Abstract
The aggregation of antitrypsin into polymers is one of the causes of neonatal hepatitis, cirrhosis, and emphysema. A similar reaction resulting in disease can occur in other human serpins, and collectively they are known as the serpinopathies. One possible therapeutic strategy involves inhibiting the conformational changes involved in antitrypsin aggregation. The citrate ion has previously been shown to prevent antitrypsin aggregation and maintain the protein in an active conformation; its mechanism of action, however, is unknown. Here we demonstrate that the citrate ion prevents the initial misfolding of the native state to a polymerogenic intermediate in a concentration-dependent manner. Furthermore, we have solved the crystal structure of citrate bound to antitrypsin and show that a single citrate molecule binds in a pocket between the A and B β-sheets, a region known to be important in maintaining antitrypsin stability.
Keywords: antitrypsin, polymerization, protein aggregation, protein unfolding, serpin
The conformational diseases, which include the serpinopathies, Alzheimer's, and Huntington's disease, result from an inappropriate conformational change leading to self-association and aggregation (Carrell and Lomas 1997). Antitrypsin (AAT) deficiency affects one in 2000 people and is linked to both liver cirrhosis and emphysema. This disease results from mutations within the AAT gene that promote aberrant conformational changes within the protein, resulting in its aggregation (Lomas et al. 1992; Devlin and Bottomley 2005). These aggregates, characterized as an ordered polymeric structure, accumulate in the endoplasmic reticulum (ER) and lead to cell damage or death, which, in turn, lead to liver damage. Emphysema is a downstream result of AAT accumulation in the ER as less AAT is secreted into circulation, thus reducing the overall inhibitory capabilities of the plasma. Target proteinases of AAT, such as elastase, then operate unhindered leading to destruction of the connective tissue within the lungs and subsequently to emphysema.
AAT aggregation involves a number of steps (James and Bottomley 1998; Dafforn et al. 1999; Devlin et al. 2002): (1) The conformational change step where the native state adopts a highly polymerogenic species termed M*, (2) M* then self-associates to form dimers, and (3) dimer–dimer self-association occurs to form long chain polymers. Preventing any of these three steps represents a plausible mechanism of inhibiting AAT aggregation. A number of approaches have shown potential in inhibiting AAT aggregation; these include chemical chaperones that stabilize the native state and peptides that block the self-association reaction (Chow et al. 2001; Devlin et al. 2001; Mahadeva et al. 2002; Zhou et al. 2004). In addition to these, it is well established that the citrate ion stabilizes AAT against aggregation through an undefined mechanism (Tew and Bottomley 2001b). In this study we reveal the molecular mechanism by which the citrate ion prevents AAT aggregation, demonstrating that it binds to the native state and dramatically slows down the first step of the aggregation pathway.
Results
Citrate stabilizes AAT against unfolding and polymerization
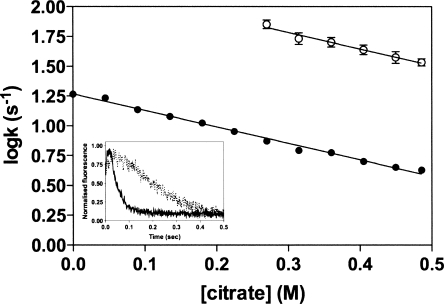
It has been demonstrated previously that sodium citrate reduces the extent of AAT aggregation by binding to and stabilizing native AAT (Tew and Bottomley 2001b). We have extended these studies by examining the unfolding kinetics of AAT in the presence of increasing concentrations of citrate in an attempt to better understand the nature of the stabilizing reaction. In these experiments, native AAT was unfolded in 5 M GdnHCl and the change in tryptophan fluorescence was monitored, over time, as a probe of conformational change. Consistent with previous studies (Pearce et al. 2000), our data reveals that in the absence of citrate, a hyperfluorescent intermediate species is formed within the dead time of the machine, which then decays to the unfolded form with an apparent relaxation rate constant of 18.5 s−1 (Fig. 1). In the presence of 0.5 M citrate, two kinetic phases are detected during unfolding: a rapid increase in fluorescence corresponding to the formation of the hyperfluorescent intermediate (kNI) and then a slow phase (kIU) corresponding to formation of the unfolded state (Fig. 1). With increasing citrate concentrations, the rate of unfolding in 5 M GdnHCl is progressively slowed and both rates display a similar dependency on citrate concentration (Fig. 1). Linear regression of the data indicated that both kNI and kIU were equally reduced by increasing citrate concentrations, with the dependency calculated as −1.38 M−1s−1 for both. This suggests that citrate binds to both the native and intermediate states of AAT. In buffer alone, or low citrate concentrations (<0.25 M), the first phase was extremely rapid and could not be determined.
