Abstract
Macromolecular crowding, a common phenomenon in the cellular environments, can significantly affect the thermodynamic and kinetic properties of proteins. A single-molecule method based on atomic force microscopy (AFM) was used to investigate the effects of macromolecular crowding on the forces required to unfold individual protein molecules. It was found that the mechanical stability of ubiquitin molecules was enhanced by macromolecular crowding from added dextran molecules. The average unfolding force increased from 210 pN in the absence of dextran to 234 pN in the presence of 300 g/L dextran at a pulling speed of 0.25 μm/sec. A theoretical model, accounting for the effects of macromolecular crowding on the native and transition states of the protein molecule by applying the scaled-particle theory, was used to quantitatively explain the crowding-induced increase in the unfolding force. The experimental results and interpretation presented could have wide implications for the many proteins that experience mechanical stresses and perform mechanical functions in the crowded environment of the cell.
Keywords: macromolecular crowding, mechanical unfolding, ubiquitin, dextran, single molecule, mechanical stability
Proteins perform their functions in environments that are crowded with various macromolecules and macromolecular assemblies, as visualized recently by high-resolution cryoelectron tomography (Medalia et al. 2002) and other techniques. While a single species of macromolecules may not be very concentrated, the total volume occupied by all macromolecules can constitute a large fraction of the interior space of a cell and intracellular compartments. As a result, proteins and other macromolecules must exist and function in a crowded environment. In order to understand the structural dynamics and functional mechanisms of protein molecules in the cell, it is necessary to consider the effects of such macromolecular crowding (Minton 2006; Zhou et al. 2008). An obvious, but important, consequence of macromolecular crowding is the excluded volume effect due to the mutual impenetrability of the macromolecules, resulting in a reduction of the accessible space for these molecules. Intrinsically unstructured proteins in dilution solutions may acquire stable structures when they are placed in crowded media (Dedmon et al. 2002; Qu and Bolen 2002). Crowding can prevent stress-induced aggregation and misfolding of proteins by altering the folding kinetics (Minton et al. 1982). The goal of the present work was to understand the effects of macromolecular crowding on the mechanical stability and unfolding kinetics of proteins at the single-molecule level using an atomic force microscopy (AFM)-based manipulation technique.
Most of our understanding on protein folding has been acquired from studies under ideal conditions with pure samples in dilute solutions. However, potentially macromolecular crowding can significantly affect the thermodynamic stability and alter the folding/unfolding kinetics of proteins. Despite a growing number of investigations on macromolecular crowding by experimental, theoretical, and computational approaches, the consequences, magnitudes, and mechanisms of crowding effects on protein folding remain controversial. For example, some theories predicted a modest effect on the thermodynamic stability by crowding (Zhou 2004) while others predicted significant stabilization (Minton 2005). Similarly, conflicting results have been reported from experimental studies. Using circular dichroism (CD) spectroscopy, fluorescence correlation spectroscopy, and NMR, Tokuriki et al. (2004) found that unfolded RNase A in 2.4 M urea at pH 3 became native when crowding agents (PEG or Ficoll) were added, and that this effect was most obvious when the “crowders” had similar sizes as the protein. Sasahara et al. (2003) reported that the thermal stability of hen egg-white lysozyme was enhanced and the acid-unfolded cytochrome c was driven to a more compact conformation by high concentrations of dextran. On the other hand, Qu and Bolen (2002) found that macromolecular crowding can enhance the folding of an intrinsically unstructured protein (TCAM), but the effect was modest. Spencer et al. (2005) observed that 180 g/L of Ficoll 70K resulted in only a modest increase of 0.5 kcal/mol in the folding stability of FKBP. Van den Berg et al. (2000) found that macromolecular crowding could accelerate a fast-track folding process, but decrease a slow-track folding process of hen lysozyme. Martin (2002) reported that some proteins (dihydrofolate reductase, enolase, and green fluorescent protein) could fold spontaneously in dilute, but not in crowded, solutions. Flaugh and Lumb (2001) found that two intrinsically disordered proteins (the C-terminal activation domain of c-Fos and the kinase-inhibition domain of p27Kip1) could not be induced into ordered structures by macromolecular crowding. These inconsistent results underscore the complexity of macromolecular crowding effects on protein folding.
The experimental studies cited above on crowding effects were all carried out by bulk techniques, which measure the average results of a large number of molecules. Over the last decade, single-molecule methods have been used in the investigation of protein folding and mechanical functions of proteins. These experiments complement bulk measurements and have helped to gain critical new insights into the mechanisms of proteins' mechanical functions (Rief et al. 1997) and the structural first dynamics and pathway heterogeneity of individual protein molecules (Yang et al. 2000; Hyeon and Thirumalai 2003; Fernandez and Li 2004). In this paper, we present the experimental study of the effects of macromolecular crowding on the mechanical stability of protein molecules using a single-molecule method. Protein molecules are known to perform various mechanical functions and experience various mechanical stresses; therefore, the elucidation of the crowding effects on the ability of protein molecules to resist mechanical stresses is important for the understanding of the structure integrity and mechanical functions of protein molecules inside the cell.
In addition to investigating the mechanical stability of proteins, there are several additional advantages in using the single-molecule approach to study the effects of macromolecular crowding on the unfolding and refolding of individual protein molecules. Experimental measurements are made in solutions with no other protein molecules except the ones under study in the polymer chain, which is in an extended conformation when the individual molecules are unfolded. This eliminates the technical difficulties caused by aggregation during certain bulk measurements. In these experiments, the mechanical force is used to induce unfolding, so that denaturants or harsh environments, such as extreme temperatures or pH, are not involved. Consequently, any complications due to changes in solution conditions are avoided. In addition, the reaction coordinate along the pulling direction is well-defined during unfolding, making modeling of the effect less arbitrary. When proteins are studied at the single-molecule level, it would be possible to distinguish and characterize macromolecular crowding effects on parallel folding pathways, such as that observed for hen lysozyme (van den Berg et al. 2000). These capacities of single-molecule methods can potentially provide unique and complementary information on macromolecular crowding effects on the energetics and kinetics of protein folding and unfolding. We investigated the force-induced unfolding of ubiquitin molecules in the presence of various concentrations of dextran from 0 to 300 g/L, which covers the range that is relevant to the crowdedness of macromolecules in cells. Ubiquitin has been used widely as a model system for protein folding studies, including single-molecule studies (Carrion-Vazquez et al. 2003; Chyan et al. 2004). Our measurements show that the forces required to unfold ubiquitin molecules are higher in crowded media than those measured in dilute solutions. In this paper, we present a complete set of experimental data and provide quantitative interpretations of the results based on the scaled-particle theory as well as discussions on the biological relevance of the observed phenomena.
