Abstract
TREX1 is the major exonuclease in mammalian cells, exhibiting the highest level of activity with a 3′→5′ activity. This exonuclease is responsible in humans for Aicardi-Goutières syndrome and for an autosomal dominant retinal vasculopathy with cerebral leukodystrophy. In addition, this enzyme is associated with systemic lupus erythematosus. TREX1 belongs to the exonuclease DEDDh family, whose members display low levels of sequence identity, while possessing a common fold and active site organization. For these exonucleases, a catalytic mechanism has been proposed that involves two divalent metal ions bound to the DEDD motif. Here we studied the interaction of TREX1 with the monovalent cations lithium and sodium. We demonstrate that these metals inhibit the exonucleolytic activity of TREX1, as measured by the classical gel method, as well as by a new technique developed for monitoring the real-time exonuclease reaction. The X-ray structures of the enzyme in complex with these two cations and with a nucleotide, a product of the exonuclease reaction, were determined at 2.1 Å and 2.3 Å, respectively. A comparison with the structures of the active complexes (in the presence of magnesium or manganese) explains that the inhibition mechanism is caused by the noncatalytic metals competing with distinct affinities for the two metal-binding sites and inducing subtle rearrangements in active centers. Our analysis also reveals that a histidine residue (His124), highly conserved in the DEDDh family, is involved in the activity of TREX1, as confirmed by mutational studies. Our results shed further light on the mechanism of activity of the DEDEh family of exonucleases.
Keywords: exonucleases, TREX1, DEDDh family, structure, enzymatic activity
The 3′→5′ exonucleases play a crucial role in many biological processes by removing nucleotides at the 3′ termini from DNA ends, which must then be remodeled in order to prevent the formation of aberrant structures. The 3′→5′ exonucleases were initially found as proofreading enzymes associated with DNA polymerases. However, in recent years several eukaryotic 3′→5′ exonucleases, such as p53, the Werner syndrome protein (WRN), and MRE11, have been reported to perform vital functions for cell survival, but are unrelated to proofreading activities (Shevelev and Hubscher 2002). Alterations in the genes encoding for these autonomous exonucleases lead to dramatic consequences, including high mutation frequencies, chromosomal instability, premature aging, cell cycle checkpoint defects, susceptibility to cancer, and even lack of viability. Despite the unquestionable evidence of the in vivo importance of autonomous exonucleases, their biological roles remain to be elucidated.
Enzymes from the DEDD 3′→5′exonuclease superfamily (also named DnaQ superfamily) are characterized by four acidic residues, three aspartates (D), and one glutamate (E), distributed in three separate sequence segments (Exo I, Exo II, and Exo III) (Moser et al. 1997). Residues from this DEDD motif comprise binding sites A and B for two divalent metal ions that are essential for catalysis (Derbyshire et al. 1995). A fifth residue, either a tyrosine or a histidine, also located at the active sites, completes the catalytic characterization represented by the DEDDy and DEDDh families, respectively. The overall structure of DEDDh family members is well preserved, although sequence similarities can be very low outside of the Exo segments (Moser et al. 1997; Zuo and Deutscher 2001). Within this family, a high structural similarity of the active-site centers suggests a common catalytic mechanism. Through this mechanism, initially proposed for the Klenow exonuclease fragment of Escherichia coli, a water molecule is activated by the conserved histidine from the motif and by the magnesium (or manganese) at site A to form a hydroxide ion, which would carry out a nucleophilic attack on the phosphorus atom of the 3′-terminal phosphodiester group, thereby contributing a 5′-monophosphate oxygen to the mononucleotide product (Brautigam et al. 1999a; Hamdan et al. 2002b).
TREX1, an exonuclease from the DEDDh family, provides the highest exonuclease activity 3′→5′ in mammals' cells (Lindahl et al. 1969; Perrino et al. 1994, 1999). TREX1 knock-out mice develop inflammatory myocarditis, resulting in progressive cardiomyopathy and a dramatic reduction in survival (Morita et al. 2004). TREX1 is also directly involved in a severe neurological brain disease, the Aicardi-Goutières syndrome (Crow et al. 2006), an autosomal dominant retinal vasculopathy with cerebral leukodystrophy (Richards et al. 2007), and is related to systemic lupus erythematosus (Lee-Kirsch et al. 2007a,b; Rice et al. 2007). TREX1 acts within the SET complex in the degradation of the DNA during granzyme A-mediated cell death (Chowdhury et al. 2006) and degrades ssDNAs that prevent chronic checkpoint activation and autoimmune disease (Yang et al. 2007). Taken together, all of the information suggests that TREX1 is required for the proper maintenance of the nonprocessed DNA intermediates produced in replication or repair events.
Here we report the inhibition of the exonucleolytic activity of TREX1 by the monovalent cations lithium and sodium. Lithium is used for the treatment of bipolar disorder and it has been recently demonstrated that it alters a number of crucial cellular responses (Shaldubina et al. 2001). The action of lithium has been well documented in phosphoryl-transfer enzymes such as kinases and phosphatases (Zhen et al. 2002; Ikeda and Kato 2003), but no effect has been previously shown in other mammalian phosphoryl-transfer enzymes such as nucleases. However, it has been proposed that lithium is toxic in yeast by inhibiting 5′→3′ exoribonucleases (Dichtl et al. 1997). On the basis of this observation, similar mechanisms may contribute to the effects of lithium in human neurobiology, or the use of lithium to treat bipolar disorder could produce a degree of toxicity or side effects. The inhibition of TREX1 exerted by sodium has not been previously reported. Several structural enzymological studies performed on DNA exonucleases have contributed to a better understanding of the catalytic mechanism of these enzymes (Brautigam and Steitz 1998; Brautigam et al. 1999a). However, no information is available on the inhibitory properties of monovalent ions, such as lithium and sodium. Here we determined the crystal structures of the exonuclease complex with these two metals at 2.1 Å and 2.3 Å, respectively. Comparison with the recently reported structures of catalytic magnesium and manganese complexes (Brucet et al. 2007), together with mutational studies, has allowed us to explain the inhibitory mechanisms of monovalent ions and identify the involvement of a new histidine residue (His124), which is highly conserved in the DEDDh family, in TREX1 activity. We developed a new technique, using SYBR Green, to monitor the exonuclease reaction in real time. Our results provide new insights into the molecular basis for the function and inhibition of exonucleases belonging to the DnaQ superfamily.
