Abstract
Most synthetic inhibitors of peptidases have been targeted to the active site for inhibiting catalysis through reversible competition with the substrate or by covalent modification of catalytic groups. Cathepsin B is unique among the cysteine peptidase for the presence of a flexible segment, known as the occluding loop, which can block the primed subsites of the substrate binding cleft. With the occluding loop in the open conformation cathepsin B acts as an endopeptidase, and it acts as an exopeptidase when the loop is closed. We have targeted the occluding loop of human cathepsin B at its surface, outside the catalytic center, using a high-throughput docking procedure. The aim was to identify inhibitors that would interact with the occluding loop thereby modulating enzyme activity without the help of chemical warheads against catalytic residues. From a large library of compounds, the in silico approach identified [2-[2-(2,4-dioxo-1,3-thiazolidin-3-yl)ethylamino]-2-oxoethyl] 2-(furan-2-carbonylamino) acetate, which fulfills the working hypothesis. This molecule possesses two distinct binding moieties and behaves as a reversible, double-headed competitive inhibitor of cathepsin B by excluding synthetic and protein substrates from the active center. The kinetic mechanism of inhibition suggests that the occluding loop is stabilized in its closed conformation, mainly by hydrogen bonds with the inhibitor, thus decreasing endoproteolytic activity of the enzyme. Furthermore, the dioxothiazolidine head of the compound sterically hinders binding of the C-terminal residue of substrates resulting in inhibition of the exopeptidase activity of cathepsin B in a physiopathologically relevant pH range.
Keywords: cysteine peptidases, inhibition, enzyme kinetics, occluding loop, docking, endopeptidase, exopeptidase
Cathepsin B, a cysteine peptidase of the papain family (EC 3.4.22.1, identifier C01.060 in the Merops database) (Rawlings et al. 2004), has been classically ranked among the lysosomal enzymes and implicated in intracellular protein digestion. Physiologically, cathepsin B is also involved in antigen processing (Matsunaga et al. 1993), in the activation of thyroglobulin, the precursor of thyroid hormones (Friedrichs et al. 2003), and in the maturation of beta-galactosidase (Okamura-Oho et al. 1997). From a pathological point of view, cathepsin B activates trypsinogen in hereditary pancreatitis (Kukor et al. 2002) and participates in apoptosis (Bröker et al. 2005), tumor progression and malignancy (Yan and Sloane 2003; Mohamed and Sloane 2006), and rheumatic diseases (Lenarcic et al. 1988; Baici et al. 1995a,b). Particular extralysosomal functions of cathepsin B are due to altered expression at the gene level and/or atypical trafficking (Müntener et al. 2003, 2004; Zwicky et al. 2003; Baici et al. 2006).
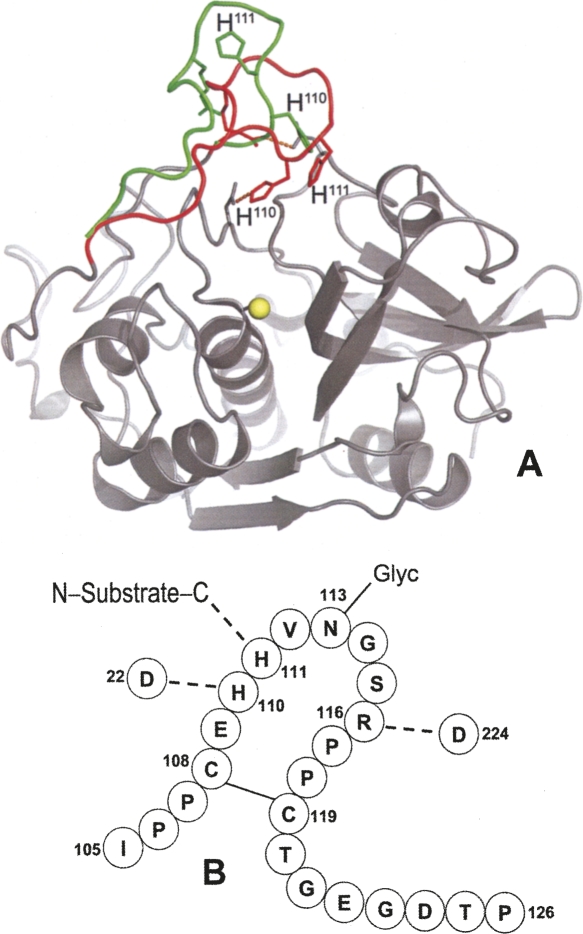
Cathepsin B is capable of endopeptidase (Mort 2004), peptidyl-dipeptidase (Aronson Jr. and Barrett 1978; Bond and Barrett 1980), and carboxypeptidase activities (Takahashi et al. 1986; Rowan et al. 1993). Among the cysteine peptidases, it owns a unique structural element, called an occluding loop, which comprises residues Ile105–Pro126 (Fig. 1; Musil et al. 1991). At low pH, two salt bridges, His110–Asp22 and Arg116–Asp224, hold the loop in a closed position over the primed subsites of the substrate binding cleft, thus preventing extended binding of polypeptides and endoproteolytic activity. The closed conformation, through the engagement of His111 in a hydrogen bond with the C-terminal carboxylate of the substrate and the loose specificity in P2′, is responsible for the peptidyl-dipeptidase activity of cathepsin B (Illy et al. 1997; Quraishi et al. 1999; Krupa et al. 2002). As mutations of the amino acids His110 and Asp22 have shown, removal of the salt bridges induces endopeptidase activity, attributed to increased flexibility of the loop (Nägler et al. 1997). This concept agrees with higher endopeptidase activity and with the competition between the occluding loop and the propeptide following deprotonation of His110 changing the pH from 4.0 to 6.0 (Quraishi et al. 1999). The flexibility of the loop was further demonstrated by the fact that cystatins A and C were able to displace the loop (Nycander et al. 1998; Pavlova et al. 2000), and deletion of residues 108–119 abolished the exopeptidase activity of cathepsin B (Illy et al. 1997).
Figure 1.
The occluding loop of human cathepsin B. (A) Closed conformation (red) in the mature enzyme (Musil et al. 1991), where a hydrogen bond is made between His110 and Asp22. The conformation seen in procathepsin B with the occluding loop lifted (green) to accommodate the propeptide, which is not shown for clarity (Turk et al. 1996). Image generated with PyMOL software (http://www.pymol.org). (B) Scheme of the occluding loop with hydrogen bonds present in the closed conformation. The symbol Glyc attached to Asn113 indicates the N-linked oligosaccharide.
A large number of synthetic compounds behave as inhibitors of cathepsin B. Most of them are either reversible or irreversible competitive inhibitors acting “inside the active center” (Otto and Schirmeister 1997; Michaud and Gour 1998; Frlan and Gobec 2006). The practical use of these inhibitors is in most cases difficult for reasons analyzed elsewhere (Baici 1998). In the present study we target cathepsin B “from the outside,” i.e., at the surface of the molecule excluding any direct interference with catalysis. Our strategy aims at proving the concept that latching the occluding loop in its closed conformation may possibly hinder the endo- and/or exopeptidase activities of the enzyme. The approach consists of computational analysis by docking a large number of compounds to the surface of cathepsin B in the occluding loop region. From 40 compounds matching the working hypothesis, 29 are commercially available and one of them inhibits at low micromolar concentration the endo- and exoproteolytic activities of cathepsin B. We analyze the kinetic mechanism of inhibition, from which we propose a model for the interaction between the compound and the enzyme.