Figure 1.
Effect of citrate concentration on rate of AAT unfolding. Plots of the logarithm of the initial rate of fluorescence increase (○) and the slower rate of fluorescence decrease (•) versus citrate concentration. The inset shows the unfolding of AAT (1 μM), monitored by rapid dilution into 5.5 M GdnHCl, in the presence (solid line) and absence (dashed line) of 0.5 M citrate. Unfolding was followed by monitoring the changes in intrinsic tryptophan fluorescence (excitation wavelength at 280 nm; emission wavelength at 330 nm).
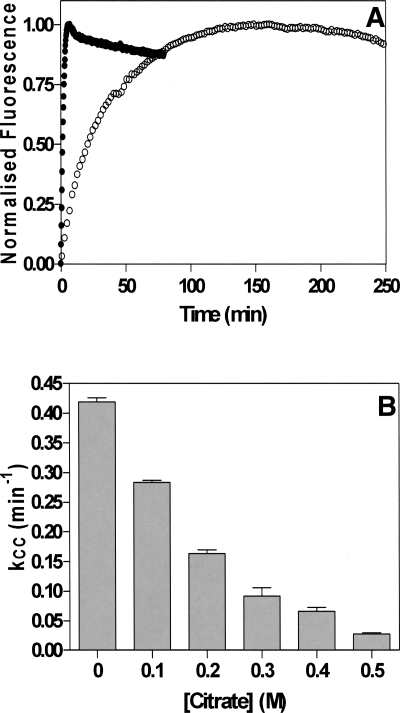
The first step of AAT polymerization requires partial unfolding of the native structure to a state termed M*, which occurs in a concentration-independent manner defined by the apparent rate constant kcc (the rate of conformational change) (Cabrita et al. 2004). Using bis-ANS, it is possible to monitor the formation of M*, as the fluorescence intensity of this probe is significantly increased in the presence of M* (James and Bottomley 1998). The second phase of aggregation, the self-association of M*, results in a decrease in bis-ANS fluorescence and is defined by the apparent rate constant kagg (the rate of aggregation) (Cabrita et al. 2004). These two phases of AAT aggregation can clearly be seen in the absence of citrate (Fig. 2A). In the presence of citrate, however, kcc is significantly reduced; for example, in 0.5 M citrate, kcc = 0.02 s−1 compared with a kcc of 0.4 s−1 in the absence of citrate (Fig. 2B). The self-association phase of AAT polymerization is extremely slow and the amplitude of the change is greatly reduced in the presence of citrate and could not be measured accurately using this assay.
Figure 2.
Polymerization of AAT in the presence of citrate. (A) AAT was incubated at 60°C in a bis-ANS solution either with (○) or without (•) 0.5 M citrate present. (B) The initial rate of conformational change, kCC, versus citrate concentration. Each bar represents the average of five separate experiments.