Results
Inhibition of TREX1 by monovalent cations
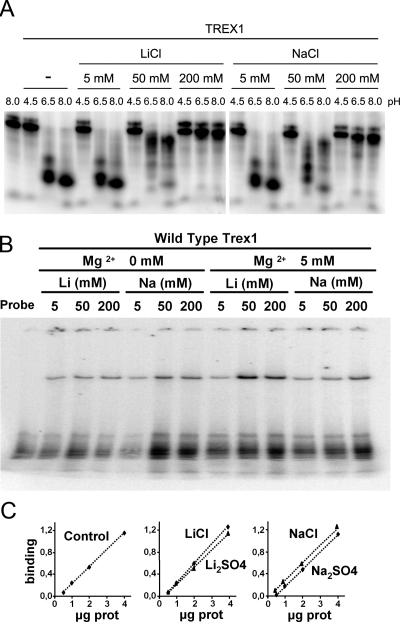
The 3′→5′ exonucleases catalyze the hydrolysis of DNA via hydrolysis of the 3′ phosphodiester bond and the release of the 3′ nucleotide. The inhibitory properties that monovalent cations, such as lithium and sodium, exert in the hydrolysis of phosphodiester bonds has been reported for other phosphoryl-transfer enzymes, such as the Mg2+-dependent phosphatases (Albert et al. 2000; Patel et al. 2002). Exonucleases and phosphatases share some catalytic properties, in particular the requirement of divalent metal cations and the initiation of the hydrolytic reaction by a nucleophilic attack on a water molecule (Brautigam et al. 1999a,b; Hamdan et al. 2002b; Patel et al. 2002). Our activity analysis shows that lithium and sodium inhibit the exonucleolytic reaction of TREX1 in a similar concentration- and pH-dependent manner, though lithium is a more effective inhibitor than sodium, in particular at neutral pH values (Fig. 1A). Inhibition effects were not detected at low ion concentrations (5 mM), while partial inhibition was clear at 50 mM, in particular for lithium, with no activity being observed at 200 mM concentrations. Similar inhibitions were obtained if we replaced LiCl and NaCl with Li2SO4 or Na2SO4. The inhibition observed in TREX1 activity in the presence of lithium or sodium was not due to a loss of DNA-binding capacity, since binding assays (EMSA) performed in the presence of 200 mM Na2SO4 or 200 mM Li2SO4 showed that the DNA-binding capacity of TREX1 was maintained under these conditions (Fig. 1B,C).
Figure 1.
TREX1 inhibition by lithium and sodium ions. (A) Exonuclease reactions for the in-gel assay were prepared with 100 pg of TREX1 enzyme and 2 nM of a 25-mer oligonucleotide and performed as described in Materials and Methods. The reactions contained increasing concentrations of lithium or sodium chloride and were buffered at a range of pH values, as indicated. (B) TREX1 binds to DNA in the presence of different concentrations of Mg2+, Li, or Na. Recombinant TREX1 was incubated with a radiolabeled probe, and the complexes DNA–protein were separated in an acrylamide gel. (C) Quantitation of TREX binding to DNA in the presence of Li or Na. EMSA assays were performed with increasing amounts of the recombinant protein in the presence of 200 mM of the indicated compounds. The quantification graph indicates the relative binding of the oligonucleotide with respect to the probe alone (arbitrary units).
Crystal structures of TREX1
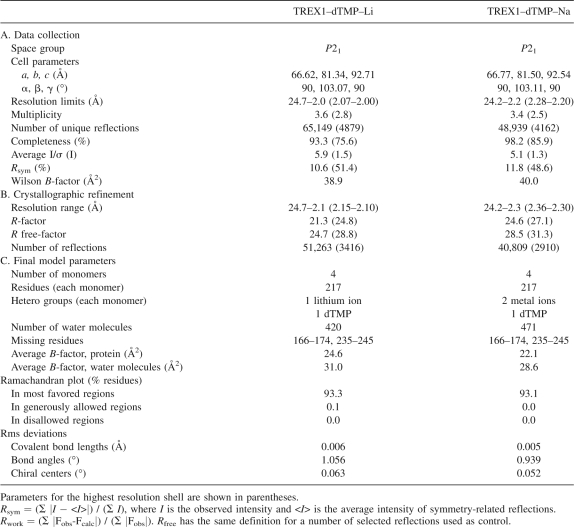
Two crystal structures of TREX1 in complex with a deoxy-thimidine-monophosphate (dTMP), a nucleotide product of the exonucleolytic reaction, were determined in the presence of either lithium or sodium (Table 1). Crystals were grown in the presence of 200 mM lithium sulfate (see Materials and Methods). For the complexes with sodium, crystals were soaked for 24 h in a solution containing an excess (200 mM) of sodium sulfate, although the presence (50 mM) of lithium sulfate was still required. The crystals were isomorphous, space group P21, and contained four protein subunits in their asymmetric unit, similarly to what was reported for the complex with magnesium (Chowdhury et al. 2006). Final models included the four subunits, with a dTMP nucleotide in each active site, and 420 or 471 solvent molecules for the lithium and sodium complexes, respectively. In the sodium and lithium complexes, TREX1 protein was organized as previously described (Mazur and Perrino 2001; Brucet et al. 2007; de Silva et al. 2007) (Fig. 2) and consisted of a molecular dimer with subunits structurally closely related to the monomeric exonucleases of the DEDDh family, despite low-sequence identities (about 20%) (1). TREX1 subunits contained a central five-stranded antiparallel β-sheet surrounded by nine α-helices. The sheet extended throughout the molecular twofold axis to the second subunit in the molecule, thereby giving a continuous 10-stranded antiparallel sheet. Within each crystal type the structures of the four subunits were very similar, with an averaged RMSD between Cα atoms of only 0.34 Å (lithium complex) and 0.24 Å (sodium complex). There was a fully disordered loop on the surface of all of the subunits, corresponding to residues Ser166–Arg174, and a partially disordered loop adjacent to a solvent-exposed proline-rich segment (residues Thr47–His53).
Table 1.