Results
Kinetic mechanism of inhibition with synthetic substrates
Z-RR↓AMC and Abz-GIVR↓AK(Dnp)-OH were used as substrates for monitoring the endo- and exoproteolytic activity of cathepsin B, respectively. Arrows in the substrate acronyms indicate the scissile bonds. Forty of 47,878 compounds screened in the docking approach were selected as candidate binders of human cathepsin B. Twenty-nine of them were commercially available and were tested as modifiers of enzyme activity, and one of them, DOFA (Fig. 2), behaved as a true inhibitor. Two compounds out of 29 were scarcely soluble in DMSO (Zinc-2005 codes 1271923 and 2990182; Supplemental Table 1). Five of them were soluble in DMSO but were prone to aggregation under the assay conditions for cathepsin B giving rise to modest and inconsistent inhibition between different assays. We considered these five compounds (958753, 1837733, 1871954, 3247330, and 1581881) as promiscuous inhibitors (Shoichet 2006). None of the compounds tested was a covalent inactivator of cathepsin B. DOFA did not inhibit human cathepsin L, and papain was only partially inhibited at millimolar concentrations (assays with Z-FR↓AMC; data not shown). A previous report of high-throughput screening classified DOFA inactive against cathepsin B (http://pubchem.ncbi.nlm.nih.gov/, CID 2078629, AID 453). Reasons can be sought in either too strict criteria or in the occasional failure of high-throughput screening methods in detecting hits (Buxser and Vroegop 2005). DOFA behaved as a reversible inhibitor of cathepsin B and manifested neither tight-binding nor slow-binding inhibition behavior. Reaction traces were linear from the very beginning of the reaction, i.e., from 3–4 ms onward, as measured with a stopped-flow apparatus. Therefore, initial velocities at variable substrate and modifier concentrations were treated as steady-state rates. In control experiments we checked the possibility that any inhibitory effect of DOFA was not due to aggregation of the compound. For this purpose 0.01% Triton X-100 was added to buffers and the DTT concentration was increased to 5 mM. In comparison with buffer not containing detergent and with DTT = 2 mM, the basal activity of cathepsin B was higher by ∼40%. However, inhibition profiles for increasing DOFA concentration, percentages of inhibition, and inhibition constants were the same.
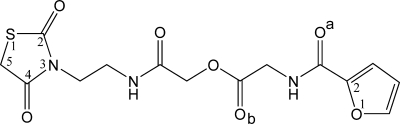
Figure 2.
Structure of the inhibitor. The IUPAC name of the compound is [2-[2-(2,4-dioxo-1,3-thiazolidin-3-yl)ethylamino]-2-oxoethyl] 2-(furan-2-carbonylamino)acetate (abbreviated DOFA; Zinc-2005 code: 2616818). Numbering and lettering used as aid for the description in the main text.
The kinetic results in this study refer to those obtained with the enzyme purified from human liver. Nevertheless, we confirmed inhibition mechanisms and kinetic parameters also with recombinant cathepsin B. The reason was to ascertain any influence of glycosylation at N113, which is located at the tip of the occluding loop, on the binding of potential inhibitors. Both kinetic and modeling results discussed below suggest that N113 glycosylation possibly occurring in the wild-type enzyme does not affect inhibition of cathepsin B by DOFA.
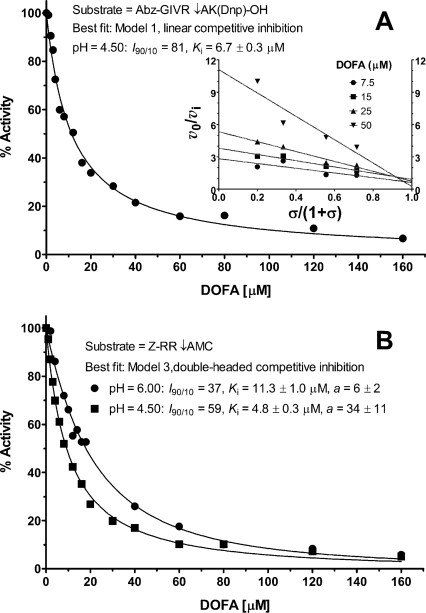
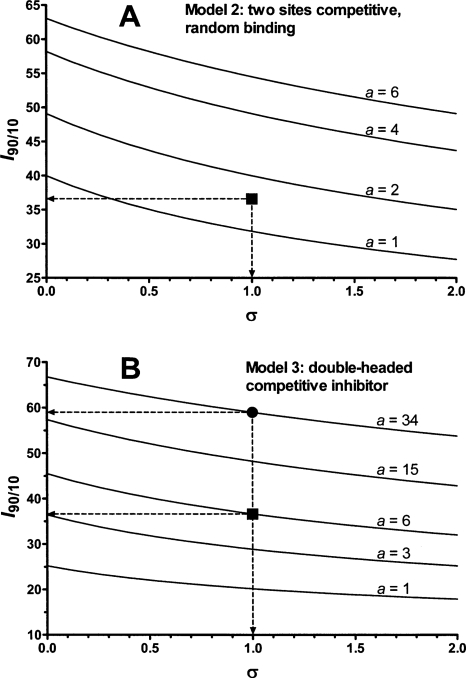
The kinetic mechanism was analyzed by a combination of graphical and regression methods to discriminate between models of enzyme–modifier interaction (Fig. 3). With the Abz-GIVR↓AK(Dnp)-OH exoproteolytic substrate at pH 4.5, the best fitting model for cathepsin B inhibition by DOFA was linear competitive inhibition with I 90/10 = 81 and K i = 6.7 μM (Fig. 4A). The specific velocity plot, typical for this mechanism, is shown in the inset of Figure 4A. With this substrate, data at pH 6.0 could not be obtained because of its scarce solubility. DOFA inhibited the endoproteolytic cathepsin B activity (Z-RR↓AMC as substrate) at pH 4.5 and 6.0. The specific velocity plot (not shown) established the linear nature of the inhibition process, i.e., enzyme activity was driven to zero at saturating inhibitor concentration, and diagnosed competitive-type inhibition. However, the specific velocity plot would not be able to discriminate between Models 1–3 in Figure 3. The activity profiles obtained at a fixed substrate concentration and variable inhibitor concentrations revealed the I 90/10 ratio to be less than 81, which can be estimated by inspection to be around 60 at pH 4.5 and around 40 at pH 6.0 (Fig. 4B). This suggests that the inhibition mechanism was not of the classical competitive type, for which I 90/10 = 81 (Model 1 in Fig. 3). On the other hand, Models 2 and 3 in the same figure predict I 90/10 values less than 81. For discriminating between models with Z-RR↓AMC as substrate, we set its concentration equal to K m so that σ = [S]/K m = 1 in the activity profiles in Figure 4B (thereby it was imperative to carefully measure the substrate concentration and K m). In Models 2 and 3, when the factor a = 1, I 90/10 depends only on σ, whereas for a ≠ 1 the I 90/10 ratio depends both on a and σ (Fig. 5A,B). Model 2 was ruled out as redundant, and Model 3 was preferred on the basis of the following quantitative and logical observations. In Model 2, two inhibitor molecules should bind to different places at the surface of the cathepsin B molecule with almost the same affinity, i.e., with the a value between 1 and 2, as shown in Figure 5A. The double-headed competitive inhibitor (Model 3 in Fig. 3) was superior also on the basis of the following quantitative observations. For Model 3, the squared symbol with dashed arrows in Figure 5B indicates the best-fit value for a = 6 at pH 6.0, which corresponds to I 90/10 = 37 and K i = 11.3 μM (Fig. 4B). Similarly, at pH 4.5, Model 3, with a = 34 (the black circle in Fig. 5B), I 90/10 = 59, and K i = 4.8 μM, produced a better fit to experimental data (Fig. 4B) than Model 2.
Figure 3.
Kinetic models for the inhibition of cathepsin B. v i and v 0 represent reaction rates in the presence and in the absence of inhibitor, respectively. σ = [S]/K m; I 90/10 = ratio of the inhibitor concentrations, which give 90% and 10% inhibition. The double-headed inhibitor possesses two distinct binding moieties that sequentially bind the enzyme.
Figure 4.
Inhibition profiles of human cathepsin B by DOFA. Main conditions and best-fit kinetic parameters are shown. (A) Inhibition profile using the FRET substrate Abz-GIVR↓AK(Dnp)-OH (pH 4.5), [S] = K m = 9.7 μM; data were obtained fluorimetrically with λex/λem at 320/420 nm. The inset shows the specific velocity plot at four substrate and four inhibitor concentrations. (B) Inhibition profiles with Z-RR↓AMC as substrate; [S] = K m = 0.47 mM at pH 6.0 (black circles) and [S] = K m = 1.54 mM at pH 4.5 (black squares). Data at pH 6.0 were collected photometrically at 360 nm and those at pH 4.5 were obtained fluorimetrically with λex/λem at 383/455 nm. Fluorescence readings in A and B were corrected for the inner filter effect.