Crystal structure of citrate bound to AAT
Our data strongly suggests that citrate reduces AAT polymerization by stabilizing and inhibiting the unfolding of native AAT. In order to understand this interaction further, we set out to determine the crystal structure of an AAT–citrate complex. Attempts to crystallize AAT in the presence of citrate were unsuccessful. This is perhaps not surprising, as the high concentrations of citrate would likely mask surface residues that have the potential to make crystal contacts. We then attempted to grow AAT crystals using the published conditions (Elliott et al. 2000) for the purpose of soaking citrate into the crystals. These attempts proved unsuccessful, so we searched for new crystallization conditions. A novel crystal form of AAT was discovered, which we call form B. These crystals were used for citrate soaking experiments. Very recently we discovered new crystals of AAT by a slight alteration of the existing form B conditions and surprisingly found that these crystals (which we call form A) appeared very similar to the published ones.
Crystal form A belongs to the C2 space group published previously (Elliott et al. 2000), while crystal form B grows in the P212121 space group. We determined the AAT structure from both crystal forms by molecular replacement and the two new apo structures were compared with the previously published apo structure (PDB id: 1QLP) (Elliott et al. 2000). The new structures are very similar to each other despite the different crystal contacts and show no significant deviations to the published structure (Cα RMS deviations of 0.3 Å and 0.4 Å, respectively) except for differences in two mobile loop regions and the presence of an oxidized cysteine (Cys232) in the new structures.
Experiments with preformed AAT crystals (crystal form B) soaked in varying concentrations of citrate indicated that the crystals only tolerated relatively low concentrations of citrate; crystals placed in solutions containing more than 10 mM citrate rapidly dissolved. At 10 mM citrate, however, the crystals proved to be stable for extended periods (more than 1 wk). Stepwise increments of citrate concentration from 10 to 100 mM were tested with citrate concentrations >10 mM, causing a loss of diffraction. The structure of the AAT–citrate complex was determined from a crystal soaked in 10 mM citrate for 8 d.
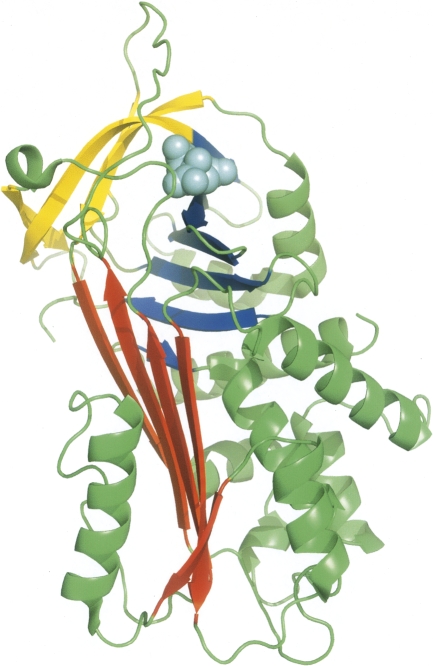
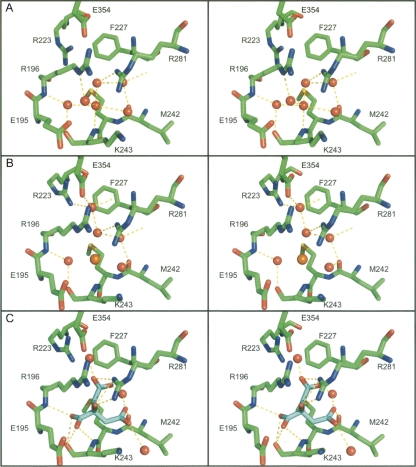
An overview of the AAT–citrate complex structure is shown in Figure 3. Introduction of the citrate ion has had minimal direct effect on the overall protein structure (Cα RMSD of 0.2 Å compared with crystal form B apo structure) or on the residue side chains surrounding the citrate ion as it occupies a void in the protein normally occupied by four water molecules between sheets B and C. The citrate molecule forms a network of hydrogen bonds and ionic interactions with the residues in the binding site (Fig. 4), not only mimicking the interactions previously generated by the displaced water molecules, but forming many new interactions. The citrate ion forms interactions directly with the side chains of two residues (Arg223 and Arg281) and the backbone of two others (Arg196 and Lys243), as well as indirectly via bridging water molecules to the side chains of Glu195 and Glu354 and backbones of Met226, Asn228, and Leu241. There are also van der Waals contacts between the citrate carbon atoms and residues Glu195, Phe227, and Met242. This extensive network of interactions bridging residues in separate sections of the B/C barrel would be expected to act as a local conformational lock, since the citrate ion would have to be ejected before any rearrangement of the pocket could occur.