Data collection, structure solution, and refinement statistics
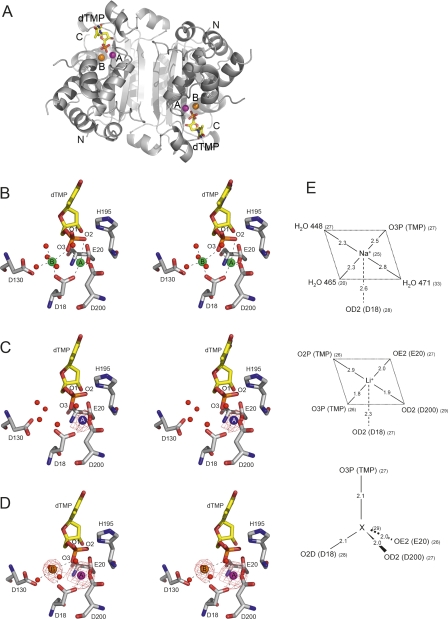
Figure 2.
TREX1–dTMP–ion complexes. (A) Overall structure of the sodium complex, representative of the organization of the three complexes (magnesium, lithium, and sodium). (B–D) Stereoviews of the metal-binding sites of the TREX1 complexes with dTMP and metal ions (B) magnesium, (C) lithium, and (D) sodium. The overall conformation of the molecular dimer of TREX1 is shown with ribbons (in gray), while dTMP molecules are represented with sticks and metal cations as spheres in green for magnesium (B), blue for lithium (C), orange and magenta for sodium and a heavier undetermined metal (X), respectively (D). The amino and carboxy ends are labeled as N and C, while the two metal-binding sites are indicated as A and B. Stereoviews of the catalytic residues from the DEDDh motif with dTMP and solvent molecules (red spheres) in the vicinity are also shown. The difference (Fo-Fc) maps at 2.5 σ are shown for the lithium and sodium structures. Coordination bonds of the cations are represented as dashed lanes. Coordinates of TREX1–dTMP–Mg were taken from the PDB code 2O4G. (E) Diagrams showing distances (in angstroms) and B-factors (in parentheses) for each ion in position A in the active center.
Active site
Comparison between the structures with lithium or sodium, or the catalytically active complexes with magnesium and manganese, indicated that differences were confined to the active-site regions (Fig. 2). The structure of the catalytic active complexes showed the binding of two magnesium (or manganese) ions and one dTMP molecule in each active site (Fig. 2A). The divalent ions were bound in the two binding sites A and B, characteristic of the DnaQ superfamily. Magnesium at site A (MgA) coordinated with oxygen atoms O2P and O3P from the dTMP phosphate group and with the carboxylate oxygen atoms from Asp18, Glu20, and Asp200 with approximate trigonal bipyramidal geometry (Brucet et al. 2007). In turn, MgB, with nearly perfect octahedral geometry, coordinated with the dTMP oxygen atom O3P, with the second carboxylate oxygen atom from Asp18 and with four water molecules, with one of the hydrogen atoms bonded to the carboxylate moiety of Asp130. Atom Nδ from His195 presented interactions with dTMP O1P and O2P, while Nε remained exposed to the solvent. Therefore, in TREX1, Asp18/Glu20/Asp130/Asp200/His195 comprised the characteristic DEDDh exonuclease motif, which presented interactions with the nucleotide similar to those determined for other members of this family (Hamdan et al. 2002b; Cheng and Patel 2004; Horio et al. 2004).
The lithium-containing complex showed, in the four crystallographically independent subunits, a peak of extra density at metal-binding site A, while no extra density was detected at site B (Fig. 2B). The extra density at site A was weak and only visible in the difference Fo-Fc map, in agreement with the low content of electrons of the lithium ion. The presence of an ion at site A could also be inferred from the proximity of the negative charges of O3P from dTMP and the carboxylate oxygen atoms from Asp18, Glu20, and Asp200. On the basis of data extracted from the Cambridge Structural Database (CSD) (Allen 2002), the geometry of these four oxygen atoms defined a tetrahedral coordination sphere well suited for a lithium ion. Confirming the presence of the lithium ion at site A, refinement proceeded smoothly when the coordinates of the ion were incorporated into the model. Therefore, site A showed some plasticity with regard to the coordination of metals of distinct sizes with either four or five ligands. This versatility may explain the inhibitory effect of lithium as resulting from competition with the catalytic metals for binding to site A. The absence of any metal ion at site B was supported by the altered geometry of this site with respect to the structure with magnesium and by the clear presence of four water molecules that occluded the space available.
Crystals soaked in the solution containing sodium sulfate showed strong peaks of extra density at metal-binding sites A and B. The ion bound at site B was coordinated to the O3P from the dTMP phosphate group, to one carboxylate oxygen atom from Asp18, and to three water molecules (Fig. 2C). Binding of sodium at site B would be in agreement with the coordination geometry for sodium, which usually presents coordination numbers of 5 or 6 (data extracted from CSD). Thermal factors in the refined model are also consistent with the presence of a sodium ion at site B. Surprisingly, the ion bound at site A showed tetra-coordinated geometry (to O3P from dTMP and to the carboxylate oxygen atoms from Asp18, Glu20, and Asp200) that is uncommon for sodium. Very low values for the thermal factors suggested the presence of an ion with a higher atomic number than sodium at site A. Since no other metal, beside sodium and lithium, were explicitly added, the binding of trace metals at site A requires affinities likely in the low nanomolar range and much higher than for lithium or sodium. Therefore, although affinity for cations at site A was always high, probably because of the concentration of negatively charged atoms around this location, its specificity was strongly modulated by the metal content at site B. With sodium at site B, the high affinity for metals with tetra-coordinated geometry at site A must compete with the binding of the catalytic cations, which would explain, at least in part, the inhibitory effects of sodium.
