Abstract
The maternal contribution of gene products enables embryos to initiate their developmental program in the absence of zygotic gene expression. In Caenorhabditis elegans, maternal CDC-25.1 levels are tightly regulated to promote early cell divisions, while stabilization of this phosphatase by gain-of-function mutations gives rise to intestinal-specific hyperplasia. To identify regulators of CDC-25.1 levels and/or function, we performed a modifier screen of the cdc-25.1(gf)-dependent hyperplasia. One of the isolated suppressor mutants possesses a donor splice site mutation in prp-8, a key splicing factor of the U5-specific snRNP. prp-8(rr40) produces aberrant prp-8 splice variants that generate C-terminal truncations at the expense of wild-type prp-8. Levels of maternal transcripts are reduced, including cdc-25.1, while zygotic transcripts appear unperturbed, suggesting a germ-line-specific role for this splicing factor in regulating the splicing, and consequently, the steady-state levels of maternal transcripts. Using a novel feeding RNAi strategy we found that only a subset of splicing factors suppress cdc-25.1(gf), suggesting that they too may play specific roles in germ-line spliceosome function. In humans, mutations in the corresponding hPrp8 C-terminal domain result in retinitis pigmentosa, a retinal-specific disorder. Intriguingly, despite affecting the general splicing apparatus, both human and C. elegans show tissue-specific defects resulting from mutations in this key splicing component. Our findings suggest that in addition to its important regulatory function in the C. elegans germ line, prp-8(rr40) may provide further insight into the etiology of this splicing-associated human disorder.
Keywords: maternal contribution, cdc-25.1, cell cycle, tissue specificity, prp-8, splicing factors, retinitis pigmentosa
INTRODUCTION
Many early embryonic events proceed under maternal control, driven by a largely unlimiting pool of mRNA and protein present in the oocyte and provided by the maternal germ line. This pool of gene products thus circumvents any requirement for the time- and energy-dependent assembly of transcription complexes and allows messages to be translated where and when necessary, while proteins can be efficiently deployed to critical regions of the developing embryo. Such maternal gene products contribute to diverse developmental processes that include axis specification, cell fate determination, and even cell cycle regulation.
From large-scale in situ hybridizations performed in the syncytial embryos of Drosophila melanogaster, it was shown that more than 65% of all genes are expressed in the maternal germ line and delivered to the developing oocyte in the form of mature transcripts, most of which are short lived and display precise localization patterns (Lecuyer et al. 2007). Microarray and EST analysis in mouse indicate that 25%–40% of all genes are deposited maternally in mature oocytes (Wang et al. 2004; Evsikov et al. 2006), while in Xenopus and most amphibians, the early embryonic developmental events rely entirely upon maternal contribution, until zygotic genes become activated at the mid-blastula transition (MBT) (Newport and Kirschner 1982; Heasman 2006). In C. elegans, the transition from maternal to zygotic developmental control occurs during embryogenesis and is subject to lineage- and gene-specific regulation (Bowerman 1998; DeRenzo and Seydoux 2004).
The early cell division cycle is controlled maternally in numerous embryos and often consists of rapid, successive S and M phases without intervening gap (G) phases (Edgar and McGhee 1988; Edgar 1994; Kipreos 2005). The lengthening of these early cell division cycles is usually triggered by the degradation of key maternal cell cycle factors, marking the transition from the maternal to the zygotic control of the cell cycle machinery (O'Farrell et al. 1989; Edgar 1994). In Drosophila, the switch from the early embryonic rapid and synchronous cell division cycles to the introduction of gap phases relies on the depletion of both the twine and string maternal gene products, two Cdc25 orthologs (Edgar and Datar 1996; Edgar and Lehner 1996; Mata et al. 2000). Likewise, in Xenopus, Cdc25A becomes down-regulated after the developmentally controlled activation of an intra-S phase checkpoint to ensure the lengthening of the cell cycle at the MBT (Kim et al. 1999; Shimuta et al. 2002).
Cdc25 family phosphatases have been shown to play pivotal roles in the decision to proceed into S or M phases of the cell cycle by removing the Wee1-mediated inhibitory phosphorylation on cyclin-dependent kinases (CDKs) (Russell and Nurse 1987; Galaktionov and Beach 1991; Mueller et al. 1995). In higher eukaryotes like Drosophila, Xenopus, or mammals, the Cdc25 family consists of related, yet functionally distinct members. C. elegans has four members of the Cdc25 phosphatase family (Ashcroft et al. 1998), with maternal CDC-25.1 being essential for germ-line proliferation, completion of meiosis, and early embryonic divisions (Ashcroft and Golden 2002). These paralogs play redundant, but also unique and characteristic, phase- and/or tissue-specific roles (O'Farrell et al. 1989; Alphey et al. 1992; Courtot et al. 1992; Busino et al. 2004; Perdiguero and Nebreda 2004; Myer et al. 2005). Destabilization of Cdc25 phosphatases is under strict cell cycle and developmental regulation. In mammals, an SCF ubiquitin E3 ligase utilizes the β-TrCP F-box protein to recognize and target Cdc25A for proteasome-mediated degradation during an unperturbed S phase, while DNA damage accelerates this process in an ATM/ATR-Chk1/Chk2-dependent manner to halt the cell cycle (Falck et al. 2001; Busino et al. 2003; Jin et al. 2003; Sorensen et al. 2003). In C. elegans, two maternal-effect, cdc-25.1 gain-of-function (gf) mutations alter residues within a destruction motif recognized by the C. elegans β-TrCP ortholog LIN-23, impairing the efficient SCFLIN-23-mediated degradation of the CDC-25.1 phosphatase during a precise developmental window during embryogenesis, triggering supernumerary cell divisions exclusively in the intestine (Bao et al. 2008; Hebeisen and Roy 2008).
To uncover additional regulatory pathways that control CDC-25.1 levels, we performed a genetic modifier screen using the cdc-25.1(rr31)gf mutant (Kostic and Roy 2002). We report here the isolation of five genes that suppress the intestinal hyperplasia associated with cdc-25.1(gf). We show that one suppressor, rr40, corresponds to a mutation, which results in a partial truncation of the C terminus of the C. elegans 220-kDa U5-specific snRNP ortholog PRP-8, an essential and key component of the spliceosome. We provide evidence that prp-8 plays important and essential roles in establishing and maintaining elevated levels of germ-line-specific gene products (maternal contribution) to sustain subsequent developmental processes throughout embryogenesis, including the proper regulation of cell division cycles. Moreover, when tested by RNAi, we show that only a subset of splicing factors act similar to prp-8 and also suppress the cdc-25.1(rr31)-associated hyperplasia, suggesting that together these factors may play a unique role during germ-line and early embryonic development. Interestingly, mutations localized in the C terminus of the ubiquitous human protein hPrp8 have been associated with autosomal dominant retinitis pigmentosa (adRP), a retinal-specific hereditary disorder (McKie et al. 2001). The intriguing parallels that exist between the human adRP and C. elegans prp-8(rr40) indicate that this mutant may be used to study the genetic and molecular events associated with adRP.
RESULTS
Isolation of mutants that reduce the tissue-specific cdc-25.1(rr31)-associated intestinal hyperplasia
The intestinal hyperplasia associated with stabilized CDC-25.1(gf) protein indicates that the timely degradation of this cell cycle regulator is crucial for the proper formation of the digestive organ. To reveal additional pathways that contribute to CDC-25.1 phosphatase levels during embryogenesis and to uncover what gene products potentiate the cdc-25.1(gf)-associated overproliferation, we performed an EMS-based suppressor screen in the cdc-25.1(rr31) gain-of-function background and selected for viable animals with a reduction of the intestinal hyperplasia (see Materials and Methods).
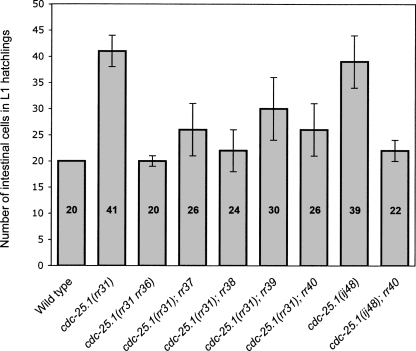
Five recessive, strict maternal effect mutations were isolated, each of which suppresses the cdc-25.1(rr31) phenotype from 50% to 100% (Fig. 1). In all cases, some animals displayed wild-type intestinal cell counts (20 cells), which is never observed in cdc-25.1(rr31) single mutants (data not shown). On the other hand, none of these suppressor mutants had less intestinal cells than the wild type, suggesting that their observed effect is not due to a general reduction in cell division, but rather to specific effects on cdc-25.1(rr31). cdc-25.1(rr31rr36) corresponds to an intragenic mutation that restores the timely degradation of CDC-25.1(rr31rr36) by affecting a potential stabilizing motif/domain of the phosphatase, thereby suppressing the cdc-25.1(rr31)-associated hyperplasia (Hebeisen and Roy 2008). Complementation analysis suggests that the four other suppressors represent individual extragenic loci (data not shown).
FIGURE 1.