Figure 5.
Ratios of inhibitor concentrations that give 90% and 10% inhibition. For Models 2 and 3 in Figure 3 the I 90/10 ratio depends on a and σ = [S]/K m. Curves were generated with Equations 4 and 6 shown in Figure 3. (A) Curves for Model 2; (B) curves for Model 3. The black squares in panels A and B correspond to the best-fit value of the coefficient a, which coincides with the value obtained by inspection of the inhibition profiles at pH 6.0 in Figure 4B. The black circle in panel B shows the best-fit value of the coefficient a (34), close to the value obtained by inspection of the inhibition profiles at pH 4.5 in Figure 4B.
Inhibition of endo- and exoproteolysis using protein substrates
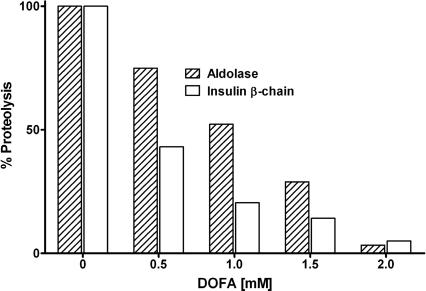
While low molecular mass oligopeptides are indispensable tools for determining inhibition mechanisms, they do not represent typical substrates for evaluating true endo- and exoproteolytic activities. Therefore, inhibition of cathepsin B endo- and exoproteolysis by DOFA was further investigated with two protein substrates: rabbit muscle aldolase and the oxidized β-chain of bovine insulin. Peptide-bond hydrolysis on both proteins at pH 6.0 was inhibited in a concentration-dependent manner by DOFA (Fig. 6) and was almost complete at saturating inhibitor concentration. As commonly observed when switching from oligopeptide to polypeptide substrates, the modifier concentrations needed for achieving inhibition were higher.
Figure 6.
Inhibition of cleavage of rabbit muscle fructose 1,6-bisphosphate aldolase and of the bovine insulin β-chain by cathepsin B. Concentration-dependent inhibition by DOFA at pH 6.0. Released peptides were quantified fluorimetrically by the fluorescamine method.
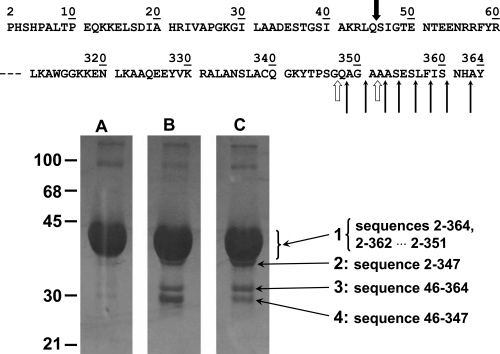
Cathepsin B has been previously reported to sequentially remove up to nine dipeptide units from the C terminus of aldolase with peptidyl-dipeptidase activity (Bond and Barrett 1980). Aldolase incubated with cathepsin B in the presence or absence of DOFA was subjected to SDS-PAGE under reducing conditions. The bands excised after electrophoretic separation were N-terminally sequenced and the masses of the polypeptides determined by mass spectrometry. The SDS gels shown in Figure 7 were purposely overloaded to reveal the less prominent bands numbered 2, 3, and 4. The N-terminal sequence of the polypeptides in bands 1 and 2 was PHSHPAL, which corresponds to positions 2–8 of aldolase. Mass analysis of the whole broad band 1 revealed the presence, besides intact aldolase with amino acids 2–364 and a mass of 39,216 Da, C-terminally degraded fragments corresponding to sequences 2–362, 2–360, and other masses down to 2–352 that were generated by the sequential removal of six dipeptide units from the C terminus. The significant presence of the 2–351 peptide indicates that aldolase degradation also comprises carboxypeptidase activity of cathepsin B. The tiny band 2 contained the polypeptide 2–347, which was the smallest fragment obtained under the described conditions and corresponded to the removal of 17 amino acids from the C terminus, i.e., 8 dipeptide units (peptidyl–dipeptidase activity) plus one amino acid (carboxypeptidase activity). Bands 3 and 4 had the common N terminus SIGTENT (positions 46–52), suggesting their generation by a minor (but very useful for the purposes of this test) endoproteolytic cleavage between Gln45 and Ser46. Band 3 corresponded to the 46–364 sequence and band 4 corresponded to sequence 46–347. Comparison of the relative intensities of bands 3 and 4 in lanes B and C (Fig. 7) suggests that DOFA inhibited the exopeptidase activity of cathepsin B on the macromolecular substrate aldolase, while the inhibitor was less or not active against the endoproteolytic activity of cathepsin B, as shown by the persistence of band 3 in lane C.
Figure 7.
Proteolysis of rabbit muscle fructose 1,6-bisphosphate aldolase by cathepsin B. Aldolase was incubated 4 h at 25°C with cathepsin B at pH 6.0, and products were separated by SDS-PAGE. (Lane A) Aldolase alone run in parallel as control; (lane B) aldolase plus cathepsin B; (lane C) aldolase with cathepsin B and DOFA. The N- and C-terminal parts of the rabbit aldolase sequence, with numbered amino acid residues, are shown at the top of the figure. Thin arrows indicate sequential peptidyl-dipeptidase cleavages by cathepsin B. The two open thick arrows show carboxypeptidase cleavages. The black thick arrow indicates a minor endoproteolytic attack site. Numbers on the left side indicate the molecular masses of markers in kilodaltons. Numbers 1–4 with long pointing arrows are assigned to the bands for referencing in the main text, and the peptides identified within the bands are shown next to the band numbers. Peptides were identified in the excised bands by N-terminal sequencing and MALDI analysis.
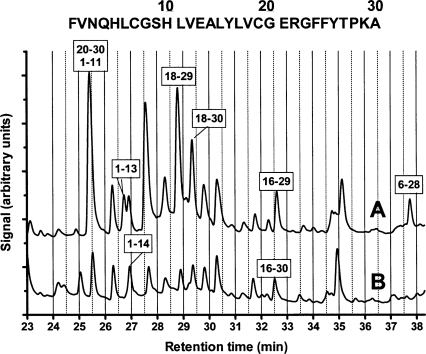
The bovine insulin β-chain is a helpful substrate for exploring the specificity of peptide-bond cleavage by peptidases, including human cathepsin B (McKay et al. 1983). The cleavage at multiple sites of the insulin β-chain by cathepsin B was almost completely inhibited by a saturating concentration of DOFA at pH 6.0 (Fig. 6). Samples of insulin β-chain incubated with cathepsin B in the presence and absence of DOFA were separated by liquid chromatography, and the eluted peaks were analyzed by mass spectrometry. We confirmed most of the cathepsin B cleavages previously described (McKay et al. 1983). Additional cleavages, which depended on the incubation time and the relative enzyme to substrate concentrations, were identified. Each of the chromatographic peaks in Figure 8 contained only one (e.g., peptides 1–13, 1–14, and 6–28) or more proteolytic fragments (e.g., peptides 20–30 plus 1–11). Only a few of them will be discussed to demonstrate the inhibitory activity of DOFA. The generation of peptides 1–11 and 20–30 by endoproteolysis was inhibited by DOFA. Fragment 1–13 was a product of carboxypeptidase activity on peptide 1–14: While DOFA completely inhibited this exoproteolytic event, endoproteolytic cleavage between residues 14 and 15 was unaffected, as shown by comparing the chromatographic profiles in the absence (Fig. 8, profile A) and in the presence of DOFA (Fig. 8, profile B). Fragment 18–29, generated by endoproteolysis at position 17–18 and exoproteolysis at position 29–30, was strongly decreased in the presence of DOFA. Comparison of the areas of fragments 18–29 and 18–30 after inhibition shows that both the exo- and the endoproteolytic events concerned with them were affected by the inhibitor. Carboxypeptidase activity inhibition is also shown by comparing the disappearance of fragment 16–29 and the appearance of fragment 16–30. Finally, the peptidyl–dipeptidase cleavage shown by fragment 6–28 disappeared in the presence of DOFA. Since the 6–30 peptide in Figure 8, profile B could not be unambiguously assigned, inhibition could be ascribed to endoproteolytic cleavage at positions 5–6, peptidyl–dipeptidase inhibition at position 28–29, or both.
