Summary
G protein-coupled receptors (GPCRs) comprise the largest family of transmembrane signaling molecules and regulate a host of physiological and disease processes. To better understand the functions of GPCRs in vivo, we quantified transcript levels of 353 non-odorant GPCRs in 41 adult mouse tissues. Cluster analysis placed many GPCRs into anticipated anatomical and functional groups and predicted novel roles for less studied receptors. From one such prediction, we showed that the Gpr91 ligand succinate can regulate lipolysis in white adipose tissue suggesting that signaling by this citric acid cycle intermediate may regulate energy homeostasis. We also showed that pairwise analysis of GPCR expression across tissues may help predict drug side effects. This resource will aid studies to understand GPCR function in vivo and may assist in the identification of therapeutic targets.
Introduction
Mammalian cells sense myriad signals in their environment via G protein-coupled receptors (GPCRs), the largest family of transmembrane signaling molecules. GPCRs can be partitioned into two groups: odorant/sensory and non-odorant. Odorant/sensory receptors are restricted to specialized cells that detect external cues — odors, tastes and pheromones — and regulate organismal behaviors such as feeding and mating. Non-odorant GPCRs are differentially expressed throughout the organism, respond to diverse endogenous ligands, and regulate a host of physiological processes including hematopoiesis, hemostasis, immune function, metabolism, neurotransmission, reproduction, cardiac function and vascular tone. Accordingly, such receptors are the targets for about one third of all approved drugs (Hopkins and Groom, 2002; Muller, 2000). Although in vivo roles have been defined for many of the approximately 370 non-odorant GPCRs in mice and humans, the expression and function of many such receptors are incompletely characterized, and a significant fraction remain orphans (Fredriksson et al., 2003; Fredriksson and Schioth, 2005; Hill et al., 2002; Joost and Methner, 2002; Vassilatis et al., 2003).
To support studies of non-odorant GPCR function, we analyzed the pattern of GPCR mRNA expression across tissues and the relative abundance for each of 353 non-odorant GPCRs in 41 tissues from adult mouse. Hierarchical clustering analysis revealed groupings of tissues and receptors that predicted physiological functions for individual receptors and receptor clusters. We tested one such prediction by examining Gpr91, a receptor for the citric acid cycle intermediate succinate (He et al., 2004). Gpr91 was grouped in the “adipose cluster”, but neither Gpr91 nor succinate was known to regulate adipocyte functions. We demonstrated that extracellular succinate can inhibit lipolysis in white adipose tissue in a manner consistent with its acting via adipocyte Gpr91. Overall, this data set provides a resource for those interested in finding new roles for GPCRs with known ligands and hints regarding the functions of orphan GPCRs and the sources of their ligands. When compared with human expression data (SymAtlas, SAGEmap), these mouse data will aid the rational use of mice to model GPCR function in human physiology and disease and may help point up new therapeutic targets and predict on-target side effects. In addition, these quantitative data describing expression of a large number of related and relatively small genes across many tissues may support studies aimed at identifying cis-acting elements and transcription factors that dictate expression in particular tissues.
Results
Tissue profiling of GPCRs by qPCR
GPCRs are usually expressed at low levels. Indeed, non-odorant GPCRs comprise about 1% of genes in the genome, but only 0.001−0.01% of expressed sequence tags (ESTs) correspond to GPCRs (Fredriksson and Schioth, 2005). Accordingly, we chose TaqMan-type quantitative real-time polymerase chain reaction (qPCR) for its high sensitivity, specificity and broad dynamic range to measure GPCR mRNA expression. Primer/probe sets were validated as described in Experimental Procedures; their sequences are provided in Supplement S1.
Transcript levels for 353 GPCRs were profiled in 41 adult tissues isolated from C57BL/6 mice. Bar graphs indicating transcript levels for each receptor in each tissue relative to internal controls (β-actin, cyclophilin, GAPDH and ribosomal protein S9) are found in Supplement S2 and at http://pdsp.med.unc.edu/apGPCRe/.
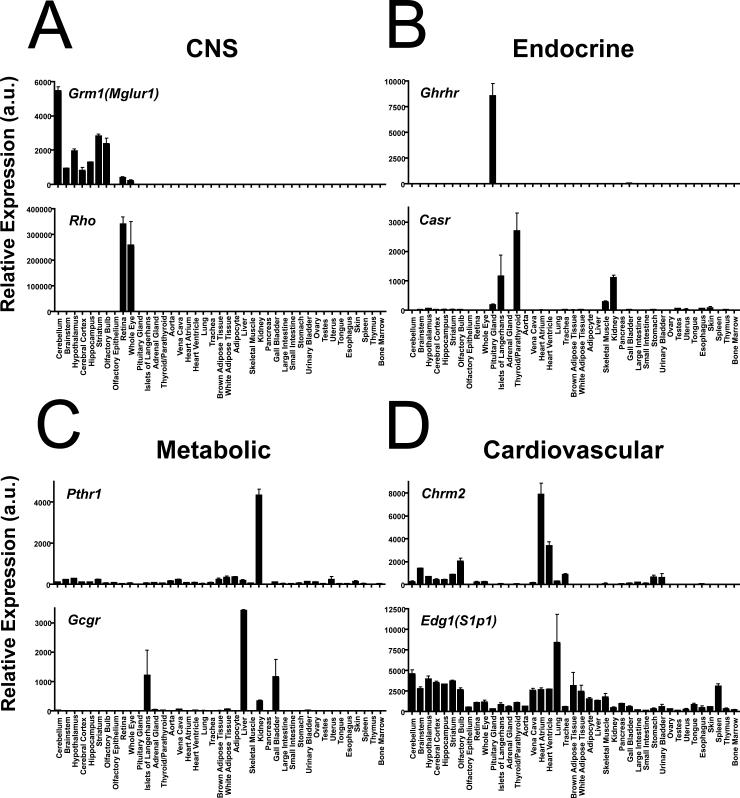
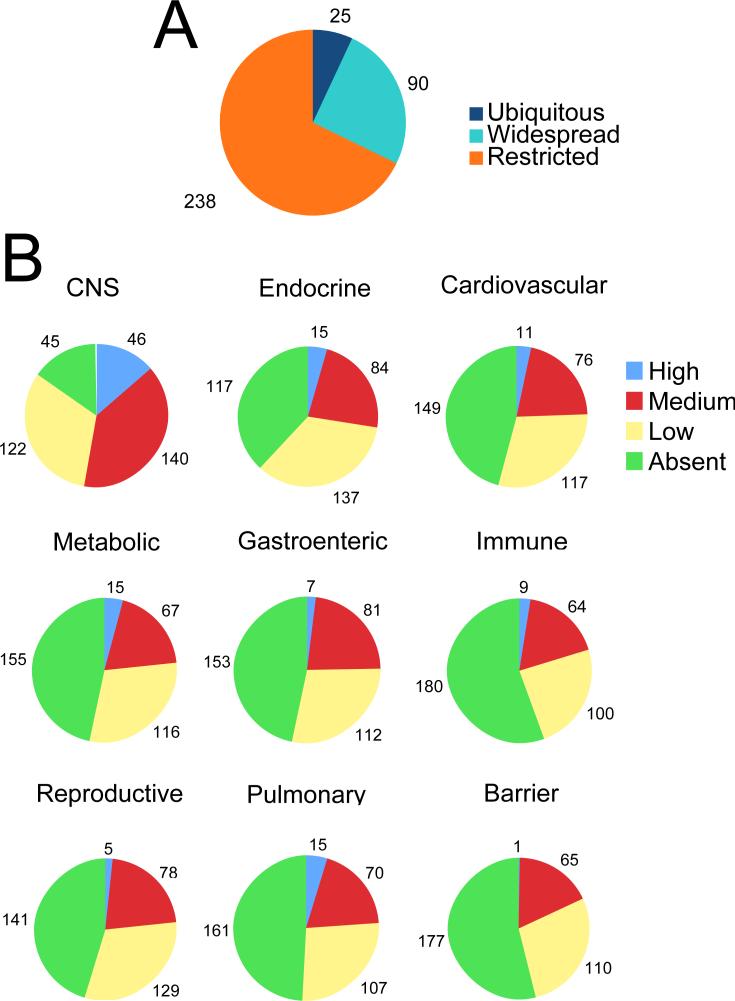
GPCR expression levels varied dramatically by tissue. Predictably, rhodopsin was the most abundantly expressed GPCR and among the most tissue-specific, present at ∼350,000 arbitrary units (a.u.) in eye but below 25 a.u. in other tissues (Fig. 2A). Because rhodopsin presumably serves no function in extraocular tissues, we adopted the convention that receptors with expression values below 25 a.u. in a given tissue were “absent”. Using this criterion, only 25 GPCRs were expressed above background in all 41 tissues assayed (Table 1); 90 were expressed in greater than half of tissues and 238 in less than half (Fig. 1A). Some ubiquitously expressed GPCRs were highly expressed in blood vessels (i.e. Edg1(S1p1), F2r(Par1), Ednra, Ptger1), perhaps accounting for their presence in all tissues (Table 1). Other ubiquitously expressed receptors, including Gpr56, Lec1(Lphn2), Lec2(Lphn1), Gpr107, Gpr108, Tm7sf1(Gpr137b), Tm7sf1l1(Gpr137), Tm7sf3 and Tpra40(Gpr175), were expressed in 5 of 5 different nonvascular cell lines tested and may indeed be expressed by most cell types in vivo (Table 1 and data not shown).