Isolation of five viable suppressors of the cdc-25.1(rr31)-associated intestinal hyperplasia. The average number of intestinal cells in L1 hatchings (sample size n > 50) is indicated for each genotype at 20°C. All animals express the intestinal-specific elt-2∷gfp marker that enables visualization and quantification of intestinal cells (Fukushige et al. 1999). Each of the suppressors (rr36–rr40) reduces the cdc-25.1(rr31)-associated intestinal hyperplasia specifically during embryogenesis, with no post-embryonic effect. rr36 is the strongest suppressor and corresponds to an intragenic mutation in the cdc-25.1 gene (Hebeisen and Roy 2008). cdc-25.1(ij48) corresponds to a second gain-of-function allele of the phosphatase that produces intestinal-specific hyperplasia similar to cdc-25.1(rr31) (Clucas et al. 2002). ij48 and rr31 both affect residues of the highly conserved DSG(X)4S destruction motif. Error bars represent standard deviation.
rr40 displays pleiotropic and temperature-sensitive effects
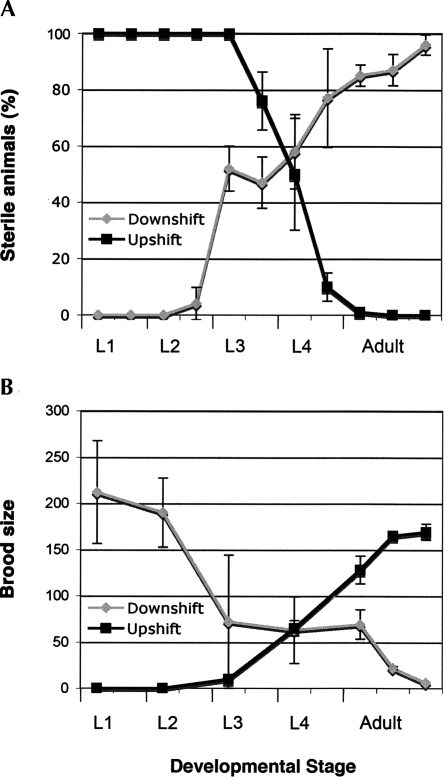
Apart from its ability to suppress the embryonic intestinal hyperplasia of cdc-25.1(rr31) mutants, rr40 displayed 26% embryonic (Emb) and early larval lethality (Lvl) (n = 2837) at 20°C together with a high incidence of male (Him) phenotype of 5.3% (n = 747), suggesting that rr40 may affect some aspects of meiotic progression or chromosome segregation. A fraction of hermaphrodites also showed sterility (Ste), while some had protruding, fragile, or multivulvae (Pvl, Rup, Muv), indicating that rr40 displays pleiotropic effects that affect animal development at different stages. The severity of these phenotypes varied with temperature. Temperature-shift experiments revealed that larvae downshifted before the L3 stage were fertile and had average brood sizes of ∼200 eggs with 20% embryonic lethality, while those downshifted after the L3 stage exhibited increasing and irreversible sterility. The few animals that produced eggs in the later downshifts had brood sizes of <10 eggs, among which ∼50% died during embryogenesis or at an early larval stage (Fig. 2, downshifts; data not shown). Complete sterility was detected in animals upshifted before the L3 stage, while penetrant embryonic/larval lethality (>95%) was observed in the progeny of fertile L4 or adult animals upshifted to the restrictive temperature (Fig. 2, upshifts; data not shown). Control cdc-25.1(rr31) mutants remain fertile and develop normally at 25°C (data not shown).
FIGURE 2.
The rr40 gene product is required from L3 to adulthood to produce and maintain a functional germ line. Reciprocal temperature-shift assays for cdc-25.1(rr31); rr40 suppressors demonstrate two major requirements for the rr40 gene product. Upshifts from 15°C (permissive) to 25°C (restrictive) are represented by black lines; downshifts from 25°C to 15°C by gray lines. (A) A first temperature-sensitive period is detected between the larval L3 and L4 stages, where rr40 is required to establish a functional gamete-producing germ line. (B) rr40 is also required continuously from the L3 stage until adulthood to maintain a normal brood size and to avoid the rapid and penetrant embryonic and early larval lethality detected in fertile adults that have been shifted at the restrictive temperature.
Thus, it appears that the rr40 gene product is required during and after the L3 stage to promote gametogenesis and throughout the subsequent stages to maintain the production of germ cells. These results, together with its maternal effect, suggest that rr40 functions mainly in the developing hermaphrodite germ line.
rr40 specifically suppresses the extra intestinal divisions observed in cdc-25.1(gf) mutants
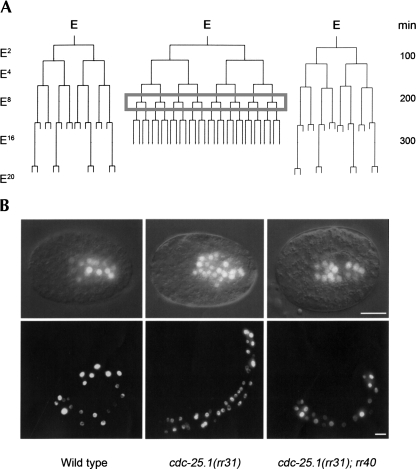
To address whether rr40 affects intestinal cell divisions during embryonic or post-embryonic development, we quantified the number of intestinal cells in cdc-25.1(rr31); rr40 hatchlings. These early L1 larvae had a reduced number of intestinal cells compared with cdc-25.1(rr31) single mutants (Fig. 1), while 32% possessed wild-type numbers of cells (n = 100) (Fig. 3B, bottom). To determine whether rr40 specifically suppresses the supernumerary divisions caused by cdc-25.1(rr31) after the E8 stage, or whether it eliminates another round of division during embryogenesis to indirectly compensate, we performed lineage analysis on cdc-25.1(rr31); rr40 embryos (Fig. 3A). We found that the supernumerary cell divisions caused by cdc-25.1(rr31) were specifically eliminated in ∼35% of the cdc-25.1(rr31); rr40 embryos (n = 16), allowing these cells to undergo the correct division pattern to produce the wild-type complement of intestinal cells (Fig. 3B, top left panel). No defect was observed in either the nuclear divisions or in the endocycles that occur after the L1 stage in the C. elegans intestine (data not shown).
FIGURE 3.
rr40 specifically suppresses the cdc-25.1(rr31)-associated supernumerary intestinal cell divisions. (A) A representative lineage map of the embryonic intestinal (E) lineage of cdc-25.1(rr31) mutants is shown alongside those of wild-type (left) and cdc-25.1(rr31); rr40 suppressors (right). E represents the founder endodermal cell specified at the embryonic 7-cell stage and the number of E descendants in wild type is indicated on the left. All animals express integrated end-3∷gfp and elt-2∷gfp transgenes, which together mark the intestinal nuclei from the embryonic E2 stage onward. The gray box highlights the supernumerary round of division that occurs during the E8 stage in cdc-25.1(rr31) and causes intestinal hyperplasia. Time scale is indicated on the right in minutes after fertilization. (B) Representative elt-2∷gfp expressing 300-min-old embryos (top) and L1 hatchlings (bottom) demonstrating the rr40-mediated suppression (right) of the cdc-25.1(rr31)-associated extra cell divisions (middle). Thirty-two percent of the cdc-25.1(rr31); rr40 suppressors were phenotypically indistinguishable from wild-type animals at 20°C (left). Scale bars, 10 μm.
To further confirm that rr40 does not simply impair cell divisions generally throughout embryogenesis, we tested whether rr40 would affect cell numbers independent of cdc-25.1(rr31) by generating rr40 single mutants. These animals consistently possessed wild-type numbers of intestinal cells and nuclei and showed the same pleiotropic spectrum of phenotypes as well as the temperature sensitivity previously described for cdc-25.1(rr31); rr40 double-mutant animals. Finally, to determine whether the rr40-mediated suppression was specific to the cdc-25.1(rr31) allele, rr40 was examined for its ability to suppress the intestinal hyperplasia of a second cdc-25.1(ij48) gain-of-function mutation that affected the same destruction motif (Clucas et al. 2002). rr40 similarly suppressed this cdc-25.1(ij48) allele (Fig. 1), suggesting that it is able to interfere with the cell cycle defect associated with these stabilized CDC-25.1(gf) proteins.
Taken together, these observations indicate that rr40 is a specific, maternal effect suppressor of the cdc-25.1(gf)-associated intestinal hyperplasia, while causing temperature-sensitive pleiotropies throughout development.
rr40 affects prp-8, a crucial and highly conserved component of the spliceosome
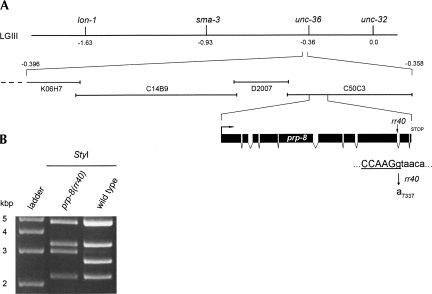
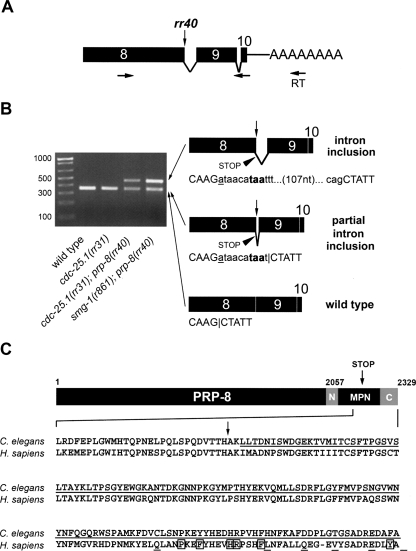
We mapped and cloned rr40 using SNP-Snip, deficiency, and 3-point mapping approaches, together with a novel hypomorphic feeding RNAi strategy (Materials and Methods; Supplemental Fig. S1). A single RNAi clone that corresponds to prp-8 (C50C3.6) fully suppressed the cdc-25.1(rr31)-associated hyperplasia and phenocopied the pleiotropic effects of rr40 (Fig. 4A; data not shown). prp-8 is an essential gene and prp-8(RNAi) causes 100% sterility or embryonic lethality (Gonczy et al. 2000; Kamath et al. 2003). Thus, prp-8(rr40) likely corresponds to a hypomorphic allele. To identify the nature of the rr40 molecular lesion, we sequenced the prp-8 gene in cdc-25.1(rr31); prp-8(rr40) suppressors. An EMS typical base-pair substitution (G7337A) was detected at the second-last donor splice site of the prp-8 genomic sequence, which was confirmed by demonstrating the formation of a StyI restriction fragment polymorphism in the mutant genomic sequence (Fig. 4B). Thus, the invariant GU nucleotides that mark the 5′ intron boundary are converted to AU in the affected prp-8(rr40) intron (Fig. 4A).
FIGURE 4.
Genetic mapping of rr40 indicates that it corresponds to an allele of the highly conserved U5-specific snRNP component prp-8. (A) Meiotic mapping in cdc-25.1(rr31) was used to place rr40 to the left of linkage group III between the visible markers unc-36 and sma-3. Feeding RNAi against the 26 genes present in this interval revealed that only C50C3.6(RNAi) phenocopied the rr40 suppression. Independent sequence analysis indicated that rr40 disrupts the second-last intron donor splice site of the prp-8 gene, converting the highly conserved GT donor sequence to AT and creating a StyI restriction polymorphism (underlined sequence). (B) StyI-digested PCR amplification products of wild-type and prp-8(rr40) genomic DNA confirm the G7337A mutation generated by rr40.