A new active residue: Histidine 124
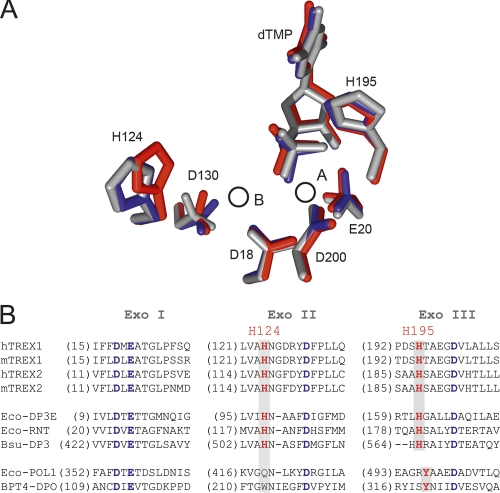
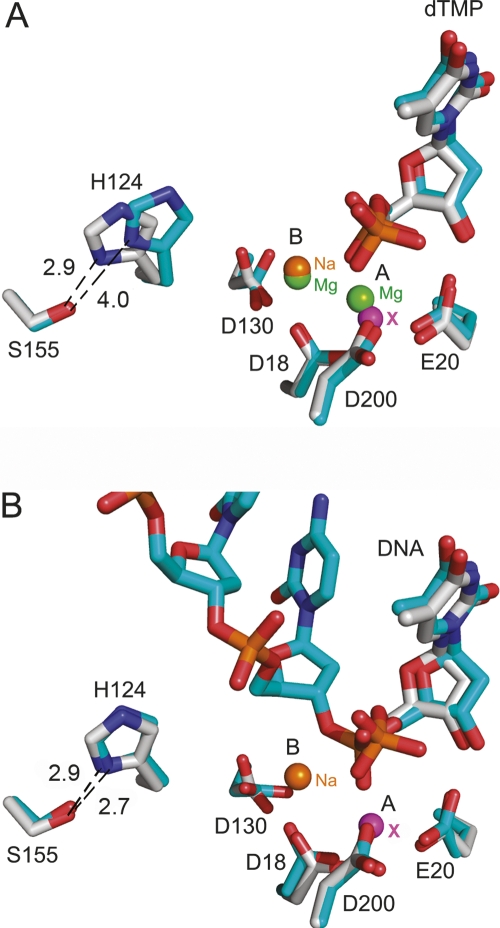
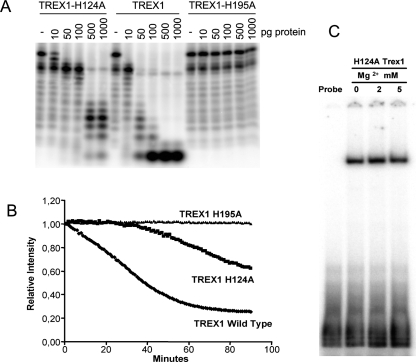
The organization of the conserved residues that comprise the DEDDh exonuclease motif was essentially identical (Fig. 3A) in the structures with magnesium (catalytically active) and with lithium or sodium (catalytically inhibited). Only Asp130 presented some variability. Asp130 interacted with the metal bound at site B only indirectly throughout coordinated water molecules, which differed depending on the complex and the cation bound. Also, the phosphate group of the dTMP was slightly closer, by about 0.7 Å, to the metal-binding site A in both the lithium and sodium complexes. However, the most conspicuous changes affected His124, a highly conserved histidine in the DEDDh family (Fig. 3B). His124 and Ser155 are highly conserved (Ser sometimes replaced by a Thr) in the DEDDh family, which is characterized by the presence of a histidine (His195 in TREX1) in the catalytic motif (Brucet et al. 2007; de Silva et al. 2007). In contrast, members of the DEDDy family, whose catalytic motif is completed by a tyrosine, lacked a histidine equivalent to His124. In the lithium and sodium structures, this histidine was displaced, with respect to its position in the magnesium complex, away from the catalytic residues. The distance between the His124 Nε atom and site B was 6.7 Å in the magnesium complex, but about 7.8 Å for both the lithium and sodium complexes, where His124 Nδ and Ser Oγ made a hydrogen bond that was absent in the magnesium complex (Fig. 4A). In the structure of TREX1 in complex with a single-stranded DNA oligonucleotide (Brucet et al. 2007), His124 presented a similar conformation, including the hydrogen bond with Ser155, to the one in the lithium or sodium structures (Fig. 4B). Therefore, during catalysis, His124 probably alternates between the two conformations, which may facilitate the release of the hydrolyzed DNA following the nonprocessive mechanism of TREX1 (Mazur and Perrino 2001). By stabilizing one of the His124 conformations, both lithium and sodium would then also interfere with the catalytic cycling. A mutant His124 to Ala was prepared to study this residue. The TREX1H124A mutant was prepared by directed mutagenesis and expressed and purified under the same conditions. The mutant protein was expressed, solubilized, and purified like the wild type, and maintained DNA-binding capacity, as shown by EMSA DNA-binding assays (data not shown). Activity analysis of the mutants H195A and H124A provided a qualitative verification of the central role of His195 and of the significant contribution of His124 to the activity of TREX1 (Fig. 5A).
Figure 3.
Histidine 124 participates in TREX1 activity. (A) Superimposition of the active site of the TREX1–dTMP–ion complexes. The conserved residues DEDDh, the His124, and the dTMP are represented as sticks and are red (magnesium complex), blue (lithium complex), or gray (sodium complex). The circles depict the positions of the metal ion binding sites A and B. (B) Sequence alignment of the exonuclease conserved motifs (Exo I, Exo II, and Exo III) of the DEDD exonucleases. Conserved residues from the DEDD motif are indicated in blue, while the catalytical histidine (His195 in TREX1) and residues corresponding to His124 in TREX1 are in red. Alignment shows that both His195 and His124 are well conserved in the DEDDh family, but absent in the DEDDy family (Eco-POL1 and BPT4-DPO). Convention used is: (m) mouse; (h) human; (Eco-DP3E) E. coli DNA pol III ε subunit; (Eco-RNT) E. coli RNase T; (Bsu-DP3) B. subtilis DNA pol III; (Eco-POL1) E. coli DNA pol I; (BPT4-DPO) bacteriophage T4 DNA pol.
Figure 4.
Histidine 124 in the inhibited and active complexes including the structure with single-stranded DNA. Active-site superimposition of the inhibited sodium complex (gray) with (A) the catalytic magnesium complex (blue) and (B) the TREX1–ssDNA complex (blue). The protein residues and the dTMP molecule are labeled and represented as atom-type sticks. The metal-binding sites A and B are also indicated, and the spheres representing the ions are labeled in green (magnesium), orange (sodium), and magenta (undetermined metal). Distances between His124 and Ser155 are clearly indicated. Coordinates of TREX1–ssDNA were taken from the PDB code 2O4I.
Figure 5.
(A) Activity of TREX1 and the variants H195A and H124A, where the exonuclease reactions were performed as described in Materials and Methods. (B) Real-time monitoring of the exonuclease reaction for TREX1, TREX1-H195A, and TREX1-H124A. DNA degradation was followed by fluorescence decay. The exonuclease reactions were performed as described in Materials and Methods. The activity was measured as the slope of each reaction curve followed by real time (product formation vs. time) vs. amount of enzyme. (C) TREX1-H124A binds to DNA in the presence of different concentrations of Mg2+.
