Abstract
VirA, a secreted effector protein from Shigella sp., has been shown to be necessary for its virulence. It was also reported that VirA might be related to papain-like cysteine proteases and cleave α-tubulin, thus facilitating intracellular spreading. We have now determined the crystal structure of VirA at 3.0 Å resolution. The shape of the molecule resembles the letter “V,” with the residues in the N-terminal third of the 45-kDa molecule (some of which are disordered) forming one clearly identifiable domain, and the remainder of the molecule completing the V-like structure. The fold of VirA is unique and does not resemble that of any known protein, including papain, although its N-terminal domain is topologically similar to cysteine protease inhibitors such as stefin B. Analysis of the sequence conservation between VirA and its Escherichia coli homologs EspG and EspG2 did not result in identification of any putative protease-like active site, leaving open a possibility that the biological function of VirA in Shigella virulence may not involve direct proteolytic activity.
Keywords: crystallography, protein crystallization, proteolysis, bacterial virulence, novel fold
Shigellosis is an infectious disease caused by Gram-negative, rod-shaped bacteria from the species Shigella. Some strains produce enterotoxin and Shiga toxin, which are very similar to the verotoxin of Escherichia coli O157:H7 (Lan and Reeves 2002). People infected with Shigella develop diarrhea, fever, and stomach cramps 2–3 d after infection. In children, severe infections accompanied by high fevers are associated with seizures. Although shigellosis is rare in the United States, one bacterial species, Shigella dysenteriae, has the potential to cause deadly epidemics in developing regions, necessitating the search for novel drugs (Dutta et al. 2003). The initial steps of Shigella infection include their attachment to and subsequent penetration of the epithelial cells of the intestinal mucosa. After infection, the bacteria multiply intracellularly and then spread to adjacent host cells. This spreading is accomplished by destabilization of the cytoplasmic network of the host and thereby results in the destruction of tissues (Parsot 2005).
Virulent species of Shigella rely on a type III secretion system (T3SS) to deliver a small number of proteins, termed effectors, into the cytosol of host cells where they subvert mechanisms that control the actin cytoskeleton so as to promote invasion and cell-to-cell spreading (Schroeder and Hilbi 2008). One of these effectors is the 45-kDa protein VirA, which creates a path that enables the bacteria to move through the dense, organized cytoplasmic network of the host cell (Ogawa et al. 2008). Shigella variants that lack a functional virA gene are unable to move through the cytoplasm, and the invasiveness of these virA mutants is attenuated, suggesting that VirA is essential for Shigella virulence (Yoshida et al. 2006). It was proposed that VirA functions as a cysteine protease that selectively degrades α-tubulin (Yoshida et al. 2006). However, how VirA may recognize and degrade α-tubulin but not β-tubulin substrates remains unclear.
The only currently known homologs of VirA are the products of espG genes found in enteropathogenic and enterohemorrhagic E. coli (EPEC and EHEC), as well as in pathogenic Citrobacter rodentium (Mundy et al. 2004). It has been shown that, like VirA, EspG disrupts microtubules in fibroblasts and nonpolarized epithelial cells (Shaw et al. 2005). Interestingly, a study conducted by Elliott et al. (2001) demonstrated that the EPEC espG gene can rescue invasion of a Shigella that contains a mutation inactivating virA. Conversely, Smollett et al. (2006) have shown that VirA complements the double-mutant EPEC ΔespG/ΔespG2.
Seeking to gain further insight into the function of VirA, including its reported ability to process α-tubulin but not β-tubulin, we determined the crystal structure of the selenomethionine (SeMet)-substituted protein and refined it with data extending to 3 Å resolution. We found the fold of full-length VirA to be novel, without any resemblance to papain-like cysteine proteases, whereas the N-terminal domain shares limited similarity to cystatin A and stefin B, protein inhibitors of cysteine proteases. Although VirA was reported to cleave α-tubulin, we have not found any structural features that resemble the active sites of known proteases. Thus, the mode of action of this unusual protein requires further study.