Fig. 2. qRT-PCR tissue distribution yields predicted patterns of expression.
Representative GPCRs that were highly expressed in distinct tissue systems were selected from Fig. 1B and Table 1 to illustrate tissue specificity, range of expression levels, and reproducibility. Examples are shown for A) CNS: metabotropic glutamate receptor 1 [Grm1(Mglur1)] and rhodopsin [Rho]. B) Endocrine system: growth hormone releasing hormone receptor [Ghrhr] and the extracellular calcium-sensing receptor [Casr]. C) Metabolic tissues: parathyroid hormone receptor 1 [Pthr1] and glucagon receptor [Gcgr]. D) Cardiovascular system: M2 muscarinic receptor [Chrm2] and the sphingosine-1-phosphate receptor 1 [Edg1(S1p1)]. Values are plotted as the mean ± SEM; n=2−5.
Table 1.
GPCRs ubiquitously and abundantly expressed in vivo.
| Ubiquitous | CNS | Endocrine | Cardiovascular |
|---|---|---|---|
| Cd97 | Adcyap1r1 | Adora1 | Agtr1(Agtr1a) |
| Ebi2 | Adora1 | Agtr1(Agtr1a) | Chrm2 |
| Edg1(S1p1) | Bai1 | Casr | Edg1(S1p1) |
| Edg3(S1p3) | Bai2 | Drd2 | Ednra |
| Ednra | Celsr2 | F2r(Par1) | Etl(Eltd1) |
| Etl(Eltd1) | Chrm1 | Ghrhr | F2r(Par1) |
| F2r(Par1) | Cnr1 | Glp1r | Fzd1 |
| Fzd1 | Drd1 | Gpr108 | Fzd2 |
| Fzd7 | Drd4 | Gpr48(Lgr4) | Fzd4 |
| Gabbr1 | Edg1(S1p1) | Gpr56 | Kiaa0758(Gpr116) |
| Gpr10(Prlhr) | Ednrb | Lec2(Lphn1) | Rdc1(Cxcr7) |
| Gpr105(P2ry14) | Fy(Darc) | Rai3(Gprc5a) | |
| Gpr107 | Fzd1 | Rdc1(Cxcr7) | |
| Gpr108 | Fzd3 | Tm7sf1(Gpr137b) | Barrier |
| Gpr19 | Gabbr1 | Tm7sf1l1(Gpr137) | Hm74(Gpr109a) |
| Gpr56 | Glp2r | ||
| Il8rb(Cxcr2) | Gpr108 | Metabolic | |
| Lec1(Lphn2) | Gpr22 | Adrb3 | Pulmonary |
| Lec2(Lphn1) | Gpr26 | Agtr1(Agtr1a) | Adrb2 |
| P2y5 | Gpr37(Paelr) | Cmkbr1l1(Ccr1l1) | Calcrl |
| Ptger1(Ep1) | Gpr37l1 | Edg1(S1p1) | Cckar |
| Tm7sf1(Gpr137b) | Gpr49(Lgr5) | Etl(Eltd1) | Cd97 |
| Tm7sf1l1(Gpr137) | Gpr51(Gabbr2) | Fzd4 | Edg1(S1p1) |
| Tm7sf3 | Gpr56 | Gcgr | Edg3(S1p3) |
| Tpra40(Gpr175) | Gprc5b(Raig2) | Gpr10(Prlhr) | Etl(Eltd1) |
| Grca(Gpr162) | Gpr48(Lgr4) | F2r(Par1) | |
| Grm1(Mglur1) | Gpr56 | Glp1r | |
| Grm3(Mglur3) | Gpr91(Sucnr1) | Gpr108 | |
| Grm4(Mglur4) | Hm74(Gpr109a) | Gpr56 | |
| Grm5(Mglur5) | Ptger3(Ep3) | Kiaa0758(Gpr116) | |
| Grm6(Mglur6) | Pthr1 | Rai3(Gprc5a) | |
| Grm7(Mglur7) | Tm7sf1(Gpr137b) | Tm7sf1(Gpr137b) | |
| Hrh3 | Tm7sf1l1(Gpr137) | ||
| Il8rb(Cxcr2) | Gastroenteric | ||
| Kiaa1828(Gpr123) | Cckar | ||
| Lec2(Lphn1) | Flj14454(Gpr128) | Immune | |
| Lec3(Lphn3) | Fzd1 | Adrb2 | |
| Ntsr2 | Glp1r | Ccr9 | |
| Opn1mw | Gpr108 | Cxcr4 | |
| Opn1sw | Ptger3(Ep3) | Edg1(S1p1) | |
| Oprl1 | Rai3(Gprc5a) | Fpr-rs2 | |
| Pgr15(Gpr165) | |||
| Pgr22(Gpr155) | Gpr18 | ||
| Rgr | Reproductive | Il8rb(Cxcr2) | |
| Rho | Agtr1(Agtr1a) | Mrga3 | |
| Tm7sf1l1(Gpr137) | Gpcr150 | P2y5 | |
| Gpr108 | |||
| Gpr19 | |||
| Tm7sf1l1(Gpr137) |
Fig. 1. Distribution of mouse GPCR mRNA expression in vivo.
A) The number of GPCRs expressed in tissues presented in pie chart form. Receptors expressed in all 41 tissues assayed are “ubiquitous”; receptors expressed in more than or less than half are “widespread” or “restricted”, respectively. A list of the “ubiquitous” receptors is included in Table 1. B) GPCR expression by tissue systems. High, medium, low and absent are defined in Results. Tissue systems are defined as: Central Nervous System (CNS: cerebellum, brainstem, hypothalamus, cerebral cortex, hippocampus, striatum, olfactory bulb, olfactory epithelium, retina, whole eye), Endocrine (pituitary gland, islets of Langerhans, adrenal gland, thyroid/parathyroid), Cardiovascular (aorta, vena cava, heart atrium, heart ventricle), Pulmonary (lung, trachea), Metabolic (brown adipose tissue, white adipose tissue, isolated adipocytes, liver, skeletal muscle, kidney), Gastroenteric (pancreas, gall bladder, large intestine, small intestine, stomach, urinary bladder), Reproductive (ovary, testes, uterus), Barrier (tongue, esophagus, skin) and Immune (spleen, thymus, bone marrow). A list of the highly expressed GPCRs in individual tissue systems is included in Table 1.
Save rhodopsin, nearly all GPCRs were expressed at levels below 10,000 a.u. To provide an overview of GPCR distribution, receptors expressed at 25−250, 250−2500, and >2500 a.u. were designated as low, medium and high expressors, respectively, and tissues were grouped by system (e.g. CNS, endocrine, cardiovascular, pulmonary, metabolic, gastrointestinal, immune, reproductive and cutaneous/barrier) (Fig. 1B). By these criteria, 15 or fewer receptors were expressed at high level in any tissue group but CNS.
As expected, receptors that were highly expressed in a given tissue included receptors established to play an important role in that tissue (Table 1). For example, light-detecting opsins were highly expressed in eye, and dopamine, gamma-aminobutyric acid (GABA) and glutamate receptors were highly expressed in CNS (Table 1 and Fig. 2A). In endocrine tissues, the extracellular calcium-sensing receptor (Casr), which regulates parathyroid hormone secretion (Ho et al., 1995), was highly expressed in parathyroid/thyroid; the growth hormone releasing hormone receptor (Ghrhr), which regulates growth hormone secretion (Lin et al., 1993), was high in pituitary; and the glucagon-like peptide receptor 1 (Glp1r), which regulates insulin secretion (Scrocchi et al., 1996), was high in islets (Table 1, Fig. 2B and Supplement S2). The β3 adrenergic receptor (Adrb3) and the niacin/ketone body receptor Hm74(Gpr109A) (Susulic et al., 1995; Tunaru et al., 2003), which regulate lipolysis, were among the most highly expressed receptors in adipose tissue (Table 1 and Fig. 4B). Glucagon receptor (Gcgr), an important regulator of glucose homeostasis (Gelling et al., 2003), was highly expressed in liver, and the parathyroid hormone receptor (Pthr1), which regulates calcium and phosphate levels and modulates the activity of 25-hydroxyvitamin D 1-hydroxylase (Amizuka et al., 1997), was highly expressed in kidney (Table 1 and Fig. 2C). In heart and blood vessels, angiotensin type 1a receptor (Agtr1), which regulates vascular tone (Ito et al., 1995; Sugaya et al., 1995), thrombin receptor (F2r; Par1) and sphingosine-1-phosphate receptor (Edg1; S1p1), which play important roles in vascular development (Connolly et al., 1996; Liu et al., 2000), and the M2 muscarinic acetylcholine receptor (Chrm2), which regulates heart rate (Gomeza et al., 1999) (Table 1 and Fig. 2D), were all highly expressed. Ccr9, Cxcr4 and Il8rb(Cxcr2), which regulate leukocyte formation and function (Nagasawa et al., 1996; Shuster et al., 1995; Wurbel et al., 2001), were abundant in immune tissues (Table 1). Taken together, these results provided confidence that high-level expression in specific tissues and tissue clusters correlates with physiological function and might predict roles for less well-characterized receptors.
