3 (Fig. 2B). This is part of a hydrogen bonding network involving the Trp L96 (Nε), Tyr P4 (OH), Glu H95 (Oε1), Gln P7 (N), and a structured water molecule filling a space between the peptide and Fab (Fig. 2C). This structured water seems critical for the binding for both its contribution to hydrogen bonding and to shape complementarity and may be the major point of action during the dissociation induced by polyols (discussed later). At the other end of the interface, the side chain of Gln P7 participates in another hydrogen bonding network with residues from CDR H1 and H2 (Fig. 2C). Arg H53 appears to contribute favorably to the binding because its long side chain bends toward the Gln P7 in one of the complexes in the asymmetric unit, acting like a “lid.” The same side chain assumes a different conformer in the other complex but still contacts with the peptide using its Cβ (Supplemental Fig. S1), suggesting a contribution of Arg H53 in delaying the dissociation.
Figure 2.
Crystal structure of P20.1 Fab in complex with peptide PRGYPGQV. (A) Ribbon diagram of the entire complex. Heavy and light chains of Fab are colored magenta and cyan, respectively. Intimate packing of the “YPGQ” motif onto the Fab surface (B) as well as the hydrogen bonding network (C) are also shown. The side chain atoms beyond Cβ of Arg P2 are omitted because of the undefined conformation. The red sphere represents a structured water molecule common to the two independent complexes in the asymmetric unit. For C, the same set of figures for another complex can be found in Supplemental Figure S1.
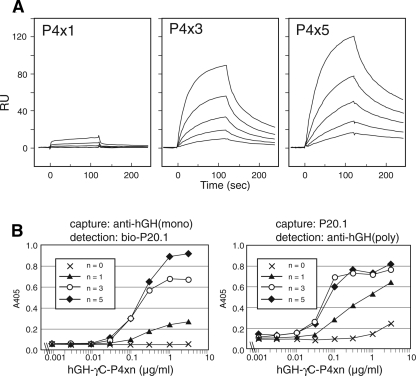
The low intrinsic affinity of P20.1 toward antigenic peptide, especially the fast dissociation rate (Fig. 1), does not seem to be compatible with its utility as an affinity purification tool. We therefore explored the possibility of increasing effective affinity by concatenating the recognition sequence. The six-residue recognition sequence (GYPGQV, referred to as P4 sequence), either alone or repeated in tandem for three or five times, was attached to the C terminus of a model fusion protein (hGH–His–γC) and tested for binding to immobilized P20.1 using Biacore. As shown in Figure 3A, the weak overall binding of hGH–His–γC–P4x1 could be substantially augmented by concatenation of the sequence, resulting in ∼80- and ∼180-fold increase in the apparent affinity for x3 and x5 peptide, respectively. This increase is caused by a delayed off rate due to the rebinding to the neighboring motifs, as confirmed by quick reversal of the association by soluble peptide (data not shown). The same set of P4xn fusion proteins were then tested for compatibility with solution-phase detection in a sandwich ELISA format. Figure 3B clearly shows that both P4x3 and x5 peptide fusions, in the concentration range of 0.03 to 1 μg/mL, can elicit concentration-dependent and saturable signals in sandwich ELISAs using P20.1 and anti-hGH antibody, while the singly tagged version (P4x1) gives unsaturable and weak signals at higher concentration. Importantly, the P20.1 mAb can be used as either capturing or detection antibody, allowing combinations with a wide variety of secondary tags.
Figure 3.
Concatenation of P4 sequence increases apparent affinity. (A) Model P4-fusion proteins were flowed over the surface immobilized with P20.1 IgG at varying concentrations (25, 12.5, 6.25, 3.13, and 1.56 nM). Apparent affinity of the binding (K d) calculated from the sensorgrams were 1470 nM, 17.5 nM, and 8.3 nM for P4x1, P4x3, and P4x5 fusion proteins, respectively. (B) Model fusion proteins with various numbers of P4 sequences were subjected to two opposite configurations of sandwich ELISA experiments described in Materials and Methods. Results are representative of three independent experiments performed in duplicate.