PRP-8 corresponds to the C. elegans U5-specific small nuclear ribonucleoparticle protein (snRNP) orthologous to the Saccharomyces cerevisiae Prp8p, the Drosophila prp8 (CG8877), and the human U5-specific 220-kDa protein hPrp8. This spliceosomal component is one of the largest and most highly conserved nuclear proteins among eukaryotes, displaying 58% identity between C. elegans and S. cerevisiae and 86% identity between C. elegans and human (Hodges et al. 1995). Prp8 was first identified in yeast in a screen for temperature-sensitive lethal mutations (Hartwell, 1967) and characterized for its role in pre-mRNA processing (Vijayraghavan et al. 1989), where it plays central roles in the 5′ and 3′ splice site recognition of pre-mRNAs in addition to its function in the extensive remodeling steps that occur during spliceosomal activation and in both catalytic transesterification reactions (Teigelkamp et al. 1995; Grainger and Beggs 2005).
prp-8(rr40) affects the correct splicing of its own intron
The accuracy of the splice site sequences is crucial for the precise recognition and removal of introns by the spliceosome. Therefore, we wondered whether the affected donor splice site in prp-8(rr40) pre-mRNA would perturb the proper splicing events of that particular intron, a process in which PRP-8 itself plays a critical role.
To reveal the splicing events that occur at the splice site affected by rr40 between exon 8 and intron 8 of prp-8(rr40), the sequences of the mRNA species that span from the middle of exon 8 to the middle of exon 10 in wild-type and prp-8(rr40) animals were amplified by semiquantitative RT–PCR (see Materials and Methods) (Fig. 5A). Instead of the single wild-type amplification product detected in control animals, two bands appeared in the cdc-25.1(rr31); prp-8(rr40) suppressor, the levels of which were reduced compared with the control (Fig. 5B). Sequence analysis of these two products indicated that intron 9 was spliced correctly in all cases, providing an internal control for spliceosomal efficiency and excluding the possibility of DNA contamination. This analysis revealed the presence of at least three different rr40 splice variants (Fig. 5B). The first sequence corresponded to the expected wild-type prp-8 message (356 nucleotides [nt]), indicating that it had been properly spliced despite its unconventional AU 5′ donor splice site. The second sequence corresponded to an aberrant message (366 nt) with a 10-nt intron inclusion produced by splicing of an unconventional UU 5′ donor splice site at intron position +10 (Fig. 5B). The third sequence (478 nt) corresponded to a second aberrant message that retained the entire 122-nt intron. The translation of both aberrant messages is predicted to produce a truncated protein that would lack the C-terminal-most 138 amino acids, removing a regulatory region shown to be very conserved among orthologs and crucial for protein interactions with other splicing factors (Fig. 5C) (Grainger and Beggs 2005; Pena et al. 2007; Zhang et al. 2007).
FIGURE 5.
Differentially spliced prp-8 mRNA species are detected in prp-8(rr40) mutants. Total mRNA was isolated from wild-type, cdc-25.1(rr31), cdc-25.1(rr31); prp-8(rr40), and smg-1(r861); prp-8(rr40) mutants and reverse transcribed (RT) with polydT primers in a controlled, semiquantitative manner (Materials and Methods). (A) Primers used for PCR amplification were located in exon 8 (sense) and exon 10 (antisense) of prp-8. The rr40 mutation is indicated by a vertical arrow. (B) Species of cDNA of varying sizes were excised from 2% agarose gels and subjected to sequence analysis (left). The sequences indicated that animals mutant for prp-8(rr40) possess wild-type and at least two aberrant forms of prp-8 mRNA, while only the wild-type prp-8 sequence was detected in both wild-type and cdc-25.1(rr31) animals. Schematic representations of the three mRNA species obtained from the prp-8(rr40) mutants are depicted on the right. Mature mRNA containing full or partial intron inclusions encode premature stop codons (arrowhead) followed by a frameshift. Uppercase letters correspond to exon sequence, lowercase letters represent intron 8. The rr40 G7337A substitution (vertical arrow) is underlined in the sequences. The nucleotides highlighted in bold represent the first stop codon encoded in the aberrant prp-8 mRNAs and the horizontal bars indicate the detected splice sites. The intron between exons 9 and 10 was correctly spliced in all mRNA species. Note that compromising the NMD pathway increases the level of aberrant prp-8(rr40) mRNA. (C) The translation of both aberrant prp-8(rr40) mRNAs is predicted to result in a truncated PRP-8(rr40) protein that lacks the C-terminal-most 138 amino acids (underlined C. elegans sequence after the STOP [vertical arrow]). The crystal structure of the peptide encompassing the N-terminal extension (N), the MPN domain and the C-terminal extension (C) was resolved for C. elegans PRP-8 (Zhang et al. 2007). The rr40-mediated truncation removes half of the PRP-8 MPN interacting domain and the entire C-terminal extension. The corresponding, highly conserved human hPrp8 sequence is represented underneath, with all hPrp8 mutations associated with human autosomal dominant retinitis pigmentosa RP13 (boxed and underlined residues represent substitution and frameshift mutations, respectively).
In summary, we show that the donor splice site mutation present in the second last prp-8(rr40) intron affects its own splicing, producing multiple prp-8 messages, two of which harbor premature termination codons (PTCs) in the C terminus due to intron inclusions and consequent frameshifts, while also producing wild-type prp-8 transcript, the levels of which are reduced.
Aberrant prp-8(rr40) transcripts are partially targeted by the NMD pathway
Our semiquantitative PCR data suggest that the wild-type prp-8 mRNA levels may be reduced, to the advantage of the mutant variants, while these aberrant mRNAs may be targeted for degradation by the nonsense-mediated mRNA decay (NMD) surveillance pathway that recognizes transcripts with PTCs (Longman et al. 2007).
To confirm the effect of prp-8(rr40) on the levels of its own transcripts, we compared the prp-8 mRNA levels in wild-type, cdc-25.1(rr31), and cdc-25.1(rr31); prp-8(rr40) animals by quantitative Northern analysis using a probe complementary to an exon sequence present in all prp-8(rr40) variants. As expected, prp-8 transcripts from control animals were detected as a single 7.4-kb band, whereas prp-8 mRNA from prp-8(rr40) mutants formed a slightly broader and more diffuse molecular weight band, the levels of which were reduced approximately twofold compared with control prp-8 levels, consistent with the semiquantitative RT–PCR results (Fig. 6A, top; data not shown).
FIGURE 6.
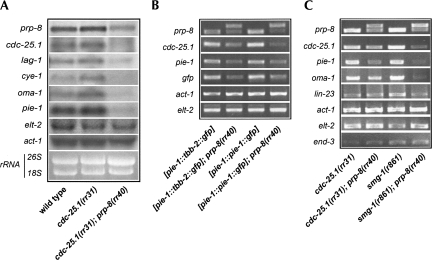
prp-8(rr40) reduces the levels of prp-8 and other maternal mRNA transcripts, without affecting zygotic transcripts. (A) Quantitative Northern analysis of the indicated mRNA performed on wild-type, cdc-25.1(rr31), and cdc-25.1(rr31); prp-8(rr40) animals. Wild-type and cdc-25.1(rr31) control animals show little variation in their mRNA levels, while prp-8 levels (top) and the levels of five other maternally provided transcripts (cdc-25.1, lag-1, cye-1, oma-1, pie-1) are substantially reduced in cdc-25.1(rr31); prp-8(rr40) suppressors compared with the rRNA loading controls and the zygotically transcribed actin (act-1) and endodermal-specific GATA factor (elt-2) genes. (B) Representative semiquantitative RT–PCRs performed on AZ244 and JH1327 animals (first and third column, respectively; see Materials and Methods for complete genotypes). These transgenic animals possess different full-length genomic, intron-containing GFP constructs exclusively in their germ line (pie-1 promoter). Both the endogenous and transgenic maternal mRNA levels are reduced in a prp-8(rr40) mutant background (second and last column, respectively), while the zygotic act-1 and elt-2 messages seem unaffected. (C) The prp-8(rr40)-associated reduction in maternal mRNA levels (cdc-25.1, pie-1, and oma-1) does not depend on the NMD of unspliced pre-mRNAs, as single RT–PCR amplicons corresponding to correctly spliced mRNAs are produced in the smg-1(r861); prp-8(rr40) double mutant, with the exception of prp-8(rr40). Every primer set encompasses one or multiple genomic introns. Representative semiquantitative RT–PCR products indicate that compromise of the NMD pathway modestly decreases the levels of the maternal mRNAs, while the levels of zygotically expressed transcripts (act-1, elt-2, end-3, and lin-23) are more stable. Note that lin-23 is contributed both maternally and zygotically.
To test whether the aberrant prp-8 transcripts are recognized and subsequently eliminated by NMD, we performed semiquantitative RT–PCR of prp-8 between exon 8 and exon 10 in prp-8(rr40); smg-1(r861) mutants that cannot elicit the NMD pathway (Hodgkin et al. 1989). While the levels of the prp-8 wild-type variant appear unaffected, blocking the NMD pathway consistently increased the population of aberrant prp-8(rr40) mRNA that retained the entire intron (Fig. 5B). Since a significant amount of aberrantly spliced mRNA was still detectable in prp-8(rr40) mutants with functional NMD pathways (Fig. 5B), we propose that these PTC-containing messages can only partially elicit a NMD response.
prp-8(rr40) affects the levels of maternal mRNA in an NMD-independent manner
PRP-8 has a highly conserved role in the formation and the catalysis of the spliceosomal complexes; thus, we reasoned that reduced levels of endogenous wild-type PRP-8 could potentially affect the splicing efficiency of every pre-mRNA. To test this possibility, we examined the mRNA levels of a number of representative transcripts by Northern analysis, including five maternally provided transcripts (oma-1, pie-1, cye-1, cdc-25.1, and lag-1) in addition to two zygotically transcribed messages (act-1 and elt-2). Surprisingly, only the maternally provided mRNA levels were reduced in prp-8(rr40) mutants, while the levels of the zygotically expressed transcripts were comparable to that of the control animals and were unaffected by the prp-8(rr40) mutation (Fig. 6A). These results were reproduced independently by semiquantitative RT–PCR for both endogenous and transgenic transcripts in various genetic backgrounds (Fig. 6B,C), confirming the differential sensitivity to prp-8(rr40) between maternal and zygotic transcripts.