Results
Effects of macromolecular crowding on secondary structures
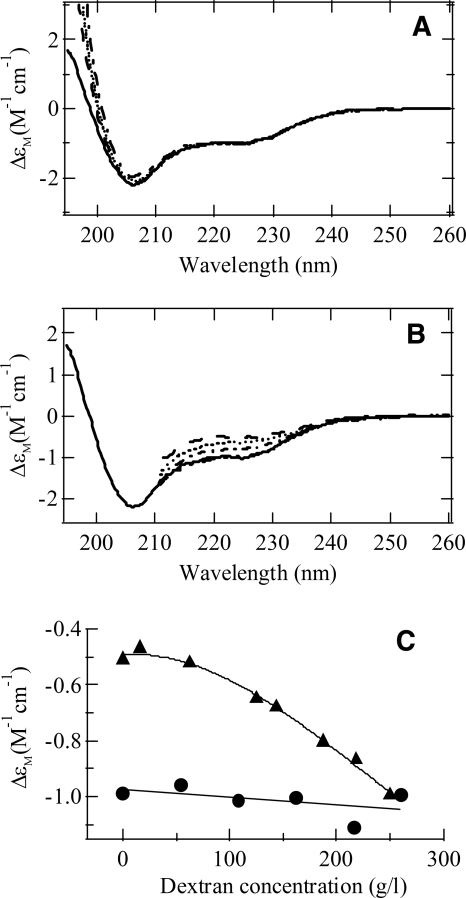
To assess the effects of crowding by dextran on the thermodynamic stability of ubiquitin molecules, CD spectroscopy was used to characterize the structural changes of ubiquitin under different conditions. As expected, the addition of various concentrations of dextran to the buffer solution did not change the CD signal when ubiquitin was in the native state, as shown in Figure 1A,C. However, the crowding condition did induce significant changes in the CD signal when the protein was in a solution containing 3.1 M GdnHCl (Fig. 1B,C). At the presence of 250 g/L dextran, the CD spectrum of ubiquitin in 3.1 M GdnHCl showed almost identical features as those in the dilute solution without GdnHCl. Our previous data showed that ubiquitin started unfolding at a GdnHCl concentration of ∼2 M, and the percentage of unfolded molecules reached 70% at 3.1 M, and 100% when the GdnHCl concentrations was >4 M (Chyan et al. 2004). Therefore, at 3.1 M GdnHCl, the addition of dextran shifted the equilibrium from a 70%:30% unfolded/folded mixture to an almost 100% folded ubiquitin population. Our data, similar to that previously reported for RNase A, lysozyme, and cytochrome c (Sasahara et al. 2003; Tokuriki et al. 2004), indicated that macromolecular crowding by dextran drove the equilibrium toward the folded state.
Figure 1.
Effect of dextran on the structure of ubiquitin. (A) CD spectra of ubiquitin in solutions (pH 5) containing various concentrations of dextran. The solid line is for ubiquitin in the absence of dextran, and the dashed, dotted, and dash-dotted lines represent the results in the presence of 110, 160, and 260 g/L dextran, respectively. (B) CD spectra of ubiquitin in solutions (pH 5) containing 3.1 M GdnHCl and in the presence of 0 (dashed line), 125 g/L (dotted line), 190 g/L (dash-dotted line), and 250 g/L (dash-dot dotted line) of dextran, respectively. The solid line is for ubiquitin in the absence of dextran and GdnHCl. (C) Molar CD extinction coefficient of ubiquitin at the wavelength of 222 nm as a function of dextran concentration at pH 5, in the absence of GdnHCl (circles) and in the presence of 3.1 M GdnHCl (triangles).
Mechanical unfolding in crowded media
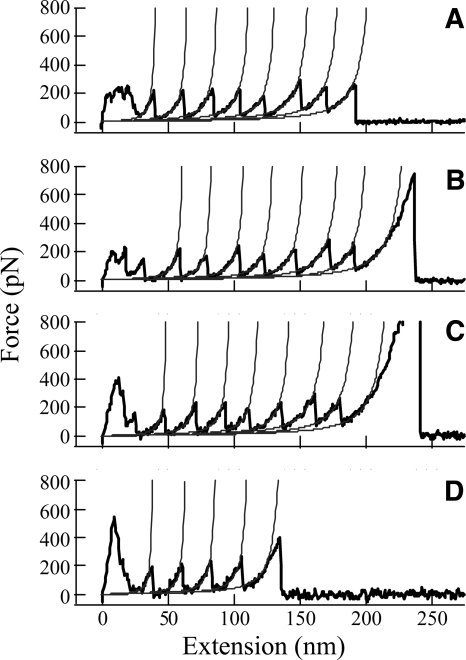
The measurements of the unfolding forces of individual protein molecules require the attachment of a protein polymer between the AFM tip and a substrate surface. A convenient and reliable method is to first deposit protein polymers on a gold surface, and then use the nonspecific interactions between the protein and the Si3N4 AFM tip to pick up a polymer from the surface and stretch it by moving the tip away (Rief et al. 1997). This has been the most widely used method in mechanical unfolding experiments. Our results show that the presence of dextran in the buffer solution in the AFM liquid chamber, even at high concentrations, did not significantly interfere with the nonspecific interactions between the tip and the protein polymers. Experiments were carried out to mechanically unfold ubiquitin molecules at dextran concentrations from 0 to 300 g/L. Figure 2 shows several mechanical unfolding “force curves” obtained by pulling ubiquitin polymers at different dextran concentrations, where each peak in the regular sawtooth patterns represents the unfolding of a single protein molecule. From these curves, several features of mechanical unfolding experiments can be observed. The beginning of each curve has irregular force peaks due to nonspecific interactions between the tip and the gold surface. The use of polymerized protein molecules circumvents the difficulties caused by the interference of these nonspecific interactions. When the tip is sufficiently far from the surface (>20 nm), the nonspecific interactions become insignificant and the unfolding of individual protein molecules can be observed. By fitting the rising part of each peak to the wormlike-chain (WLC) model (Fig. 2), the contour length increase from an unfolding event can be obtained. A comparison of this contour length increment with that calculated from the structure of the protein serves as one of the parameters to verify that the observed peaks are indeed from unfolding of individual protein molecules. The unfolding force for each ubiquitin molecule is not the same, because thermal fluctuation plays an important role in such single-molecule experiments. The average and the distribution of the unfolding forces are determined by the free-energy barrier height and position of the transition state (Carrion-Vazquez et al. 1999; Li et al. 2000). Using Monte Carlo simulations based on a two-state model for protein unfolding, force curves of the force-induced unfolding of ubiquitin molecules were generated. The unfolding rates in the absence of an applied force and the distance between the native state and transition state along the pulling direction were determined (Fig. 3).
Figure 2.
Force versus extension curves obtained from stretching ubiquitin polymers in PBS buffer containing: (A) 0, (B) 100, (C) 200, and (D) 300 g/L of dextran. The pulling speed used for these curves was 0.25 μm/sec. The rising parts of the force peaks are fitted to the WLC model. The persistence length p used in the fitting was 0.4 nm. The contour length increment ΔL between adjacent peaks, i.e., the increase in the polymer length upon each unfolding event, was found to be ΔL = 24.9 ± 2.4 nm (n = 676). This value is consistent with the expected value of 24.4 nm from the structure of ubiquitin (Chyan et al. 2004). The value of ΔL was found to be independent on the dextran concentration, with values of 25.2 ± 2.5, 24.7 ± 2.5, 24.6 ± 2.1, and 25.0 ± 2.6 nm for dextran concentrations of 0, 100, 200, and 300 g/L, respectively. With an automatic procedure to fit the data by varying both the persistence length and the contour length, it was found that the persistence length did not depend on dextran concentration.