The successful engineering of an epitope tag that shows good solution binding behaviors prompted us to test if P20.1 can be used to capture and release tagged proteins from crude mixtures. Despite the slightly lower affinity compared to the P4x5 sequence, we chose P4x3 for this practical application due to the smaller size (18 res vs. 30 res). Thus, the model fusion protein (hGH–His–γC–P4x3) was expressed in 293T cells, and the ability of P20.1-coupled Sepharose to capture and release the target protein from the culture supernatant of transfected cells was assessed. As shown in Figure 4, the tagged protein was specifically captured and completely eluted from the resin by a buffer containing >100 μg/mL 8-mer free peptide (lanes 1, 2). It was also confirmed that >85% of the tagged protein was captured during the 1-h incubation at 4°C (data not shown). The eluted fraction contained almost exclusively the tagged protein alone, even though the starting material contained 10% fetal calf serum and the expressed protein constituted ∼0.01% of the protein present in the culture supernatant. The P20.1–Sepharose thus achieved 10,000-fold purification in just one step, which is in sharp contrast with the eluted material from the Ni-NTA resin (lane 8) that shows the presence of several serum-derived proteins even after extensive washing steps.
Figure 4.
Specific binding and elution of tagged protein. Model protein (hGH–His–γC–P4x3) secreted into culture medium from transiently transfected HEK293T cells was captured to P20.1–Sepharose (lanes 1–7) or Ni-NTA agarose (lane 8). Resins were washed four times with TBS and eluted in the following conditions: TBS containing 100 μg/mL (lane 1) or 1 mg/mL (lane 2) PRGYPGQV peptide, 50 mM glycine-HCl, pH 2.3 (lane 3), 50 mM triethylamine, pH 11.5 (lane 4), 2 M potassium iodide (lane 5), 40% (v/v) propylene glycol + 1 M NaCl (lane 6), TBS alone (lane 7), and 250 mM imidazole (lane 8). Eluted materials were concentrated, analyzed with reducing SDS-PAGE, and Coomassie-stained for visualization.
We next explored whether other elution conditions can be employed. For reversible (i.e., nondenaturing) elution of antigens from the immunoaffinity resins, conditions such as extreme pH and chaotropic ions are frequently used. In the case of P20.1–Sepharose, however, acidic conditions (pH 2.3) or 2 M potassium iodide did not cause any elution of the protein, and high pH buffer (pH 11.5) resulted in partial elution (Fig. 4, lanes 3–5). Some monoclonal antibodies are known to be sensitive to the high concentrations of polyols such as propylene glycol for their antigen recognition (Burgess and Thompson 2002), and we decided to test the possibility that P20.1–P4 peptide interaction is polyol-sensitive. Indeed, a buffer containing 40% propylene glycol and 1 M NaCl completely eluted tagged protein from the P20.1–Sepharose (Fig. 4, lane 6). Further experiments revealed that complete elution can be achieved by >40% propylene glycol or other water-miscible organic solvents such as ethylene glycol and DMSO, without adding high concentrations of salt (Supplemental Fig. S2). These solvents are known to destroy the hydration structure of the solute, and the effective elution may be attributed to their ability to replace the interfacial water involved in the hydrogen bonding network (Fig. 2C).
As propylene glycol is not likely to have a denaturing effect on protein structure and can be easily removed from the sample by dialysis, P4x3-tagging seems ideal for the protein production/purification for structural studies. We applied the system to a crystallization project conducted in our lab, a 145-residue fragment of an extracellular protein F-spondin corresponding to its N-terminal reeler domain. Figure 5A shows the successful one-step purification of the F-spondin fragment from the culture media of transiently transfected 293T cells using P20.1–Sepharose and propylene glycol elution. Furthermore, the protein, after the cleavage of the tag by TEV protease, crystallized immediately in multiple conditions (Fig. 5B, inset), which diffracted up to 1.45 Å (Fig. 5B). Importantly, the structure determination was completed within only 3 wk after the completion of the DNA construction, underscoring the very high efficiency of affinity purification using the P20.1/P4x3 tag.