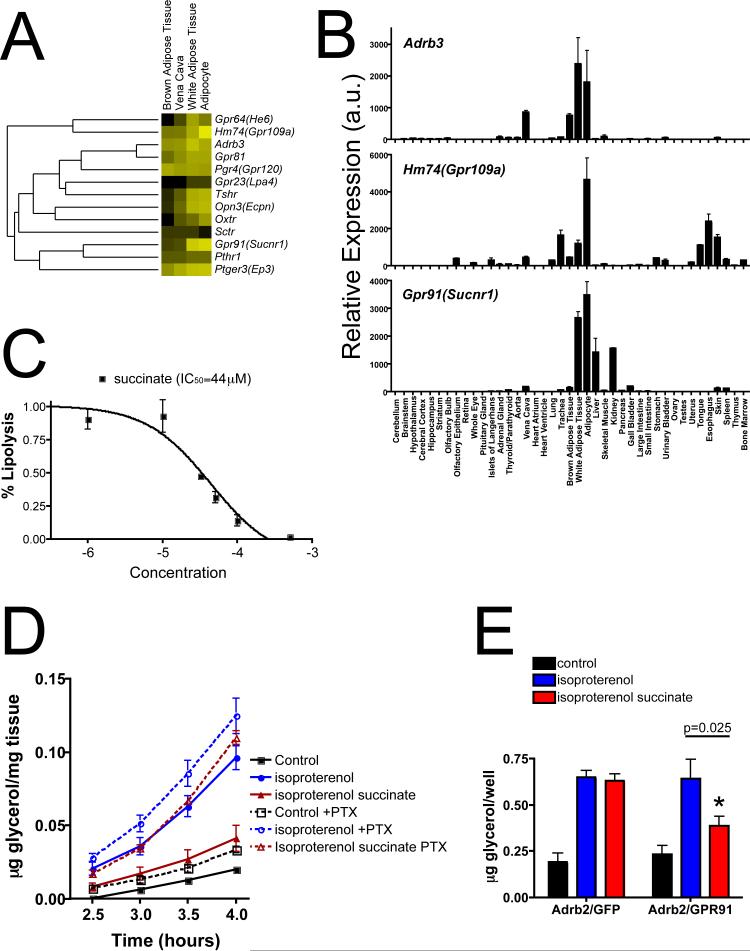
Fig. 4. Gpr91(Sucnr1) and its ligand, succinate, can inhibit lipolysis in adipocytes.
A) The “adipose” cluster (blue circle, lower part Fig. 3A) contained numerous regulators of adipocyte function. B) Adrb3, Hm74(Gpr109a) and Gpr91(Sucnr1) were expressed at similarly high levels in white adipose tissues (WAT) and isolated adipocytes. C) Isolated WAT was treated with isoproterenol (20nM) to stimulate lipolysis, as measured by glycerol release at 3 hours. Succinate added concurrently with isoproterenol inhibited lipolysis in a concentration-dependent manner with an apparent IC50 of approximately 44μM. D) Isolated WAT was pretreated either with KRH/BSA or KRH/BSA containing pertussis toxin (PTX; 100 ng/mL) for 3 hours prior to exposure to the indicated conditions. Isoproterenol (20nM)-stimulated glycerol release was inhibited by the addition of 80μM succinate. Pretreatment of WAT with PTX abrogated this effect, suggesting succinate's inhibition of lipolysis occurred in a Gi/o-dependent manner, consistent with Gpr91 activation. E) Differentiated 3T3-L1 cells, which do not express Gpr91 mRNA, were transfected with mammalian expression vectors containing β2 adrenergic receptor (Adrb2) and control (green fluorescent protein; GFP) or Adrb2 and GPR91. Succinate (100μM) inhibited isoproterenol (10nM)-induced lipolysis in cells co-transfected with GPR91, but not GFP. The data shown are mean ± SD, n=3; p=0.025, unpaired student's t-test. This experiment was done three times with similar results.
Hierarchical clustering of GPCR expression
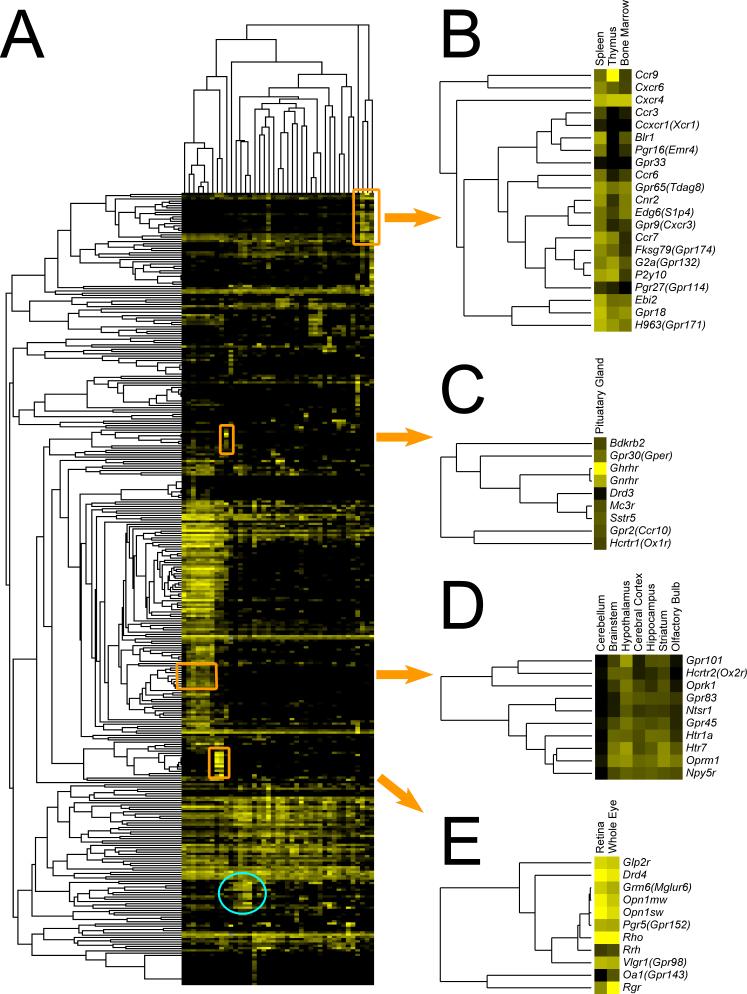
GPCR qPCR results were analyzed by unsupervised, hierarchical clustering to further explore potential relationships between receptor expression and tissue function. The resulting dendograms for both the tissue and receptor axes showed functional clusters (Fig. 3A for a thumbnail image and supplemental S3 for a full size form). CNS tissues cerebellum, brainstem, hypothalamus, cerebral cortex, hippocampus, striatum, olfactory bulb, retina and whole eye clustered together as did the immune/hematopoietic tissues spleen, thymus and bone marrow. The steroidogenic organs adrenal gland and ovary clustered, but testes showed a very distinct pattern of GPCR expression. Liver, kidney and gall bladder formed a group, as did large intestine, small intestine, pancreas and stomach. Skin, esophagus and tongue also formed a cluster, perhaps related to their common barrier function. Cardiac atrium and ventricle, skeletal muscle, aorta and urinary bladder formed a cluster, perhaps in part due to their sharing a relative abundance of muscle cells. Brown adipose tissue (BAT), white adipose tissue (WAT), isolated adipocytes and vena cava formed an “adipose” cluster. The presence of vena cava in this cluster likely represents the incomplete removal of surrounding fat from the samples.
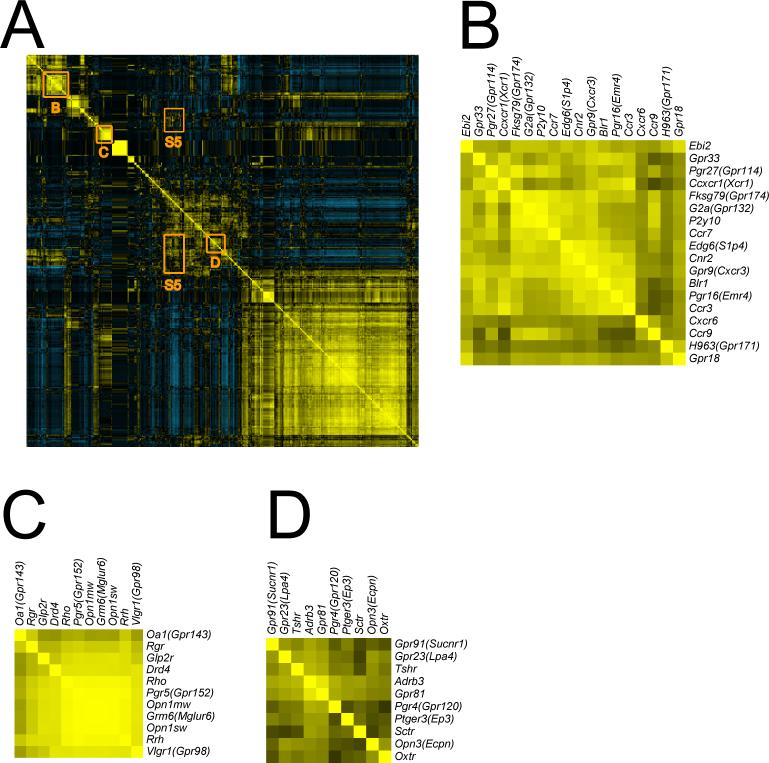
Fig. 3. Unsupervised hierarchical clustering of GPCR expression across tissues.