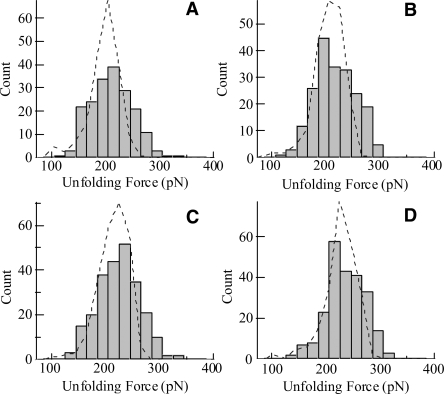
Figure 3.
Distribution of the measured unfolding force of ubiquitin obtained in PBS buffer containing (A) 0, (B) 100, (C) 200, and (D) 300 g/L of dextran at a pulling speed of 0.25 μm/sec. The dashed lines are the results from Monte Carlo simulations. The numbers of data points are 189, 202, 242, and 232, respectively, and the forces are (average ± standard deviation): 210 ± 30, 215 ± 37, 224 ± 39, and 234 ± 35 pN, respectively, for the dextran concentration of 0, 100, 200, and 300 g/L. The Monte Carlo simulation results, scaled by the number of data points, were obtained with the parameters of Δx u = 0.225 nm and k u0 = 5.0 × 10−5, 4.3 × 10−5, 2.6 × 10−5, and 1.3 × 10−5 sec−1, respectively, for the dextran concentrations listed above. The simulation results have narrower distributions than the experimental data because of experimental noises.
Dependence of unfolding forces on dextran concentration
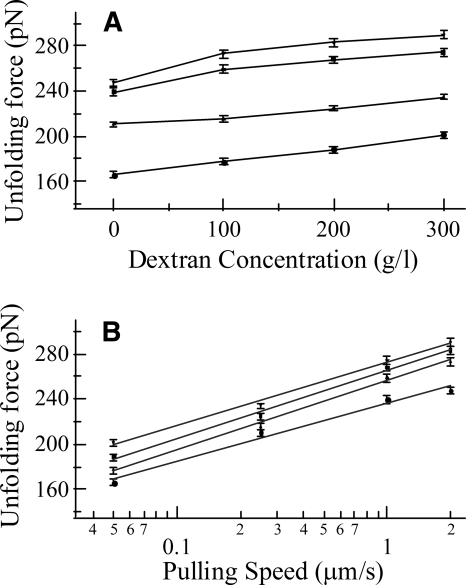
The effects of macromolecular crowding on the mechanical unfolding of ubiquitin were determined by measuring the unfolding forces when the protein molecules were in solutions with different concentrations of dextran. Figure 3 shows the distribution of the unfolding forces when the protein was in solutions with 0, 100, 200, and 300 g/L of dextran, respectively. These force histograms show that the average unfolding force increased from 210 pN to 234 pN as the dextran concentration was increased from 0 to 300 g/L (see also Fig. 4A), while the widths of the distributions did not change appreciably. The data suggest that the crowding conditions caused a decrease in the zero-force unfolding rates, k u0, of the ubiquitin molecules but did not change the distance between the native state and the transition state, Δx u, since the distribution width is primarily determined by Δx u as shown by Monte Carlo simulations (G. Yang, unpubl.). Figure 4A shows that the experimental data obtained using different pulling speeds all exhibited the same trend of increase in unfolding forces as the solution became more crowded with dextran. Within the range of dextran concentrations used in the experiments, the unfolding forces increased nearly linearly with the dextran concentration. Furthermore, when the unfolding forces are plotted as a function of the pulling speed, the unfolding forces are found to increase linearly with the logarithm of the speed (Fig. 4B), indicating that the force-induced unfolding of the ubiquitin molecules followed the two-state model at each dextran concentration (Rief et al. 1997). The straight lines in the plot are nearly parallel to each other, which again indicate that crowding reduces k u0 but does not change Δx u, as suggested by our Monte Carlo simulation results. In presenting the data in Figure 4, the standard error of the mean is used to quantify the uncertainties of the experimental results because this value provides an estimate of how well the measured average values represent the protein population, while the standard deviation reflects the distribution of individual data points around the mean.
Figure 4.
(A) Dependence of the unfolding forces of ubiquitin on dextran concentration at four different pulling speeds. From top to bottom, the pulling speeds were: 2.0 μm/sec, 1.0 μm/sec, 0.25 μm/sec, and 0.05 μm/sec, respectively. Each point in the plot is the average of a number of data points (from 166 to 290). The error bars represent the standard errors of mean (standard deviation divided by the square root of the number of data points). The unfolding force changes almost linearly with dextran concentration with an average slope of 0.11 pN/(g/L). (B) The dependence of the unfolding forces on the pulling speeds for four different dextran concentrations. From top to bottom, the dextran concentrations were 300 g/L, 200 g/L, 100 g/L, and 0 g/L, respectively. The lines are linear fits to the data for each dextran concentration, as predicted by the two-state model for unfolding (Rief et al. 1997). The error bars are the standard errors of the mean.
Effect of macromolecular crowding on the unfolding rate
Previous published work has shown that macromolecular crowding can stabilize the native state of a protein (Sasahara et al. 2003; Spencer et al. 2005) and enhances the folding rate (van den Berg et al. 1999; Ellis 2001), but there are few reports on the effects on the unfolding rates. In a recent study, Ai et al. (2006) used 15N NMR spin relaxation dispersion to investigate the effects of crowding on the folding and unfolding rates of Rd-apocyt b 562. The presence of 85 g/L PEG 20K increased the folding rate by a modest 50% and had no significant effect on the unfolding rate. Cheung et al. (2005) and Cheung and Thirumalai (2007) used an off-lattice model to simulate macromolecular crowding effects on the folding thermodynamics and kinetics of an all-β-sheet WW domain. The simulation also found that crowding increased the folding stability, and this increase was mostly the result of an increased folding rate. A continuous three-dimensional polymer model based on the concept of depletion force was used to qualitatively explain the crowding-induced increase in protein's mechanical stability based on our initial results (Ping et al. 2006). Here, a quantitative model is used to interpret the experimental observations, which provides the first evidence that the unfolding rate (at zero external force; k u0) is lowered in a crowded solution.
When a protein molecule is subjected to an external force, its unfolding rate changes as (Bell 1978; Evans and Ritchie 1999):
 |
where F is the applied force, Δx u is the distance between the native state and the transition state along the pulling direction, and k B T is the thermal energy. At the same pulling speed, the observed unfolding rate k u is the same; therefore, a higher force F can be the result of a lower k u0, a smaller Δx u or both. As discussed above, the data show that Δx u (0.225 nm, see Fig. 3, caption) does not change when the solution is crowded with dextran. Consequently, the observed unfolding force increase in crowded solutions, as shown in Figure 4A, suggests that the zero-force unfolding rate k u0 is lowered by macromolecular crowding, according to Equation 1. Here it should be pointed out that a very small decrease in Δx u, not discernible in the data, could still contribute to the observed unfolding force increase since the unfolding force depends sensitively on Δx u.