To determine whether the reduction in maternal mRNA levels was due to splicing defects that would result in the NMD of incorrectly or unspliced pre-mRNA, we performed semiquantitative RT–PCR of these transcripts in smg-1(r861); prp-8(rr40) double mutants. Similar to what we found in prp-8(rr40), single RT–PCR products were detected in smg-1(r861); prp-8(rr40) for all maternal and zygotic transcripts tested (except for prp-8 itself), corresponding to the expected sizes for each properly spliced mRNA (Fig. 6B,C). These data suggest that splicing per se is functional in prp-8(rr40) mutants and that the observed decrease in the maternal mRNAs occurs independently of the NMD pathway.
To assess whether the spectrum of phenotypes associated with prp-8(rr40) are due exclusively to the decreased levels of functional full-length PRP-8, or if the truncated prp-8(rr40) gene products may contribute to these defects, potentially in a dominant-negative manner, we analyzed the degree of the cdc-25.1(rr31)-dependent intestinal hyperplasia in cdc-25.1(rr31); prp-8(rr40); smg-1(RNAi) animals, in which the truncated forms of prp-8 are more abundant (Fig. 5B). The RNAi-mediated knockdown of smg-1 had a minimal effect on the intestinal cell numbers observed in cdc-25.1(rr31); prp-8(rr40) hatchlings (27.5 ± 5.5 [n = 64] in cdc-25.1(rr31); prp-8(rr40); smg-1(RNAi) animals versus 25.8 ± 5.3 [n = 80] in cdc-25.1(rr31); prp-8(rr40)), suggesting that loss of the NMD pathway does not significantly modify the prp-8(rr40)-associated suppression of the intestinal hyperplasia. The intestinal cell counts in both smg-1(r861) and wild-type animals subjected to smg-1(RNAi) looked completely wild type (20 intestinal cells). In addition, we also quantified the penetrance of the prp-8(rr40)-associated Emb and Him phenotypes in smg-1(r861); prp-8(rr40) double mutants (31% Emb, n = 1425 and 8.4% Him, n = 986) and found that they were only marginally enhanced compared with prp-8(rr40) single mutants (26% Emb, n = 2837 and 5.3% Him, n = 747), while the frequency of Emb and Him phenotypes in control smg-1(r861) were essentially indistinguishable from wild-type animals (Emb, 1.2%, n = 1870, and 0.8% Him, n = 1848). Thus, it seems that the truncated variant of PRP-8(rr40) does not appear to act dominant negatively when permitted to accumulate in a Smg background, as it does not significantly contribute to the prp-8(rr40)-associated defects. However, since blocking the NMD pathway in prp-8(rr40) mutants reduced the levels of maternal mRNAs, albeit only marginally (compare second and last column in Fig. 6C), we cannot formally exclude that the truncated PRP-8(rr40) protein is also capable of assembling into the spliceosome, which might consequently reduce its efficiency.
Taken together, we found that maternally loaded transcripts are very sensitive to the prp-8(rr40) mutation, while the levels of the zygotic transcripts appear less perturbed by these changes. Moreover, the decline in maternal mRNAs was not dependent on NMD of unspliced pre-mRNA. Our results imply that the low level of wild-type PRP-8 protein synthesized in prp-8(rr40) mutants meets or surpasses a functional threshold that is sufficient for the splicing of pre-mRNAs, allowing most of these animals to survive, while the truncated prp-8(rr40) gene products contribute only minimally, if at all, to the various prp-8(rr40)-associated phenotypic traits.
Low PRP-8 levels suppress a second maternal effect gain-of-function mutation
To determine whether the prp-8(rr40)-associated differences in maternally loaded mRNA levels were physiologically relevant, we investigated whether reduced levels of PRP-8 could affect a previously characterized mutation in oma-1, an essential maternal gene that coordinates maternal protein degradation during early embryogenesis (Lin 2003). OMA-1 becomes phosphorylated by MBK-2, which is required both for its activation in the one-cell embryo and its subsequent degradation after the first mitosis (Lin 2003; Nishi and Lin 2005; Shirayama et al. 2006; Stitzel et al. 2006). oma-1(zu405) is a gain-of-function mutant, wherein its mbk-2-dependent destruction site becomes stabilized, similar to what occurs with the CDC-25.1(rr31) protein (Kostic and Roy 2002; Nishi and Lin 2005). This perturbation in OMA-1(zu405) destabilization affects the timely degradation of PIE-1, POS-1, MEX-1, MEX-5, and SKN-1 among others, causing these maternal factors to perdure, triggering embryonic arrest (Lin 2003; Shirayama et al. 2006).
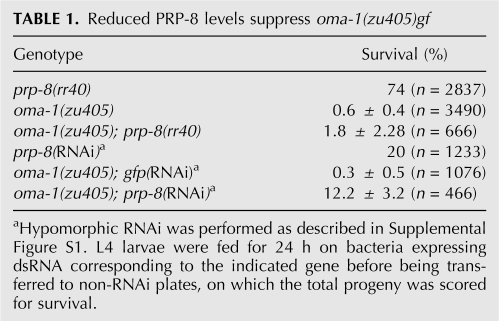
If PRP-8 plays a general role in maintaining adequate levels of maternal transcripts, then prp-8(rr40) or prp-8(RNAi) should reduce the levels of the proteins stabilized in oma-1(zu405) and attenuate the severity of its embryonic lethal phenotype. Consistent with this, we found that a reduction in prp-8 expression levels reproducibly increased the number of viable embryos in the oma-1(zu405); prp-8(rr40) double mutants (threefold), while oma-1(zu405); prp-8 (hypomorphic RNAi) resulted in a 50-fold increase in the number of survivors compared with oma-1(zu405) animals fed on control gfp(RNAi)-inducing bacteria (Table 1).
TABLE 1.
Reduced PRP-8 levels suppress oma-1(zu405)gf
Therefore, the reduction in prp-8 expression levels suppresses another stabilizing maternal-effect gf mutation, lending further genetic support for an important role of PRP-8 in the germ line in maintaining the levels of maternally provided transcripts.
Only a subset of splicing factors comprising U2- and U5-specific snRNPs suppress the cdc-25.1(gf) embryonic phenotype
Because of the essential role PRP-8 plays in the spliceosomal reactions and based on our previous results, it is plausible that limiting PRP-8 activity would mainly affect maternal mRNA levels through affecting overall splicing efficiency.
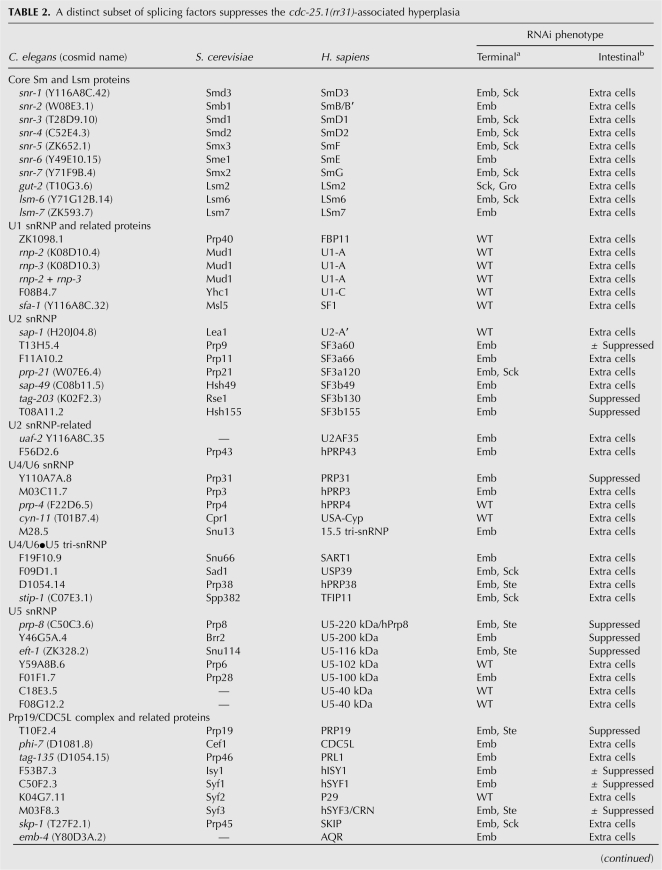
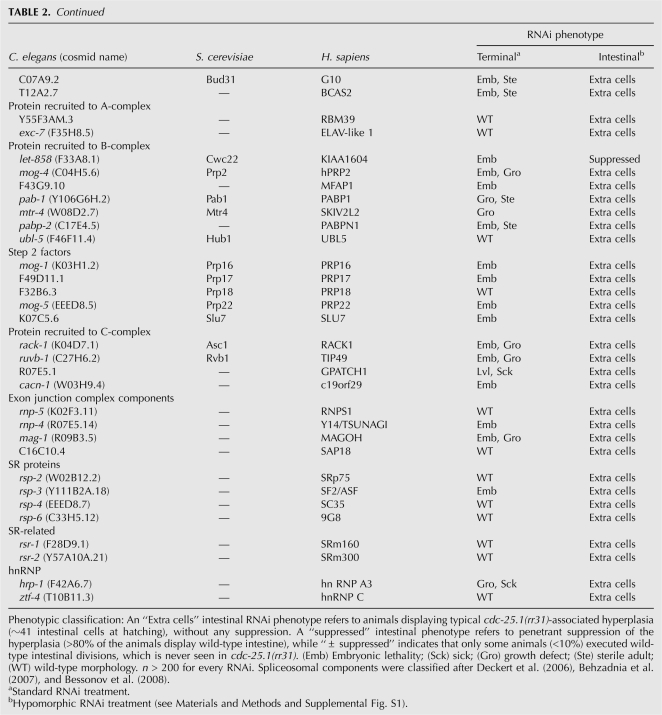
To determine whether a general reduction of splicing activity may account for our findings, we tested whether decreasing the expression levels of other splicing components by RNAi would suppress the cdc-25.1(rr31)-associated intestinal hyperplasia in a manner similar to prp-8(rr40) or prp-8(RNAi). A total of 81 dsRNA-expressing RNAi clones, all representing genes implicated in the splicing process or associated splicing events were fed to cdc-25.1(rr31) animals (Table 2). Surprisingly, only a subset of the clones that induce embryonic lethality as a terminal phenotype gave rise to significant and reproducible suppression of the cdc-25.1(rr31)-associated cell cycle defect. These genes corresponded to essential spliceosomal components and were primarily members of the U2- and U5-specific snRNP proteins, or components of the PRP19 complex, which is also transiently associated with the U5 snRNP in a larger 35S complex (Makarov et al. 2002; Table 2).