A) Transformed qRT-PCR data for the 353 GPCRs assayed in the 41 tissues was evaluated by unsupervised hierarchical clustering with average linkage using Cluster 3.0 and visualized using Java TreeView (see Experimental Procedures). A thumbnail image is shown here, a full size version is available in Supplement S3. Multiple clusters and subclusters were seen, 5 were chosen for further analysis. The fifth cluster (blue circle towards the bottom) is discussed in Fig. 4. B) A portion of the “immune/hematopoietic” cluster is shown; note the abundance of chemokine receptors. C) The “pituitary” cluster contains many well-documented regulators of pituitary function, including Ghrhr and Gnrhr. D) A small portion of the “CNS” cluster, by far the largest. This portion contains receptors for important neurotransmitters including serotonin, neuropeptide Y, orexin and opiates. E) The “eye/retinal” cluster contains light-sensing opsins as well as other receptors known to regulate vision.
A number of receptor axis clusters were easily recognized (Fig. 3). A portion of the “immune/hematopoietic” cluster is shown in Fig. 3B. Included are Ccr9, Cxcr6, Cxcr4, Ccr3, Ccxcr1(Xcr1), Blr1, Pgr16(Emr4), Gpr33, Ccr6, Gpr65(Tdag8), Cnr2, Edg6(S1p4), Gpr9(Cxcr3), Ccr7, Fksg79(Gpr174), G2a(Gpr132), P2Y10, Pgr27(Gpr114), Ebi2, Gpr18 and H963(Gpr171). Most of these receptors are known to be expressed in and/or to play a role in immune cells(Birkenbach et al., 1993; Forster et al., 1996; Forster et al., 1999; Humbles et al., 2004; Karsak et al., 2007; Kim et al., 2001; Le et al., 2001; Malone et al., 2004; Nagasawa et al., 1996; Rao et al., 1999; Soto et al., 1998; Stacey et al., 2002; Varona et al., 2001; Wurbel et al., 2001), but no role for Fksg79(Gpr174), Pgr27(Gpr114) and H963(Gpr171) in this context has been described.
The pituitary cluster (Fig. 3C) includes Bdkrb2, Gpr30(Gper), Ghrhr, Gnrhr, Drd3, Mc3r, Sstr5, Gpr2(Ccr10) and Hcrtr1(Ox1r); among these, Gpr2(Ccr10) has not been previously implicated in pituitary gland function (Brailoiu et al., 2007; Date et al., 2000; Kumar et al., 1997; Lin et al., 1993; Lorsignol et al., 1999; Tsutsumi et al., 1992).
The CNS cluster was by far the largest. Our analysis (Fig. 1B and Fig. 3), as well as others (Vassilatis et al., 2003), suggests that >80% of all non-odorant GPCRs are expressed in CNS. In Fig. 3D we show a small portion of the CNS cluster that includes Gpr101, Hcrtr2(Ox2r), Oprk1, Gpr83, Ntsr1, Gpr45, Htr1a, Htr7, Oprm1 and Npy5r, all of which have been implicated in regulation of neuronal function (Bates et al., 2006; Filliol et al., 2000; Lovenberg et al., 1993; Marsh et al., 1998; Mazella et al., 1996; Popova et al., 2007; Simonin et al., 1995; Willie et al., 2003).
Finally Fig. 2E shows the “eye/retina” cluster which contains Glp2r, Drd4, Grm6(Mglur6), Opn1mw, Opn1sw, Pgr5(Gpr152), Rho, Rrh, Vlgr1(Gpr98), Oa1(Gpr143) and Rgr. Save Glp2r and Pgr5(Gpr152), all are known to function in the eye (Chen et al., 2001; Chiu et al., 1994; Cohen et al., 1992; Dryja et al., 2005; Incerti et al., 2000; McGee et al., 2006; Nathans and Hogness, 1983; Sun et al., 1997a; Sun et al., 1997b).
Taken together, the results outlined above reveal that tissues cluster into largely expected functional groups based purely on their GPCR repertoires, and both expected and unique groups of receptors cluster by tissue function. These data identify sets of receptors involved in specific aspects of physiology and should prove useful in providing clues regarding in vivo roles for orphan GPCRs and new roles for receptors with known ligands. To test the latter prediction, we sought a possible role for a recently de-orphanized receptor, Gpr91, found in the adipose cluster.
Extracellular succinate inhibits lipolysis, likely via Gpr91
Thirteen GPCRs defined an adipose cluster (Fig. 4A and circled area in Fig. 3A): Gpr64(He6), Hm74(Gpr109a), Adrb3, Gpr81, Pgr4(Gpr120), Gpr23(Lpa4), Tshr, Opn3, Oxtr, Sctr, Gpr91(Suncr1), Pthr1 and Ptger3(Ep3). Ligands for most of these receptors are known to affect adipocyte function (Butcher and Carlson, 1970; Gotoh et al., 2007; Moskowitz and Fain, 1969; Rodbell, 1964; Sinha et al., 1976; Susulic et al., 1995; Tunaru et al., 2003; Valet et al., 1998), but to our knowledge, no role for Gpr64(He6), Gpr81, Opn3 or Gpr91(Suncr1) in adipose tissue has been reported. He et al. demonstrated that the citric acid cycle intermediate succinate can activate Gpr91 and that Gpr91, presumably in kidney, mediates elevation of plasma renin levels and blood pressure in response to exogenous succinate (He et al., 2004). Like Adrb3 and Hm74(Gpr109a), Gpr91(Suncr1) mRNA was most highly expressed in white adipose tissue (WAT) and was abundant in purified adipocytes (Fig. 4B). Gpr91 is at least partially Gi-coupled (He et al., 2004), and Gi-coupled GPCRs are known to inhibit lipolysis in adipose tissues (Moreno et al., 1983). Accordingly, we examined the effect of succinate on isoproterenol-induced lipolysis in isolated WAT (Isoproterenol has relatively low affinity for Adrb3 and most likely acts via Adrb2 in this context).
Succinate inhibited lipolysis in a dose-dependent manner with an apparent IC50 of 44μM (Fig. 4C), a concentration similar to the EC50 for succinate activation of Gpr91 heterologously expressed in 293 cells (He et al., 2004). Inhibition of lipolysis by succinate was ablated by pertussis toxin pretreatment. 3T3-L1 adipocyte-like cells, which do not express Gpr91 mRNA (unpublished observation) and were not sensitive to the anti-lipolytic effects of succinate, became succinate-sensitive when transfected with a mammalian expression vector for GPR91 (Fig. 4E). These data strongly suggest that succinate inhibits lipolysis in WAT via a Gi-coupled GPCR, presumably Gpr91.
Pair-wise analysis of GPCR co-expression may help identify multiple roles for individual GPCRs
To explore whether co-expression of GPCRs might provide additional clues to function, we compared the expression pattern of each GPCR to that of every other to generate the map shown in Fig. 5. Positive and negative correlation of expression patterns are indicated by yellow and blue colors, respectively (Fig. 5A and Supplemental S4). Such correlation maps are a powerful tool for organizing and analyzing gene-gene and protein-protein interactions on a global scale (Collins et al., 2007a; Collins et al., 2007b; Krogan et al., 2006; Schuldiner et al., 2005; Segre et al., 2005).
Fig. 5. Hierarchical-Cluster analysis of pairwise GPCR expression reveals additional levels of interaction.
A) Pearson correlation r coefficients were calculated for interactions between each GPCR with all others based on tissue expression patterns. The resulting data set was further analyzed using Cluster 3.0 with complete linkage and visualized using TreeView. A thumbnail image is shown here, a full size image is available in Supplemental S4. Receptors with similar distributions are shown in yellow; distinct distributions are shown in blue; X- and Y-axis are mirror images of one another. The diagonal represents each receptor interacting with itself (perfect similarity in distribution). B) Clustering of receptors by similarity of expression reveals a immune/hematopoietic grouping very similar to that in Fig. 3B; C) an eye/retinal cluster similar to that in Fig. 3E; and D) an adipose cluster similar to that in Fig. 4A. However, analysis of GPCR interaction clusters off the diagonal suggested receptor functions outside of their most obvious physiological roles. This might aid in understanding and predicting on-target drug side effects (see Discussion and supplementary figure S5).
Not surprisingly, tissue-specific receptor clusters similar to those seen in Fig. 3 were, for the most part, recapitulated as distinct blocks along the diagonal. Examples are shown in Fig. 5A, B and C, which largely reproduce the immune/hematopoietic, eye/retina, and adipose clusters of Fig. 3B and E and 4A, respectively. Off-diagonal blocks drew attention to possible roles for receptors in other contexts. By pointing up possible roles for receptors outside of their main physiological cluster, such analysis may be useful in understanding and predicting on-target drug side effects (See Discussion).
Discussion
We have quantitated mRNA levels for the non-odorant G protein-coupled receptors encoded in the mouse genome in 41 tissues and provide this data set as a resource for predicting roles for incompletely characterized GPCRs, exploring tissue-specific gene expression, and other purposes. The fact that tissues that comprise classical physiological systems (cardiovascular, gastrointestinal, etc.) were clustered together simply on the basis of their GPCR repertoires speaks to the key roles that GPCRs play in homeostatic regulation.