Figure 5.
Purification of a recombinant protein using P4x3 tag/P20.1 system yields high-quality structural data. (A) SDS-PAGE analysis of fractions eluted from P20.1–Sepharose during the purification of N-terminally P4x3-tagged F-spondin fragment. Despite the presence of numerous serum-derived proteins in the original sample as well as in the wash fractions (lanes 1–3), the eluate fractions (lanes 4–7) contained tagged protein (arrowhead), which was essentially contaminant-free. This protein, after the removal of the tag, was crystallized (B, inset) and led to a determination of 1.45 Å resolution structure (Nagae et al. 2008). A diagram of weighted 2|F obs| − |F calc| electron density map is shown in B.
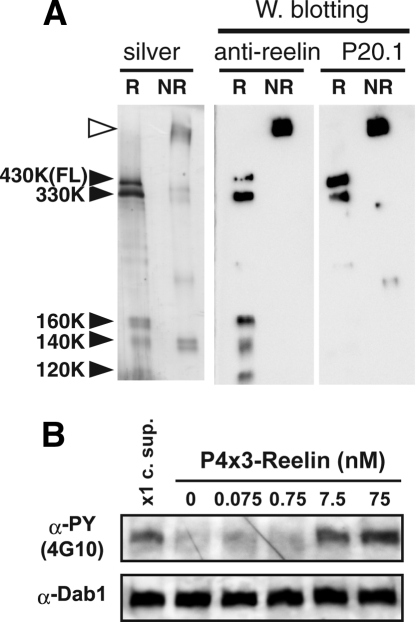
Efficient capture of low abundance proteins and mild elution by propylene glycol also made it possible to purify a giant and labile secreted glycoprotein reelin from culture supernatant. Purification of reelin, a 3461-residue protein secreted from a class of neuronal cells that plays critical roles in brain development (Cooper 2008), has been notoriously difficult because of its very high molecular weight, oligomeric state, the lack of good monoclonal antibodies, and the presence of proteolytic cleavage that removes the C-terminal part of the protein (Kubo et al. 2002; Jossin et al. 2007). By appending the P4x3 tag sequence after the signal peptide, full-length reelin could be purified from the culture supernatant to homogeneity (Fig. 6A). Furthermore, the purified reelin was biologically active in inducing Dab1 phosphorylation in cultured neurons (Fig. 6B). Retention of full activity is postulated by comparison of the concentration dependence of activity to that of partially purified reelin reported previously (Benhayon et al. 2003).
Figure 6.
Purification of biologically active full-length reelin using P4x3 tag/P20.1 system. (A) Full-length reelin purified on a P20.1–Sepharose column was separated on 7.5% SDS-PAGE gel under reducing (R) or nonreducing (NR) conditions. Western blotting using anti-reelin antibody directed against N-terminal part reveals that all five bands (black arrowheads) that appear in the silver-stained gel under reducing condition correspond to intact full-length reelin (430 kDa) or protease-processed fragments originated from it (330, 160, 140, and 120 kDa), confirming the high purity of the sample. Furthermore, lack of the P20.1 reactivity in the 120–160 kDa fragments shows that they are devoid of the very N terminus. Under the nonreducing condition, reelin migrates as a disulfide-bonded high molecular weight multimer (open arrowhead) as reported previously. (B) Mouse cortical neurons were incubated for 20 min with the purified full-length reelin at the indicated concentration or with culture supernatant (x1 c. sup.). Total cell lysate was prepared, subjected to SDS-PAGE, and phosphorylated, and total Dab1 were detected by Western blotting using 4G10 and anti-Dab1, respectively.