To monitor the exonuclease in real time, we developed a rapid and sensitive technique based on SYBR Green staining of the DNA probes. This technique allows the simultaneous monitoring of a broad range of conditions without using radioactivity (Fig. 5B). In the assay, a duplex probe was used to detect DNA degradation. SYBR Green bound to the probe, which resulted in high fluorescence intensity with a maximum peak at 540 nm. When the TREX1 reaction advanced, the DNA was hydrolyzed and the SYBR Green released, resulting in the decay of the fluorescence intensity. When TREX1 was inactive, no decay in fluorescence was detected (Fig. 5B). For H195A (where His195 is mutated to Ala), fluorescence remained constant, indicating that the mutated enzyme was unable to degrade DNA. For the H124A variant, the decay of the fluorescence intensity was slower than for wild-type TREX1. This observation indicates a loss of activity and thus confirms the involvement of His124 in the activity of TREX1. We used this technique to quantify the exonuclease activity of TREX1 and the mutated TREX1-H195A and H124A proteins. The relative activity of the mutated enzymes with respect to the active TREX1 was calculated from the data extracted by real-time monitoring (see Materials and Methods) and is shown in Table 2. The H195A mutation presented a dramatic change in enzymatic activity, the TREX1-H195A enzyme being inactive under the experimental conditions. The H124A mutation had a sevenfold reduction in activity compared with the wild-type protein, thereby indicating the participation of His124 in the enzymatic activity, but not an essential role for this residue in catalysis. The inhibition of TRE1-H124A activity was not due to a loss of DNA-binding capacity (Fig. 5C).
Table 2.
Relative activity of TREX1 proteins
Discussion
Here we demonstrate that the monovalent cations, lithium and sodium, inhibit the exonucleolytic activity of TREX1, an exonuclease belonging to the DnaQ superfamily. The inhibitory effects of these two cations followed a similar concentration- and pH-dependent pattern, though lithium showed greater inhibitory capacity than sodium. We developed a new technique to monitor the exonuclease activity of TREX1. Previous techniques have been based on end-point radioactive methods. The new method we propose is rapid, inexpensive, and sensitive, and is based on SYBR Green dye fluorescence detection, which allows exonuclease activity to be followed simultaneously in real time and allows for a wide range of conditions. The technique can be used for double-stranded DNA (dsDNA) and also with 3′ mispaired or overhanging dsDNA, as SYBR Green binds only to dsDNA but not to single-stranded DNA (ssDNA). A method also based on SYBR Green dying, named ExoIII-Dye-based assay, has been successfully developed for detecting sequence-specific DNA-binding proteins (Chen et al. 2006), in which fluorescence can be related to Exo III protection by a particular binding-protein bound to a specific DNA.
Comparison of the crystal structures of TREX1 with both lithium and sodium to the available structures of TREX1 (PDB codes 2O4G and 2O4I) with the catalytic divalent cations indicate subtle rearrangements only in the active site, in particular in the vicinity of metal-binding sites A and B. Our results show that site A has high affinity for metals, probably as a result of the concentration of negatively charged atoms around this location, and the site was always found occupied by a cation. However, affinity at site A showed some flexibility with regard to coordinating metals of distinct sizes with either four or five ligands. This versatility might explain that inhibition by lithium results directly from the competition of this cation with the catalytic metals for binding to site A. Sodium shows a preference to occupy site B only, even though this binding strongly affects the coordination preferences at site A, which is then capable of binding to a trace metal. Therefore, inhibition by sodium would affect site A indirectly, which might explain the weaker inhibitory effects of this cation than lithium. The presence of other metals, such as zinc, at site A has been described, although with magnesium at site B, for the Klenow exonuclease fragment of E. coli DNA polymerase III (Brautigam and Steitz 1998; Brautigam et al. 1999b), and has been related to catalytic states of the enzyme. Inhibition of TREX1 by lithium and sodium is consistent with alterations in the two-metal-ion phosphoryl-transfer mechanism proposed for the DnaQ family (Brautigam et al. 1999a; Hamdan et al. 2002b). According to this hypothesis, the metal ion located at site A would activate an hydroxyl nucleophile from a water molecule, via reducing the pKa of the water, and the ion located at site B would stabilize the transition state intermediate and the leaving group (Steitz and Steitz 1993; Hamdan et al. 2002b). Thus, A and B sites play crucial roles in catalysis, and the chemical properties of divalent cations are essential in phosphoryl-transfer reactions. An appropriate acid hydrolysis constant is required for the cation to participate in the abstraction of a proton from a water molecule to form a nucleophile. The acid hydrolysis constants are greater for lithium and sodium than for magnesium or manganese, thereby preventing these monovalent ions from participating in forming the nucleophile. Moreover, the charge density of magnesium allows it to more readily stabilize the transition state of phosphoryl hydrolysis, while monovalent ions do not exert this stability. Recently, the currently accepted model has been discussed (Pingoud et al. 2005) and other roles have been proposed for the divalent ions, including participation in substrate recognition and product release and a more active role of site-B metal in catalysis (Yang et al. 2006). For such roles, proper size and coordination geometry of the ions is crucial for the reaction to take place. Other atoms than divalent ions magnesium or manganese, such as monovalent ions, show distinct atomic radii and a variable number of ligands. These features may explain why sodium and lithium cannot support catalysis by the two-metal-ion mechanism.
Furthermore, subtle rearrangements in the exonuclease active center caused by the presence of the monovalent cations may also contribute to inhibition. In the inhibited lithium and sodium complexes, the oxygen atoms from the dTMP-phosphate group approach the carboxylate oxygen atoms from residue Glu20. The proximity of these charged groups should alter the pKs of neighboring residues, in particular of the catalytic His195, thereby inhibiting the reaction in a similar way to what occurs at low pH. A pH dependence for other DEDDh exonucleases, such as the Klenow fragment (Brautigam and Steitz 1998), the ε186 subunit from E. coli DNA polymerase III (Hamdan et al. 2002a), and TREX2 (Mazur and Perrino 2001), has also been reported. This dependency has been related to the protonation state of the imidazole ring of the catalytic histidine (corresponding to His195 in TREX1). On the basis of the accepted catalytic mechanism for DEDDh exonucleases (Brautigam et al. 1999a; Hamdan et al. 2002b), the histidine initiates the reaction by acting as a base to deprotonate a water molecule, which will initiate a nucleophilic attack on the phosphodiester bond of the DNA substrate. Protonation of this histidine at low pH or altered pK values, induced by the monovalent cations, would impede His195 to act as a base.