Our anatomic expression profiling yielded a large amount of information consistent with known physiology, and high-level expression of a GPCR in a particular tissue cluster or specific tissue correlated well with its physiological role. While this result is not surprising, it does provide confidence that roles for orphan receptors or GPCRs not known to play a role in a particular physiological process might be predicted by presence in a given cluster. Our demonstration that Gpr91 expression pointed to a role for extracellular succinate in regulating lipolysis in adipocytes validates this notion and is also of intrinsic interest.
The concentration of succinate in plasma has been reported at 5−125 μM, a range that surrounds the EC50 for Gpr91 activation (He et al., 2004) and the IC50 for inhibition of lipolysis in WAT (Fig. 4). Succinate concentrations increase during exercise and metabolic acidosis and, in rodents, in hyperglycemic metabolic states (Forni et al., 2005; Hochachka and Dressendorfer, 1976; Krebs, 1950; Kushnir et al., 2001; Nordmann and Nordmann, 1961; Sadagopan et al., 2007). Thus, excursions in the levels of extracellular succinate do occur and might regulate adipocyte function in vivo. Adipocyte function was not investigated in mice lacking Gpr91, which are grossly healthy (He et al., 2004). Overall, a physiological role for succinate in regulating adipocyte metabolism is plausible, but when and how such a system might be important and/or redundant with other systems that govern adipocyte function remains unknown.
49 of the 353 GPCRs profiled were expressed in only one or two of the 41 tissues examined (see Table 2 in the Supplement). Such confined expression might point up targets of pharmaceutical interest. For example, testes showed a GPCR expression pattern very distinct from that of other tissues. Gpr150, Gpr66 and Gpr15, Mtnr1a, Pgr23 were almost perfectly specific to testes. Whether such receptors play a role in spermatogenesis or other testicular functions and their potential utility as targets for drugs aimed at controlling fertility is unknown.
A comparison of each receptor's expression pattern with that of every other (Fig. 5) provided a means of pointing up possible roles for a given receptor outside its main physiological cluster. Hm74(Gpr109a), the ketone body receptor that is activated therapeutically by niacin to treat dyslipidemias (Soga et al., 2003; Tunaru et al., 2003; Wise et al., 2003) provides an interesting example. By traditional clustering analysis Hm74(Gpr109a) is placed in the “adipose” cluster (Fig. 4A), and activation of adipocyte Hm74 likely mediates the anti-lipolytic actions of niacin (Tunaru et al., 2003). It was recently shown that the skin flushing side effect of niacin (Carlson, 2005) is mediated by Hm74 expression by bone-marrow derived epidermal Langerhans cells that release of vasodilatory prostanoids (Benyo et al., 2006; Benyo et al., 2005). By quantitative profiling across tissues, Hm74 was noted to be expressed relatively highly in skin and other ”barrier-cluster” tissues as well as adipose (Fig. 4B). By expression correlation analysis, Hm74 was not found with the adipose cluster on the diagonal but instead clustered with receptors with more widespread expression (Supplemental Figures S4 & S5). A search of off-diagonal interactions revealed that Hm74 interacts not only with the adipose cluster, but also with receptors in both immune and barrier clusters (Supplemental Figure S4 & S5). Thus, analysis of GPCR expression data from these different perspectives may generate hypotheses regarding on-target side effects of drugs.
GPCR genes are usually relatively small, often intronless, and range from closely to distantly related. These features plus the availability of quantitative expression data across multiple tissues for hundreds of related genes that show clusters based on shared tissue-specific patterns may provide a resource for those interested in identifying the combinations of cis-acting elements that specify gene expression in a given cell type.
Some GPCRs are thought to function as heterodimers, and cluster analysis for co-expression might point to potential receptor pairs. For example, Gabbr1 is currently thought to function as an obligate heterodimer with Gpr51(Gabbr2) (Jones et al., 1998; Kaupmann et al., 1998; Kuner et al., 1999; White et al., 1998). Our data (Supplement S2) and that of others (Calver et al., 2000) demonstrate similar expression patterns for these receptors in CNS and divergent expression in the periphery. These results are consistent with Gabbr1 and Gabbr2 heterodimer formation in the CNS but raise the possibility that in the periphery, expression of one or the other partner is regulated, that these receptors may use other partners, and/or that heterodimerization may not be required in all settings (Cheng et al., 2007).
Several caveats should be stated regarding interpreting our expression data. 1) Relative mRNA levels, of course, will not always reflect relative protein expression levels nor the relative importance of a particular receptor in a particular tissue. Indeed, some important receptors such as adrenergic receptors were expressed at relatively low levels. 2) Tissues are comprised of multiple cell types, and receptor expression can be restricted to a minority cell type. In the extreme, receptor expression in a minority population can be missed by whole tissue analysis. For example, neither our qPCR data (supplemental S2) nor those of others (Liberles and Buck, 2006) detected trace amine associated receptor 1 (Taar1) expression in brain, but a Taar 1 Lac-Z knock-in mouse revealed Taar1 expression in discrete neuronal populations and Taar1-dependent regulation of dopaminergic activity (Lindemann et al., 2008). 3) The presence of a receptor within a particular anatomical cluster does not exclude important functions in tissues outside that cluster. For example, the D2 dopamine receptor (Drd2) is recognized as the major dopamine receptor subtype in pituitary (Kelly et al., 1997; Saiardi et al., 1997), but Drd2 was in the CNS cluster while the D3 dopamine receptor (Drd3) was in the pituitary cluster (Figure 3C). The cluster analysis presented here used the Pearson correlation, which normalizes expression levels (see Methods) to focus on gaining information from the pattern of gene expression at the cost of ignoring absolute levels of receptor expression, and the Drd cluster results are presumably a result of the fact that Drd2 is expressed at moderate levels in numerous CNS structures while Drd3 is expressed at very low levels in only a few structures (supplemental S2). However, our quantitative expression data (supplemental S2) reveals that Drd2 is expressed approximately 100-fold higher than Drd3 in pituitary, consistent with the dominant role of Drd2 in pituitary function. Thus, this resource is best utilized when the data are analyzed from several perspectives and should be viewed as a means of generating hypotheses to be tested experimentally. Raw expression data for the 353 GPCRs in the 41 organ samples are available online at http://pdsp.med.unc.edu/apGPCRe/. for those who wish to perform their own analyses.
Lastly, expression of individual GPCRs in specific tissues can be different in human and mouse. When used together with GPCR expression patterns in human (SymAtlas, SAGEmap), our dataset should facilitate rational use of mouse to model the roles of GPCRs in human physiology and disease.
Materials and Methods
qRT-PCR
10 week old C56BL/6 mice (Jackson Labs) were housed in UCSF animals facilities for 2 weeks prior to organ harvest. Mice were anesthetized with ketamine/xylazine and transcardially-perfused with saline to remove blood. Organs were dissected, rapidly frozen in liquid nitrogen and stored at −80°C until time of RNA isolation. Corresponding organs from 2 male and 2 female mice were pooled to yield a single tissue RNA sample (i.e., for each separate replicate, organs of 4 mice were pooled), with the exception of sex-specific organs, which were pooled from 2 mice. Pancreas was removed and stored for 24 hours in RNAlater at 4°C prior to storage at −80°C. Islets of Langerhans were isolated from 20 male C57BL/6 males by the UCSF islet isolation core. Primary adipocytes were isolated from sex organ fats pads as described (Rodbell, 1964). Tissues were first homogenized and crude RNA extracted with Trizol (Invitrogen). RNA was further purified using RNeasy columns (Qiagen) with on-column DNAse I digestion. RNA samples were DNAse I-treated a second time and concentrated using the Zymo Research DNA-free RNA kit. First strand synthesis was performed using the iScript kit (Biorad). qPCR assays were performed in a 384-well format using an ABI7900HT, Platinum qPCR mix (Invitrogen) and 10μL reactions. Cycle threshold (Ct) values were collected at 0.2 for all samples; Ct values for individual GPCRs were compared to Ct values for 4 internal controls (β-actin, cyclophilin, GAPDH and ribosomal protein S9) for all tissues. Taqman primer/probes have been described (Regard et al., 2007) and sequences are available in supplemental data (S1). There were 22 mouse GPCRs for which we were unable to generate primer/probe sets that produced a signal above background in any tissue. A lack of data should be viewed as “no data” for these receptors, which are listed in Supplemental Table S3. PCR efficiencies were calculated as described (Peirson et al., 2003). 2(CtGPCR –Ctcontrol) multiplied by 215 was used for graphical representation of qPCR data. Repeated qPCR analysis of a given tissue RNA sample, i.e., technical replicates, yielded variations of less than 10%. 2 to 5 independently prepared RNA samples from separate tissue isolations were analyzed to assess in vivo expression variability, which was also generally less than 10%. Data presented in Supplement 2 and elsewhere represent the mean ± SEM (n=2−5).