The P4x3 tag system in combination with immobilized P20.1 as the purification tool has great advantages over existing technologies. First, the antibody has high specificity toward the concatenated P4 sequence and therefore only artificially tagged proteins are bound, enabling one-step purification of target proteins that are present in low abundance in the starting material. This is particularly useful when high-level recombinant production cannot be expected, such as in the case of the so-called “difficult-to-express proteins” or membrane proteins. We have so far applied this tagging system to the production of nine soluble proteins (four C-terminally and five N-terminally tagged proteins) and one single-pass membrane protein (tagged at its intracellular tail), all using a mammalian expression system, and experienced no negative effect on the expression level or the protein stability, indicating that it is a very robust system compatible with a wide range of applications. Second, the use of water-miscible organic solvents in the elution step offers a mild, complete, and inexpensive way to achieve one-step purification. The benefit of this mild elution condition is twofold; it maintains the structural and functional integrity of the protein to be purified, and at the same time it causes minimum damage to the antibody, leading to an extended lifetime of the affinity column. Binding/elution cycles can be repeated more than 10 times without any loss of binding capacity (data not shown). When the addition of organic solvents are undesirable, such as in the case of weakly interacting protein complexes or multispan membrane proteins, one can still use peptide elution followed by column regeneration with 40% propylene glycol. The eight-residue peptide used for the competitive elution (PRGYPGQV) is freely soluble in various buffers and can be synthesized with high yield probably because of the presence of prolines, adding other advantages to this system. Thirdly, this system will become widely available to basic researchers. We deposited the P20.1 hybridoma to a public cell bank (Riken BioResource Center, http://www.brc.riken.jp/lab/cell/english/) so that researchers in nonprofit organizations will have access to the antibody. Also, mammalian expression vectors with either an N- or C-terminally added P4x3 tag sequence, followed or preceded by a TEV cleavage site (Supplemental Fig. S3), are available upon request for direct cloning or PCR-based transfer to any desirable vectors. Finally, and most importantly, an atomic resolution structure is available for P4–P20.1 interaction, opening a possibility for further improvement of the affinity and/or selectivity by modifying either the peptide or the antibody. Availability of the scFv fragment of P20.1 adds a further advantage to such attempts. As the core binding interface is comprised of only four residues from the peptide side with high shape and hydrogen bonding complementarity, engineering of CDR residues involved in this interface will be difficult. Therefore, mutations outside this core interface should be the primary target for the engineering. We have introduced an additional negative charge by mutating both Thr L28 and Ser L30 to Glu in the P20.1 scFv, hoping that it would increase the affinity by attracting Arg P2 that was highly mobile in the current structure (Supplemental Fig. S4). Unfortunately, this design did not cause any increase in the affinity (Supplemental Fig. S4), suggesting a need for more elaborate exploration of interface residues on both antibody and the peptide, possibly using a high-throughput combinatorial screening. Nevertheless, the fact that wild-type P20.1–peptide interaction uses small interface with relatively low affinity makes it an ideal starting system or “scaffold” for the engineering of specialized antibody peptide pairs by incorporating additional interactions. Such derivatives will be most efficiently developed when the system is publicly available.
Materials and Methods
Proteins and constructs
Establishment of mouse hybridoma cell line secreting P20.1 antibody (IgG1, λ) and its characterization have been described previously (Sangawa et al. 2008). Nucleotide sequence encoding the variable region of P20.1 was determined as described in the Supplemental materials. Expression construct for the scFv contained an initiation Met followed by a VH region, a (GGGGS)3 linker, a VL region, a hexahistidine tag, and a Myc tag, cloned into NdeI-BamHI site of pET11c vector. The resultant vector was used to transform BL21 Escherichia coli, and scFv was purified from the inclusion body of the IPTG-induced bacteria by Ni-NTA chromatography in the presence of 6 M guanidium hydrochloride and refolded by sequential dialysis (Kipriyanov et al. 1996).