Results and Discussion
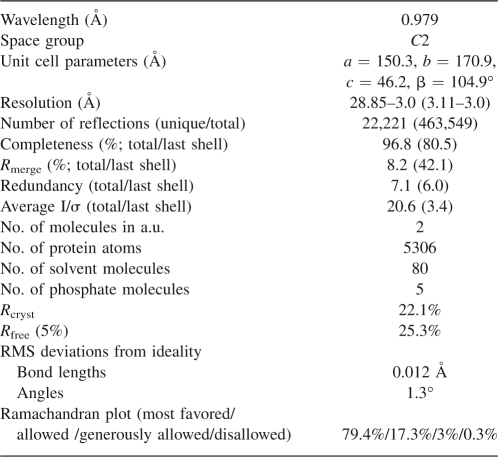
The structure of recombinant VirA was solved by single-wavelength anomalous diffraction (SAD) of a single crystal of SeMet-substituted protein. The enzyme was expressed in E. coli, purified to homogeneity, and crystallized using vapor-diffusion technique. The crystals belong to the monoclinic space group C2, with unit cell parameters a = 150.3 Å, b = 170.9 Å, c = 46.2 Å, β = 104.9°, and diffract to 3 Å resolution. Each asymmetric unit contains two molecules of VirA, resulting in a Matthews coefficient V m (Matthews 1968) of 3.2 Å3/Da and a solvent content of 61%. Due to the presence of considerable disorder and to limited resolution of diffraction data, we were able to trace only 684 residues out of 800 expected in the dimeric protein. The residues modeled in each of the crystallographically independent molecules are 1–13, 58–310, 314–328, and 339–399. Assignment of the sequence to the traced chain was facilitated by identifying the locations of all SeMet residues, based on the anomalous signal of the selenium atoms. However, the identification of the N-terminal 13 residues that form a short helix is only tentative since their sequence does not contain strong markers. Nevertheless, despite the relatively low resolution of the diffraction data, we consider the tracing of the visible parts of the polypeptide chain (with the exception of the N terminus) to be unambiguous.
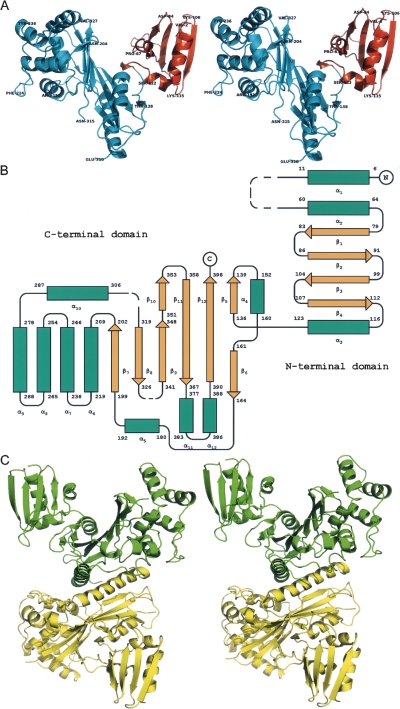
The refined structure reveals two independently folded domains that resemble the letter “V” (Fig. 1A). The partially disordered N-terminal domain includes residues 1–130, whereas the larger C-terminal domain encompasses residues 131 through 400. The current model consists of 12 α-helices and 12 β-strands, including a four-stranded antiparallel β-sheet located in the N-terminal domain and a six-stranded antiparallel β-sheet located in the C-terminal domain (Fig. 1B). A noteworthy feature of the structure is a long, serine-rich helix α10 (287–306) that appears to stabilize the dimer in the crystal (and, presumably, also in solution, since VirA appears to migrate as a dimer on native gels and on size-exclusion columns). That helix is responsible for most of the interactions in the dimer (Fig. 1C). The area buried upon dimer formation is 2487 Å2 per subunit.
Figure 1.
The structure of VirA. (A) Ribbon diagram showing the structure of a monomer of VirA. The N-terminal domain is colored gold; the C-terminal domain, blue. Location of selected residues are marked. (B) A topology diagram of the secondary structure of VirA, showing α-helices as cylinders, β-strands as arrows, and irregular structure as lines. Broken lines mark the missing residues. Secondary structure elements are labeled as discussed in the text. (C) A dimer of VirA, with the two molecules shown in green and yellow. All images representing structures were created in PyMOL (DeLano Scientific).