Figure 3.
Diagrammatic representation of the three-dimensional structure of the AAT–citrate complex. The protein is shown in ribbon style, with sheet A in blue, sheet B in red, sheet C in yellow, the reactive center loop in green, and the citrate ion is shown as cyan-colored spheres.
Figure 4.
Analysis of the citrate-binding pocket. Stereoscopic figure of residues contributing to the citrate-binding pocket in (A) crystal form A, (B) crystal form B, and (C) the citrate complex. In all three, the protein is shown as sticks colored by atom type (carbon: green, oxygen: red, nitrogen: blue, sulfur: yellow), with water molecules shown as red spheres. In crystal form B, the chloride ion is shown as an orange sphere (B) while in the citrate complex (B) the citrate is shown as sticks colored by atom type, except that the carbon atoms are in cyan. Comparison of the three sites shows that minimal rearrangement of the site has occurred to adapt to citrate binding.
In the published structure of AAT, the citrate binding site is filled with a network of six water molecules (PDB id: 1QLP) (Elliott et al. 2000). Five of these water molecules are found in our form A structure, and four of them are conserved in our form B structure. One of the water molecules seen in the published and form A structures is replaced by a chloride ion in form B. The potential role of cavities in the stability and folding of AAT has been discussed previously (Elliott et al. 2000; Lee et al. 2001; Parfrey et al. 2003). The citrate site, however, has not been identified in these earlier works; for example, Elliott and colleagues focused on four sites (not including the citrate site) that are conserved between different serpin structures (Elliott et al. 2000), while Parfrey and coworkers targeted a specific site between sheet A and helices D and E for mutagenic analysis (Parfrey et al. 2003).
Citrate binds to a pocket in the B/C barrel of AAT
This work indicated that citrate may exert its protective effects on AAT via binding to specific sites within the B/C barrel of AAT. A number of studies have shown that this region is proposed to play a crucial role in establishing and maintaining serpin stability (Seo et al. 2000; Tew and Bottomley 2001a). The structures described here suggested a number of residues that may be responsible for binding the citrate ion as well as identifying residues that make up the binding pocket. To confirm this, we made the following single point mutations (R196A, R223A, M226A, F227A, R281A, and R281E) to test the importance of the pocket for AAT folding and stability. All of these proteins were expressed as inclusion bodies, and despite repeated attempts using a number of different methods, we were unable to refold three of the proteins (R223A, M226A, and F227A) to a monomeric native state.
The other three proteins (R281A, R281E, and R196A) were fully active and retained their susceptibility to the stabilizing effects of citrate (data not shown). The result for the R196A mutant confirms that the side chain is neither critical for stabilizing the pocket, nor is involved in binding to the citrate ion, thus confirming the crystal structure result that suggested that the interaction between bound citrate and Arg196 is solely through the backbone amide of the residue. The lack of a change in either of the Arg281 mutants is surprising, as the residue is one of those that directly interact with citrate in the bound complex structure. Inspection of the AAT structure reveals that Arg281 is a fully exposed surface residue and that a minor rearrangement of the Arg196 side chain, impossible in the presence of Arg281, could compensate for any interactions lost in substitution of Arg281.
The failure of many of the mutants to refold properly highlights the sensitivity of the citrate-binding pocket for AAT folding and stability. Combinations of two or more mutations in this site were also made in an attempt to abrogate the citrate effect; in no construct with multiple substitutions could we obtain properly folded protein.