Lipolysis
C57BL6 mice were euthanized by cervical dislocation. Sex organ fat pads were dissected and finely minced and washed 4 times with Krebs-Ringer Hepes buffer containing 4% fatty acid free BSA, 5mM glucose, 0.1mM ascorbic acid (KRH-BSA). 25−30 mg of WAT mincate was placed in the upper well of transwells in a 24-well plate and placed in a tissue culture incubator at 37°C with the specified cocktail for the indicated amount of time. KRH-BSA samples were taken from the wells at the specified time to quantify glycerol release and divided by the weight of the mincate. Pertussis toxin (Calbiochem) pretreatment was performed for 4 hours in a tissue culture incubator with mild shaking. 3T3-L1 cells (ATCC) were cultured and differentiated using standard protocols. Fully differentiated 3T3-L1 cells were transfected (Amaxa, Nucleofection) with mammalian expression vectors containing β2-adrenergic receptor (Adrb2 – generously provided by Dr. J. Silvio Gutkind) and GPR91 (Missouri S&T) and allowed to recover 48−72 hours prior to lipolysis assays. Free glycerol was quantified using the Free Glycerol reagent (Sigma). IC50 for succinate inhibition of isoproterenol-induced lipolysis was calculated using Prism4.
Hierarchical Clustering
All analysis of the data was performed using Python, Scipy and Excel. For pair-wise correlation in Figure 5, Pearson correlation r values were computed between the expression vectors of every pair of genes in the data set. Transformed data was hierarchically clustered using Cluster 3.0 (Eisen et al., 1998) and visualized using Java TreeView (Saldanha, 2004).
Supplementary Material
Acknowledgements
We thank the Coughlin lab for valuable discussions; Ivo Cornelissen, Gerard Honig, and Grant Li for technical assistance in isolating mouse tissues and Stuart Peirson for sharing qPCR efficiency transformations. We also thank Dale Webster and Joseph Derisi for help with informatics, Silvio Gutkind for providing the Adrb2 construct and Bryan Roth for critical reading of the manuscript. This work was supported by the NIH (S.R.C) and Sandler Family Foundation (J.B.R.). The authors have declared that no conflict of interest exists.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amizuka N, Lee HS, Kwan MY, Arazani A, Warshawsky H, Hendy GN, Ozawa H, White JH, Goltzman D. Cell-specific expression of the parathyroid hormone (PTH)/PTH-related peptide receptor gene in kidney from kidney-specific and ubiquitous promoters. Endocrinology. 1997;138:469–481. doi: 10.1210/endo.138.1.4845. [DOI] [PubMed] [Google Scholar]
- Bates B, Zhang L, Nawoschik S, Kodangattil S, Tseng E, Kopsco D, Kramer A, Shan Q, Taylor N, Johnson J, et al. Characterization of Gpr101 expression and G-protein coupling selectivity. Brain Res. 2006;1087:1–14. doi: 10.1016/j.brainres.2006.02.123. [DOI] [PubMed] [Google Scholar]
- Benyo Z, Gille A, Bennett CL, Clausen BE, Offermanns S. Nicotinic acid-induced flushing is mediated by activation of epidermal langerhans cells. Mol Pharmacol. 2006;70:1844–1849. doi: 10.1124/mol.106.030833. [DOI] [PubMed] [Google Scholar]
- Benyo Z, Gille A, Kero J, Csiky M, Suchankova MC, Nusing RM, Moers A, Pfeffer K, Offermanns S. GPR109A (PUMA-G/HM74A) mediates nicotinic acid-induced flushing. J Clin Invest. 2005;115:3634–3640. doi: 10.1172/JCI23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbach M, Josefsen K, Yalamanchili R, Lenoir G, Kieff E. Epstein-Barr virus-induced genes: first lymphocyte-specific G protein-coupled peptide receptors. J Virol. 1993;67:2209–2220. doi: 10.1128/jvi.67.4.2209-2220.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brailoiu E, Dun SL, Brailoiu GC, Mizuo K, Sklar LA, Oprea TI, Prossnitz ER, Dun NJ. Distribution and characterization of estrogen receptor G protein-coupled receptor 30 in the rat central nervous system. J Endocrinol. 2007;193:311–321. doi: 10.1677/JOE-07-0017. [DOI] [PubMed] [Google Scholar]
- Butcher RW, Carlson LA. Effects of secretin on fat mobilizing lipolysis and cyclic AMP levels in rat adipose tissue. Acta Physiol Scand. 1970;79:559–563. doi: 10.1111/j.1748-1716.1970.tb04758.x. [DOI] [PubMed] [Google Scholar]
- Calver AR, Medhurst AD, Robbins MJ, Charles KJ, Evans ML, Harrison DC, Stammers M, Hughes SA, Hervieu G, Couve A, et al. The expression of GABA(B1) and GABA(B2) receptor subunits in the cNS differs from that in peripheral tissues. Neuroscience. 2000;100:155–170. doi: 10.1016/s0306-4522(00)00262-1. [DOI] [PubMed] [Google Scholar]
- Carlson LA. Nicotinic acid: the broad-spectrum lipid drug. A 50th anniversary review. J Intern Med. 2005;258:94–114. doi: 10.1111/j.1365-2796.2005.01528.x. [DOI] [PubMed] [Google Scholar]
- Chen P, Hao W, Rife L, Wang XP, Shen D, Chen J, Ogden T, Van Boemel GB, Wu L, Yang M, et al. A photic visual cycle of rhodopsin regeneration is dependent on Rgr. Nat Genet. 2001;28:256–260. doi: 10.1038/90089. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Tu C, Rodriguez L, Chen TH, Dvorak MM, Margeta M, Gassmann M, Bettler B, Shoback D, Chang W. Type B gamma-aminobutyric acid receptors modulate the function of the extracellular Ca2+− sensing receptor and cell differentiation in murine growth plate chondrocytes. Endocrinology. 2007;148:4984–4992. doi: 10.1210/en.2007-0653. [DOI] [PubMed] [Google Scholar]
- Chiu MI, Zack DJ, Wang Y, Nathans J. Murine and bovine blue cone pigment genes: cloning and characterization of two new members of the S family of visual pigments. Genomics. 1994;21:440–443. doi: 10.1006/geno.1994.1292. [DOI] [PubMed] [Google Scholar]
- Cohen AI, Todd RD, Harmon S, O'Malley KL. Photoreceptors of mouse retinas possess D4 receptors coupled to adenylate cyclase. Proc Natl Acad Sci U S A. 1992;89:12093–12097. doi: 10.1073/pnas.89.24.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007a;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007b;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Connolly AJ, Ishihara H, Kahn ML, Farese RV, Jr., Coughlin SR. Role of the thrombin receptor in development and evidence for a second receptor. Nature. 1996;381:516–519. doi: 10.1038/381516a0. [DOI] [PubMed] [Google Scholar]
- Date Y, Mondal MS, Matsukura S, Ueta Y, Yamashita H, Kaiya H, Kangawa K, Nakazato M. Distribution of orexin/hypocretin in the rat median eminence and pituitary. Brain Res Mol Brain Res. 2000;76:1–6. doi: 10.1016/s0169-328x(99)00317-4. [DOI] [PubMed] [Google Scholar]
- Dryja TP, McGee TL, Berson EL, Fishman GA, Sandberg MA, Alexander KR, Derlacki DJ, Rajagopalan AS. Night blindness and abnormal cone electroretinogram ON responses in patients with mutations in the GRM6 gene encoding mGluR6. Proc Natl Acad Sci U S A. 2005;102:4884–4889. doi: 10.1073/pnas.0501233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filliol D, Ghozland S, Chluba J, Martin M, Matthes HW, Simonin F, Befort K, Gaveriaux-Ruff C, Dierich A, LeMeur M, et al. Mice deficient for delta- and mu-opioid receptors exhibit opposing alterations of emotional responses. Nat Genet. 2000;25:195–200. doi: 10.1038/76061. [DOI] [PubMed] [Google Scholar]
- Forni LG, McKinnon W, Lord GA, Treacher DF, Peron JM, Hilton PJ. Circulating anions usually associated with the Krebs cycle in patients with metabolic acidosis. Crit Care. 2005;9:R591–595. doi: 10.1186/cc3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster R, Mattis AE, Kremmer E, Wolf E, Brem G, Lipp M. A putative chemokine receptor, BLR1, directs B cell migration to defined lymphoid organs and specific anatomic compartments of the spleen. Cell. 1996;87:1037–1047. doi: 10.1016/s0092-8674(00)81798-5. [DOI] [PubMed] [Google Scholar]
- Forster R, Schubel A, Breitfeld D, Kremmer E, Renner-Muller I, Wolf E, Lipp M. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Schioth HB. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol. 2005;67:1414–1425. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- Gelling RW, Du XQ, Dichmann DS, Romer J, Huang H, Cui L, Obici S, Tang B, Holst JJ, Fledelius C, et al. Lower blood glucose, hyperglucagonemia, and pancreatic alpha cell hyperplasia in glucagon receptor knockout mice. Proc Natl Acad Sci U S A. 2003;100:1438–1443. doi: 10.1073/pnas.0237106100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomeza J, Shannon H, Kostenis E, Felder C, Zhang L, Brodkin J, Grinberg A, Sheng H, Wess J. Pronounced pharmacologic deficits in M2 muscarinic acetylcholine receptor knockout mice. Proc Natl Acad Sci U S A. 1999;96:1692–1697. doi: 10.1073/pnas.96.4.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh C, Hong YH, Iga T, Hishikawa D, Suzuki Y, Song SH, Choi KC, Adachi T, Hirasawa A, Tsujimoto G, et al. The regulation of adipogenesis through GPR120. Biochem Biophys Res Commun. 2007;354:591–597. doi: 10.1016/j.bbrc.2007.01.028. [DOI] [PubMed] [Google Scholar]