TABLE 2.
A distinct subset of splicing factors suppresses the cdc-25.1(rr31)-associated hyperplasia
That the compromise of other splicing components also suppresses the cdc-25.1(rr31)-associated intestinal hyperplasia is consistent with PRP-8(rr40) triggering its effects through reducing spliceosomal efficiency. However, only a limited subset of the essential splicing factors tested, namely, those associated with U2, U5, and/or PRP19, are capable of altering the intestinal hyperplasia of cdc-25.1(rr31) mutants like prp-8(rr40), suggesting that in C. elegans these factors/complexes may play specialized and limiting roles in regulating spliceosome efficiency in a tissue/germ-line-specific manner.
DISCUSSION
The early developmental program of C. elegans is tightly regulated by gene products that are expressed within the maternal germ line. The expression levels of such genes would be expected to be very high in order to support the developmental events that unravel during embryogenesis, while the localization, activity, and degradation of these gene products has to be tightly regulated and coordinated with the emerging zygotic gene-expression program. The isolation of a strict maternal effect gain-of-function mutation in cdc-25.1 that triggers tissue-specific embryonic hyperplasia by interfering with the timely proteolysis of the CDC-25.1(gf) phosphatase indicates that the regulation of the early embryonic cell divisions can be perturbed by changes in the levels of crucial maternal factors. The importance of Cdc25A expression and degradation during animal development prompted us to look for genes that would regulate the levels of this phosphatase, using suppression of the cdc-25.1(gf)-associated intestinal hyperplasia as a readout. With this strategy, we identified unexpected tissue-specific maternal roles for the general splicing factor prp-8.
prp-8(rr40) causes incomplete cryptic splicing, decreasing its own expression level
We found that the prp-8(rr40) allele contains a mutant donor splice site, which induces cryptic splicing and affects the efficient removal of the affected intron. In prp-8(rr40) mutants, wild-type prp-8 transcripts are detectable but at reduced levels, while the additional prp-8 mRNA variants that are generated are partially eliminated by NMD.
This incomplete targeting of the prp-8(rr40) PTC by the NMD is likely due to the positional effect that has been described for NMD response in C. elegans. Unlike in mammals, PTC recognition seems to be independent of the formation of a downstream exon junction complex (EJC) at splice site boundaries, but mainly dependent on the distance of the PTCs from the natural 3′ end of the transcript (Longman et al. 2007). Consistent with this, the partial NMD response associated with aberrant prp-8(rr40) transcripts may reflect the close proximity of their PTCs to the natural stop, similar to what has been reported for other incompletely degraded PTC-containing transcripts (Dibb et al. 1985; Bejsovec and Anderson 1988; Longman et al. 2007).
On the other hand, the ability of the spliceosome to recognize and properly process unconventional AU or UU donor splice sites, as detected in prp-8(rr40) mRNAs, has already been noticed in other C. elegans splice site mutants (Aroian et al. 1993; Rushforth and Anderson 1996). This phenomenon reveals the intriguing plasticity of the spliceosomal macrocomplex in identifying splice site boundaries and indicates that the information required in cis to mark the splice site boundaries is not exclusively encrypted in the highly conserved donor and acceptor sequences.
PRP-8 plays a predominant role in the germ line
In C. elegans, the germ line represents the only tissue with sustained cell division and growth, enabling germ cells to proliferate after every somatic cell division is terminated. Furthermore, cellularization ensures that oocytes are loaded with the maternal factors required for oogenesis and early embryonic development. We show that the mRNA levels of transcripts that are expressed in the maternal germ line are reduced in an NMD-independent manner when wild-type PRP-8 levels are reduced by rr40, while the levels of zygotically transcribed messages seem less affected. This phenomenon highlights an intrinsic difference in the splicing and/or expression requirement between the germ line and the somatic tissues in C. elegans. High levels of functional PRP-8 protein are likely necessary in the germ line to support its important splicing requirement, and thus, a reduction in PRP-8 levels below a critical threshold would predominantly affect mechanisms that rely on maternal factors. Consistent with this, the maternal effect of the prp-8(rr40) mutation, its associated Ste and Emb phenotypes, together with its temperature-sensitive period and its effect on cdc-25.1(rr31) and oma-1(zu405), all pinpoint toward a significant role for PRP-8 in the germ line. Remarkably, available in situ hybridization data indicates that prp-8 is almost exclusively expressed in the germ line, with almost no detectable signal in the somatic tissues (http://nematode.lab.nig.ac.jp/db2/ShowGeneInfo.php?celk=CELK00510).
It is surprising that a gene product presumed to be required in all cells for the splicing of every transcript would be restricted to the germ line of an organism. Thus, it seems most likely that prp-8 is abundantly expressed in the developing germ line to enable the high splicing requirement of maternal transcripts in early embryos, and that later on, this maternal prp-8 contribution (in the form of stable mRNA and/or protein) and possibly limited zygotic prp-8 expression, would suffice for the lower splicing requirement necessary for the limited and periodic developmental pulses of zygotic expression. Consistent with the important requirement for RNA metabolism in the germ line and early embryos, maternal transcripts corresponding to genes involved in splicing (including prp8) and translation were enriched in Drosophila embryos and detected to be the most stable of all mRNA (Lecuyer et al. 2007).
Only a subset of splicing factors suppress intestinal hyperplasia
By using the feeding RNAi library, we noticed that only a subset of the 81 tested splicing factors was able to suppress the cdc-25.1(rr31)-associated hyperplasia (Table 2). Surprisingly, essential splicing factors like the U1 to U5 core snRNP components snr-1 to snr-7 that give rise to complete embryonic lethality as a terminal phentoype did not show any suppression of the hyperplasia following RNAi treatment. Since our feeding RNAi protocol guarantees a reproducible RNAi effect by triggering the strongest viable hypomorphic phenotype possible for any clone resulting in 100% lethality (Supplemental Fig. S1; Materials and Methods), this result suggests that a general reduction of splicing efficiency cannot fully account for the observed suppression.
The suppressors reported in Table 2 correspond mainly to members of the U2 snRNP, U5 snRNP, or associated factors thereof, like the Prp19 complex (NTC) members that transiently associate with U5 proteins (Makarov et al. 2002). Intriguingly, the C-terminal fragment that is deleted in the truncated variants of PRP-8(rr40) corresponds to a protein-interaction domain that enables interaction with its own N-terminal domain and with at least two other U5-specific splicing factors, namely Brr2 and Snu114 (Pena et al. 2007; Zhang et al. 2007). Remarkably, we found that RNAi-mediated knockdown of their C. elegans orthologs completely suppressed the cdc-25.1(rr31)-associated hyperplasia, similar to prp-8 (Table 2). Consistent with the high homology of Prp8 proteins between species, both at the molecular and functional levels, it is very likely that C. elegans PRP-8 also interacts with these factors through its C-terminal domain to form a multisubunit complex. We suggest that the factors that suppress the cdc-25.1(rr31)-associated hyperplasia represent a specific class of spliceosomal components, the function and/or level of which may be particularly important in the C. elegans germ line to regulate the levels of mature maternal mRNAs that have to be loaded into the developing oocytes to control embryonic cell division cycles, in addition to other developmental processes.
Links between splicing eventsand cell cycle regulation
Links between splicing events and cell cycle regulation that involve Prp8 and other splicing components have been reported in budding and fission yeast, while in Drosophila, orthologs of the splicing factors that suppress cdc-25.1(rr31) affect cell growth, supporting the idea that the links between splicing and cell cycle control may have been conserved from yeast to higher eukaryotes (Shea et al. 1994; Lundgren et al. 1996; Potashkin et al. 1998; Ben-Yehuda et al. 2000a,b; Russell et al. 2000; Habara et al. 2001; Coelho et al. 2005).
It seems unlikely that PRP-8 [or the other cdc-25.1(rr31) suppressors] plays any kind of direct role in cell cycle regulation, mainly because of the pleiotropic phenotypes associated with prp-8(rr40). Consistent with this, recent reports provide growing evidence that in yeast, the cell cycle phenotypes arise through splicing-dependent mechanisms that indirectly affect the expression of proteins involved in the regulation of cell cycle and/or growth (Burns et al. 2002; Dahan and Kupiec 2002, 2004; Rasheva et al. 2006; Kaplan and Kupiec 2007). Since compromise of maternal cdc-25.1 or cye-1 (cyclin E) can suppress the cdc-25.1(rr31)-associated hyperplasia (Kostic and Roy 2002), it is more plausible that the prp-8(rr40)-mediated suppression is caused, at least in part, by the reduction of these two important maternally contributed cell cycle regulators, the levels of which would become limiting for the supernumerary intestinal divisions, resulting in a tissue-specific suppression of the hyperplasia.
Tissue-specific defects from mutationsin general splicing factors
In metazoans, null mutations in spliceosomal components are most often embryonic lethal, owing to their essential functions at the cellular level. Interestingly, mutations that disrupt the assembly or function of snRNPs have been associated with several tissue-specific diseases in humans (Faustino and Cooper 2003). Haploinsufficent and/or dominant-negative mutations in human hPrp8, along with mutations in three other general and ubiquitous splicing factors (hPrp31, hPrp3, and Pap1) involved in the formation of U4/U6·U5 tri-snRNP (Makarova et al. 2002), have been linked with various forms of the hereditary autosomal dominant retinal-specific disorder retinitis pigmentosa (McKie et al. 2001; Vithana et al. 2001; Chakarova et al. 2002; Maita et al. 2004, 2005). This neurodegenerative syndrome causes severe visual impairment as a consequence of photoreceptor apoptosis and eventually leads to other pathological symptoms in the retina that often result in complete blindness (Kennan et al. 2005; Mordes et al. 2006). Remarkably, all hPrp8 mutations that give rise to retinitis pigmentosa (RP13) affect residues concentrated within its last exon at the end of the hPrp8 C-terminal binding domain (Fig. 5C) (Grainger and Beggs 2005). A hPrp8 fragment lacking these residues completely failed to interact with its N terminus domain, with hBrr2 or hSnu114 (Pena et al. 2007), while these interactions were altered in yeast Prp8p variants that mimicked RP13 point mutations (Boon et al. 2007). The resulting splicing efficiency decrease due to impairment in protein–protein interactions together with a possible disruption in the structure or folding kinetics of the C-terminal binding domain are believed to underlie the molecular basis of the RP13 disorder, but the reasons for the associated retinal-specific phenotype still remain controversial (Boon et al. 2007; Pena et al. 2007; Zhang et al. 2007).