Conclusion
We show that citrate binds to both the native and intermediate conformations of AAT, and in so doing can prevent unfolding and aggregation. Crystallography of AAT with the protective molecule citrate bound reveals that this is mediated through residues within the B/C barrel interacting with the citrate molecule. Our data indicate that the contacts formed in the native state are maintained in the intermediate conformation. These data suggest that the B/C barrel remains intact in both the native and intermediate states, which is consistent with previous studies suggesting that this region forms a stable platform upon which all conformational changes, inhibitory and misfolding, take place.
Materials and Methods
Mutagenesis and purification of AAT
Variants of AAT were generated using the Stratagene Quikchange site-directed mutagenesis method. Recombinant AAT was purified as previously described (Bottomley and Stone 1998; Bird et al. 2004).
Unfolding kinetics
AAT (10 μM) was rapidly mixed at a 10:1 ratio with 5.5 M GdnHCl. Both protein and denaturant were prepared in varying concentrations of sodium citrate, between 0 and 0.5 M. The final concentrations of denaturant and protein in the reaction mixture were 5 M and 1 μM, respectively. Samples were rapidly mixed and the change in fluorescence measured using an Applied Photophysics SV18 stopped flow fluorimeter at 22°C. Excitation and emission wavelengths were 280 nm and 330 nm, respectively, with slit widths of 3 mm used. The experimental data were fit to a double exponential equation, using the manufacturer's software.
Polymerization assays
For each polymerization assay the buffer (50 mM Tris-Hcl , pH 7.4, 100 mM NaCl) was preheated to 60°C and the reaction started by the addition of AAT. Polymerization was followed using bis-ANS as previously described (James and Bottomley 1998; Cabrita et al. 2004).
Crystallography of AAT
Recombinant AAT was crystallized by the hanging-drop vapor diffusion method. The C2 crystal form (crystal form A) was generated from an initial protein solution of 50 mM Tris-HCl pH 8.0, 75 mM NaCl at a protein concentration of 3 mg/mL. Crystals grew in 29% (w/v) PEG 3350 and 400 mM NaF at room temperature with rod-shaped crystals appearing after 1 d, growing to full size (0.2 mm × 0.2 mm × 0.5 mm) within 2 wk. Crystals were cryoprotected prior to data collection by brief transfer to buffer containing 10% (v/v) glycerol, 30% (w/v) PEG 3350, and 367 mM NaF. Data were collected at 100 K on an ADSC CCD detector on the BioCARS beamline 14-BM-C at the Advance Photon Source.
For crystal form B, the initial protein solution consisted of 50 mM Tris-HCl pH 8.0, 75 mM NaCl at a protein concentration of 8 mg/mL. The best crystals grew in 27% (w/v) PEG 4000, 0.2 M sodium acetate and 100 mM Tris-HCl pH 8.4 at room temperature (22°C) with rod-shaped crystals appearing after 1 d and grew to full size (up to 0.1 mm × 0.08 mm × 0.5 mm) within 5 d.