Figure 8.
Proteolysis of the bovine insulin β-chain by cathepsin B. Incubation at pH 5.5 and 25°C in the absence (A) and in the presence (B) of DOFA. Peptides released by enzymatic digestion were characterized by LC/MALDI/MS. The sequence at the top of the figure shows the sequence of bovine insulin, β-chain with amino acid numbering. Boxed numbers, e.g., 6–28, indicate peptides released by the endo- and exoproteolytic action of cathepsin B.
Discussion
The docking approach used in this study aimed at identifying compounds that bind to the occluding loop of human cathepsin B and thereby modulate its enzymatic activity. Considering that no other means are available for selecting small molecules for a rather flexible protein segment, the most diverse compound library was used. There is a fundamental difference between our methodology and previous approaches to inhibitors that might interact with parts of the occluding loop of cathepsin B (Murata et al. 1991; Michaud and Gour 1998; Cathers et al. 2002). These studies exploited the properties of the fungal inhibitor E-64 (Hanada et al. 1978) and the crystal structure of cathepsin B (Musil et al. 1991). New inhibitors, such as CA-074, were designed by modification of the E-64 structure to create interactions with the primed sites of the substrate binding cleft and thus with the occluding loop (Murata et al. 1991). In this approach the epoxy moiety of the modifiers acted as a warhead against the active site cysteine to produce potent irreversible inhibition (Matsumoto et al. 1999). Conversely, we did not target the active site, and the molecules selected by docking were devoid of chemically active warheads. The finding of only one active compound out of 29 substances is in line with previous studies, which showed that candidate inhibitors predicted by docking algorithms are not necessarily active in vitro (Huang et al. 2005).
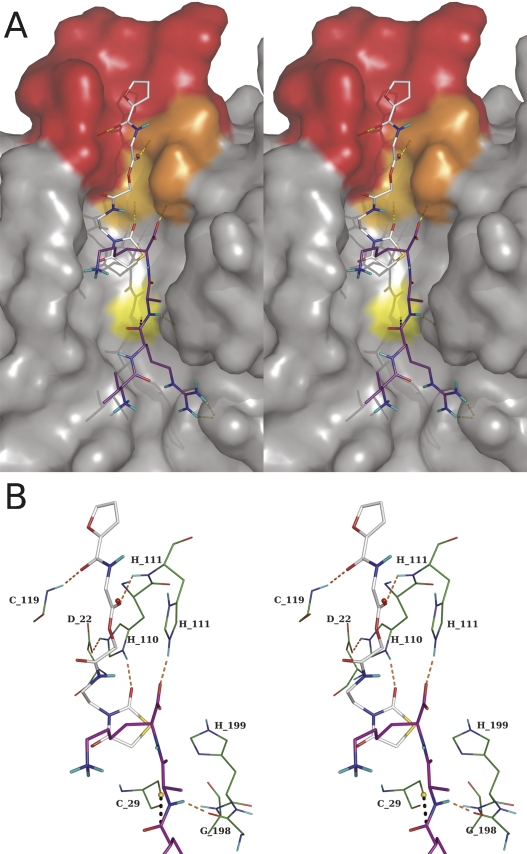
In agreement with published data (Illy et al. 1997; Nägler et al. 1997; Quraishi et al. 1999; Krupa et al. 2002) our results support the notion that the occluding loop of cathepsin B is a highly flexible segment. Short episodes of opening and closing govern endo- and exoproteolysis, which thus occur intermittently in the pH range 4.5–6.0. The compound identified by docking, DOFA, behaves as a reversible competitive inhibitor of the exo- and endopeptidase activities of cathepsin B with no involvement of a substrate-like binding mode in the active center of the enzyme. It can inhibit endoproteolysis most likely by forcing the loop in the closed conformation, and exoproteolysis by disturbing the accommodation of the C terminus of polypeptides in the primed side of the binding cleft. Docking calculations suggest that the dioxothiazolidine moiety of DOFA is located in the binding pocket of the side chain of the substrate C-terminal residue when the loop is closed. It makes a hydrogen bond with Nδ1 of His110 and sterically hinders substrate binding for exoproteolytic activity (Fig. 9A,B). In this figure, the structures of DOFA and the four amino acids VRAK, which are part of the FRET substrate used for measuring exoproteolytic activity, were modeled on the known structure of human cathepsin B merely to show their spatial relationship. In fact, such a ternary ESI complex does not exist according to the kinetic mechanism; only the ES and EI complexes exist. This structural arrangement is consistent with the kinetic mechanism of inhibition: At pH 4.5 the exoproteolytic activity of cathepsin B is inhibited in a linear competitive manner by virtue of the dioxothiazolidine ring. Docking suggests that DOFA binds at the surface of the occluding loop and is involved in three hydrogen bonds with the enzyme (Fig. 9A). Of particular interest is that the carbonyl oxygen involved in a hydrogen bond with the amide of His111 occupies the place of the oxygen of a conserved water molecule present in the crystal structures of human cathepsin B. This position is shown as a red sphere in Figure 9B. Together with the favorable van der Waals interactions, three hydrogen bonds stabilize the loop in the closed conformation (Fig. 9B). These structural properties are reflected in the kinetic mechanism observed with Z-RR↓AMC. Despite not being a complete model for endoproteolysis, the dipeptide is unquestionably cleaved by cathepsin B in the endo-, not in the exoproteolytic mode. The double-headed competitive kinetic mechanism diagnosed for DOFA on Z-RR↓AMC, and the changes in the kinetic parameters observed when going from pH 4.5 to 6.0, insinuate cooperation between the furan and the dioxothiazolidine moieties. Kinetic measurements alone would be unable to identify which element makes the stronger contribution to binding energy. However, with the support of structural information from modeling discussed above, we suggest that the dioxothiazolidine moiety takes the leader function. In the double-headed mechanism at pH 4.5, the K i value of 4.8 μM is thus ascribed to the dioxothiazolidine part of the molecule and K i ≈ 160 μM to the furan part (factor a = 34 in Model 3, Fig. 3). The increase of pH to 6.0 disfavors the K i of the dioxothiazolidine portion, which becomes 11.3 μM and favors binding of the furan moiety, decreasing its K i to 68 μM.
Figure 9.
Stereoview of inhibitor and substrate within the structure of human cathepsin B. (A) The protein surface is shown with the occluding loop in red, His110 and His111 in orange, and the catalytic Cys29 in yellow. The substrate (carbon atoms in purple) is shown as residues VR↓AK, which is part of the FRET substrate used in this study to monitor peptidyl–dipeptidase activity. The Dnp group, which is exposed to the solvent, was omitted for clarity. The pose of DOFA obtained by docking (carbon atoms in gray) is visible in the upper part with the furan moiety in the occluding loop and the dioxothiazolidine ring sterically interfering with the lysine side chain of the substrate. (B) Details of the presumed interactions between DOFA and cathepsin B residues (carbon atoms in green). The inhibitor makes three hydrogen bonds with residues of the occluding loop: carbonyl group labeled “a” in Figure 2 with amide nitrogen of Cys119, carbonyl group labeled “b” in Figure 2 with amide nitrogen of His111, and 2-oxo group of the dioxothiazolidine ring with Nδ1 of His110. The hydrogen bonds of the occluding loop in the closed conformation, His110–Asp22 and His111 with the C-terminal carbonyl of the substrate, are also shown. The red sphere indicates the position of a water molecule in the cathepsin B crystal structure. Images generated with PyMOL software (see Fig. 1).
The measurements with protein substrates, aldolase and the insulin β-chain, confirm the results just discussed with synthetic substrates, as well as the previously recorded ability of cathepsin B to exert peptidyl–dipeptidase and carboxypeptidase activity in the pH range 5.5–6.0 (Mort et al. 1998). While efficiently inhibiting these two exoproteolytic activities, DOFA was not able to inhibit all endoproteolytic events. For instance, the minor endoproteolytic action at position 45–46 of rabbit muscle aldolase was only moderately or not affected by DOFA, as was not position 14–15 of the insulin β-chain. Yet, other endoproteolytic attacks on the insulin β-chain were efficiently inhibited.