- He W, Miao FJ, Lin DC, Schwandner RT, Wang Z, Gao J, Chen JL, Tian H, Ling L. Citric acid cycle intermediates as ligands for orphan G-protein-coupled receptors. Nature. 2004;429:188–193. doi: 10.1038/nature02488. [DOI] [PubMed] [Google Scholar]
- Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, Chrystal MA, Cravchik A, Collins FH, Robertson HM, Zwiebel LJ. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- Ho C, Conner DA, Pollak MR, Ladd DJ, Kifor O, Warren HB, Brown EM, Seidman JG, Seidman CE. A mouse model of human familial hypocalciuric hypercalcemia and neonatal severe hyperparathyroidism. Nat Genet. 1995;11:389–394. doi: 10.1038/ng1295-389. [DOI] [PubMed] [Google Scholar]
- Hochachka PW, Dressendorfer RH. Succinate accumulation in man during exercise. Eur J Appl Physiol Occup Physiol. 1976;35:235–242. doi: 10.1007/BF00423282. [DOI] [PubMed] [Google Scholar]
- Hopkins AL, Groom CR. The druggable genome. Nature reviews. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- Humbles AA, Lloyd CM, McMillan SJ, Friend DS, Xanthou G, McKenna EE, Ghiran S, Gerard NP, Yu C, Orkin SH, et al. A critical role for eosinophils in allergic airways remodeling. Science. 2004;305:1776–1779. doi: 10.1126/science.1100283. [DOI] [PubMed] [Google Scholar]
- Incerti B, Cortese K, Pizzigoni A, Surace EM, Varani S, Coppola M, Jeffery G, Seeliger M, Jaissle G, Bennett DC, et al. Oa1 knock-out: new insights on the pathogenesis of ocular albinism type 1. Hum Mol Genet. 2000;9:2781–2788. doi: 10.1093/hmg/9.19.2781. [DOI] [PubMed] [Google Scholar]
- Ito M, Oliverio MI, Mannon PJ, Best CF, Maeda N, Smithies O, Coffman TM. Regulation of blood pressure by the type 1A angiotensin II receptor gene. Proc Natl Acad Sci U S A. 1995;92:3521–3525. doi: 10.1073/pnas.92.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA, Borowsky B, Tamm JA, Craig DA, Durkin MM, Dai M, Yao WJ, Johnson M, Gunwaldsen C, Huang LY, et al. GABA(B) receptors function as a heteromeric assembly of the subunits GABA(B)R1 and GABA(B)R2. Nature. 1998;396:674–679. doi: 10.1038/25348. [DOI] [PubMed] [Google Scholar]
- Joost P, Methner A. Phylogenetic analysis of 277 human G-protein-coupled receptors as a tool for the prediction of orphan receptor ligands. Genome Biol. 2002;3:RESEARCH0063. doi: 10.1186/gb-2002-3-11-research0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsak M, Gaffal E, Date R, Wang-Eckhardt L, Rehnelt J, Petrosino S, Starowicz K, Steuder R, Schlicker E, Cravatt B, et al. Attenuation of allergic contact dermatitis through the endocannabinoid system. Science. 2007;316:1494–1497. doi: 10.1126/science.1142265. [DOI] [PubMed] [Google Scholar]
- Kaupmann K, Malitschek B, Schuler V, Heid J, Froestl W, Beck P, Mosbacher J, Bischoff S, Kulik A, Shigemoto R, et al. GABA(B)-receptor subtypes assemble into functional heteromeric complexes. Nature. 1998;396:683–687. doi: 10.1038/25360. [DOI] [PubMed] [Google Scholar]
- Kelly MA, Rubinstein M, Asa SL, Zhang G, Saez C, Bunzow JR, Allen RG, Hnasko R, Ben-Jonathan N, Grandy DK, et al. Pituitary lactotroph hyperplasia and chronic hyperprolactinemia in dopamine D2 receptor-deficient mice. Neuron. 1997;19:103–113. doi: 10.1016/s0896-6273(00)80351-7. [DOI] [PubMed] [Google Scholar]
- Kim CH, Kunkel EJ, Boisvert J, Johnston B, Campbell JJ, Genovese MC, Greenberg HB, Butcher EC. Bonzo/CXCR6 expression defines type 1-polarized T-cell subsets with extralymphoid tissue homing potential. J Clin Invest. 2001;107:595–601. doi: 10.1172/JCI11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs HA. Chemical composition of blood plasma and serum. Annu Rev Biochem. 1950;19:409–430. doi: 10.1146/annurev.bi.19.070150.002205. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kumar U, Laird D, Srikant CB, Escher E, Patel YC. Expression of the five somatostatin receptor (SSTR1−5) subtypes in rat pituitary somatotrophes: quantitative analysis by double-layer immunofluorescence confocal microscopy. Endocrinology. 1997;138:4473–4476. doi: 10.1210/endo.138.10.5566. [DOI] [PubMed] [Google Scholar]
- Kuner R, Kohr G, Grunewald S, Eisenhardt G, Bach A, Kornau HC. Role of heteromer formation in GABAB receptor function. Science. 1999;283:74–77. doi: 10.1126/science.283.5398.74. [DOI] [PubMed] [Google Scholar]
- Kushnir MM, Komaromy-Hiller G, Shushan B, Urry FM, Roberts WL. Analysis of dicarboxylic acids by tandem mass spectrometry. High-throughput quantitative measurement of methylmalonic acid in serum, plasma, and urine. Clin Chem. 2001;47:1993–2002. [PubMed] [Google Scholar]
- Le LQ, Kabarowski JH, Weng Z, Satterthwaite AB, Harvill ET, Jensen ER, Miller JF, Witte ON. Mice lacking the orphan G protein-coupled receptor G2A develop a late-onset autoimmune syndrome. Immunity. 2001;14:561–571. doi: 10.1016/s1074-7613(01)00145-5. [DOI] [PubMed] [Google Scholar]
- Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442:645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- Lin SC, Lin CR, Gukovsky I, Lusis AJ, Sawchenko PE, Rosenfeld MG. Molecular basis of the little mouse phenotype and implications for cell type-specific growth. Nature. 1993;364:208–213. doi: 10.1038/364208a0. [DOI] [PubMed] [Google Scholar]
- Lindemann L, Meyer CA, Jeanneau K, Bradaia A, Ozmen L, Bluethmann H, Bettler B, Wettstein JG, Borroni E, Moreau JL, et al. Trace amine-associated receptor 1 modulates dopaminergic activity. J Pharmacol Exp Ther. 2008;324:948–956. doi: 10.1124/jpet.107.132647. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wada R, Yamashita T, Mi Y, Deng CX, Hobson JP, Rosenfeldt HM, Nava VE, Chae SS, Lee MJ, et al. Edg-1, the G protein-coupled receptor for sphingosine-1-phosphate, is essential for vascular maturation. J Clin Invest. 2000;106:951–961. doi: 10.1172/JCI10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsignol A, Vande Vijver V, Ramaekers D, Vankelecom H, Denef C. Detection of melanocortin-3 receptor mRNA in immature rat pituitary: functional relation to gamma3-MSH-induced changes in intracellular Ca2+ concentration? J Neuroendocrinol. 1999;11:171–179. doi: 10.1046/j.1365-2826.1999.00305.x. [DOI] [PubMed] [Google Scholar]
- Lovenberg TW, Baron BM, de Lecea L, Miller JD, Prosser RA, Rea MA, Foye PE, Racke M, Slone AL, Siegel BW, et al. A novel adenylyl cyclase-activating serotonin receptor (5-HT7) implicated in the regulation of mammalian circadian rhythms. Neuron. 1993;11:449–458. doi: 10.1016/0896-6273(93)90149-l. [DOI] [PubMed] [Google Scholar]
- Malone MH, Wang Z, Distelhorst CW. The glucocorticoid-induced gene tdag8 encodes a pro-apoptotic G protein-coupled receptor whose activation promotes glucocorticoid-induced apoptosis. J Biol Chem. 2004;279:52850–52859. doi: 10.1074/jbc.M408040200. [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Hollopeter G, Kafer KE, Palmiter RD. Role of the Y5 neuropeptide Y receptor in feeding and obesity. Nat Med. 1998;4:718–721. doi: 10.1038/nm0698-718. [DOI] [PubMed] [Google Scholar]
- Mazella J, Botto JM, Guillemare E, Coppola T, Sarret P, Vincent JP. Structure, functional expression, and cerebral localization of the levocabastine-sensitive neurotensin/neuromedin N receptor from mouse brain. J Neurosci. 1996;16:5613–5620. doi: 10.1523/JNEUROSCI.16-18-05613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee J, Goodyear RJ, McMillan DR, Stauffer EA, Holt JR, Locke KG, Birch DG, Legan PK, White PC, Walsh EJ, et al. The very large G-protein-coupled receptor VLGR1: a component of the ankle link complex required for the normal development of auditory hair bundles. J Neurosci. 2006;26:6543–6553. doi: 10.1523/JNEUROSCI.0693-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno FJ, Mills I, Garcia-Sainz JA, Fain JN. Effects of pertussis toxin treatment on the metabolism of rat adipocytes. J Biol Chem. 1983;258:10938–10943. [PubMed] [Google Scholar]