The N-terminal domain of the VirA molecule starts with a stretch of 13 residues disconnected from the rest of the model, putatively assigned as 1–13, that includes small helix α1 (residues 6–11). A perpendicularly oriented second helix α2 leads, through an extended linker region, to a four-stranded antiparallel β-sheet (β1–β4), followed by helix α3, which ends the first domain. The C-terminal domain begins with a short strand β5 followed by helix α4, which is partially shielded by the six-stranded antiparallel β-sheet. The loop extending from helix α4 wraps around this sheet and connects to helix α5, which is followed by α6, α7, and α8, forming a small helical bundle. Helix α9 protrudes through the back end of the bundle and loops around to the long α10 helix situated behind the main sheet and concludes with a short helix α11, followed by the last helix α12.
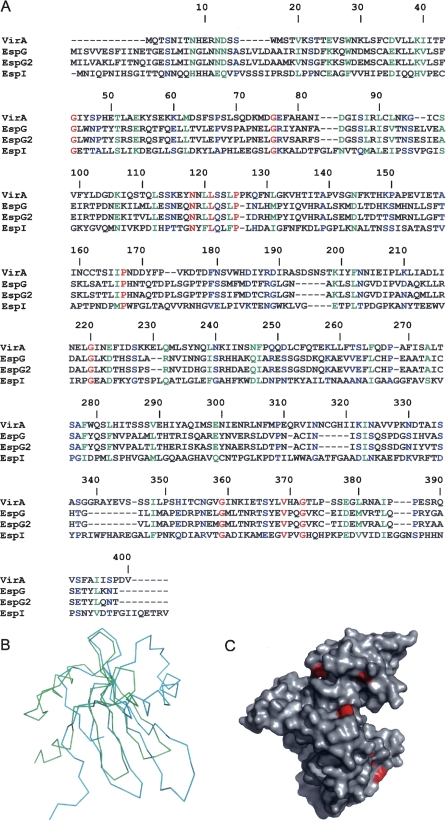
One of the reasons cited by Yoshida et al. (2006) for postulating cysteine protease-like activity of VirA was the loss of the apparent enzymatic activity when Cys34 (Fig. 2A) was mutated to a serine. The region surrounding this putative active site is disordered in our current model, and no other part of the protein bears any resemblance to the structure of papain. The N-terminal regions of proteins exported by the T3SS are frequently disordered (Galan and Wolf-Watz 2006), since these areas provide export signals, and a similar situation might exist in the case of VirA.
Figure 2.
Conservation of the sequence in the VirA/EspG family. (A) Sequence alignment of VirA, EspG, EspG2, and EspI proteins performed using ClustalW. Red denotes identical, green denotes strongly similar, and blue denotes weakly similar conservation. (B) Superposition of the Cα coordinates of the N-terminal domain of VirA and the inhibitor stefin B from the complex with papain (PDB code 1STF). VirA is traced in green; stefin B, in cyan. (C) Surface diagram of VirA in which the residues that are strictly conserved among VirA, EspG, EspG2, and EspI are colored red.
By creating a series of truncated proteins, a previous study identified the tubulin-binding region of VirA to contain residues 224–315 (Yoshida et al. 2002). Structurally, these residues comprise the helices α7–α10. However, as mentioned above, helix α10 plays a major role in dimer formation, thus it may not participate in binding tubulin but rather be necessary for maintaining VirA in a dimeric state. Some of these residues line the inner surface of the “V” structure, and a large cleft present between the two domains is of sufficient size to postulate that it could accommodate a tubulin dimer. It is surprising, though, that the N-terminal domain was not implicated in tubulin binding, although it forms the other arm of the letter “V” and any plausible binding mode would necessitate interactions with both domains.