The pooled experimental evidence from the above described properties of DOFA inhibition strengthens the notion that not only pH dependent, intermittent closing and opening of the occluding loop, but also substrate structure determine together the susceptibility of particular peptide bonds. The binding affinity of DOFA is possibly not high enough to compete with a substrate, which optimally binds at both unprimed and primed sites. It appears that the occluding loop of cathepsin B can be assisted in assuming its closed or open conformation by particular polypeptides with an induced fit mechanism that promotes either exo- or endoproteolysis. This observation forms the basis of cathepsin B inhibition by its own propeptide (Quraishi et al. 1999) and its resistance to cystatin A and C inhibition (Nycander et al. 1998; Pavlova et al. 2000).
The above results draw attention to the physiopathological role of cathepsin B, which exerts its action in a relatively broad range of pH values, with exoproteolysis in the range 5–6 and endoproteolysis in the range 5–7.4. The pH in early endosomes is ∼6.2 and drops to 5.0–5.5 in late endosomes and lysosomes (Gu and Gruenberg 2000). Extracellularly, the pH can vary from relatively acidic, e.g., at the periphery of solid tumors (Rofstad et al. 2006), to 7.4 in blood. Contrary to commonplace textbook information, proteolysis within phagolysosomes does not quite occur in a very acidic environment. For instance, 5 min after initiation of phagocytosis in human polymorphonuclear leukocytes, the relatively acidic milieu of the lysosomes (pH ≈ 5) increases in the phagocytic vacuoles to a value of 7.8. It falls then to 7.4 after 15 min, to 6.4 after 30 min, and 5.7 after 60 min (rounded values from Cech and Lehrer [1984]). Various hydrolases are expected to find their optimal conditions of action during this forth and back excursion of pH (Baici et al. 1996).
It has been suggested that “targeting of the occluding loop of cathepsin B may be a poor inhibitor design strategy if the enzyme environment has a pH greater than 5.5” (Cathers et al. 2002). This statement refers to the above discussed approach “from inside.” Since endo- and exoproteolysis by cathepsin B occurs in the physiologically and pathologically relevant pH range of 5–6, our approach of targeting the occluding loop from the enzyme surface may represent an alternative starting point for the development of clinically active compounds.
Materials and Methods
Materials and general methods
Recombinant human cathepsin B was prepared from Escherichia coli inclusion bodies (Kuhelj et al. 1995). Wild-type cathepsin B from human liver was obtained from Calbiochem. The synthetic substrates Z-FR↓AMC2 and Z-RR↓AMC were purchased from Bachem and the internally quenched fluorogenic substrate Abz-GIVR↓AK(Dnp)OH from Merck. Rabbit muscle fructose-1,6-bisphosphate aldolase (EC 4.1.2.13) and fluorescamine were from Sigma-Aldrich Chemie and the oxidized bovine insulin β-chain from Serva Feinbiochemica. The putative binders/modifiers of cathepsin B activity determined by docking analysis and their sources are listed in Supplemental Table 1. The compound studied in detail, [2-[2-(2,4-dioxo-1,3-thiazolidin-3-yl)ethylamino]-2-oxoethyl] 2-(furan-2-carbonylamino)acetate (IUPAC name, here abbreviated DOFA; Fig. 2), Zinc-2005 code 2616818 (Irwin and Shoichet 2005) was obtained from Enamine. The compounds were dissolved as concentrated stock solutions in dimethyl sulphoxide, stored at 4°C, and diluted into the appropriate buffer at the moment of the assay. Photometric measurements were carried out with a Cary50 UV-visible spectrophotometer, and fluorescence was measured with an Aminco SPF-500 fluorimeter operating in the ratio mode. Reversed phase HPLC was performed in a Hewlett Packard Series 1100 apparatus with a Nucleosil 120-5 C18 column (4.0 × 250 mm). Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) and liquid chromatography followed by mass analysis, LC-MALDI/MS, was performed at the Functional Genomics Center in Zurich.
Docking
The A and B chains of the human cathepsin B structure, entry 1HUC in the Brookhaven database, were considered and all water molecules were removed. Hydrogens were added to side chains and termini of the protein to simulate conditions at pH 6–7. Cys29 was in the reduced form, and His110 was protonated at both imidazole nitrogen atoms. CHARMm atom types and force field parameters were assigned and used for both protein and ligands (Momany and Rone 1992), and hydrogens were minimized with the program CHARMM (Brooks et al. 1983). The search for a putative binding site focused on the occluding loop of the enzyme, for which no inhibitor has been reported to date. A benzene molecule was used as a probe with the program SEED (Solvation Energy for Exhaustive Docking) to identify possible binding pockets whose main interaction component is supposed to be hydrophobicity (Majeux et al. 1999). The surface tested by SEED consists of cathepsin B residues within a 9 Å distance from His110, which locks the occluding loop in a closed position. The benzenes with the most favorable free energy of binding, excluding those in the catalytic site, were used to define the binding site for docking, which included all the 42 residues of the protein that had >50% of their atoms within 5 Å from the optimal poses of benzenes.
About 3 million compounds in the ZINC 2005 library (Irwin and Shoichet 2005) were clustered using a 17-field fingerprint by Decomposition and Identification of Molecules (Kolb and Caflisch 2006). Molecules with more than nine rotatable bonds were neglected. The resulting 48,026 cluster representatives were docked as in previous in silico campaigns (Huang et al. 2005, 2006; Kolb et al. 2008). For each molecule docked by the program FFLD (Fragment-based Flexible Ligand Docking) (Budin et al. 2001; Cecchini et al. 2004), the 300 most favorable poses were clustered with the leader algorithm yielding 764,776 poses (∼17 poses per compound, 44,527 compounds). The cluster representatives were minimized in the rigid protein using CHARMM.
It is necessary to weed out unlikely poses to prevent a high number of false positives (Kolb et al. 2008). Suitable filters are cutoffs in the total van der Waals interaction energy and in the van der Waals efficiency (defined as the ratio between van der Waals energy and molecular mass). A cutoff of −30 kcal/mol for intermolecular van der Waals energy was selected by plotting a histogram of the van der Waals energies (Supplemental Fig. 1). Analogously, a cutoff of −0.09 kcal/g was chosen for the van der Waals efficiency (Supplemental Fig. 2). A total of 106,502 poses of 10,709 compounds survived both filters. Poses closer than 4 Å to Cys29 of the catalytic site and those with no hydrogen bond to the protein were neglected, yielding 25,178 remaining poses (5678 compounds).
The electrostatic contribution to the binding free energy was evaluated by the finite difference Poisson approach (module PBEQ of CHARMM) (Im et al. 1998). Poses were ranked for visual inspection according to most favorable van der Waals energy, van der Waals efficiency, electrostatics, and sum of van der Waals and electrostatic energy. Poses with high ranking in two lists or more were selected and visually inspected. Forty poses belonging to 40 different compounds were selected in this way. According to the predicted binding modes, three regions of the protein were exploited for binding: one on the occluding loop and one each on immediately adjacent regions. Twenty-nine out of the 40 candidate compounds were commercially available.