A mammalian expression vector pSGHV0 (Leahy et al. 2000) that carries hGH minigene followed by a Hisx8 and a tobacco etch virus (TEV) protease cleavage site have been used to construct an expression plasmid for human fibrinogen γ chain C-terminal domain (γC) (Xiao et al. 2004). At the C-terminal of this construct (hGH–His–γC), various numbers of P4 sequences (i.e., GYPGQV) were appended using extension PCR to construct vectors for P4xn-tagged model fusion proteins. These plasmids were used to transiently transfect 293T cells and the corresponding fusion proteins were purified from the culture supernatants using Ni-NTA-agarose chromatography. For N-terminally tagged secreted proteins, pcDNA3.1 vector (Invitrogen) was modified to incorporate signal sequence from mouse nidogen (Takagi et al. 2003) followed by three concatenations of P4 sequence and a TEV site. The resultant vector (pENP4x3) was used to construct expression plasmids for F-spondin fragment and full-length reelin.
Structure determination of P20.1 Fab in complex with P4 peptide
Fab fragment was prepared by papain treatment of P20.1 antibody, which was purified from culture supernatants of hybridoma. Fab fragment was mixed with the eight-residue synthetic peptide (NH2-PRGYPGQV-COOH) at a molar ratio of 1:10 and subjected to crystallization. Diffraction quality crystals were grown in a crystallization buffer containing 20%–23% (wt/vol) PEG 3000 and 0.1 M sodium acetate (pH 4.5). Crystals were cryoprotected with a harvesting buffer containing glycerol and used for data collection at SPring-8 BL-44XU. The crystal diffracted X-ray to 1.8 Å resolution, and it belongs to the triclinic space group P1 with cell dimensions of a = 40.05 Å, b = 65.27 Å, c = 85.03 Å, α = 99.93°, β = 93.50°, and γ = 96.46°. Initial phases were determined by a molecular replacement method using the mouse antibody Fab structure containing the λ chain (PDB accession code 1NC4) as a search model. After the phase improvement, the structure of the P4 peptide was clearly visualized in the electron density map. As a result of iterative model fitting and refinement, the crystallographic R-factor and free R-factor were reduced to 17.2% and 20.4% at 1.8 Å resolution, respectively. Details of experimental procedures and calculations are described in the Supplemental material, and statistics for data collection and structure refinement are summarized in Supplemental Tables 1 and 2.
Binding measurements
Two different sandwich ELISA conditions were established to detect P4xn-tagged protein in solution. In the first condition, an anti-hGH monoclonal antibody (HGH-B, American Type Culture Collection) was coated on microtiter wells (NUNC Maxisorp) and incubated with various concentrations of purified model proteins diluted in 20 mM Tris, 150 mM NaCl, pH 7.5 (TBS) containing 1 mg/mL BSA. This capturing step was conducted at room temperature for 3 h, washed three times with TBS, and probed with biotinylated P20.1 in combination with peroxidase-streptavidin conjugate. In the second condition, an inverted configuration was employed where capture and detection antibodies were replaced with P20.1 and rabbit anti-hGH antibody (Biodesign), respectively. Surface plasmon resonance experiments were performed using Biacore 2000 and Biacore X100 equipments (GE Healthcare) as described previously (Sangawa et al. 2008).
Purification and characterization of P4x3-tagged proteins
Coupling of P20.1 IgG to CNBr-activated Sepharose 4 Fast Flow (GE Healthcare) was performed according to the protocol provided by the manufacturer and routinely yielded 2∼3 mg IgG/mL gel. The expression plasmid coding for an N-terminal reeler domain of human F-spondin with N-terminal P4x3 tag was used to transiently transfect 293T cells plated in 15-cm culture dishes, and ∼400 mL of culture supernatant was harvested 6 d after the transfection. Cleared supernatant was directly loaded to P20.1–Sepharose (2.5 mL bed vol) equilibrated with TBS. After washing the column with 20 mL of TBS, the bound protein was eluted by 40% (v/v) propylene glycol in TBS. Eluted fractions were combined and dialyzed against TBS, at which step the volume of the dialyzate increased by ∼150%. The eluted material (∼0.6 mg protein) was concentrated and treated with TEV protease to cleave off the tag, followed by a final purification by MonoQ column (GE Healthcare) to remove the protease and the cleaved tag. The purified protein was concentrated to 4.5 mg/mL using VIVASPIN6 (Sartorius), and subjected to the crystallization. The entire purification took only 4 d, and diffraction-quality crystalls were obtained in <1 d. The detailed characterization and structure determination of F-spondin reeler domain will be published elsewhere (Nagae et al. 2008).