To determine if the structure of VirA resembles any other proteins with known structure, including the microtubule-destabilizing proteins, we searched the Protein Data Bank (PDB) for its structural homologs using the DALI server (Holm and Sander 1993). Although no overall strong similarity to any known proteins was found, the N-terminal domain of VirA exhibits detectable similarity (Z-score 4.1) to cysteine protease inhibitors such as stefin B (PDB accession code 1stf) or cystatin A (1gd3). The structure of stefin B was solved in a complex with papain (Stubbs et al. 1990), and 55 out of a total of 98 residues of the inhibitor can be superimposed on residues within the sequence 57–130 of VirA with an root-mean-square (RMS) deviation of 2.4 Å (Fig. 2B). However, the sequence identity is extremely low at 7%. When the N-terminal domain of VirA is superimposed on the stefin B molecule complexed with papain, the remaining part of VirA does not clash with the papain molecule, making it possible to postulate that the function of VirA might be to create a scaffold for a papain-like cysteine protease, rather than function as an enzyme by itself.
VirA is a member of the EspG family, which also includes E. coli EspG, C. rodentium EspI, as well as E. coli EspG2 (Fig. 2A). Sequence similarity between VirA and EspG is ∼40% (21% identity), strongly suggestive of similar three-dimensional structures. In addition, these proteins also appear to share functional similarity, since both EspG and EspG2 have been shown to disrupt the microtubule network of intestinal epithelial cells (Elliott et al. 2001). The similarity between VirA, EspG, EspI, and EspG2 suggests a shared fold and mechanism of action within this family.
It has been postulated that all members of the EspG family exhibit cysteine protease-like activity (Tomson et al. 2005; Smollett et al. 2006; Yoshida et al. 2006). In a similar assay, we investigated the ability of VirA to degrade α/β-tubulin. However, we were unable to detect any proteolytic activity using the purified protein, and the sample of VirA purified by us did not exhibit any microtubule-severing activity in a microscopic assay using rhodamine microtubules (A. Roll-Mecak, pers. comm.). Even the alignment of the primary structures of these proteins does not provide a clear indication that this should be the case. The putative catalytic residue in VirA, Cys34, cannot be aligned well with its closest equivalent, Cys50 in EspG, whereas the corresponding region of EspG2 completely lacks any cysteine residues (Fig. 2A). No other cysteine throughout the whole sequence is conserved among these proteins. Although a number of serine residues are conserved (Ser14, Ser22, Ser123, Ser227, Ser276, and Ser366), none of them is close to either a conserved histidine or a lysine, which would be necessary to create the minimum catalytic dyad found in serine proteases (Dodson and Wlodawer 1998). Ser14 and Ser22 are located within the disordered fragment of the N-terminal domain, Ser276 is buried, and the remaining conserved serine residues, although found on the surface, do not have partners which could activate them for catalysis. None of the lysines and only a single histidine (His291) is conserved, with the latter residue situated at the beginning of the long stabilizing helix, α10, which is on the opposite side of the putative tubulin-binding cleft. Very few conserved residues are found on the protein surface (Fig. 2C), and those that are do not create a plausible active site, raising a possibility that the reported proteolytic activity of this protein might be due to a so-far undetected protease for which it would act as a binding partner.
Conclusions
Although the structure of VirA was solved at a resolution of 3 Å and the quality of the refined model is only moderate, we have full confidence that the fold of the protein is described correctly. With the exception of the N-terminal domain that topologically resembles several inhibitors of cysteine proteases, the fold of VirA is otherwise novel. Neither the structure by itself nor sequence comparisons with the related proteins EspG and EspG2 support the hypothesis that VirA is a cysteine protease. In particular, the part of the N-terminal domain that contains Cys34, previously postulated to be the catalytic residue, is completely disordered, making it unlikely that it could form a part of the active site. However, the puzzling similarity of the fold of the N-terminal domain of VirA to the inhibitors of cysteine proteases raises a remote possibility that this virulence effector might function as a scaffold rather than an enzyme. Although this hypothesis currently lacks experimental proof, further investigation of the role of VirA in the virulence of Shigella is clearly warranted.