Explanation of the decrease in k u0 using scaled-particle theory
The zero-force unfolding rate can be written in the form:
 |
where ΔG ≠ u0 is the unfolding free-energy barrier in the absence of force and A is a pre-exponential factor determined by the internal dynamics of the protein molecule as well as the solution conditions. The reduction in the unfolding rate k u0 by macromolecular crowding can be due to either an increase in the energy barrier ΔG ≠ u0 or a decrease in the prefactor A. The transition state (TS) for unfolding consists of protein conformations that are more expanded than those in the native state, and hence will be more disfavored entropically by the presence of crowders, leading to an increase in the unfolding free-energy barrier height. The crowders also serve as obstacles for—and therefore slow down—the overall diffusion motion of the protein molecule and its internal dynamics. In the presence of a high concentration of dextran, the solution becomes more viscous (at 300 g/L, the solution viscosity is ∼25 cP). Ansari et al. (1992) showed that, using the Kramers theory, the pre-exponential factor in the rate equation for conformational change of proteins was inversely proportional to the sum of two contributions: One relates to the solution viscosity and the other to the internal viscosity of the protein from the motions of protein atoms relative to each other. Monte Carlo simulations indicated no large scale conformational changes on reaching the transition state during the mechanical unfolding of ubiquitin, and the rate-limiting step mostly involved residues shielded from the solvent (Kleiner and Shakhnovich 2007), therefore the crowding agent is expected to have minimal effects on the internal viscosity of the protein. The effect of solution viscosity on the pre-exponential factor depends on the fraction of protein atoms in contact with solvent molecules (Ansari et al. 1992). Due to the large size of the dextran molecules relative to ubiquitin, the friction experienced by the protein molecules is much less than that expected from the macroscopic viscosity of the solution (Jas et al. 2001). From these considerations, the pre-exponential factor for the unfolding rate expression is treated as unchanged by the presence of the crowding agents in the discussions below.
For the effect of crowding on ΔG ≠ u0, we model the crowder as a sphere (with radius R c) for simplicity. We also model the folded protein molecule as a sphere, with radius a F, and the TS also as another sphere, with a somewhat enlarged radius a TS. A similar treatment of the TS was used by Hayer-Hartl and Minton (2006) in their calculations of the effect of confinement on the folding rate. The crowders occupy volumes that are inaccessible to the protein. The excluded volumes serve to increase the free energy of the protein. According to the scaled-particle theory (Lebowitz et al. 1965), the increase in the free energy of a protein modeled as a sphere is given by
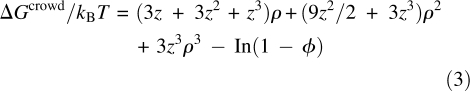 |
where z = a/R c (with a = a F for the folded state and a TS for the TS), φ = 4πR c 3 c/3 is the volume fraction of the crowders (c: number concentration), and ρ = φ/(1 – φ). The change in the unfolding free-energy barrier by the crowders is then
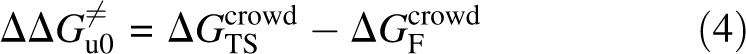 |
The change in k u0 due to macromolecular crowding can be calculated once the radii of the folded protein, the TS, and the crowder are specified. Based on the structure of ubiquitin, an estimate of a F = 1.8 nm can be given. The TS can be assigned a radius of ∼1.9 nm based on the magnitude of Δx u. Using the data published by Weiss et al. (2004) and the molecule weight of the dextran (40 kDa) used in our experiments, the crowder can be assigned a radius of 3.2 nm. The solid line in Figure 5 represents the calculation result using these parameters, assuming that the prefactor A is unaffected by crowding. Compared with the experimental results in Figure 3, the calculations show the correct trend but underestimate the decrease in k u0 at dextran concentrations ∼200 g/L.
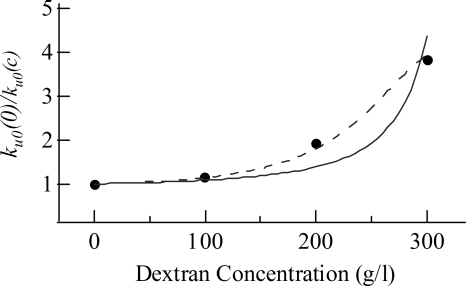
Figure 5.
Macromolecular crowding-induced decrease in the zero-force unfolding rate k u0. The ordinate is the ratio of k u0 in the absence of crowding agent to that at various dextran concentrations. The experimental values (filled circles) of k u0 are those listed in the caption of Figure 3. The calculated values (solid and dashed lines) were obtained using Equations 2 to 4. The solid line was obtained by assuming a fixed radius of 3.2 nm for the dextran molecules, and a fixed radius of 1.9 nm for the TS of ubiquitin. The dashed line was obtained by assuming that the radii of the dextran molecules and the TS of ubiquitin are both reduced at high concentrations of dextran, with R c (nm) = 4.0 − 1.84 ± 10−3 c − 8.75 × 10−7 c 2, and a TS (nm) = 1.9 − 5.0 × 10−5 c − 5.0 × 10−7 c 2, where the dextran concentration c is in g/L.
There are several possibilities that can account for this underestimate, with the most obvious being the treatments of dextran and the TS as spheres of fixed sizes. Dextran is a flexible polymer with a very short persistence length (Rief et al. 1998); therefore, it is not unreasonable to assume a spherical shape for the molecules in solution, especially when present in high concentrations. (In a very dilute solution, it is perhaps more appropriate to model dextran as a random-flight polymer chain. This and other models would predict a qualitatively similar result [Berg 1990; Minton 2005; Zhou 2008]. That is, the TS with an expanded volume relative to the folded state will be more adversely affected by macromolecular crowding than the folded state. Again, an increase in unfolding free-energy barrier by crowding is predicted.) However, the radius of dextran molecules of a specific molecular weight has not been determined unequivocally, with reported values ranging from 3 to 5 nm for the dextran molecules with an average molecular weight of 40 kDa (Ioan et al. 2000; Weiss et al. 2004). In addition, the volume of dextran molecules shrinks as the concentration increases (Ogston and Preston 1979). The volume of the protein in transition state may also be reduced by crowding. Theoretically, the TS is expected to become more compact under crowded conditions since some of the more open conformations in the TS will be eliminated by the crowding agent due to volume exclusion (Zhou 2004; Cheung et al. 2005). In particular, a unstructured region known to be present in the transition state of ubiquitin (Went and Jackson 2005) could be affected by crowding. By assuming that crowding does induce a volume reduction of the dextran molecules and the ubiquitin TS (see Fig. 5, caption), the experimentally observed changes in k u0 due to macromolecular crowding can be well reproduced by the scaled-particle theory, as shown by the dashed line in Figure 5.
Discussion
Biological implications of the experimental results
Generation and transformation of mechanical forces are involved in many biological processes (Bustamante et al. 2004; Gao et al. 2006; Forman and Clarke 2007). Motor proteins convert chemical energy into mechanical work for muscle contraction, protein and vesicle transport, and chromosome separation during mitosis and meiosis, and cell motility. F1 ATP synthase uses mechanical torques as the energy source for ATP synthesis (Yoshida et al. 2001). Mechanosensory proteins convert the stimulus from mechanical forces into an electrical or biochemical signal (Hamill and Martinac 2001). During chaperonin-assisted protein folding, mechanical forces are involved in unfolding the misfolded protein (Brinker et al. 2001). Many proteins with functions to withstand mechanical stresses have modular architectures, i.e., they are composed of tandem-linked domains that are individually folded. The force-induced unfolding/refolding and deformation of these domains regulate the tensions in the protein molecules as required by the cellular processes, such as titin during muscle stretch (Granzier and Labeit 2004), fibronectin in the assembly and cell attachment of the extracellular matrix (Erickson 2002; Oberhauser et al. 2002), and spectrin in the deformation of erythrocytes (Vera et al. 2005).