Kinetics
The peptide Z-RR↓AMC with 7-amino-4-methlycoumarin (AMC) as leaving group is a handy substrate for measuring the endopeptidase activity of cathepsin B (Barrett 1980), in which the two arginine residues correspond to the P1 and P2 positions (Schechter and Berger 1967). We preferred this substrate over Z-FR↓AMC for its slower hydrolysis that allowed long measuring times with low substrate consumption. The buffer for measurements at pH 6.00 was 50 mM sodium phosphate containing 2 mM EDTA and 2 mM dithiothreitol (DTT), hereafter referred to as pH 6.0 buffer. For measurements at pH 4.50, 0.1 M sodium acetate, 0.2 M NaCl, 2 mM EDTA, and 2.5 mM DTT were used (hereafter referred to as pH 4.5 buffer). In control experiments aimed at ascertaining any aggregation phenomena on the part of the putative inhibitors, the DTT concentration was increased to 5 mM and the buffer included 0.01% (v/v) of hydrogenated Triton X-100, which does not absorb light in the ultraviolet range and is not fluorescent. The buffers were prepared and used at the same temperature of the kinetic assays, 25 ± 1°C. The final concentration of dimethyl sulphoxide, used for preparing stock solutions of the compounds, was 0.2% (v/v) in experiments with synthetic substrates and 1% (v/v) in those with protein substrates. Reaction progress with Z-RR↓AMC at pH 4.5 and 6.0 was followed by either measuring the change in absorbance at 360 nm, with Δε = 11,400 M−1cm−1 as the difference absorption coefficient between free and bound AMC or fluorimetrically with excitation and emission wavelengths, λex and λem, set at 383 and 455 nm, respectively. Exopeptidase activity was continuously monitored fluorimetrically with the Förster resonance energy transfer (FRET) labeled substrate Abz-GIVR↓AK(Dnp)-OH (Cotrin et al. 2004). Appropriate corrections for inner filter effects were taken into account in fluorescence measurements considering the absorbencies of the substrates and of the tested compounds at λex and λem (Palmier and Van Doren 2007). With the AMC-based substrate, λex at 383 nm allowed high substrate concentrations to be used with small inner filter corrections. Since with the FRET substrate inner filter effects were more pronounced, λex and λem were selected according to the compound being tested. The combination λex/λem 320/420 nm, often used in assays with this type of substrate, could not be utilized for the strong absorbance at 320 nm of one of the compounds (ZINC-2005 code 1797383), which simulated strong enzyme inhibition, whereas the combination λex/λem 370/430 nm revealed no inhibition. Any modifier-independent contribution to enzyme activity loss by denaturation, adherence to the vessel walls, or other causes was monitored in time-course assays by the Selwyn method (Selwyn 1965). Screening and primary analysis of the kinetic inhibition mechanism of the compounds was performed on initial velocity data obtained at various concentrations of substrate and compounds. The specific velocity plot, a sensitive tool for detecting mixed-type and/or hyperbolic mechanisms was routinely used in this phase (Baici 1981). The kinetic inhibition models considered and their related rate equations are shown in Figure 3. In Models 2 and 3, a is a coefficient, which is equal to 1 when the first binding process does not influence the second one. If the equilibrium constant of the vacant site is influenced after binding to the first site, a ≠ 1. Kinetic analysis was performed by fitting the equations in Figure 3 to experimental data using GraphPad Prism version 5.00 for Windows. The runs test and analysis of residuals were performed to monitor deviations from a model, and discrimination between mechanisms was made by analysis of variance of the difference between the sum of squares (extra sum-of-squares test) and calculation of F ratios and p values. According to Occam's razor principle, the simplest mechanism describing experimental data was considered superior to a more complex (redundant) mechanism. Additionally, a powerful and decisive tool for model discrimination was the I 90/10 ratio, i.e., the ratio of inhibitor concentration necessary to achieve 90% and 10% inhibition. This parameter was determined experimentally and compared with values calculated from Equations 4 and 6 in Figure 3.
Protein substrates
The activity of cathepsin B on rabbit muscle aldolase was quantified by measuring primary amino groups derived from peptide-bond hydrolysis with fluorescamine (Schwabe 1975). Aldolase was dissolved in pH 6.0 buffer without DTT, incubated 3 h at 37°C with cathepsin B, which was pre-activated with DTT, and DOFA under vigorous shaking to prevent precipitation. Final concentrations were: aldolase = 3.5 mg/mL = 21.7 μM, cathepsin B = 2.3 μM, DOFA = 0.5, 1.0, 1.5, and 2.0 mM. The reaction was stopped with trichloroacetic acid, 5% (w/v) final concentration, and centrifuged. We added 0.1 mL of the clear supernatant to 2.0 mL 0.2 M sodium borate buffer (pH 8.50), and 1.0 mL of fluorescamine solution (15 mg/100 mL acetone). The fluorescence of labeled peptides was measured with λex/λem = 390/480 nm. Blanks were subtracted and controls compared with the samples containing the compounds. Measurements with the oxidized β-chain of bovine insulin as substrate were performed in pH 6.0 buffer using the same procedure. Final concentrations were: insulin β-chain = 72 μg/mL = 20.6 μM, cathepsin B = 2.3 μM, DOFA = 0.5, 1.0, 1.5, and 2.0 mM.
Endo- and exoproteolysis of protein substrates
Reaction products resulting from incubation of aldolase with cathepsin B and DOFA were also assessed by denaturing gel electrophoresis. Final concentrations in incubation mixtures at 25°C under continuous shaking for 4 h were: aldolase 3.5 mg/mL (21.7 μM) in pH 6.0 buffer without DTT, 2.3 μM of preactivated cathepsin B, and 1.0 mM DOFA. After centrifugation, the clear supernatant was analyzed by sodium dodecyl sulphated, polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions with 12.5% polyacrylamide and silver stained. For N-terminal sequencing by Edman degradation, the bands were transferred to a polyvinylidene fluoride membrane. Peptides in the excised bands were also identified by MALDI analysis.
The oxidized β-chain of bovine insulin was incubated at a final concentration of 1.0 mg/mL (286 μM) in 0.1 M sodium acetate buffer containing 0.2 mM NaCl, 2 mM EDTA, and 2.5 mM DTT (pH 5.50) with 1.2 μM cathepsin B (control) or cathepsin B plus 1.0 mM DOFA. After 20 h at 25°C under shaking, solutions were centrifuged and the solvent removed in a Speed Vac concentrator. The residue was resuspended in 60 μL of deionized water containing 0.1% trifluoroacetic acid and centrifuged before analysis by LC/MALDI/MS.
Electronic supplemental material
Supplemental Table 1 shows the identification codes and the sources of the cathepsin B inhibitors. Supplemental Figure 1 presents the van der Waals interaction energies distribution used in the docking procedure and shows the cutoff of −30 kcal/mol for intermolecular van der Waals energy. Analogously, Supplemental Figure 2 shows the van der Waals interaction energy efficiencies distribution.
Acknowledgments
Docking calculations were performed on Matterhorn, a Beowulf Linux cluster at the University of Zurich, and we thank C. Bolliger, T. Steenbock, and A. Godknecht for computer support. We thank A. Widmer (Novartis Pharma, Basel, Switzerland) for providing the molecular modeling program Wit!P, which was used for preparing the structures. We also thank the staff at the Functional Genomics Center in Zurich for expert assistance in LC/MALDI/MS measurements. This work was supported by grant 31-113345/1 of the Swiss National Foundation and by the Albert-Böni Foundation (to A.B.), and by a grant of the Hartmann-Müller Foundation (to A.C.).
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Antonio Baici, Department of Biochemistry, University of Zurich, Winterthurerstrasse 190, CH-8057 Zurich, Switzerland; e-mail: abaici@bioc.uzh.ch; fax: 41-44-6356805.
Abbreviations: Abz, ortho-aminobenzoyl; AMC, 7-amino-4-methylcoumarin; DTT, dithiothreitol; Z, benzyloxycarbonyl; Dnp, Nε-2,4-dinitrophenyl; FRET, Förster resonance energy transfer; DOFA, [2-[2-(2,4-dioxo-1,3-thiazolidin-3-yl)ethylamino]-2-oxoethyl] 2-(furan-2-carbonylamino)acetate; MALDI, matrix-assisted laser desorption/ionization; MS, mass spectrometry; LC, liquid chromatography; SDS-PAGE, sodium dodecyl sulfate, polyacrylamide gel electrophoresis.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.037341.108.