Materials and Methods
Cloning, expression, and purification
The open reading frame (ORF) of Shigella flexneri VirA was amplified from genomic DNA (American Type Culture Collection, Manassas, VA) by the polymerase chain reaction (PCR) using the following oligonucleotide primers: 5′-GAGAACCTGTACTTCCAGGGTATGCAGACATCAAACATAACTAACC-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTATTAAACATCAGGAGATATGATGG-3′ (primer R). The PCR amplicon was subsequently used as template for a second PCR with the following primers: 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCGGAGAACCTGTACTTCCAG-3′ and primer R (above). The amplicon from the second PCR was inserted by recombinational cloning into the entry vector pDONR201 (Invitrogen), and the nucleotide sequence was confirmed experimentally. The ORF of VirA (M1–V400), now with a recognition site (ENLYFQ/G) for tobacco etch virus (TEV) protease fused in-frame to its N terminus, was moved by recombinational cloning into the destination vector pDEST-HisMBP (Tropea et al. 2007) to produce pDZ1952. pDZ1952 directs the expression of a maltose-binding protein (MBP)-VirA fusion protein with an intervening TEV protease recognition site. The MBP contains an N-terminal hexahistidine tag for affinity purification by immobilized metal affinity chromatography (IMAC). The fusion protein was expressed in the E. coli strain BL21 (DE3) CodonPlus-RIL (Stratagene). Cells containing the expression vector were grown to mid-log phase (OD600 ∼ 0.5) at 37°C in Luria broth containing 100 μg mL−1 ampicillin, 30 μg mL−1 chloramphenicol, and 0.2% glucose. Overproduction of the fusion protein was induced with isopropyl-β-D-thiogalactopyranoside at a final concentration of 1 mM for 4 h at 30°C. The cells were pelleted by centrifugation and stored at −80°C.
All purification procedures were performed at 4°C–8°C. Ten grams of E. coli cell paste was suspended in ice-cold 50 mM sodium phosphate (pH 7.5), 200 mM NaCl, 25 mM imidazole buffer (buffer A) containing 2 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride (AEBSF, Roche Molecular Biochemicals). The cells were lysed with an APV-1000 homogenizer (Invensys) at 10,000 psi and centrifuged at 30,000g for 30 min. The supernatant was filtered through a 0.22-μm polyethersulfone membrane and applied to a 12 mL Ni-NTA superflow column (Qiagen) equilibrated in buffer A. The column was washed to baseline with buffer A and then eluted with a linear gradient of imidazole to 250 mM. Fractions containing recombinant His6-MBP-VirA were pooled, concentrated using an Amicon YM30 membrane (Millipore Corporation), diluted with 50 mM sodium phosphate (pH 7.5), 200 mM NaCl buffer to reduce the imidazole concentration to ∼25 mM, and digested overnight at 4°C with His6-tagged TEV protease (Kapust et al. 2001). The digest was applied to a second Ni-NTA superflow column equilibrated in buffer A and the VirA emerged in the column effluent. The column effluent was incubated overnight with 10 mM dithiothreitol (DTT), concentrated using an Amicon YM10 membrane, and applied to a HiPrep 26/60 Sephacryl S-100 HR column (Amersham Biosciences) equilibrated in 25 mM Tris, 150 mM NaCl, 2 mM Tris(2-carboxyethyl) phosphine hydrochloride buffer. The peak fractions containing recombinant VirA were pooled and concentrated to 15–20 mg mL−1 (estimated at 280 nm using a molar extinction coefficient of 36,900 M−1 cm−1). Aliquots were flash-frozen in liquid nitrogen and stored at −80°C. The final product was judged to be >95% pure by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The molecular weight of VirA was confirmed by electrospray mass spectroscopy.
Crystallization and data collection
Prior to crystallization, VirA was concentrated to a final concentration of 20 mg/mL in 25 mM Tris pH 7.2, 150 mM NaCl, 2 mM Tris (2-carboxyethyl) phosphine hydrochloride buffer. The crystals were grown in hanging drops composed of an equal volume of VirA and 0.8 M sodium/potassium phosphate, pH 6.1 at 22°C. The crystals were dipped into 0.8 M sodium/potassium phosphate, pH 6.1 + 30% glycerol to cryo-protect before flash-freezing in liquid nitrogen. The diffraction data were collected at the SER-CAT 22-ID beamline (Advanced Photon Source, Argonne National Laboratory). After a fluorescence scan was performed, the wavelength was tuned to the selenium absorption peak and data were collected at 0.97923 Å wavelength on a MAR300CCD detector. A 3.0 Å SAD data set was collected and then indexed and scaled with the HKL2000 program suite (Otwinowski and Minor 1997). The statistics of diffraction data and refinement are given in Table 1.