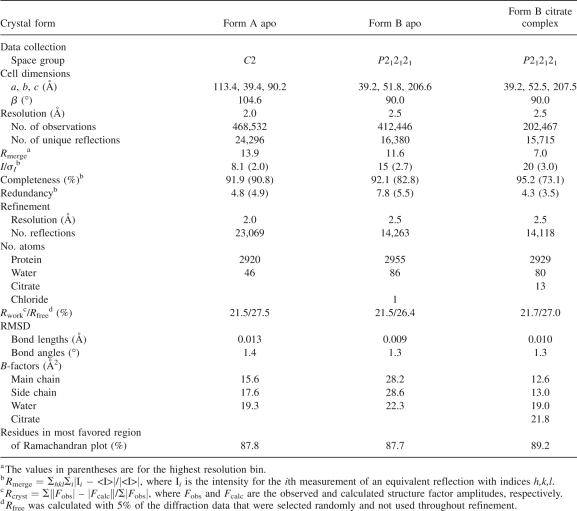
Citrate was introduced into preformed form B crystals by soaking in 10 mM sodium citrate, 27% (w/v) PEG 4000, 0.2 M sodium acetate, and 100 mM Tris pH 8.4 for 8 d. Prior to data collection, the crystals were briefly (∼5 sec) transferred to a cryoprotectant buffer containing 5% (v/v) glycerol, 27% (w/v) PEG 4000, 0.2 M sodium acetate, and 100 mM Tris-Cl pH 8.4. The X-ray diffraction data from apo form B crystals were collected on BioCARS beamline 14-BM-D at the Advanced Photon Source from a single frozen crystal at 100 K. The X-ray diffraction data for the citrate-soaked form B crystals were collected at 100 K using a MARResearch imaging plate detector with CuK α X-rays generated by a Rigaku RU-200 rotating anode X-ray generator. Diffraction data were processed and analyzed using the HKL (Otwinowski and Minor 1997) and Collaborative Computational Project, Number 4 (1994) packages. Data collection statistics are shown in Table 1.
Table 1.
Data collection and refinement statistics
Structure determination
Refinement of the structure began with the published apo model of AAT (PDB id: 1QLP) (Elliott et al. 2000) with water molecules omitted. For crystal form A (which has the same C2 space group and cell dimensions of the published structure), initial phases were determined by molecular replacement with AMoRe (Navaza 2001). Rigid body refinement of the starting model in REFMAC (Murshudov et al. 1997) was used to compensate for changes in the crystal packing environment, and then the model was subject to restrained refinement. Waters and cysteine-S-dioxide (Cys232) were added using COOT (Emsley and Cowtan 2004) before further rounds of refinement. Tightly restrained individual B-factor refinement was used and a bulk solvent correction applied. Using TLS refinement (Winn et al. 2001; Painter and Merritt 2006) in REFMAC, the model has been refined to a crystallographic R factor of 21.5% (R free of 27.5%) for all data to 2.0 Å. The final model includes residues 24–392 and 46 water molecules.
The initial phases for crystal form B were obtained by molecular replacement with AMoRe (Navaza 2001) as described for crystal form A. Rigid body refinement of the starting model in REFMAC (Murshudov et al. 1997) was used to compensate for changes in the crystal packing environment, and then the model was subject to restrained refinement. Waters, cysteine-S-dioxide (Cys232), a chloride ion (for the apo-protein), and citrate (for the citrate complex) were added using COOT (Emsley and Cowtan 2004) before further rounds of refinement as described for crystal form A. A chloride ion was built into the citrate-binding pocket of the apo-protein structure to improve the fit for density originally attributed to a water molecule. The identity of the chloride was suggested by examination of the position and geometry of the site; this ion is not apparent in crystal form A. The chloride ion binds to the backbone amide of Lys243 and the side chain of Arg281. After final TLS refinement in REFMAC, the R factor for the apo-protein in crystal form B was 21.5% (R free of 26.4%) for all data to 2.5 Å, and for the citrate complex was 21.7% (R free of 27.0%) for all data to 2.5 Å. The final apo-protein model includes residues 24–394, one chloride ion, and 86 water molecules, while the final citrate-complex structure encompasses residues 24–393 and 81 water molecules. The position of the citrate molecule was confirmed by examination of an omit map in the region surrounding the citrate binding site.
Refinement statistics for all three structures are shown in Table 1. The structures were assessed for stereochemical quality with PROCHECK (Laskowski et al. 1993) and gave results in the range expected for well-refined structures at their respective resolutions. In all three structures, all residues lie within the allowed regions of the Ramachandran plot, with only two residues in the generously allowed region for both crystal form B structures. The two apo structures superpose closely with the published structure with overall RMSDs of 0.3 and 0.4 Å for all Cα atoms in crystal forms A and B, respectively. The major outliers (>1.5 Å) occur in two regions of the protein, between residues 107 and 109, and in the mobile reactive center loop and between residues 344 and 349. These are regions of relatively poor electron density that reflects local disorder within the structures.