The active center rearrangements of TREX1 also affect His124, a highly conserved residue in the DEDDh family, which we have confirmed, by mutational analysis, participates in exonucleolytic activity. In the complex of TREX1 with a single-stranded DNA oligonucleotide (Brucet et al. 2007), His124 present a similar conformation to those in the inhibited structures with lithium or sodium and a distinct conformation to that found in magnesium complex. Therefore, lithium and sodium stabilize only one of the several conformations that His124 requires, likely during a catalytic cycle and consequently, interfere with activity. However, other possibilities need to be taken into account such as (1) that the histidine helps in binding the ssDNA substrate; (2) that the bulky histidine side chain is required to stabilize the conformation of its backbone O atom, which makes a hydrogen bond with an inner-sphere ligand of metal ion B; and (3) that the histidine side chain is required to stabilize the conformation of the loop between b4 and a5, which could be relevant for the positioning of catalytic residue E130 and/or substrate binding.
These results provide new insights into the function and inhibition of exonucleases of the DnaQ superfamily. In particular, our findings demonstrate: (1) the central catalytic role of metal ion at site A and its strong modulation by the occupancy at site B, and (2) the significant active role of His124, which, in parallel with Ser155 (or Thr in some cases), is highly conserved in the DEDDh family.
Materials and Methods
Protein preparation
The murine wild-type TREX1 cDNA construct (pETM-10-TREX1 vector, corresponding to residues 9-245) was produced in E. coli and purified as described (Brucet et al. 2007). The cDNA for the mutants TREX1-H195A and H124A were produced from the pETM-10-TREX1 vector, using PCR site-directed mutagenesis strategy with the QuikChange Site-Directed Mutagenesis kit (Stratagene). The mutant proteins were purified by the same protocol.
Crystallization, data collection, structure determination, and refinement
Purified TREX1 protein solution (wild-type TREX1), at 8 mg/mL, was mixed with 2 mM deoxythymidine-5′-monophosphate (dTMP), left O/N at 4°C, and used for crystallization trials. For the lithium complexes, 1 μL of this solution was mixed with 1 μL of reservoir solution (22% PEG3350, 100 mM MES pH 6.0, 200 mM Li2SO4) and crystallization was carried out by the hanging-drop vapor-diffusion method at 20°C. For the sodium complexes, crystals were soaked for 24 h with the same crystallization solution, except that it contained 200 mM Na2SO4 and 50 mM Li2SO4 instead of 200 mM Li2SO4. Crystals were flash-cooled with 30% ethylene glycol as a cryoprotectant. Diffraction data were collected at the European Synchrotron Radiation Facility (Grenoble, France), and processed using DENZO and SCALEPACK (Otwinowski and Minor 1997). The first models for both complexes were obtained by molecular replacement from the TREX1–dTMP–Mg structure (PDB code 2O4G) (Brucet et al. 2007), using the TREX1 protein without any heterogroup. The models were completed and refined using programs from the CCP4 package and the graphic programs Turbo Frodo. Residues 166–174 were disordered and were therefore excluded from the model. Analysis of metal peaks using distinct types of different Fourier maps ([2Fobs-Fcalc], [Fobs-Fcalc], Omitt maps) was performed. Ramachandran plots calculated using the PROCHECK program (Laskowski et al. 1993) show that >93% of all residues in the models had Φ and Ψ angles in the most preferred regions, with no residues in the disallowed regions. Statistics for diffraction data and model refinement are summarized in Table 1. Structure representations were done with the PyMOL program (DeLano Scientific).
Radioactive exonuclease activity assay
A 25-mer single-stranded DNA (5′-GCTAGGCAGGAACCCCTCCTCCCCT-3′) was labeled with 32P, and then annealed with its complementary oligonucleotide at 1:1 molar ratio (final volume 100 μL) by heating at 95°C for 10 min and then decreasing the temperature to RT. Exonuclease reactions were performed in a final volume of 10 μL containing 20 mM Tris-HCl pH 7.5, 5 mM MgCl2, 2 mM DTT, 100 μg/mL BSA, 2 nM 5′-32P-labeled 25-mer DNA, and the indicated amount of TREX1 or mutated TREX1 enzymes (10–1000 pg, corresponding to 0.037–3.7 nM final concentration). TREX1 dilutions were prepared at 4°C in reaction buffer (20 mM Tris-HCl pH 7.5, 5 mM MgCl2, 2 mM DTT, 100 μg/mL BSA). Reactions were initiated by the addition of the enzyme, performed at RT for 30 min, and quenched by the addition of 30 μL of cold 95% ethanol. Samples were dried in vacuum, resuspended in 5 μL of formamide, and heated at 95°C for 5 min. These reaction products (all of the volume) were resolved on 22% PAGE–TBE gels containing 8 M urea. Radiolabeled bands were visualized by phosphorimaging (Molecular Dynamics). Oligonucleotides, used as substrates, were 5′-end labeled using T4 Polynucleotide Kinase (USB Corporation). For the pH-dependence studies, the conditions were the same, except for the buffering agents: 20 mM of TrisHCl (pH 6.5 or 8.0) or sodium acetate (pH 4.5) were used instead of TrisHCl pH 7.5. For the inhibition assays, 1 μL of 10× LiCl, NaCl, Li2SO4, or Na2SO4 was added to the 10-μL final reaction volume. Sodium acetate at 20 mM was used in the reaction as buffering agent. At this concentration sodium does not affect hydrolysis, as checked in previous experiments (data not shown).