We report here that the residues mutated in RP13 are missing from the corresponding C. elegans PRP-8(rr40) variants that retain intronic sequences (Fig. 5C), while loss of function in the ortholog of hPrp31, which triggers RP11 due to haploinsufficiency (Rio Frio et al. 2008) similarly suppresses the intestinal hyperplasia in our RNAi assay (Table 2). hPrp3 triggers RP most probably in a dominant-negative manner (Gonzalez-Santos et al. 2008), and loss-of-function in its C. elegans ortholog did not suppress cdc-25.1(rr31).
The differential sensitivity to a mutation in a general component of the spliceosome between tissues of a same organism indicates that the intrinsic nature and dynamics of a given tissue play a major role in the expressivity of a phenotypic trait. Since the retina constitutes a tissue with sustained expression of genes and high turnover of proteins (especially rhodopsin) involved in maintaining the light-sensitive properties of its photoreceptors, it has been proposed that a general reduction in expression would primarily affect this type of tissue (Kennan et al. 2005). Thus, it appears that metabolically active tissues like the human retina, or the C. elegans germ line, are more susceptible than others when the splicing efficiency of crucial components drops below a threshold, because the cells constituting these specialized tissues rely on high expression levels to efficiently execute their respective physiological functions. The striking parallels that exist between the gene identities, the molecular lesions, the effects of dosage, and the tissue-specific effects seen in the phenotypes of hPprp8/hPrp31-linked retinitis pigmentosa and the prp-8(rr40)-associated germ-line defect suggest that this model may provide molecular insight as to how these mutations manifest in a single tissue.
MATERIALS AND METHODS
Strains
All C. elegans strains were cultured at 20°C according to standard procedures unless stated otherwise (Brenner 1974). The following strains, rearrangements, and transgenes were used: wild-type Bristol (N2); Hawaiian ecotype CB4856 (Hodgkin and Doniach 1997). MR156 rrIs01[elt-2∷GFP; unc-119(+)]. MR142 cdc-25.1(rr31); rrIs01. MR183 cdc-25.1(rr31rr36); rrIs01. MR184 cdc-25.1(rr31); rr37; rrIs01. MR185 cdc-25.1(rr31); rr38; rrIs01. MR186 cdc-25.1(rr31); rr39; rrIs01. MR187 cdc-25.1(rr31); rr40; rrIs01. MR246 rrEx17[elt-2∷GFP] in CB4856. MR320 cdc-25.1(rr31); lon-1(e1137) unc-32(e189); rrIs01. MR372 cdc-25.1(rr31); unc-32(e189) sma-3(e491); rrIs01. MR475 cdc-25.1(rr31); sma-3(e491) unc-36(e251); rrIs01. MR375 cdc-25.1(rr31) unc-13(e450); lon-1(e1137) rr40; rrIs01. MR349 cdc-25.1(ij48); rrIs01. MR396 cdc-25.1(ij48); lon-1(e1137) rr40; rrIs01. MR625 smg-1(r861); rr40; rrIs01. TR1331 smg-1(r861). JR2274 wIs137(end-3∷END-3[P202L]∷GFP; pRF4) (gift from M. Maduro, University of California Riverside). AZ244 unc-119(ed3); ruIs57([pAZ147: pie-1 promoter/β-tubulin∷GFP; unc-119(+)]) (Praitis et al. 2001). JH1327 axEx73(pJH3.92: pie-1 promoter/pie-1∷GFP; rol-6(su1006)) (Reese et al. 2000). DP38 unc-119(ed3). TX20 oma-1(zu405). BC4634 dpy-17(e164) sDf125 unc-32(e189); sDp3. BC4638 dpy-17(e164) sDf127(s2428) unc-32(e189); sDp3. BC4696 dpy-17(e164) sDf128(s2786) unc-32(e189); sDp3. BC4677 dpy-17(e164) sDf135(s2767) unc-32(e189); sDp3. CX2914 nDf16/dpy-17(e164) unc-32(e189). MT3022 nDf20/sma-2(e502) unc-32(e189).
Semiclonal screen for maternal-effect suppressors of cdc-25.1(rr31)
MR142 cdc-25.1(rr31); rrIs01 animals were mutagenized at the L4 stage with 40 mM ethylmethanesulfonate (EMS) as previously described (Brenner 1974). Mutagenized worms were transferred to 100-mm plates (30–40 per plate) and allowed to produce progeny. The L4 stage F1 progeny were picked to 35-mm plates (two per plate). The gravid adult F2 progeny were washed off half of each plate, pooled, subjected to alkaline/hypochlorite, and the resultant F3 generation was synchronized at the L1 stage. F3 animals were grown to the L4 stage and screened under a fluorescent dissecting microscope for suppression of the cdc-25.1(rr31)-associated supernumerary divisions. Candidate mutants were singled and their progeny re-evaluated. A total of 5456 haploid genomes were analyzed and five suppressors were isolated.
Mapping and cloning of rr40
MR187 cdc-25.1(rr31); rr40; rrIs01 hermaphrodites were first crossed with Hawaiian CB4856 males carrying an extrachromosomal rrEx17(elt-2∷gfp) transgene to allow for efficient visualization of the intestinal lineage in the cross progeny. A total of 75 suppressed F2 and 75 nonsuppressed F2 adults were isolated and lysed, and SNP-snip mapping using bulk segregant analysis was performed on the respective pools (Wicks et al. 2001), placing rr40 in the middle of linkage group (LG) III. Subsequent deficiency and meiotic mapping refined the rr40 locus between sma-3 and unc-36, tightly linked and to the left of unc-36, between –0.396 and –0.355 map units. This 107-kb interval is covered by four cosmids and contains a total of 26 predicted genes. Systematic feeding RNAi of genes present in the mapping interval (Kamath et al. 2003) was then used to find candidates that would suppress the cdc-25.1(rr31)-associated hyperplasia and phenocopy the rr40-mediated suppression. Under standard RNAi conditions (see below), none of the clones tested suppressed cdc-25.1(rr31), but four clones caused penetrant sterility (Ste) and 100% embryonic lethality (Emb). To test for their suppression potential, we developed a novel alternative feeding RNAi protocol that ensures the strongest viable hypomorphic phenotype possible for any given clone triggering complete Ste and/or Emb. Using this alternative RNAi strategy, one of the four clones consistently suppressed the cdc-25.1(rr31)-mediated hyperplasia. Independent PCRs and sequencing analysis of this gene in rr40 mutants revealed a single, EMS-typical base-pair substitution in prp-8 (C50C3.6).
Standard and hypomorphic feeding RNAi
Standard feeding RNAi
Bacterial colonies from the feeding RNAi library were grown overnight at 37°C and dsRNA synthesis was induced on NGM plates containing 50 μg/μL ampicillin and 1 mM IPTG for 24 h at room temperature. L4 stage MR142 cdc-25.1(rr31); rrIs01 larvae were fed for 16 h on these plates and transferred for an additional day to a second RNAi plate, on which the progeny was scored for suppression of the cdc-25.1(rr31)-associated hyperplasia.
Hypomorphic feeding RNAi
dsRNA expression from bacteria containing the clones of interest was induced as described for standard feeding RNAi. L4 animals were allowed to feed on these RNAi plates for a precise and empirically determined period of time. This feeding period was determined as the one triggering the terminal Ste/Emb phenotype for around 24 h after the transfer of the animals from the RNAi plate to a non-RNAi standard nematode growing medium (NGM) plate seeded with OP50 bacteria, and typically varied from 8 to 30 h (Supplemental Fig. S1). Embryos laid by these transferred adults that eventually hatch possess minimal, but viable levels of the targeted gene product to survive embryogenesis, and can be scored for the strongest post-embryonic hypomorphic phenotypes. This protocol was successfully used to test the multiple lethal RNAi clones of the spliceosomal machinery (Table 2).
Lineage analysis
MR379 wIs137; rrIs01 and MR380 cdc-25.1(rr31); wIs137; rrIs01 were generated by crossing JR2274 hermaphrodites with MR142 males. MR389 cdc-25.1(rr31); prp-8(rr40); wIs137; rrIs01 was generated by crossing MR380 hermaphrodites with MR187 males. Embryos were dissected from gravid MR379, MR380, or MR389 hermaphrodites and mounted on 2% agarose pads. The GFP expressed of the end-3 promoter is visible from the E2 (two intestinal cells) to the E8 stage (Maduro et al. 2005), while the elt-2∷gfp marks intestinal nuclei from the E8 stage to adulthood (Fukushige et al. 1999). Intestinal cell divisions were monitored from the appearance of the intestinal precursor cell E onward using both DIC and epifluorescence.
RT–PCR and Northern blots
Total RNA was isolated from synchronized adults with a Trizol-based extraction method (Invitrogen). The RNA concentration was precisely quantified and adjusted to 200 μg/μL using a Nano-Drop ND-1000 spectrophotometer. First-strand cDNA was performed on 1 mg of RNA using polydT primers and Expand Reverse Transcriptase (Roche) following the manufacturer's protocol. For each primer pair, parameters were optimized for semiquantitative RT–PCR (Marone et al. 2001). The cryptic splice sites and intron inclusions produced by the prp-8(rr40) mutation were detected by sequence analysis of the two bands detected in RT–PCRs (with polydT or sequence specific reverse primers) across the rr40 locus in MR187 animals and comparison with the current genomic C. elegans database accessible through Wormbase (www.wormbase.org).
DIG-labeled antisense RNA probes generated from prp-8 [probe is complementary to a sequence present in all prp-8(rr40) variants], cdc-25.1, cye-1, oma-1, pie-1, elt-2, and act-1 cDNA were used to perform Northern analysis on total RNA according to the manufacturer's protocol (Roche). Probes were detected chromogenically using NBT/BCIP. Detailed description of all primer sequences and/or cloning strategies are available upon request.
Microscopy and image processing
Light microscopy, image analysis, and processing were performed essentially as previously described (Kostic and Roy 2002).