References
- Aronson N.N., Jr, Barrett A.J. The specificity of cathepsin B. Hydrolysis of glucagon at the C-terminus by a peptidyldipeptidase mechanism. Biochem. J. 1978;171:759–765. doi: 10.1042/bj1710759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baici A. The specific velocity plot. A graphical method for determining inhibition parameters for both linear and hyperbolic enzyme inhibitors. Eur. J. Biochem. 1981;119:9–14. doi: 10.1111/j.1432-1033.1981.tb05570.x. [DOI] [PubMed] [Google Scholar]
- Baici A. Inhibition of extracellular matrix-degrading endopeptidases: Problems, comments, and hypotheses. Biol. Chem. 1998;379:1007–1018. [PubMed] [Google Scholar]
- Baici A., Hörler D., Lang A., Merlin C., Kissling R. Cathepsin B in osteoarthritis: Zonal variation of enzyme activity in human femoral head cartilage. Ann. Rheum. Dis. 1995a;54:281–288. doi: 10.1136/ard.54.4.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baici A., Lang A., Hörler D., Kissling R., Merlin C. Cathepsin B in osteoarthritis: Cytochemical and histochemical analysis of human femoral head cartilage. Ann. Rheum. Dis. 1995b;54:289–297. doi: 10.1136/ard.54.4.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baici A., Szedlacsek S.E., Früh H., Michel B.A. pH-Dependent hysteretic behaviour of human myeloblastin (leucocyte proteinase 3) Biochem. J. 1996;317:901–905. doi: 10.1042/bj3170901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baici A., Müntener K., Willimann A., Zwicky R. Regulation of human cathepsin B by alternative mRNA splicing: Homeostasis, fatal errors and cell death. Biol. Chem. 2006;387:1017–1021. doi: 10.1515/BC.2006.125. [DOI] [PubMed] [Google Scholar]
- Barrett A.J. Fluorimetric assays for cathepsin B and cathepsin H with methylcoumarylamide substrates. Biochem. J. 1980;187:909–912. doi: 10.1042/bj1870909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J.S., Barrett A.J. Degradation of fructose-1,6-bisphosphate aldolase by cathepsin B. A further example of peptidyldipeptidase activity of this proteinase. Biochem. J. 1980;189:17–25. doi: 10.1042/bj1890017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröker L.E., Kruyt F.A.E., Giaccone G. Cell death independent of caspases: A review. Clin. Cancer Res. 2005;11:3155–3162. doi: 10.1158/1078-0432.CCR-04-2223. [DOI] [PubMed] [Google Scholar]
- Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- Budin N., Majeux N., Caflisch A. Fragment-based flexible ligand docking by evolutionary optimization. Biol. Chem. 2001;382:1365–1372. doi: 10.1515/BC.2001.168. [DOI] [PubMed] [Google Scholar]
- Buxser S., Vroegop S. Calculating the probability of detection for inhibitors in enzymatic or binding reactions in high-throughput screening. Anal. Biochem. 2005;340:1–13. doi: 10.1016/j.ab.2005.01.034. [DOI] [PubMed] [Google Scholar]
- Cathers B.E., Barrett C., Palmer J.T., Rydzewski R.M. pH Dependence of inhibitors targeting the occluding loop of cathepsin B. Bioorg. Chem. 2002;30:264–275. doi: 10.1016/s0045-2068(02)00009-3. [DOI] [PubMed] [Google Scholar]
- Cecchini M., Kolb P., Majeux N., Caflisch A. Automated docking of highly flexible ligands by genetic algorithms: A critical assessment. J. Comput. Chem. 2004;25:412–422. doi: 10.1002/jcc.10384. [DOI] [PubMed] [Google Scholar]
- Cech P., Lehrer R.I. Phagolysosomal pH of human neutrophils. Blood. 1984;63:88–95. [PubMed] [Google Scholar]
- Cotrin S.S., Puzer L., Judice W.A.D., Juliano L., Carmona A.K., Juliano M.A. Positional-scanning combinatorial libraries of fluorescence resonance energy transfer peptides to define substrate specificity of carboxydipeptidases: Assays with human cathepsin B. Anal. Biochem. 2004;335:244–252. doi: 10.1016/j.ab.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Friedrichs B., Tepel C., Reinheckel T., Deussing J., von Figura K., Herzog V., Peters C., Saftig P., Brix K. Thyroid functions of mouse cathepsins B, K, and L. J. Clin. Invest. 2003;111:1733–1745. doi: 10.1172/JCI15990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frlan R., Gobec S. Inhibitors of cathepsin B. Curr. Med. Chem. 2006;13:2309–2327. doi: 10.2174/092986706777935122. [DOI] [PubMed] [Google Scholar]
- Gu F., Gruenberg J. ARF1 regulates pH-dependent COP functions in the early endocytic pathway. J. Biol. Chem. 2000;275:8154–8160. doi: 10.1074/jbc.275.11.8154. [DOI] [PubMed] [Google Scholar]
- Hanada K., Tamai M., Yamagishi M., Ohmura S., Sawada J., Tanaka I. Isolation and characterization of E-64, a new thiol protease inhibitor. Agric. Biol. Chem. 1978;42:523–528. [Google Scholar]
- Huang D.Z., Lüthi U., Kolb P., Edler K., Cecchini M., Audetat S., Barberis A., Caflisch A. Discovery of cell-permeable non-peptide inhibitors of β-secretase by high-throughput docking and continuum electrostatics calculations. J. Med. Chem. 2005;48:5108–5111. doi: 10.1021/jm050499d. [DOI] [PubMed] [Google Scholar]
- Huang D.Z., Lüthi U., Kolb P., Cecchini M., Barberis A., Caflisch A. In silico discovery of β-secretase inhibitors. J. Am. Chem. Soc. 2006;128:5436–5443. doi: 10.1021/ja0573108. [DOI] [PubMed] [Google Scholar]
- Illy C., Quraishi O., Wang J., Purisima E., Vernet T., Mort J.S. Role of the occluding loop in cathepsin B activity. J. Biol. Chem. 1997;272:1197–1202. doi: 10.1074/jbc.272.2.1197. [DOI] [PubMed] [Google Scholar]
- Im W., Beglov D., Roux B. Continuum solvation model: Computation of electrostatic forces from numerical solutions to the Poisson–Boltzmann equation. Comput. Phys. Commun. 1998;111:59–75. [Google Scholar]
- Irwin J.J., Shoichet B.K. ZINC—a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb P., Caflisch A. Automatic and efficient decomposition of two-dimensional structures of small molecules for fragment-based high-throughput docking. J. Med. Chem. 2006;49:7384–7392. doi: 10.1021/jm060838i. [DOI] [PubMed] [Google Scholar]
- Kolb P., Huang D., Dey F., Caflisch A. Discovery of kinase inhibitors by high-throughput docking and scoring based on a transferable linear interaction energy model. J. Med. Chem. 2008;51:1179–1188. doi: 10.1021/jm070654j. [DOI] [PubMed] [Google Scholar]
- Krupa J.C., Hasnain S., Nägler D.K., Menard R., Mort J.S. S2′ substrate specificity and the role of His110 and His111 in the exopeptidase activity of human cathepsin B. Biochem. J. 2002;361:613–619. doi: 10.1042/0264-6021:3610613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhelj R., Dolinar M., Pungercar J., Turk V. The preparation of catalytically active human cathepsin B from its precursor expressed in Escherichia coli in the form of inclusion bodies. Eur. J. Biochem. 1995;229:533–539. doi: 10.1111/j.1432-1033.1995.0533k.x. [DOI] [PubMed] [Google Scholar]
- Kukor Z., Mayerle J., Krüger B., Tóth M., Steed P.M., Halangk W., Lerch M.M., Sahin-Tóth M. Presence of cathepsin B in the human pancreatic secretory pathway and its role in trypsinogen activation during hereditary pancreatitis. J. Biol. Chem. 2002;277:21389–21396. doi: 10.1074/jbc.M200878200. [DOI] [PubMed] [Google Scholar]
- Lenarcic B., Gabrijelcic D., Rozman B., Drobnic-Kosorok M., Turk V. Human cathepsin B and cysteine proteinase inhibitors (CPIs) in inflammatory and metabolic joint diseases. Biol. Chem. Hoppe Seyler. 1988;369:257–261. [PubMed] [Google Scholar]