Table 1.
Data collection and refinement statistics
Structure determination and refinement
The self-rotation function calculated with POLARRFN (CCP4 1994) revealed a prominent peak for κ = 180°, indicating the presence of a noncrystallographic twofold symmetry axis. XPREP (Sheldrick 2001) was used to extract anomalous differences, and SHELXD (Sheldrick 2008) was employed to locate the positions of selenium sites. HA_NCS (Terwilliger 2002) also located a twofold NCS axis from the anomalous sites independently. The NCS from anomalous sites had the same orientation as that from the self-rotation function, which verified the validity of the selenium sites. The identified anomalous scattering sites were input for SAD phasing to several programs, e.g., SHELXE (Sheldrick 2008), SOLVE (Terwilliger and Berendzen 1999), and PHASER (McCoy et al. 2007). However, the electron-density maps after density modification were largely uninterpretable, and only some helical fragments and turns could be automatically assigned with the program RESOLVE (Terwilliger 2003). Because the anomalous sites were verified by the self-rotation function, the residue clusters built by RESOLVE at least contained some useful phasing information to improve the map. Thus, the modeled fragments and selenium sites were fed back to PHASER again to combine the incomplete but partially correct atomic model and the anomalous signal. After several cycles, the combined phase information became stronger, allowing the missing selenium atoms to be located and additional secondary structure elements to be identified. The resulting electron density map was largely interpretable and almost a complete model was built with the program BUCCANEER (Cowtan 2006). The structure was subsequently refined with PHENIX.REFINE (Adams et al. 2002) using NCS, experimental phases, and stereochemical information as restraints (Table 1). Solvent molecules were added with COOT (Emsley and Cowtan 2004), in Fo-Fc electron density peaks close to polar atoms of the protein. The quality of the final model was evaluated with the program PROCHECK (Laskowski et al. 1993) and was found to be acceptable, given the limited resolution of the diffraction data (all outliers of the Ramachandran plot are located in the poorly defined loop regions). The coordinates and structure factors have been submitted to the PDB under accession code 3ee1.
Note added in proof
The structure of an N-terminally truncated VirA, crystallized in a space group different from the full-length protein described here, appeared in press when this manuscript was under review (Germane et al. 2008). The results presented here, in particular concerning the novel fold of VirA and the lack of detectable enzymatic activity, are fully consistent with their observations.
Acknowledgments
We thank Dr. Antonina Roll-Mecak for testing the microtubule-severing activity of VirA. Diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) beamline 22-ID, located at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31-109-Eng-38. This project was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and in part with Federal funds from the National Cancer Institute, NIH, under contract no. NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Reprint requests to: Alexander Wlodawer, Protein Structure Section, Macromolecular Crystallography Laboratory, NCI-Frederick, PO Box B, Frederick, MD 21702, USA; e-mail: wlodawea@mail.nih.gov; fax: (301) 846-6322.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.037978.108.