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank all the members of the Roy laboratory, particularly Patrick Narbonne and Jimmy Ouellet for ideas and discussion. We are grateful to Rueyling Lin, Morris Maduro, and the Caenorhabditis Genetic Center, which is founded by the NIH National Center for Research Resources (NCRR), for C. elegans strains and reagents, and to the Kohara laboratory for making expression data available to the community. This work was supported by a Research Award from the Canadian Cancer Society. R.R. is a CIHR New Investigator.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1168408.
REFERENCES
- Alphey L., Jimenez J., White-Cooper H., Dawson I., Nurse P., Glover D.M. twine, a cdc25 homolog that functions in the male and female germ line of Drosophila . Cell. 1992;69:977–988. doi: 10.1016/0092-8674(92)90616-k. [DOI] [PubMed] [Google Scholar]
- Aroian R.V., Levy A.D., Koga M., Ohshima Y., Kramer J.M., Sternberg P.W. Splicing in Caenorhabditis elegans does not require an AG at the 3′ splice acceptor site. Mol. Cell. Biol. 1993;13:626–637. doi: 10.1128/mcb.13.1.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft N., Golden A. CDC-25.1 regulates germ line proliferation in Caenorhabditis elegans . Genesis. 2002;33:1–7. doi: 10.1002/gene.10083. [DOI] [PubMed] [Google Scholar]
- Ashcroft N.R., Kosinski M.E., Wickramasinghe D., Donovan P.J., Golden A. The four cdc25 genes from the nematode Caenorhabditis elegans . Gene. 1998;214:59–66. doi: 10.1016/s0378-1119(98)00228-5. [DOI] [PubMed] [Google Scholar]
- Bao Z., Zhao Z., Boyle T.J., Murray J.I., Waterston R.H. Control of cell cycle timing during C. elegans embryogenesis. Dev. Biol. 2008;318:65–72. doi: 10.1016/j.ydbio.2008.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadnia N., Golas M.M., Hartmuth K., Sander B., Kastner B., Deckert J., Dube P., Will C.L., Urlaub H., Stark H., et al. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 2007;26:1737–1748. doi: 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejsovec A., Anderson P. Myosin heavy-chain mutations that disrupt Caenorhabditis elegans thick filament assembly. Genes & Dev. 1988;2:1307–1317. doi: 10.1101/gad.2.10.1307. [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S., Dix I., Russell C.S., McGarvey M., Beggs J.D., Kupiec M. Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae . Genetics. 2000a;156:1503–1517. doi: 10.1093/genetics/156.4.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S., Russell C.S., Dix I., Beggs J.D., Kupiec M. Extensive genetic interactions between PRP8 and PRP17/CDC40, two yeast genes involved in pre-mRNA splicing and cell cycle progression. Genetics. 2000b;154:61–71. doi: 10.1093/genetics/154.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonov S., Anokhina M., Will C.L., Urlaub H., Luhrmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- Boon K.L., Grainger R.J., Ehsani P., Barrass J.D., Auchynnikava T., Inglehearn C.F., Beggs J.D. prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast. Nat. Struct. Mol. Biol. 2007;14:1077–1083. doi: 10.1038/nsmb1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B. Maternal control of pattern formation in early Caenorhabditis elegans embryos. Curr. Top. Dev. Biol. 1998;39:73–117. doi: 10.1016/s0070-2153(08)60453-6. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C.G., Ohi R., Mehta S., O'Toole E.T., Winey M., Clark T.A., Sugnet C.W., Ares M., Jr, Gould K.L. Removal of a single α-tubulin gene intron suppresses cell cycle arrest phenotypes of splicing factor mutations in Saccharomyces cerevisiae . Mol. Cell. Biol. 2002;22:801–815. doi: 10.1128/MCB.22.3.801-815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L., Donzelli M., Chiesa M., Guardavaccaro D., Ganoth D., Dorrello N.V., Hershko A., Pagano M., Draetta G.F. Degradation of Cdc25A by β-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- Busino L., Chiesa M., Draetta G.F., Donzelli M. Cdc25A phosphatase: Combinatorial phosphorylation, ubiquitylation, and proteolysis. Oncogene. 2004;23:2050–2056. doi: 10.1038/sj.onc.1207394. [DOI] [PubMed] [Google Scholar]
- Chakarova C.F., Hims M.M., Bolz H., Abu-Safieh L., Patel R.J., Papaioannou M.G., Inglehearn C.F., Keen T.J., Willis C., Moore A.T., et al. Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum. Mol. Genet. 2002;11:87–92. doi: 10.1093/hmg/11.1.87. [DOI] [PubMed] [Google Scholar]
- Clucas C., Cabello J., Bussing I., Schnabel R., Johnstone I.L. Oncogenic potential of a C.elegans cdc25 gene is demonstrated by a gain-of-function allele. EMBO J. 2002;21:665–674. doi: 10.1093/emboj/21.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho C.M., Kolevski B., Walker C.D., Lavagi I., Shaw T., Ebert A., Leevers S.J., Marygold S.J. A genetic screen for dominant modifiers of a small-wing phenotype in Drosophila melanogaster identifies proteins involved in splicing and translation. Genetics. 2005;171:597–614. doi: 10.1534/genetics.105.045021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtot C., Fankhauser C., Simanis V., Lehner C.F. The Drosophila cdc25 homolog twine is required for meiosis. Development. 1992;116:405–416. doi: 10.1242/dev.116.2.405. [DOI] [PubMed] [Google Scholar]
- Dahan O., Kupiec M. Mutations in genes of Saccharomyces cerevisiae encoding pre-mRNA splicing factors cause cell cycle arrest through activation of the spindle checkpoint. Nucleic Acids Res. 2002;30:4361–4370. doi: 10.1093/nar/gkf563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan O., Kupiec M. The Saccharomyces cerevisiae gene CDC40/PRP17 controls cell cycle progression through splicing of the ANC1 gene. Nucleic Acids Res. 2004;32:2529–2540. doi: 10.1093/nar/gkh574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert J., Hartmuth K., Boehringer D., Behzadnia N., Will C.L., Kastner B., Stark H., Urlaub H., Luhrmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRenzo C., Seydoux G. A clean start: Degradation of maternal proteins at the oocyte-to-embryo transition. Trends Cell Biol. 2004;14:420–426. doi: 10.1016/j.tcb.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Dibb N.J., Brown D.M., Karn J., Moerman D.G., Bolten S.L., Waterston R.H. Sequence analysis of mutations that affect the synthesis, assembly and enzymatic activity of the unc-54 myosin heavy chain of Caenorhabditis elegans . J. Mol. Biol. 1985;183:543–551. doi: 10.1016/0022-2836(85)90170-6. [DOI] [PubMed] [Google Scholar]
- Edgar B.A. Cell-cycle control in a developmental context. Curr. Biol. 1994;4:522–524. doi: 10.1016/s0960-9822(00)00113-5. [DOI] [PubMed] [Google Scholar]
- Edgar L.G., McGhee J.D. DNA synthesis and the control of embryonic gene expression in C. elegans . Cell. 1988;53:589–599. doi: 10.1016/0092-8674(88)90575-2. [DOI] [PubMed] [Google Scholar]
- Edgar B.A., Datar S.A. Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila's early cell cycle program. Genes & Dev. 1996;10:1966–1977. doi: 10.1101/gad.10.15.1966. [DOI] [PubMed] [Google Scholar]
- Edgar B.A., Lehner C.F. Developmental control of cell cycle regulators: A fly's perspective. Science. 1996;274:1646–1652. doi: 10.1126/science.274.5293.1646. [DOI] [PubMed] [Google Scholar]
- Evsikov A.V., Graber J.H., Brockman J.M., Hampl A., Holbrook A.E., Singh P., Eppig J.J., Solter D., Knowles B.B. Cracking the egg: Molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo. Genes & Dev. 2006;20:2713–2727. doi: 10.1101/gad.1471006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J., Mailand N., Syljuasen R.G., Bartek J., Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410:842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- Faustino N.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes & Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Fukushige T., Hendzel M.J., Bazett-Jones D.P., McGhee J.D. Direct visualization of the elt-2 gut-specific GATA factor binding to a target promoter inside the living Caenorhabditis elegans embryo. Proc. Natl. Acad. Sci. 1999;96:11883–11888. doi: 10.1073/pnas.96.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaktionov K., Beach D. Specific activation of cdc25 tyrosine phosphatases by B-type cyclins: Evidence for multiple roles of mitotic cyclins. Cell. 1991;67:1181–1194. doi: 10.1016/0092-8674(91)90294-9. [DOI] [PubMed] [Google Scholar]
- Gonczy P., Echeverri C., Oegema K., Coulson A., Jones S.J., Copley R.R., Duperon J., Oegema J., Brehm M., Cassin E., et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Santos J.M., Cao H., Duan R.C., Hu J. Mutation in the splicing factor Hprp3p linked to retinitis pigmentosa impairs interactions within the U4/U6 snRNP complex. Hum. Mol. Genet. 2008;17:225–239. doi: 10.1093/hmg/ddm300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger R.J., Beggs J.D. Prp8 protein: At the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habara Y., Urushiyama S., Shibuya T., Ohshima Y., Tani T. Mutation in the prp12+ gene encoding a homolog of SAP130/SF3b130 causes differential inhibition of pre-mRNA splicing and arrest of cell-cycle progression in Schizosaccharomyces pombe . RNA. 2001;7:671–681. doi: 10.1017/s1355838201001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J. Bacteriol. 1967;93:1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J. Patterning the early Xenopus embryo. Development. 2006;133:1205–1217. doi: 10.1242/dev.02304. [DOI] [PubMed] [Google Scholar]
- Hebeisen M., Roy R. CDC-25.1 stability is regulated by distinct domains to restrict cell division during embryogenesis in C. elegans . Development. 2008;135:1259–1269. doi: 10.1242/dev.014969. [DOI] [PubMed] [Google Scholar]