SYBR Green-based exonuclease activity assay
We used the same dsDNA substrate as in the radioactive exonuclease activity assay. Exonuclease reactions were performed in a 96-well plate. First, 10× of the double-stranded DNA oligonucleotide (1 μM) was incubated with 10x SYBR Green (Invitrogen), heated for 10 min at 95°C, and left for 30 min at RT, so as to anneal the DNA probe and to allow the SYBR Green to incorporate into the DNA (the concentration of both DNA and SYBR Green was 5x). For each well, the reaction solution contained 20 mM Tris-HCl pH 7.5, 5 mM MgCl2, 2 mM DTT, and 100 μg/mL BSA and 10 ng of enzyme (24.5 nM final concentration). A mix of the reaction solution was prepared, and 12 μL of the mix was added to each well. We then added 3 μL of the SYBR Green-DNA mix to the wall of each well and, after one spin of the plate, the reaction was followed in real time with an ABIPrism 7700 sequence detection system (Applied Biosystems) (Program: 25°C 5 min, 25°C 1 min for 90 times, 4°C 2 min). TREX1 dilutions were prepared at 4°C in reaction buffer (20 mM Tris-HCl pH 7.5, 5 mM MgCl2, 2 mM DTT, 100 μg/mL BSA). The activity was measured as the slope of each reaction curve followed by real time (product formation vs. time) vs. amount of enzyme. Relative activity was calculated as the relation between the activities of each enzyme. For the pH dependence and the inhibition studies, the buffers and the inhibitors used were the same as for the assay performed in gel.
Electrophoretic mobility shift assays (EMSA)
These assays were performed as described (Casals et al. 2007). Briefly, binding reactions were prepared with 2 μg of recombinant TREX1 or the H124 mutant TREX1 and 60,000 cpm 32P-labeled probe in the presence of 2 μg of poly(dI-dC), in a final volume of 15 μL containing 3× binding buffer (12 mM HEPES, pH 7.9; 60 mM KCl; 12% glycerol, 0.12 mM EDTA; 0.3 mM PMSF, and 0.3 mM DTT) supplemented with Mg2+, Li or Na. An 8-min preincubation of extracts and poly(dI-dC) was performed. The radiolabeled probe was then added and incubated for an additional 15 min at RT. Samples were loaded onto 6% acrylamide gel containing 5% glycerol and 0.25% TBE, and electrophoresed at 4°C. Band-shift gels were dried and bands were visualized using the Phosphorimager (Molecular Dynamics). For competition experiments, 100-fold excess of unlabeled primers were included in the binding reaction. The oligonucleotides used as probes in the assay were 5′-end-labeled using T4 Polynucleotide Kinase (USB Corp.). The sequence of the used oligonucleotide is the same as that described in the SYBR Green-based exonuclease activity assay.
Protein Data Bank deposition
Coordinates have been deposited in the Protein Data Bank database with accession codes 3B6O (TREX1–dTMP–Li complex) and 3B6P (TREX1–dTMP–Na complex).
Acknowledgments
This work was supported by grant BFU2005-08686-C02-01 to I.F. and BFU2004-05725/BMC and BFU2007-63712/BMC to A.C. from the Spanish Ministerio de Educación y Cultura (MEC). We thank the Scientific and Technical Services (Genomics Unit) of the “Universitat de Barcelona” for helping in the developing of the SYBR Green assay. The technical help of Rosa Perez-Luque is highly appreciated. We also thank Tanya Yates for editing the manuscript.
Footnotes
Reprint requests to: Antonio Celada, Macrophage Biology Group (IBMB-UB), Parc Científic de Barcelona, Josep Samitier 1-5, 08028 Barcelona, Spain; e-mail: acelada@ub.edu; fax: 34-93-4034747.
Abbreviations: Exo, exonuclease motive; CSD, the Cambridge Structural Database; TREX1, three-prime repair exonuclease 1; dTMP, deoxithymidine-5′-monophosphate; MES, 4-morpholineethanesulfonic acid.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.036426.108.
References
- Albert A., Yenush L., Gil-Mascarell M.R., Rodriguez P.L., Patel S., Martinez-Ripoll M., Blundell T.L., Serrano R. X-ray structure of yeast Hal2p, a major target of lithium and sodium toxicity, and identification of framework interactions determining cation sensitivity. J. Mol. Biol. 2000;295:927–938. doi: 10.1006/jmbi.1999.3408. [DOI] [PubMed] [Google Scholar]
- Allen F.H. The Cambridge Structural Database: A quarter of a million crystal structures and rising. Acta Crystallogr. B. 2002;58:380–388. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- Brautigam C.A., Steitz T.A. Structural principles for the inhibition of the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates. J. Mol. Biol. 1998;277:363–377. doi: 10.1006/jmbi.1997.1586. [DOI] [PubMed] [Google Scholar]
- Brautigam C.A., Aschheim K., Steitz T.A. Structural elucidation of the binding and inhibitory properties of lanthanide (III) ions at the 3′–5′ exonucleolytic active site of the Klenow fragment. Chem. Biol. 1999a;6:901–908. doi: 10.1016/s1074-5521(00)80009-5. [DOI] [PubMed] [Google Scholar]
- Brautigam C.A., Sun S., Piccirilli J.A., Steitz T.A. Structures of normal single-stranded DNA and deoxyribo-3′-S-phosphorothiolates bound to the 3′–5′ exonucleolytic active site of DNA polymerase I from Escherichia coli . Biochemistry. 1999b;38:696–704. doi: 10.1021/bi981537g. [DOI] [PubMed] [Google Scholar]
- Brucet M., Querol-Audi J., Serra M., Ramirez-Espain X., Bertlik K., Ruiz L., Lloberas J., Macias M.J., Fita I., Celada A. Structure of the dimeric exonuclease TREX1 in complex with DNA displays a proline-rich binding site for WW domains. J. Biol. Chem. 2007;282:14547–14557. doi: 10.1074/jbc.M700236200. [DOI] [PubMed] [Google Scholar]
- Casals C., Barrachina M., Serra M., Lloberas J., Celada A. Lipopolysaccharide up-regulates MHC class II expression on dendritic cells through an AP-1 enhancer without affecting the levels of CIITA. J. Immunol. 2007;178:6307–6315. doi: 10.4049/jimmunol.178.10.6307. [DOI] [PubMed] [Google Scholar]