References
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- Dodson G., Wlodawer A. Catalytic triads and their relatives. Trends Biochem. Sci. 1998;23:347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- Dutta S., Dutta S., Dutta P., Matsushita S., Bhattacharya S.K., Yoshida S. Shigella dysenteriae serotype 1, Kolkata, India. Emerg. Infect. Dis. 2003;9:1471–1474. doi: 10.3201/eid0911.020652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott S.J., Krejany E.O., Mellies J.L., Robins-Browne R.M., Sasakawa C., Kaper J.B. EspG, a novel type III system-secreted protein from enteropathogenic Escherichia coli with similarities to VirA of Shigella flexneri . Infect. Immun. 2001;69:4027–4033. doi: 10.1128/IAI.69.6.4027-4033.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Galan J.E., Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- Germane K.L., Ohi R., Goldberg M.B., Spiller B.W. Structural and functional studies indicate that Shigella VirA is not a protease and does not directly destabilize microtubules. Biochemistry. 2008;47:10241–10243. doi: 10.1021/bi801533k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Kapust R.B., Tözsér J., Fox J.D., Anderson D.E., Cherry S., Copeland T.D., Waugh D.S. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- Lan R., Reeves P.R. Escherichia coli in disguise: Molecular origins of Shigella . Microbes Infect. 2002;4:1125–1132. doi: 10.1016/s1286-4579(02)01637-4. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: Program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Matthews B.W. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallograhic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy R., Petrovska L., Smollett K., Simpson N., Wilson R.K., Yu J., Tu X., Rosenshine I., Clare S., Dougan G., et al. Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infect. Immun. 2004;72:2288–2302. doi: 10.1128/IAI.72.4.2288-2302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Handa Y., Ashida H., Suzuki M., Sasakawa C. The versatility of Shigella effectors. Nat. Rev. Microbiol. 2008;6:11–16. doi: 10.1038/nrmicro1814. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Parsot C. Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors. FEMS Microbiol. Lett. 2005;252:11–18. doi: 10.1016/j.femsle.2005.08.046. [DOI] [PubMed] [Google Scholar]
- Schroeder G.N., Hilbi H. Molecular pathogenesis of Shigella spp.: Controlling host cell signaling, invasion, and death by type III secretion. Clin. Microbiol. Rev. 2008;21:134–156. doi: 10.1128/CMR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw R.K., Smollett K., Cleary J., Garmendia J., Straatman-Iwanowska A., Frankel G., Knutton S. Enteropathogenic Escherichia coli type III effectors EspG and EspG2 disrupt the microtubule network of intestinal epithelial cells. Infect. Immun. 2005;73:4385–4390. doi: 10.1128/IAI.73.7.4385-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G.M. XPREP program. 6.14 Bruker Nonius, Inc; Madison, WI: 2001. [Google Scholar]
- Sheldrick G.M. A short history of SHELX. Acta Crystallogr. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Smollett K., Shaw R.K., Garmendia J., Knutton S., Frankel G. Function and distribution of EspG2, a type III secretion system effector of enteropathogenic Escherichia coli . Microbes Infect. 2006;8:2220–2227. doi: 10.1016/j.micinf.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Stubbs M.T., Laber B., Bode W., Huber R., Jerala R., Lenarcic B., Turk V. The refined 2.4 Å X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: A novel type of proteinase inhibitor interaction. EMBO J. 1990;9:1939–1947. doi: 10.1002/j.1460-2075.1990.tb08321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. Rapid automatic NCS identification using heavy-atom substructures. Acta Crystallogr. D Biol. Crystallogr. 2002;58:2213–2215. doi: 10.1107/S0907444902016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. SOLVE and RESOLVE: Automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C., Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomson F.L., Viswanathan V.K., Kanack K.J., Kanteti R.P., Straub K.V., Menet M., Kaper J.B., Hecht G. Enteropathogenic Escherichia coli EspG disrupts microtubules and in conjunction with Orf3 enhances perturbation of the tight junction barrier. Mol. Microbiol. 2005;56:447–464. doi: 10.1111/j.1365-2958.2005.04571.x. [DOI] [PubMed] [Google Scholar]
- Tropea J.E., Cherry S., Nallamsetty S., Bignon C., Waugh D.S. A generic method for the production of recombinant proteins in Escherichia coli using a dual hexahistidine-maltose-binding protein affinity tag. Methods Mol. Biol. 2007;363:1–19. doi: 10.1007/978-1-59745-209-0_1. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Katayama E., Kuwae A., Mimuro H., Suzuki T., Sasakawa C. Shigella deliver an effector protein to trigger host microtubule destabilization, which promotes Rac1 activity and efficient bacterial internalization. EMBO J. 2002;21:2923–2935. doi: 10.1093/emboj/cdf319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Handa Y., Suzuki T., Ogawa M., Suzuki M., Tamai A., Abe A., Katayama E., Sasakawa C. Microtubule-severing activity of Shigella is pivotal for intercellular spreading. Science. 2006;314:985–989. doi: 10.1126/science.1133174. [DOI] [PubMed] [Google Scholar]