- Hodges P.E., Jackson S.P., Brown J.D., Beggs J.D. Extraordinary sequence conservation of the PRP8 splicing factor. Yeast. 1995;11:337–342. doi: 10.1002/yea.320110406. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Doniach T. Natural variation and copulatory plug formation in Caenorhabditis elegans . Genetics. 1997;146:149–164. doi: 10.1093/genetics/146.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., Papp A., Pulak R., Ambros V., Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans . Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Shirogane T., Xu L., Nalepa G., Qin J., Elledge S.J., Harper J.W. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes & Dev. 2003;17:3062–3074. doi: 10.1101/gad.1157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kaplan Y., Kupiec M. A role for the yeast cell cycle/splicing factor Cdc40 in the G1/S transition. Curr. Genet. 2007;51:123–140. doi: 10.1007/s00294-006-0113-y. [DOI] [PubMed] [Google Scholar]
- Kennan A., Aherne A., Humphries P. Light in retinitis pigmentosa. Trends Genet. 2005;21:103–110. doi: 10.1016/j.tig.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Li C., Maller J.L. A maternal form of the phosphatase Cdc25A regulates early embryonic cell cycles in Xenopus laevis . Dev. Biol. 1999;212:381–391. doi: 10.1006/dbio.1999.9361. [DOI] [PubMed] [Google Scholar]
- Kipreos E.T. C. elegans cell cycles: Invariance and stem cell divisions. Nat. Rev. Mol. Cell Biol. 2005;6:766–776. doi: 10.1038/nrm1738. [DOI] [PubMed] [Google Scholar]
- Kostic I., Roy R. Organ-specific cell division abnormalities caused by mutation in a general cell cycle regulator in C. elegans . Development. 2002;129:2155–2165. doi: 10.1242/dev.129.9.2155. [DOI] [PubMed] [Google Scholar]
- Lecuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T.R., Tomancak P., Krause H.M. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lin R. A gain-of-function mutation in oma-1, a C. elegans gene required for oocyte maturation, results in delayed degradation of maternal proteins and embryonic lethality. Dev. Biol. 2003;258:226–239. doi: 10.1016/s0012-1606(03)00119-2. [DOI] [PubMed] [Google Scholar]
- Longman D., Plasterk R.H., Johnstone I.L., Caceres J.F. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes & Dev. 2007;21:1075–1085. doi: 10.1101/gad.417707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren K., Allan S., Urushiyama S., Tani T., Ohshima Y., Frendewey D., Beach D. A connection between pre-mRNA splicing and the cell cycle in fission yeast: cdc28+ is allelic with prp8+ and encodes an RNA-dependent ATPase/helicase. Mol. Biol. Cell. 1996;7:1083–1094. doi: 10.1091/mbc.7.7.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M.F., Hill R.J., Heid P.J., Newman-Smith E.D., Zhu J., Priess J.R., Rothman J.H. Genetic redundancy in endoderm specification within the genus Caenorhabditis . Dev. Biol. 2005;284:509–522. doi: 10.1016/j.ydbio.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Maita H., Kitaura H., Keen T.J., Inglehearn C.F., Ariga H., Iguchi-Ariga S.M. PAP-1, the mutated gene underlying the RP9 form of dominant retinitis pigmentosa, is a splicing factor. Exp. Cell Res. 2004;300:283–296. doi: 10.1016/j.yexcr.2004.07.029. [DOI] [PubMed] [Google Scholar]
- Maita H., Kitaura H., Ariga H., Iguchi-Ariga S.M. Association of PAP-1 and Prp3p, the products of causative genes of dominant retinitis pigmentosa, in the tri-snRNP complex. Exp. Cell Res. 2005;302:61–68. doi: 10.1016/j.yexcr.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Makarov E.M., Makarova O.V., Urlaub H., Gentzel M., Will C.L., Wilm M., Lührmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- Makarova O.V., Makarov E.M., Liu S., Vornlocher H.P., Lührmann R. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 2002;21:1148–1157. doi: 10.1093/emboj/21.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marone M., Mozzetti S., De Ritis D., Pierelli L., Scambia G. Semiquantitative RT-PCR analysis to assess the expression levels of multiple transcripts from the same sample. Biol. Proced. Online. 2001;3:19–25. doi: 10.1251/bpo20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J., Curado S., Ephrussi A., Rorth P. Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell. 2000;101:511–522. doi: 10.1016/s0092-8674(00)80861-2. [DOI] [PubMed] [Google Scholar]
- McKie A.B., McHale J.C., Keen T.J., Tarttelin E.E., Goliath R., van Lith-Verhoeven J.J., Greenberg J., Ramesar R.S., Hoyng C.B., Cremers F.P., et al. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13) Hum. Mol. Genet. 2001;10:1555–1562. doi: 10.1093/hmg/10.15.1555. [DOI] [PubMed] [Google Scholar]
- Mordes D., Luo X., Kar A., Kuo D., Xu L., Fushimi K., Yu G., Sternberg P., Jr, Wu J.Y. Pre-mRNA splicing and retinitis pigmentosa. Mol. Vis. 2006;12:1259–1271. [PMC free article] [PubMed] [Google Scholar]
- Mueller P.R., Coleman T.R., Kumagai A., Dunphy W.G. Myt1: A membrane-associated inhibitory kinase that phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science. 1995;270:86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- Myer D.L., Bahassi E.M., Stambrook P.J. The Plk3-Cdc25 circuit. Oncogene. 2005;24:299–305. doi: 10.1038/sj.onc.1208278. [DOI] [PubMed] [Google Scholar]
- Newport J., Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Nishi Y., Lin R. DYRK2 and GSK-3 phosphorylate and promote the timely degradation of OMA-1, a key regulator of the oocyte-to-embryo transition in C. elegans . Dev. Biol. 2005;288:139–149. doi: 10.1016/j.ydbio.2005.09.053. [DOI] [PubMed] [Google Scholar]
- O'Farrell P.H., Edgar B.A., Lakich D., Lehner C.F. Directing cell division during development. Science. 1989;246:635–640. doi: 10.1126/science.2683080. [DOI] [PubMed] [Google Scholar]
- Pena V., Liu S., Bujnicki J.M., Luhrmann R., Wahl M.C. Structure of a multipartite protein–protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol. Cell. 2007;25:615–624. doi: 10.1016/j.molcel.2007.01.023. [DOI] [PubMed] [Google Scholar]
- Perdiguero E., Nebreda A.R. Regulation of Cdc25C activity during the meiotic G2/M transition. Cell Cycle. 2004;3:733–737. [PubMed] [Google Scholar]
- Potashkin J., Kim D., Fons M., Humphrey T., Frendewey D. Cell-division-cycle defects associated with fission yeast pre-mRNA splicing mutants. Curr. Genet. 1998;34:153–163. doi: 10.1007/s002940050381. [DOI] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans . Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheva V.I., Knight D., Bozko P., Marsh K., Frolov M.V. Specific role of the SR protein splicing factor B52 in cell cycle control in Drosophila . Mol. Cell. Biol. 2006;26:3468–3477. doi: 10.1128/MCB.26.9.3468-3477.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese K.J., Dunn M.A., Waddle J.A., Seydoux G. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol. Cell. 2000;6:445–455. doi: 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- Rio Frio T., Wade N.M., Ransijn A., Berson E.L., Beckmann J.S., Rivolta C. Premature termination codons in PRPF31 cause retinitis pigmentosa via haploinsufficiency due to nonsense-mediated mRNA decay. J. Clin. Invest. 2008;118:1519–1531. doi: 10.1172/JCI34211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushforth A.M., Anderson P. Splicing removes the Caenorhabditis elegans transposon Tc1 from most mutant pre-mRNAs. Mol. Cell. Biol. 1996;16:422–429. doi: 10.1128/mcb.16.1.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P., Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 1987;49:559–567. doi: 10.1016/0092-8674(87)90458-2. [DOI] [PubMed] [Google Scholar]
- Russell C.S., Ben-Yehuda S., Dix I., Kupiec M., Beggs J.D. Functional analyses of interacting factors involved in both pre-mRNA splicing and cell cycle progression in Saccharomyces cerevisiae . RNA. 2000;6:1565–1572. doi: 10.1017/s1355838200000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea J.E., Toyn J.H., Johnston L.H. The budding yeast U5 snRNP Prp8 is a highly conserved protein which links RNA splicing with cell cycle progression. Nucleic Acids Res. 1994;22:5555–5564. doi: 10.1093/nar/22.25.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimuta K., Nakajo N., Uto K., Hayano Y., Okazaki K., Sagata N. Chk1 is activated transiently and targets Cdc25A for degradation at the Xenopus midblastula transition. EMBO J. 2002;21:3694–3703. doi: 10.1093/emboj/cdf357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Soto M.C., Ishidate T., Kim S., Nakamura K., Bei Y., van den Heuvel S., Mello C.C. The conserved kinases CDK-1, GSK-3, KIN-19, and MBK-2 promote OMA-1 destruction to regulate the oocyte-to-embryo transition in C. elegans . Curr. Biol. 2006;16:47–55. doi: 10.1016/j.cub.2005.11.070. [DOI] [PubMed] [Google Scholar]
- Sorensen C.S., Syljuasen R.G., Falck J., Schroeder T., Ronnstrand L., Khanna K.K., Zhou B.B., Bartek J., Lukas J. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell. 2003;3:247–258. doi: 10.1016/s1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- Stitzel M.L., Pellettieri J., Seydoux G. The C. elegans DYRK kinase MBK-2 marks oocyte proteins for degradation in response to meiotic maturation. Curr. Biol. 2006;16:56–62. doi: 10.1016/j.cub.2005.11.063. [DOI] [PubMed] [Google Scholar]
- Teigelkamp S., Newman A.J., Beggs J.D. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 1995;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan U., Company M., Abelson J. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae . Genes & Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- Vithana E.N., Abu-Safieh L., Allen M.J., Carey A., Papaioannou M., Chakarova C., Al-Maghtheh M., Ebenezer N.D., Willis C., Moore A.T., et al. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11) Mol. Cell. 2001;8:375–381. doi: 10.1016/s1097-2765(01)00305-7. [DOI] [PubMed] [Google Scholar]
- Wang Q.T., Piotrowska K., Ciemerych M.A., Milenkovic L., Scott M.P., Davis R.W., Zernicka-Goetz M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev. Cell. 2004;6:133–144. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- Wicks S.R., Yeh R.T., Gish W.R., Waterston R.H., Plasterk R.H. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Zhang L., Shen J., Guarnieri M.T., Heroux A., Yang K., Zhao R. Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants. Protein Sci. 2007;16:1024–1031. doi: 10.1110/ps.072872007. [DOI] [PMC free article] [PubMed] [Google Scholar]