The protocol for expression and purification of N-terminally P4x3-tagged full-length reelin was the same as that for F-spondin fragment, except that the yield was much lower because of the inefficient expression. About 100 μg of purified reelin was obtained from ∼2 L of culture supernatant. The biological activity of purified reelin was assessed by Dab1 phosphorylation assay using mouse cortical neurons (Yasui et al. 2007).
Protein Data Bank deposition
Coordinates have been deposited in the Protein Data Bank with the accession code 2ZPK.
Acknowledgments
We thank Emiko Mihara and Maiko Nampo for their excellent technical assistance. We also thank Atsushi Nakagawa, Mamoru Suzuki, Eiki Yamashita, and Masato Yoshimura of SPring-8 BL-44XU for providing the data collection facilities and for support, and Dan Leahy for critical reading of the manuscript. This work was supported in part by the “Target Proteins Research Program (TPRP)” grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) and by the Protein 3000 Project grant from MEXT.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Junichi Takagi, Laboratory of Protein Synthesis and Expression, Institute for Protein Research, Osaka University, 3-2 Yamadaoka, Suita, Osaka 565-0871, Japan; e-mail: takagi@protein.osaka-u.ac.jp; fax: 81-6-6879-8609.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.038299.108.
References
- Arnau J., Lauritzen C., Petersen G.E., Pedersen J. Current strategies for the use of affinity tags and tag removal for the purification of recombinant proteins. Protein Expr. Purif. 2006;48:1–13. doi: 10.1016/j.pep.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Benhayon D., Magdaleno S., Curran T. Binding of purified reelin to ApoER2 and VLDLR mediates tyrosine phosphorylation of disabled-1. Brain Res. Mol. Brain Res. 2003;112:33–45. doi: 10.1016/s0169-328x(03)00032-9. [DOI] [PubMed] [Google Scholar]
- Boldicke T., Struck F., Schaper F., Tegge W., Sobek H., Villbrandt B., Lankenau P., Bocher M. A new peptide-affinity tag for the detection and affinity purification of recombinant proteins with a monoclonal antibody. J. Immunol. Methods. 2000;240:165–183. doi: 10.1016/s0022-1759(00)00167-8. [DOI] [PubMed] [Google Scholar]
- Burgess R.R., Thompson N.E. Advances in gentle immunoaffinity chromatography. Curr. Opin. Biotechnol. 2002;13:304–308. doi: 10.1016/s0958-1669(02)00340-3. [DOI] [PubMed] [Google Scholar]
- Chang I.F. Mass spectrometry-based proteomic analysis of the epitope-tag affinity purified protein complexes in eukaryotes. Proteomics. 2006;6:6158–6166. doi: 10.1002/pmic.200600225. [DOI] [PubMed] [Google Scholar]
- Cooper J.A. A mechanism for inside-out lamination in the neocortex. Trends Neurosci. 2008;31:113–119. doi: 10.1016/j.tins.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Coughlin S.R. Thrombin signalling and protease-activated receptors. Nature. 2000;407:258–264. doi: 10.1038/35025229. [DOI] [PubMed] [Google Scholar]
- Crusius K., Finster S., McClary J., Xia W., Larsen B., Schneider D., Lu H.T., Biancalana S., Xuan J.A., Newton A., et al. Tab2, a novel recombinant polypeptide tag offering sensitive and specific protein detection and reliable affinity purification. Gene. 2006;380:111–119. doi: 10.1016/j.gene.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Einhauer A., Jungbauer A. The FLAG peptide, a versatile fusion tag for the purification of recombinant proteins. J. Biochem. Biophys. Methods. 2001;49:455–465. doi: 10.1016/s0165-022x(01)00213-5. [DOI] [PubMed] [Google Scholar]