One common feature in the aforementioned biological processes is that they involve protein molecules in deformed or unfolded conformations. Such conformations present expanded excluded volumes relative to those in the folded state, and hence are particularly open to the effects of macromolecular crowding when the processes occur in environments crowded with other macromolecules. Our experimental results demonstrate that the mechanical stability of these proteins can be enhanced by macromolecular crowding. Since the number and species of macromolecules present in various locations and during distinct stages of cell development are different, it is likely that varying degrees enhancement in mechanical stability by macromolecular crowding can serve as a means to fine-tune protein functions and regulate the biomolecular processes.
Comparison between theory and experiment
The observed increase in the unfolding force for ubiquitin by the presence of dextran can be explained by a crowding-induced increase in the unfolding free-energy barrier as calculated from the scaled-particle theory. This theory has recently been used to account for the macromolecular crowding effects on the polymerization of sickle hemoglobin (Liu et al. 2008). Though the theoretical calculations involve several approximations and simplifications, the model probably captures the essential physics of the enhancement of the protein's mechanical stability due to macromolecular crowding. Refinement of the theoretical model will be possible when more experimental data are obtained at different temperatures and using crowding agents of different sizes and species.
The zero-force unfolding rate k u0 was obtained from Monte Carlo simulations based on the Bell model (Bell 1978) for a two-state protein (see Equation 1). This is the most widely used approach to analyze data from mechanical stretching experiments due to its simplicity and its applicability to various biopolymers (Rief et al. 1998). Schlierf and Rief used the mechanical unfolding data of the protein ddFLN4 to demonstrate that Kramer's diffusion model could fit the measured unfolding force data better than the Bell model for proteins with broad free-energy barriers (Schlierf and Rief 2006). Since ubiquitin has a relatively narrow unfolding transition state, as indicated by the value of Δx u, the Bell model is still valid for the description of the system. Zinober et al. (2002) showed that the distribution and magnitude of the unfolding forces of the protein domain titin I27 were influenced by many other variables in addition to the intrinsic properties of the proteins, such as the unfolding event number, the length of the tethered protein polymer, and the spring constant of the cantilever. Some of these variables, such as the cantilever spring constant and the varying force loading rate during pulling, can be integrated into the Monte Carlo simulations, while others, such as the length of the tethered polymer, vary for each pulling action and cannot be reproduced reliably in the simulations. Williams et al. (2003), using mutant forms of titin I27, showed that the transition state structure and energy of the protein depended on the force loading rate; therefore, the protein's unfolding characteristics at low loading rates could not be obtained from direct extrapolation based on the data obtained at high loading rates. These subtle factors complicate the data interpretation and the comparison of data from different laboratories. In this paper, the Bell model is used because the primary conclusion of the work, i.e., macromolecular crowding enhances the mechanical stability of protein molecules, does not depend on the specific models for data interpretation. In addition, the calculated ratio k u0(0)/k u0(c) presented in Figure 5 does not change significantly when different models are used.
Dextran as a crowding agent
The ideal crowding agents should provide environments that mimic the interior conditions of a cell. One approach would be using cell extracts as mimetics of macromolecular crowding. However, the heterogeneity in the chemical, geometrical, and physical properties of cell extracts makes it difficult to collect the relevant experimental data and to use theoretical models for interpretation of the results. Therefore, purified macromolecules have been used as crowding agents in most experimental studies of the crowding effects, and dextran is one of the most commonly used macromolecules since it is inert, highly soluble in water, and available in various sizes and large quantities. The persistence length of dextran, determined by single-molecule measurements, was found to be 0.4 nm (at low force regime) (Rief et al. 1998), indicating that dextran is a very flexible polymer which should exist as a random coil with a spherical shape in aqueous solutions, especially when present in high concentrations. In previous studies of macromolecular crowding on protein folding stability and kinetics, crowding agents have included dextran (Sasahara et al. 2003), Ficoll (Tokuriki et al. 2004; Spencer et al. 2005), polyethylene glycol (Tokuriki et al. 2004; Ai et al. 2006), and bovine serum albumin (Qu and Bolen 2002; Zhou et al. 2004).
Effects of viscous drag on the AFM cantilever
A solution with a high concentration of dextran molecules becomes very viscous. The viscosity increases exponentially with the concentration of dextran molecules. For dextran with a molecular weight of 40 kDa, the viscosity of the solution increases from 1 cP to ∼25 cP at room temperature when dextran concentration is increased from 0 to 300 g/L (http://www.dextran.nu/dex1.html). The measured forces are underestimated due to the viscous drag on the AFM cantilever, and this underestimation is more severe at higher dextran concentrations and higher pulling speeds. Oden et al. (1996) investigated the effects of viscous drag on high frequency vibrations of cantilevers, by modeling the system as a damped one-dimensional harmonic oscillator. Alcaraz et al. (2002) developed a scaled spherical model to assess the effect of the hydrodynamic drag on AFM cantilevers for microrheological measurements in liquid at low Reynolds numbers. Janovjak et al. (2005) used this model to measure the viscous drag on the unfolding forces of a multidomain protein in fast pulling experiments with pulling speeds in the range of 5–140 μm/sec. These authors showed that the unfolding forces could be underestimated by >20% due to viscous drag at a pulling speed of 30 μm/sec.
In contrast to the experiments discussed above, our measurements were carried out at relatively low pulling speeds of 0.05–2 μm/sec, and the unfolding force underestimation due to viscous drag on the cantilever was found not to be significant. In the experiments, the pulling motion is realized by moving the sample away from the stationary cantilever, and the cantilever does not experience a viscous drag when it is not bending. As shown in Figure 2, after the tethered protein polymer becomes detached (or when no polymer is tethered), the cantilever signal remains constant although the sample surface continues to move away from the cantilever. The cantilever signal recorded on approaching the surface is also constant over this distance range, indicating the absence of a force on the cantilever. When the cantilever is bending, the drag force can be estimated from the Stokes law. The cantilever used in the experiments is triangular in shape, with a length of 180 μm and a width of 18 μm for each arm. When the cantilever is pulled, it bends in a nonlinear fashion (Sader 1995) with the tip-bearing end moving to a maximum distance of ∼3 nm (the cantilevers used in the experiments have spring constants ∼80 pN/nm). The tip moves at a slower speed than that of the sample since the tip is linked to the sample through the flexible protein chain. From the data shown in Figure 2, it can be estimated that, when the sample is pulled at 2 μm/sec (the highest speed used), the maximum speed of the tip will be ∼0.8 μm/sec, which occurs near the top of the peaks in a force curve. To estimate the viscous drag on the cantilever when the tip is pulled, we consider a circular disc (due to the lack of viscous drag equation for rectangular and more complicated shapes) of radius 18 μm at the end of the cantilever moving at a speed of 0.8 μm/sec, since the rest of the cantilever moves much less than the end. The viscous drag on the disc is F v = 16ηRv, where η is the viscosity of the solution, R is the radius of the disc, and v is the speed. Using the values cited above, F v can be estimated at 5.7 pN. This is the maximum viscous force at the highest dextran concentration (300 g/L) and fastest pulling speed (2 μm/sec), and it acts on the cantilever for only a brief period of time when the bending approaches the maximum value, i.e., near the top of the force peaks. Under other conditions, the viscous drag is much reduced. In addition, the shape of the peaks in the force curves resembles those reported by Janovjak et al. (2005) after correction for viscous drag, and the rising parts of the peaks can be fitted well by the WLC model (Fig. 2), indicating that the viscous drag effect was not significant. If the viscous drag effect were taken into account, the curves in Figure 4A would shift up slightly at the high dextran concentrations.