- Majeux N., Scarsi M., Apostolakis J., Ehrhardt C., Caflisch A. Exhaustive docking of molecular fragments with electrostatic solvation. Proteins. 1999;37:88–105. [PubMed] [Google Scholar]
- Matsumoto K., Mizoue K., Kitamura K., Tse W.C., Huber C.P., Ishida T. Structural basis of inhibition of cysteine proteases by E-64 and its derivatives. Biopolymers. 1999;51:99–107. doi: 10.1002/(SICI)1097-0282(1999)51:1<99::AID-BIP11>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Matsunaga Y., Saibara T., Kido H., Katunuma N. Participation of cathepsin B in processing of antigen presentation to MHC class II. FEBS Lett. 1993;324:325–330. doi: 10.1016/0014-5793(93)80144-j. [DOI] [PubMed] [Google Scholar]
- McKay M.J., Offermann M.K., Barrett A.J., Bond J.S. Action of human liver cathepsin B on the oxidized insulin B chain. Biochem. J. 1983;213:467–471. doi: 10.1042/bj2130467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud S., Gour B.J. Cathepsin B inhibitors as potential anti-metastatic agents. Expert Opin. Ther. Pat. 1998;8:645–672. [Google Scholar]
- Mohamed M.M., Sloane B.F. Cysteine cathepsins: Multifunctional enzymes in cancer. Nat. Rev. Cancer. 2006;6:764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- Momany F.A., Rone R. Validation of the general purpose QUANTA®3.2/CHARMm® force field. J. Comput. Chem. 1992;13:888–900. [Google Scholar]
- Mort J.S. Cathepsin B. In: Barrett A.J., Rawlings N.D., Woessner J.F. Jr, editors. Handbook of proteolytic enzymes. 2nd ed. Elsevier; London, UK: 2004. pp. 1079–1086. [Google Scholar]
- Mort J.S., Magny M.C., Lee E.R. Cathepsin B: An alternative protease for the generation of an aggrecan “metalloproteinase” cleavage neoepitope. Biochem. J. 1998;335:491–494. doi: 10.1042/bj3350491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müntener K., Zwicky R., Csucs G., Baici A. The alternative use of exons 2 and 3 in cathepsin B mRNA controls enzyme trafficking and triggers nuclear fragmentation in human cells. Histochem. Cell Biol. 2003;119:93–101. doi: 10.1007/s00418-002-0487-y. [DOI] [PubMed] [Google Scholar]
- Müntener K., Zwicky R., Csucs G., Rohrer J., Baici A. Exon skipping of cathepsin B: Mitochondrial targeting of a lysosomal peptidase provokes cell death. J. Biol. Chem. 2004;279:41012–41017. doi: 10.1074/jbc.M405333200. [DOI] [PubMed] [Google Scholar]
- Murata M., Miyashita S., Yokoo C., Tamai M., Hanada K., Hatayama K., Towatari T., Nikawa T., Katunuma N. Novel epoxysuccinyl peptides. Selective inhibitors of cathepsin B, in vitro. FEBS Lett. 1991;280:307–310. doi: 10.1016/0014-5793(91)80318-w. [DOI] [PubMed] [Google Scholar]
- Musil D., Zucic D., Turk D., Engh R.A., Mayr I., Huber R., Popovic T., Turk V., Towatari T., Katunuma N., et al. The refined 2.15 Å X-ray crystal structure of human liver cathepsin B: The structural basis for its specificity. EMBO J. 1991;10:2321–2330. doi: 10.1002/j.1460-2075.1991.tb07771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nägler D.K., Storer A.C., Portaro F.C.V., Carmona E., Juliano L., Menard R. Major increase in endopeptidase activity of human cathepsin B upon removal of occluding loop contacts. Biochemistry. 1997;36:12608–12615. doi: 10.1021/bi971264+. [DOI] [PubMed] [Google Scholar]
- Nycander M., Estrada S., Mort J.S., Abrahamson M., Björk I. Two-step mechanism of inhibition of cathepsin B by cystatin C due to displacement of the proteinase occluding loop. FEBS Lett. 1998;422:61–64. doi: 10.1016/s0014-5793(97)01604-9. [DOI] [PubMed] [Google Scholar]
- Okamura-Oho Y., Zhang S.Q., Callahan J.W., Murata M., Oshima A., Suzuki Y. Maturation and degradation of β-galactosidase in the post-Golgi compartment are regulated by cathepsin B and a non-cysteine protease. FEBS Lett. 1997;419:231–234. doi: 10.1016/s0014-5793(97)01461-0. [DOI] [PubMed] [Google Scholar]
- Otto H.H., Schirmeister T. Cysteine proteases and their inhibitors. Chem. Rev. 1997;97:133–171. doi: 10.1021/cr950025u. [DOI] [PubMed] [Google Scholar]
- Palmier M.O., Van Doren S.R. Rapid determination of enzyme kinetics from fluorescence: Overcoming the inner filter effect. Anal. Biochem. 2007;371:43–51. doi: 10.1016/j.ab.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova A., Krupa J.C., Mort J.S., Abrahamson M., Björk I. Cystatin inhibition of cathepsin B requires dislocation of the proteinase occluding loop. Demonstration by release of loop anchoring through mutation of His110. FEBS Lett. 2000;487:156–160. doi: 10.1016/s0014-5793(00)02337-1. [DOI] [PubMed] [Google Scholar]
- Quraishi O., Nägler D.K., Fox T., Sivaraman J., Cygler M., Mort J.S., Storer A.C. The occluding loop in cathepsin B defines the pH dependence of inhibition by its propeptide. Biochemistry. 1999;38:5017–5023. doi: 10.1021/bi981950o. [DOI] [PubMed] [Google Scholar]
- Rawlings N.D., Tolle D.P., Barrett A.J. MEROPS: The peptidase database (http://merops.sanger.ac.uk) Nucleic Acids Res. 2004;32:D160–D164. doi: 10.1093/nar/gkh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rofstad E.K., Mathiesen B., Kindem K., Galappathi K. Acidic extracellular pH promotes experimental metastasis of human melanoma cells in athymic nude mice. Cancer Res. 2006;66:6699–6707. doi: 10.1158/0008-5472.CAN-06-0983. [DOI] [PubMed] [Google Scholar]
- Rowan A.D., Feng R., Konishi Y., Mort J.S. Demonstration by electrospray mass spectrometry that the peptidylpeptidase activity of cathepsin B is capable of rat cathepsin B C-terminal processing. Biochem. J. 1993;294:923–927. doi: 10.1042/bj2940923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter I., Berger A. On the size of the active sites in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Schwabe C. A fluorescence assay for proteolytic enzymes. Anal. Biochem. 1975;53:484–490. doi: 10.1016/0003-2697(73)90098-5. [DOI] [PubMed] [Google Scholar]
- Selwyn M.J. A simple test for inactivation of an enzyme during assay. Biochim. Biophys. Acta. 1965;105:193–195. doi: 10.1016/s0926-6593(65)80190-4. [DOI] [PubMed] [Google Scholar]
- Shoichet B.K. Screening in a spirit haunted world. Drug Discov. Today. 2006;11:607–615. doi: 10.1016/j.drudis.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Dehdarani A.H., Yonezawa S., Tang J. Porcine spleen cathepsin B is an exopeptidase. J. Biol. Chem. 1986;261:9375–9381. [PubMed] [Google Scholar]
- Turk D., Podobnik M., Kuhelj R., Dolinar M., Turk V. Crystal structures of human procathepsin B at 3.2 and 3.3 Å resolution reveal an interaction motif between a papain-like cysteine protease and its propeptide. FEBS Lett. 1996;384:211–214. doi: 10.1016/0014-5793(96)00309-2. [DOI] [PubMed] [Google Scholar]
- Yan S.Q., Sloane B.F. Molecular regulation of human cathepsin B: Implication in pathologies. Biol. Chem. 2003;384:845–854. doi: 10.1515/BC.2003.095. [DOI] [PubMed] [Google Scholar]
- Zwicky R., Müntener K., Csucs G., Goldring M.B., Baici A. Exploring the role of 5′-alternative splicing and of the 3′-untranslated region of cathepsin B mRNA. Biol. Chem. 2003;384:1007–1018. doi: 10.1515/BC.2003.113. [DOI] [PubMed] [Google Scholar]