- Jossin Y., Gui L., Goffinet A.M. Processing of reelin by embryonic neurons is important for function in tissue but not in dissociated cultured neurons. J. Neurosci. 2007;27:4243–4252. doi: 10.1523/JNEUROSCI.0023-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipriyanov S.M., Little M., Kropshofer H., Breitling F., Gotter S., Dubel S. Affinity enhancement of a recombinant antibody: Formation of complexes with multiple valency by a single-chain Fv fragment-core streptavidin fusion. Protein Eng. 1996;9:203–211. doi: 10.1093/protein/9.2.203. [DOI] [PubMed] [Google Scholar]
- Kubo K., Mikoshiba K., Nakajima K. Secreted reelin molecules form homodimers. Neurosci. Res. 2002;43:381–388. doi: 10.1016/s0168-0102(02)00068-8. [DOI] [PubMed] [Google Scholar]
- Leahy D.J., Dann C.E., III, Longo P., Perman B., Ramyar K.X. A mammalian expression vector for expression and purification of secreted proteins for structural studies. Protein Expr. Purif. 2000;20:500–506. doi: 10.1006/prep.2000.1331. [DOI] [PubMed] [Google Scholar]
- Lichty J.J., Malecki J.L., Agnew H.D., Michelson-Horowitz D.J., Tan S. Comparison of affinity tags for protein purification. Protein Expr. Purif. 2005;41:98–105. doi: 10.1016/j.pep.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Nagae M., Nishikawa K., Yasui N., Yamasaki M., Nogi T., Takagi J. Structure of F-spondin reeler domain reveals a unique β-sandwich fold with a deformable disulfide-bonded loop. Acta Cryst. D. 2008;64:1138–1145. doi: 10.1107/S0907444908028308. [DOI] [PubMed] [Google Scholar]
- O'Hare H.M., Johnsson K., Gautier A. Chemical probes shed light on protein function. Curr. Opin. Struct. Biol. 2007;17:488–494. doi: 10.1016/j.sbi.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Sangawa T., Nogi T., Takagi J. A murine monoclonal antibody that binds N-terminal extracellular segment of human protease activated receptor-4. Hybridoma. 2008 doi: 10.1089/hyb.2008.0027. (in press) [DOI] [PubMed] [Google Scholar]
- Shimada M., Chen X., Cvrk T., Hilfiker H., Parfenova M., Segre G.V. Purification and characterization of a receptor for human parathyroid hormone and parathyroid hormone-related peptide. J. Biol. Chem. 2002;277:31774–31780. doi: 10.1074/jbc.M204166200. [DOI] [PubMed] [Google Scholar]
- Takagi J., Yang Y., Liu J.-H., Wang J.-H., Springer T.A. Complex between nidogen and laminin fragments reveals a paradigmatic β-propeller interface. Nature. 2003;424:969–974. doi: 10.1038/nature01873. [DOI] [PubMed] [Google Scholar]
- Terpe K. Overview of tag protein fusions: From molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2003;60:523–533. doi: 10.1007/s00253-002-1158-6. [DOI] [PubMed] [Google Scholar]
- Waugh D.S. Making the most of affinity tags. Trends Biotechnol. 2005;23:316–320. doi: 10.1016/j.tibtech.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Xiao T., Takagi J., Coller B.S., Wang J.H., Springer T.A. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui N., Nogi T., Kitao T., Nakano Y., Hattori M., Takagi J. Structure of a receptor-binding fragment of reelin and mutational analysis reveal a recognition mechanism similar to endocytic receptors. Proc. Natl. Acad. Sci. 2007;104:9988–9993. doi: 10.1073/pnas.0700438104. [DOI] [PMC free article] [PubMed] [Google Scholar]