Conclusion
In this work, a single-molecule method has been applied, for the first time, to the investigation of macromolecular crowding effects on the mechanical unfolding of protein molecules. This approach has the benefits of avoiding using denaturants, extreme pH, or high temperatures to induce folding/unfolding transitions. A simple model based on the scaled-particle theory was used to provide physical insights into the effects of macromolecular crowding on the unfolding kinetics. Our observation that the mechanical stability of ubiquitin is enhanced by macromolecular crowding suggests that proteins can withstand higher mechanical stress in vivo than that measured in dilute solutions. In addition to proteins with specific mechanical functions, all proteins experience mechanical stress one way or the other inside the cell; therefore, the reported results can help in understanding the mechanisms of the macromolecular crowding effects on the stability and function of proteins. Our findings may also help to improve the performance of engineered proteins intended for withstanding high mechanical stresses by tuning environmental conditions.
Materials and Methods
Protein samples
To facilitate the measurements using AFM, the ubiquitin molecules were made into polymers via protein engineering. The details of the polymer synthesis were reported previously (Chyan et al. 2004). Briefly, the clone containing the ubiquitin (UBI) gene was first amplified with PCR, and the PCR product was purified and ligated into a vector. After verification, this construct was used to create dimeric, tetrameric, and octameric UBI genes via iterative cloning. The doubly digested octameric UBI gene was subcloned into a pET30a expression vector and transformed into Escherichia coli strain BL21(DE3). The polymerized ubiquitin molecules (octamers) were then isolated and purified. The octamer ubiquitin thus synthesized contains AMADIGS residues on the N terminus, eight repeats of ubiquitin with RS residues in between each repeat, and two cysteine residues at the C terminus. CD measurements showed that polymerization did not change the structure and stability of the ubiquitin molecules significantly (Chyan et al. 2004).
Crowding agent
Dextran molecules, with an average molecular weight of 40 kDa (purchased from Sigma-Aldrich Inc.), were used as the crowding agent. The hydrodynamic radii of the dextran molecules have an average value of 3.5 nm (Weiss et al. 2004). In the experiments, dextran was dissolved in PBS (phosphate buffered saline, containing 126 mM NaCl, 7.2 mM Na2HPO4, 3 mM NaH2PO4, pH 7.0) buffer to a desired concentration, and the solution was then injected into the AFM liquid chamber. Electric conductivity, pH, and dynamic light scattering of the buffer solutions containing various concentrations of dextran molecules were checked to ensure that the dextran powder did not contain significant amount of small molecules.
Circular dichroism studies
CD spectra were recorded on a Jasco J-715 CD spectrometer. CD spectra were collected using a cylindrical quartz cuvette with a 1-mm path length. The step resolution was 0.2 nm with 1.0 nm bandwidth at a scan speed of 50 nm/min. Each CD spectrum was averaged over 16 measurements and corrected for the appropriate buffer baseline. All spectra are presented as the molar CD absorption coefficient (Δε M). Concentrations of ubiquitin were determined by UV absorption using the extinction coefficient at 276 nm, 1450 M−1 cm−1 (on the basis of ubiquitin monomer). Sample concentrations ranged from 4 to 20 μM. The contents of secondary structures were calculated using the software package CDPro.
Mechanical unfolding measurements
The mechanical unfolding experiments were performed using a modified commercial Nanoscope IIIa scanning probe microscope (Digital Instruments/Veeco). The cantilevers used in the experiments were triangular Si3N4 cantilevers with a nominal spring constant of 50 pN/nm. The value of the spring constant of each cantilever was calibrated individually using the method of thermal energy equipartition (Hutter and Bechhofer 1993). The polymerized ubiquitin molecules were dissolved in PBS buffer to a protein concentration of 50 μg/mL. The specimen for the mechanical unfolding experiments was prepared by depositing 20 μL of the protein solution onto a fresh gold surface and allowing the molecules to adsorb for 10 min. After washing off the unbound molecules with PBS, the sample was placed in the liquid chamber of the AFM. The experiments were carried out with both the sample and the tip immerged in the buffer. While protein molecules remained on the surface, dextran solutions of specified concentrations were injected into the liquid chamber as desired. To minimize errors caused by other factors, each sample was studied over the entire range of concentrations from 0 to 300 g/L.
Data analysis
The raw data was first screened for curves showing multiple unfolding events with the characteristic sawtooth pattern. To evaluate the structural changes upon the unfolding of a ubiquitin molecule in the polymer, the force versus extension relationship was fitted to the WLC model (Bustamante et al. 1994):
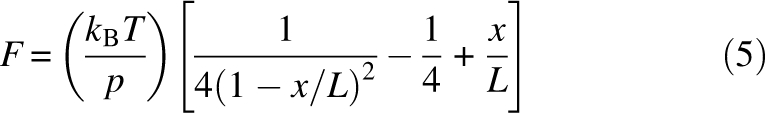 |
where L is the contour length, x is the end-to-end distance of the chain, and p is the persistence length.
Monte Carlo simulations
Monte Carlo simulations were performed to determine the unfolding rate of the protein. In the simulations, force versus extension curves were generated by assuming the polymer to be a WLC chain, and the cantilever to be a linear spring. To determine if a folded protein molecule will unfold at a particular force, a probability is calculated according to the theory developed by Bell (1978) and elaborated by Evans and Ritchie (1999) for two-state unfolding:
where k u is the unfolding rate constant when an external force F is applied to the protein, k u0 is the unfolding rate constant in the absence of external force, Δt is the time interval over which the force F is acting on the protein, Δx u is the distance between the native state and the transition state along the pulling direction, and k B T is the thermal energy. At each force level, every folded ubiquitin monomer in the polymer is polled for unfolding by comparing the unfolding probability with a randomly generated number, before the chain is pulled further. In the simulations, 50 force curves were generated for each set of parameters on pulling octameric ubiquitin chains, thus 400 points were acquired for each data set. The values of k u0 and Δx u for the protein were obtained as adjustable parameters that best fitted the simulation results to the experimental data. The fitting was performed for the distributions of the unfolding forces, and for the dependence of the unfolding forces on the pulling speed.
Acknowledgments
We thank Len Finegold for critical comments, Meihong Su and Yao Yang for technical assistance, and Zenghui Liu for helpful discussions. This work was supported by National Institutes of Health Grants R01-GM071793 (to G.Y.) and R01-GM058187 (to H.-X.Z.).