Protein Data Bank accession numbers
Atomic coordinates and structure factors (accession codes 2QUG, 3CWL, and 3CWM for the apo crystal form A, apo crystal form B, and citrate complexes, respectively) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org).
Acknowledgments
We thank Alistair Draffan, John Lambert, and Simon Tucker from Biota Holdings Limited for advice and encouragement. We also thank Harry Tong and other BioCARS staff for their help at the Advanced Photon Source. This work was supported by the Australian Synchrotron Research Program, which is funded by the Commonwealth of Australia under the Major National Research Facilities Program. Use of the Advanced Photon Source was supported by the U.S. DOE, Basic Energy Sciences, Office of Energy Research. This work was also supported by grants from the Australian Research Council (ARC) and the National Health and Medical Research Council (NHMRC). S.C.F. was supported by a NHMRC Industry Fellowship. M.W.P. is an ARC Federation Fellow and an NHMRC Honorary Fellow. S.P.B. is a NHMRC Senior Fellow.
Footnotes
Reprint requests to: Stephen P. Bottomley, Department of Biochemistry and Molecular Biology, PO Box 13D, Monash University, Clayton, Victoria 3800, Australia; e-mail: steve.bottomley@med.monash.edu.au; fax: 61-3-99054699.
Abbreviations: AAT, α1-antitrypsin; GdnHC1, guanidinium hydrochloride; M*, partial unfolded state of AAT.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.037234.108.
References
- Bird P.I., Pak S.C., Worrall D.M., Bottomley S.P. Production of recombinant serpins in Escherichia coli . Methods. 2004;32:169–176. doi: 10.1016/s1046-2023(03)00208-1. [DOI] [PubMed] [Google Scholar]
- Bottomley S.P., Stone S.R. Protein engineering of chimeric serpins: An investigation into effects of the serpin scaffold and reactive centre loop length. Protein Eng. 1998;11:1243–1247. doi: 10.1093/protein/11.12.1243. [DOI] [PubMed] [Google Scholar]
- Cabrita L.D., Dai W., Bottomley S.P. Different conformational changes within the F-helix occur during serpin folding, polymerization, and proteinase inhibition. Biochemistry. 2004;43:9834–9839. doi: 10.1021/bi0491346. [DOI] [PubMed] [Google Scholar]
- Carrell R.W., Lomas D.A. Conformational disease. Lancet. 1997;350:134–138. doi: 10.1016/S0140-6736(97)02073-4. [DOI] [PubMed] [Google Scholar]
- Chow M.K., Devlin G.L., Bottomley S.P. Osmolytes as modulators of conformational changes in serpins. Biol. Chem. 2001;382:1593–1599. doi: 10.1515/BC.2001.194. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Dafforn T.R., Mahadeva R., Elliott P.R., Sivasothy P., Lomas D.A. A kinetic mechanism for the polymerization of α1-antitrypsin. J. Biol. Chem. 1999;274:9548–9555. doi: 10.1074/jbc.274.14.9548. [DOI] [PubMed] [Google Scholar]
- Devlin G.L., Bottomley S.P. A protein family under “stress”—serpin stability, folding and misfolding. Front. Biosci. 2005;10:288–299. doi: 10.2741/1528. [DOI] [PubMed] [Google Scholar]
- Devlin G.L., Parfrey H., Tew D.J., Lomas D.A., Bottomley S.P. Prevention of polymerization of M and Z α1-antitrypsin (α1-AT) with trimethylamine N-oxide. Implications for the treatment of α1-AT deficiency. Am. J. Respir. Cell Mol. Biol. 2001;24:727–732. doi: 10.1165/ajrcmb.24.6.4407. [DOI] [PubMed] [Google Scholar]