- Chen Z., Ji M., Hou P., Lu Z. Exo-dye-based assay for rapid, inexpensive, and sensitive detection of DNA-binding proteins. Biochem. Biophys. Res. Commun. 2006;345:1254–1263. doi: 10.1016/j.bbrc.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Patel D.J. Crystallographic structure of the nuclease domain of 3′hExo, a DEDDh family member, bound to rAMP. J. Mol. Biol. 2004;343:305–312. doi: 10.1016/j.jmb.2004.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D., Beresford P.J., Zhu P., Zhang D., Sung J.S., Demple B., Perrino F.W., Lieberman J. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol. Cell. 2006;23:133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Crow Y.J., Hayward B.E., Parmar R., Robins P., Leitch A., Ali M., Black D.N., van Bokhoven H., Brunner H.G., Hamel B.C., et al. Mutations in the gene encoding the 3′–5′ DNA exonuclease TREX1 cause Aicardi-Goutières syndrome at the AGS1 locus. Nat. Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- Derbyshire V., Pinsonneault J.K., Joyce C.M. Structure-function analysis of 3′→5′-exonuclease of DNA polymerases. Methods Enzymol. 1995;262:363–385. doi: 10.1016/0076-6879(95)62030-3. [DOI] [PubMed] [Google Scholar]
- de Silva U., Choudhury S., Bailey S.L., Harvey S., Perrino F.W., Hollis T. The crystal structure of TREX1 explains the 3′ nucleotide specificity and reveals a polyproline II helix for protein partnering. J. Biol. Chem. 2007;282:10537–10543. doi: 10.1074/jbc.M700039200. [DOI] [PubMed] [Google Scholar]
- Dichtl B., Stevens A., Tollervey D. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 1997;16:7184–7195. doi: 10.1093/emboj/16.23.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan S., Bulloch E.M., Thompson P.R., Beck J.L., Yang J.Y., Crowther J.A., Lilley P.E., Carr P.D., Ollis D.L., Brown S.E., et al. Hydrolysis of the 5′-p-nitrophenyl ester of TMP by the proofreading exonuclease (ε) subunit of Escherichia coli DNA polymerase III. Biochemistry. 2002a;41:5266–5275. doi: 10.1021/bi0159480. [DOI] [PubMed] [Google Scholar]
- Hamdan S., Carr P.D., Brown S.E., Ollis D.L., Dixon N.E. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure. 2002b;10:535–546. doi: 10.1016/s0969-2126(02)00738-4. [DOI] [PubMed] [Google Scholar]
- Horio T., Murai M., Inoue T., Hamasaki T., Tanaka T., Ohgi T. Crystal structure of human ISG20, an interferon-induced antiviral ribonuclease. FEBS Lett. 2004;577:111–116. doi: 10.1016/j.febslet.2004.09.074. [DOI] [PubMed] [Google Scholar]
- Ikeda A., Kato T. Biological predictors of lithium response in bipolar disorder. Psychiatry Clin. Neurosci. 2003;57:243–250. doi: 10.1046/j.1440-1819.2003.01112.x. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Lee-Kirsch M.A., Chowdhury D., Harvey S., Gong M., Senenko L., Engel K., Pfeiffer C., Hollis T., Gahr M., Perrino F.W., et al. A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. J. Mol. Med. 2007a;85:531–537. doi: 10.1007/s00109-007-0199-9. [DOI] [PubMed] [Google Scholar]
- Lee-Kirsch M.A., Gong M., Chowdhury D., Senenko L., Engel K., Lee Y.A., de Silva U., Bailey S.L., Witte T., Vyse T.J., et al. Mutations in the gene encoding the 3′–5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat. Genet. 2007b;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Gally J.A., Edelman G.M. Properties of deoxyribonuclease 3 from mammalian tissues. J. Biol. Chem. 1969;244:5014–5019. [PubMed] [Google Scholar]
- Mazur D.J., Perrino F.W. Excision of 3′ termini by the Trex1 and TREX2 3′→5′ exonucleases. Characterization of the recombinant proteins. J. Biol. Chem. 2001;276:17022–17029. doi: 10.1074/jbc.M100623200. [DOI] [PubMed] [Google Scholar]
- Morita M., Stamp G., Robins P., Dulic A., Rosewell I., Hrivnak G., Daly G., Lindahl T., Barnes D.E. Gene-targeted mice lacking the Trex1 (DNase III) 3′→5′ DNA exonuclease develop inflammatory myocarditis. Mol. Cell. Biol. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M.J., Holley W.R., Chatterjee A., Mian I.S. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Patel S., Martinez-Ripoll M., Blundell T.L., Albert A. Structural enzymology of Li+-sensitive/Mg2+-dependent phosphatases. J. Mol. Biol. 2002;320:1087–1094. doi: 10.1016/s0022-2836(02)00564-8. [DOI] [PubMed] [Google Scholar]
- Perrino F.W., Miller H., Ealey K.A. Identification of a 3′→5′-exonuclease that removes cytosine arabinoside monophosphate from 3′ termini of DNA. J. Biol. Chem. 1994;269:16357–16363. [PubMed] [Google Scholar]
- Perrino F.W., Mazur D.J., Ward H., Harvey S. Exonucleases and the incorporation of aranucleotides into DNA. Cell Biochem. Biophys. 1999;30:331–352. doi: 10.1007/BF02738118. [DOI] [PubMed] [Google Scholar]
- Pingoud A., Fuxreiter M., Pingoud V., Wende W. Type II restriction endonucleases: Structure and mechanism. Cell. Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- Rice G., Newman W.G., Dean J., Patrick T., Parmar R., Flintoff K., Robins P., Harvey S., Hollis T., O'Hara A., et al. Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutières Syndrome. Am. J. Hum. Genet. 2007;80:811–815. doi: 10.1086/513443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A., van den Maagdenberg A.M., Jen J.C., Kavanagh D., Bertram P., Spitzer D., Liszewski M.K., Barilla-Labarca M.L., Terwindt G.M., Kasai Y., et al. C-terminal truncations in human 3′–5′ DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nat. Genet. 2007;39:1068–1070. doi: 10.1038/ng2082. [DOI] [PubMed] [Google Scholar]
- Shaldubina A., Agam G., Belmaker R.H. The mechanism of lithium action: State of the art, ten years later. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2001;25:855–866. doi: 10.1016/s0278-5846(01)00154-3. [DOI] [PubMed] [Google Scholar]
- Shevelev I.V., Hubscher U. The 3′–5′ exonucleases. Nat. Rev. Mol. Cell Biol. 2002;3:364–376. doi: 10.1038/nrm804. [DOI] [PubMed] [Google Scholar]
- Steitz T.A., Steitz J.A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Lee J.Y., Nowotny M. Making and breaking nucleic acids: Two-Mg2+-ion catalysis and substrate specificity. Mol. Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Yang Y.G., Lindahl T., Barnes D.E. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Zhen X., Torres C., Friedman E. Lithium regulates protein tyrosine phosphatase activity in vitro and in vivo. Psychopharmacology. 2002;162:379–384. doi: 10.1007/s00213-002-1126-y. [DOI] [PubMed] [Google Scholar]
- Zuo Y., Deutscher M.P. Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]