Footnotes
Reprint requests to: Guoliang Yang, Department of Physics, Drexel University, 3141 Chestnut Street, Philadelphia, PA 19104, USA; e-mail: gyang@drexel.edu; fax: (215) 895-5934.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.037325.108.
References
- Ai X., Zhou Z., Bai Y., Choy W.Y. 15N NMR spin relaxation dispersion study of the molecular crowding effects on protein folding under native conditions. J. Am. Chem. Soc. 2006;128:3916–3917. doi: 10.1021/ja057832n. [DOI] [PubMed] [Google Scholar]
- Alcaraz J., Buscemi L., Puig-de-Morales M., Colchero J., Baro A., Navajas D. Correction of microrheological measurements of soft samples with atomic force microscopy for the hydrodynamic drag on the cantilever. Langmuir. 2002;18:716–721. [Google Scholar]
- Ansari A., Jones C.M., Henry E.R., Hofrichter J., Eaton W.A. The role of solvent viscosity in the dynamics of protein conformational changes. Science. 1992;256:1796–1798. doi: 10.1126/science.1615323. [DOI] [PubMed] [Google Scholar]
- Bell G.I. Models of the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- Berg O.G. The influence of macromolecular crowding on thermodynamic activity: Solubility and dimerization constants for spherical and dumbbell-shaped molecules in a hard-sphere mixture. Biopolymers. 1990;30:1027–1037. doi: 10.1002/bip.360301104. [DOI] [PubMed] [Google Scholar]
- Brinker A., Pfeifer G., Kerner M.J., Naylor D.J., Hartl F.U., Hayer-Hartl M. Dual function of protein confinement in chaperonin-assisted protein folding. Cell. 2001;107:223–233. doi: 10.1016/s0092-8674(01)00517-7. [DOI] [PubMed] [Google Scholar]
- Bustamante C., Marko J.F., Siggia E.D., Smith S. Entropic elasticity of λ-phage DNA. Science. 1994;265:1599–1601. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- Bustamante C., Chemla Y.R., Forde N.R., Izhaky D. Mechanical processes in biochemistry. Annu. Rev. Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- Carrion-Vazquez M., Oberhauser A.F., Fowler S.B., Marszalek P.E., Broedel S.E., Clarke J., Fernandez J.M. Mechanical and chemical unfolding of a single protein: A comparison. Proc. Natl. Acad. Sci. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrion-Vazquez M., Li H., Lu H., Marszalek P.E., Oberhauser A.F., Fernandez J.M. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Biol. 2003;10:738–743. doi: 10.1038/nsb965. [DOI] [PubMed] [Google Scholar]
- Cheung M.S., Thirumalai D. Effects of crowding and confinement on the structures of the transition state ensemble in proteins. J. Phys. Chem. B. 2007;111:8250–8257. doi: 10.1021/jp068201y. [DOI] [PubMed] [Google Scholar]
- Cheung M.S., Klimov D., Thirumalai D. Molecular crowding enhances native state stability and refolding rates of globular proteins. Proc. Natl. Acad. Sci. 2005;102:4753–4758. doi: 10.1073/pnas.0409630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyan C.-L., Lin F.-C., Peng H., Yuan J.-M., Chang C.-H., Lin S.-H., Yang G. Reversible mechanical unfolding of single ubiquitin molecules. Biophys. J. 2004;87:3995–4006. doi: 10.1529/biophysj.104.042754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedmon M.M., Patel C.N., Young G.B., Pielak G.J. FlgM gains structure in living cells. Proc. Natl. Acad. Sci. 2002;99:12681–12684. doi: 10.1073/pnas.202331299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R.J. Macromolecular crowding: Obvious but underappreciated. Trends Biochem. Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- Erickson H.P. Stretching fibronectin. J. Muscle Res. Cell Motil. 2002;23:575–580. doi: 10.1023/a:1023427026818. [DOI] [PubMed] [Google Scholar]
- Evans E., Ritchie K. Strength of a weak bond connecting flexible polymer chains. Biophys. J. 1999;76:2439–2447. doi: 10.1016/S0006-3495(99)77399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J.M., Li H. Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science. 2004;303:1674–1678. doi: 10.1126/science.1092497. [DOI] [PubMed] [Google Scholar]
- Flaugh S.L., Lumb K.J. Effects of macromolecular crowding on the intrinsically disordered proteins c-Fos and p27Kip1. Biomacromolecules. 2001;2:538–540. doi: 10.1021/bm015502z. [DOI] [PubMed] [Google Scholar]
- Forman J.R., Clarke J. Mechanical unfolding of proteins: Insights into biology, structure and folding. Curr. Opin. Struct. Biol. 2007;17:58–66. doi: 10.1016/j.sbi.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Gao M., Sotomayor M., Villa E., Lee E.H., Schulten K. Molecular mechanisms of cellular mechanics. Phys. Chem. Chem. Phys. 2006;8:3692–3706. doi: 10.1039/b606019f. [DOI] [PubMed] [Google Scholar]
- Granzier H.L., Labeit S. The giant protein titin: A major player in myocardial mechanics, signaling, and disease. Circ. Res. 2004;94:284–295. doi: 10.1161/01.RES.0000117769.88862.F8. [DOI] [PubMed] [Google Scholar]
- Hamill O.P., Martinac B. Molecular basis of mechanotransduction in living cells. Physiol. Rev. 2001;81:685–740. doi: 10.1152/physrev.2001.81.2.685. [DOI] [PubMed] [Google Scholar]
- Hayer-Hartl M., Minton A.P. A simple semiempirical model for the effect of molecular confinement upon the rate of protein folding. Biochemistry. 2006;45:13356–13360. doi: 10.1021/bi061597j. [DOI] [PubMed] [Google Scholar]
- Hutter J.L., Bechhofer J. Calibration of atomic force microscope tips. Rev. Sci. Instrum. 1993;64:1868–1873. [Google Scholar]
- Hyeon C., Thirumalai D. Can energy landscape roughness of proteins and RNA be measured by using mechanical unfolding experiments? Proc. Natl. Acad. Sci. 2003;100:10249–10253. doi: 10.1073/pnas.1833310100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioan C.E., Aberle T., Burchard W. Structure properties of dextran. 2. Dilute solution. Macromolecules. 2000;33:5730–5739. [Google Scholar]
- Janovjak H., Struckmeier J., Muller D.J. Hydrodynamic effects in fast AFM single-molecule force measurements. Eur. Biophys. J. 2005;34:91–96. doi: 10.1007/s00249-004-0430-3. [DOI] [PubMed] [Google Scholar]
- Jas G.S., Eaton W.A., Hofrichter J. Effect of viscosity on the kinetic of α-helix and β-hairpin formation. J. Phys. Chem. B. 2001;105:261–272. [Google Scholar]
- Kleiner A., Shakhnovich E. The mechanical unfolding of ubiquitin through all-atom Monte Carlo simulation with a go-type potential. Biophys. J. 2007;92:2054–2061. doi: 10.1529/biophysj.106.081257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebowitz J.L., Helfand E., Praestgaard E. Scaled particle theory of fluid mixtures. J. Chem. Phys. 1965;43:774–779. [Google Scholar]