- Devlin G.L., Chow M.K., Howlett G.J., Bottomley S.P. Acid Denaturation of α1-antitrypsin: Characterization of a novel mechanism of serpin polymerization. J. Mol. Biol. 2002;324:859–870. doi: 10.1016/s0022-2836(02)01088-4. [DOI] [PubMed] [Google Scholar]
- Elliott P.R., Pei X.Y., Dafforn T.R., Lomas D.A. Topography of a 2.0 Å structure of α1-antitrypsin reveals targets for rational drug design to prevent conformational disease. Protein Sci. 2000;9:1274–1281. doi: 10.1110/ps.9.7.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- James E.L., Bottomley S.P. The mechanism of α1-antitrypsin polymerization probed by fluorescence spectroscopy. Arch. Biochem. Biophys. 1998;356:296–300. doi: 10.1006/abbi.1998.0751. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Lee C., Sun E.J., Yu M.-H. Role of the connectivity of secondary structure segments in the folding of α1-antitrypsin. Biochem. Biophys. Res. Commun. 2001;287:636–641. doi: 10.1006/bbrc.2001.5638. [DOI] [PubMed] [Google Scholar]
- Lomas D.A., Evans D.L., Finch J.T., Carrell R.W. The mechanism of Z α1-antitrypsin accumulation in the liver. Nature. 1992;357:605–607. doi: 10.1038/357605a0. [DOI] [PubMed] [Google Scholar]
- Mahadeva R., Dafforn T.R., Carrell R.W., Lomas D.A. 6-Mer peptide selectively anneals to a pathogenic serpin conformation and blocks polymerization. Implications for the prevention of Z α1-antitrypsin-related cirrhosis. J. Biol. Chem. 2002;277:6771–6774. doi: 10.1074/jbc.C100722200. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in the oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Painter J., Merritt E.A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- Parfrey H., Mahadeva R., Ravenhill N.A., Zhou A., Dafforn T.R., Foreman R.C., Lomas D.A. Targeting a surface cavity of α1-antitrypsin to prevent conformational disease. J. Biol. Chem. 2003;278:33060–33066. doi: 10.1074/jbc.M302646200. [DOI] [PubMed] [Google Scholar]
- Pearce M.C., Rubin H., Bottomley S.P. Conformational change and intermediates in the unfolding of α1-antichymotrypsin. J. Biol. Chem. 2000;275:28513–28518. doi: 10.1074/jbc.M004310200. [DOI] [PubMed] [Google Scholar]
- Seo E.J., Im H., Maeng J.S., Kim K.E., Yu M.H. Distribution of the native strain in human α1-antitrypsin and its association with protease inhibitor function. J. Biol. Chem. 2000;275:16904–16909. doi: 10.1074/jbc.M001006200. [DOI] [PubMed] [Google Scholar]
- Tew D.J., Bottomley S.P. Probing the equilibrium denaturation of the serpin α1-antitrypsin with single tryptophan mutants; evidence for structure in the urea unfolded state. J. Mol. Biol. 2001a;313:1161–1169. doi: 10.1006/jmbi.2001.5104. [DOI] [PubMed] [Google Scholar]
- Tew D.J., Bottomley S.P. The citrate ion increases the conformational stability of antitrypsin. Biochim. Biophys. Acta. 2001b;1481:11–17. doi: 10.1016/s0167-4838(00)00118-7. [DOI] [PubMed] [Google Scholar]
- Winn M.D., Isupov M.N., Murshudov G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Zhou A., Stein P.E., Huntington J.A., Sivasothy P., Lomas D.A., Carrell R.W. How small peptides block and reverse serpin polymerisation. J. Mol. Biol. 2004;342:931–941. doi: 10.1016/j.jmb.2004.07.078. [DOI] [PubMed] [Google Scholar]