- Moskowitz J, Fain JN. Hormonal regulation of lipolysis and phosphorylase activity in human fat cells. J Clin Invest. 1969;48:1802–1808. doi: 10.1172/JCI106146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller G. Towards 3D structures of G protein-coupled receptors: a multidisciplinary approach. Curr Med Chem. 2000;7:861–888. doi: 10.2174/0929867003374534. [DOI] [PubMed] [Google Scholar]
- Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- Nathans J, Hogness DS. Isolation, sequence analysis, and intronexon arrangement of the gene encoding bovine rhodopsin. Cell. 1983;34:807–814. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- Nordmann J, Nordmann R. Organic acids in blood and urine. Adv Clin Chem. 1961;4:53–120. doi: 10.1016/s0065-2423(08)60035-9. [DOI] [PubMed] [Google Scholar]
- Peirson SN, Butler JN, Foster RG. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003;31:e73. doi: 10.1093/nar/gng073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova NK, Naumenko VS, Plyusnina IZ. Involvement of brain serotonin 5-HT1A receptors in genetic predisposition to aggressive behavior. Neurosci Behav Physiol. 2007;37:631–635. doi: 10.1007/s11055-007-0062-z. [DOI] [PubMed] [Google Scholar]
- Rao S, Garrett-Sinha LA, Yoon J, Simon MC. The Ets factors PU.1 and Spi-B regulate the transcription in vivo of P2Y10, a lymphoid restricted heptahelical receptor. J Biol Chem. 1999;274:34245–34252. doi: 10.1074/jbc.274.48.34245. [DOI] [PubMed] [Google Scholar]
- Regard JB, Kataoka H, Cano DA, Camerer E, Yin L, Zheng YW, Scanlan TS, Hebrok M, Coughlin SR. Probing cell type-specific functions of G(i) in vivo identifies GPCR regulators of insulin secretion. J Clin Invest. 2007 doi: 10.1172/JCI32994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodbell M. Metabolism of Isolated Fat Cells. I. Effects of Hormones on Glucose Metabolism and Lipolysis. J Biol Chem. 1964;239:375–380. [PubMed] [Google Scholar]
- Sadagopan N, Li W, Roberds SL, Major T, Preston GM, Yu Y, Tones MA. Circulating Succinate is Elevated in Rodent Models of Hypertension and Metabolic Disease. Am J Hypertens. 2007;20:1209–1215. doi: 10.1016/j.amjhyper.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Bozzi Y, Baik JH, Borrelli E. Antiproliferative role of dopamine: loss of D2 receptors causes hormonal dysfunction and pituitary hyperplasia. Neuron. 1997;19:115–126. doi: 10.1016/s0896-6273(00)80352-9. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Segre D, Deluna A, Church GM, Kishony R. Modular epistasis in yeast metabolism. Nat Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- Shuster DE, Kehrli ME, Jr., Ackermann MR. Neutrophilia in mice that lack the murine IL-8 receptor homolog. Science. 1995;269:1590–1591. doi: 10.1126/science.7667641. [DOI] [PubMed] [Google Scholar]
- Simonin F, Gaveriaux-Ruff C, Befort K, Matthes H, Lannes B, Micheletti G, Mattei MG, Charron G, Bloch B, Kieffer B. kappa-Opioid receptor in humans: cDNA and genomic cloning, chromosomal assignment, functional expression, pharmacology, and expression pattern in the central nervous system. Proc Natl Acad Sci U S A. 1995;92:7006–7010. doi: 10.1073/pnas.92.15.7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha TK, Thajchayapong P, Queener SF, Allen DO, Bell NH. On the lipolytic action of parathyroid hormone in man. Metabolism. 1976;25:251–260. doi: 10.1016/0026-0495(76)90083-4. [DOI] [PubMed] [Google Scholar]
- Soga T, Kamohara M, Takasaki J, Matsumoto S, Saito T, Ohishi T, Hiyama H, Matsuo A, Matsushime H, Furuichi K. Molecular identification of nicotinic acid receptor. Biochem Biophys Res Commun. 2003;303:364–369. doi: 10.1016/s0006-291x(03)00342-5. [DOI] [PubMed] [Google Scholar]
- Soto H, Wang W, Strieter RM, Copeland NG, Gilbert DJ, Jenkins NA, Hedrick J, Zlotnik A. The CC chemokine 6Ckine binds the CXC chemokine receptor CXCR3. Proc Natl Acad Sci U S A. 1998;95:8205–8210. doi: 10.1073/pnas.95.14.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey M, Chang GW, Sanos SL, Chittenden LR, Stubbs L, Gordon S, Lin HH. EMR4, a novel epidermal growth factor (EGF)-TM7 molecule up-regulated in activated mouse macrophages, binds to a putative cellular ligand on B lymphoma cell line A20. J Biol Chem. 2002;277:29283–29293. doi: 10.1074/jbc.M204306200. [DOI] [PubMed] [Google Scholar]
- Sugaya T, Nishimatsu S, Tanimoto K, Takimoto E, Yamagishi T, Imamura K, Goto S, Imaizumi K, Hisada Y, Otsuka A, et al. Angiotensin II type 1a receptor-deficient mice with hypotension and hyperreninemia. J Biol Chem. 1995;270:18719–18722. doi: 10.1074/jbc.270.32.18719. [DOI] [PubMed] [Google Scholar]
- Sun H, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J. Peropsin, a novel visual pigment-like protein located in the apical microvilli of the retinal pigment epithelium. Proc Natl Acad Sci U S A. 1997a;94:9893–9898. doi: 10.1073/pnas.94.18.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Macke JP, Nathans J. Mechanisms of spectral tuning in the mouse green cone pigment. Proc Natl Acad Sci U S A. 1997b;94:8860–8865. doi: 10.1073/pnas.94.16.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susulic VS, Frederich RC, Lawitts J, Tozzo E, Kahn BB, Harper ME, Himms-Hagen J, Flier JS, Lowell BB. Targeted disruption of the beta 3-adrenergic receptor gene. J Biol Chem. 1995;270:29483–29492. doi: 10.1074/jbc.270.49.29483. [DOI] [PubMed] [Google Scholar]
- Tsutsumi M, Zhou W, Millar RP, Mellon PL, Roberts JL, Flanagan CA, Dong K, Gillo B, Sealfon SC. Cloning and functional expression of a mouse gonadotropin-releasing hormone receptor. Mol Endocrinol. 1992;6:1163–1169. doi: 10.1210/mend.6.7.1324422. [DOI] [PubMed] [Google Scholar]
- Tunaru S, Kero J, Schaub A, Wufka C, Blaukat A, Pfeffer K, Offermanns S. PUMA-G and HM74 are receptors for nicotinic acid and mediate its anti-lipolytic effect. Nat Med. 2003;9:352–355. doi: 10.1038/nm824. [DOI] [PubMed] [Google Scholar]
- Valet P, Pages C, Jeanneton O, Daviaud D, Barbe P, Record M, Saulnier-Blache JS, Lafontan M. Alpha2-adrenergic receptor-mediated release of lysophosphatidic acid by adipocytes. A paracrine signal for preadipocyte growth. J Clin Invest. 1998;101:1431–1438. doi: 10.1172/JCI806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varona R, Villares R, Carramolino L, Goya I, Zaballos A, Gutierrez J, Torres M, Martinez AC, Marquez G. CCR6-deficient mice have impaired leukocyte homeostasis and altered contact hypersensitivity and delayed-type hypersensitivity responses. J Clin Invest. 2001;107:R37–45. doi: 10.1172/JCI11297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, Brown A, Rodriguez SS, Weller JR, Wright AC, et al. The G protein-coupled receptor repertoires of human and mouse. Proc Natl Acad Sci U S A. 2003;100:4903–4908. doi: 10.1073/pnas.0230374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JH, Wise A, Main MJ, Green A, Fraser NJ, Disney GH, Barnes AA, Emson P, Foord SM, Marshall FH. Heterodimerization is required for the formation of a functional GABA(B) receptor. Nature. 1998;396:679–682. doi: 10.1038/25354. [DOI] [PubMed] [Google Scholar]
- Willie JT, Chemelli RM, Sinton CM, Tokita S, Williams SC, Kisanuki YY, Marcus JN, Lee C, Elmquist JK, Kohlmeier KA, et al. Distinct narcolepsy syndromes in Orexin receptor-2 and Orexin null mice: molecular genetic dissection of Non-REM and REM sleep regulatory processes. Neuron. 2003;38:715–730. doi: 10.1016/s0896-6273(03)00330-1. [DOI] [PubMed] [Google Scholar]
- Wise A, Foord SM, Fraser NJ, Barnes AA, Elshourbagy N, Eilert M, Ignar DM, Murdock PR, Steplewski K, Green A, et al. Molecular identification of high and low affinity receptors for nicotinic acid. J Biol Chem. 2003;278:9869–9874. doi: 10.1074/jbc.M210695200. [DOI] [PubMed] [Google Scholar]
- Wurbel MA, Malissen M, Guy-Grand D, Meffre E, Nussenzweig MC, Richelme M, Carrier A, Malissen B. Mice lacking the CCR9 CC-chemokine receptor show a mild impairment of early T- and B-cell development and a reduction in T-cell receptor gammadelta(+) gut intraepithelial lymphocytes. Blood. 2001;98:2626–2632. doi: 10.1182/blood.v98.9.2626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.