- Li H., Oberhauser A.F., Fowler S.B., Clarke J., Fernandez J.M. Atomic force microscopy reveals the mechanical design of a modular protein. Proc. Natl. Acad. Sci. 2000;97:6527–6531. doi: 10.1073/pnas.120048697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Weng W., Bookchin R.M., Lew V.L., Ferrone F.A. Free energy of sickle hemoglobin polymerization: A scaled-particle treatment for use with dextran as a crowding agent. Biophys. J. 2008;94:3629–3634. doi: 10.1529/biophysj.107.117465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. Requirement for GroEL/GroES-dependent protein folding under nonpermissive conditions of macromolecular crowding. Biochemistry. 2002;41:5050–5055. doi: 10.1021/bi015925l. [DOI] [PubMed] [Google Scholar]
- Medalia O., Weber I., Frangakis A.S., Nicastro D., Gerisch G., Baumeister W. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298:1209–1213. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- Minton A.P. Models for excluded volume interaction between an unfolded protein and rigid macromolecular co-solutes: Macromolecular crowding and protein stability revisited. Biophys. J. 2005;88:971–985. doi: 10.1529/biophysj.104.050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton A.P. How can biochemical reactions within cells differ from those in test tubes? J. Cell Sci. 2006;119:2863–2869. doi: 10.1242/jcs.03063. [DOI] [PubMed] [Google Scholar]
- Minton K.W., Karmin P., Hahn G.M., Minton A.P. Nonspecific stabilization of stress-susceptible proteins by stress-resistant proteins: A model for the biological role of heat shock proteins. Proc. Natl. Acad. Sci. 1982;79:7107–7111. doi: 10.1073/pnas.79.23.7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhauser A.F., Badilla-Fernandez C., Carrion-Vazquez M., Fernandez J.M. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J. Mol. Biol. 2002;319:433–447. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]
- Oden P.I., Chen G.Y., Steele R.A., Warmack R.J., Thundat T. Viscous drag measurements utilizing microfabricated cantilevers. Appl. Phys. Lett. 1996;68:3814–3816. [Google Scholar]
- Ogston A.G., Preston B.N. The molecular compression of dextran. Biochem. J. 1979;183:1–9. doi: 10.1042/bj1830001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping G., Yang G., Yuan J.-M. Depletion force from macromolecular crowding enhances mechanical stability of protein molecules. Polymer (Guildf.) 2006;47:2564–2570. [Google Scholar]
- Qu Y., Bolen W. Efficacy of macromolecular crowding in forcing proteins to fold. Biophys. Chem. 2002;101–102:155–165. doi: 10.1016/s0301-4622(02)00148-5. [DOI] [PubMed] [Google Scholar]
- Rief M., Gautel M., Oesterheld F., Fernandez J., Gaub H. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- Rief M., Fernandez J.M., Gaub H.E. Elastically coupled two-level systems as a model for biopolymer extensibility. Phys. Rev. Lett. 1998;81:4764–4767. [Google Scholar]
- Sader J.E. Parallel beam approximation for V-shaped atomic force microscope cantilevers. Rev. Sci. Instrum. 1995;66:4583–4587. [Google Scholar]
- Sasahara K., McPhie P., Minton A.P. Effect of dextran on protein stability and conformation attributed to macromolecular crowding. J. Mol. Biol. 2003;326:1227–1237. doi: 10.1016/s0022-2836(02)01443-2. [DOI] [PubMed] [Google Scholar]
- Schlierf M., Rief M. Single-molecule unfolding force distributions reveal a funnel-shaped energy landscape. Biophys. J. 2006;90:L33–L35. doi: 10.1529/biophysj.105.077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer D.S., Xu K., Logan T.M., Zhou H.-X. Effects of pH, salt, and macromolecular crowding on the stability of FK506-binding protein: An integrated experimental and theoretical study. J. Mol. Biol. 2005;351:219–232. doi: 10.1016/j.jmb.2005.05.029. [DOI] [PubMed] [Google Scholar]
- Tokuriki N., Kinjo M., Negi S., Hoshino M., Goto Y., Urabe I., Yomo T. Protein folding by the effects of macromolecular crowding. Protein Sci. 2004;13:125–133. doi: 10.1110/ps.03288104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg B., Ellis R.J., Dobson C.M. Effects of macromolecular crowding on protein folding and aggregation. EMBO J. 1999;18:6927–6933. doi: 10.1093/emboj/18.24.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg B., Wain R., Dobson C.M., Ellis R.J. Macromolecular crowding perturbs protein refolding kinetics: Implications for folding inside the cell. EMBO J. 2000;19:3870–3875. doi: 10.1093/emboj/19.15.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera C., Skelton R., Bossens F., Sung L.A. 3-D nanomechanics of an erythrocyte junctional complex in equibiaxial and anisotropic deformations. Ann. Biomed. Eng. 2005;33:1387–1404. doi: 10.1007/s10439-005-4698-y. [DOI] [PubMed] [Google Scholar]
- Weiss M., Elsner M., Kartberg F., Nilssony T. Anomalous subdiffusion is a measure for cytoplasmic crowding in living cells. Biophys. J. 2004;87:3518–3524. doi: 10.1529/biophysj.104.044263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Went H.M., Jackson S.E. Ubiquitin folds through a highly polarized transition state. Protein Eng. Des. Sel. 2005;18:229–237. doi: 10.1093/protein/gzi025. [DOI] [PubMed] [Google Scholar]
- Williams P.M., Fowler S.B., Best R.B., Toca-Herrera J.L., Scott K.A., Steward A., Clarke J. Hidden complexity in the mechanical properties of titin. Nature. 2003;422:446–449. doi: 10.1038/nature01517. [DOI] [PubMed] [Google Scholar]
- Yang G., Cecconi C., Baase W.A., Vetter I.R., Breyer W.A., Haack J.A., Matthews B.W., Dahlquist F.W., Bustamante C. Solid-state synthesis and mechanical unfolding of polymers of T4 lysozyme. Proc. Natl. Acad. Sci. 2000;97:139–144. doi: 10.1073/pnas.97.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Muneyuki E., Hisabori T. ATP synthase—a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2001;2:669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- Zhou H.-X. Protein folding and binding in confined spaces and in crowded solutions. J. Mol. Recognit. 2004;17:368–375. doi: 10.1002/jmr.711. [DOI] [PubMed] [Google Scholar]
- Zhou H.-X. Protein folding in confined and crowded environments. Arch. Biochem. Biophys. 2008;469:76–82. doi: 10.1016/j.abb.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.-R., Liang Y., Du F., Zhou Z., Chen J. Mixed macromolecular crowding accelerates the oxidative refolding of reduced, denatured lysozyme. J. Biol. Chem. 2004;279:55109–55116. doi: 10.1074/jbc.M409086200. [DOI] [PubMed] [Google Scholar]
- Zhou H.-X., Rivas G., Minton A.P. Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinober R.C., Brockwell D.J., Beddard G.S., Blake A.W., Olmsted P.D., Radford S.E., Smith D.A. Mechanically unfolding proteins: The effect of unfolding history and the supramolecular scaffold. Protein Sci. 2002;11:2759–2765. doi: 10.1110/ps.0224602. [DOI] [PMC free article] [PubMed] [Google Scholar]