Abstract
Gene expression programs can become activated in response to extracellular signals. One evolutionarily conserved example is binding of Wnt glycoproteins to their receptor, which triggers a signal transduction cascade that stabilizes cytoplasmic β-catenin protein, allowing it to translocate into the nucleus. There, β-catenin binds to TCF/Lef family transcription factors and promotes the expression of target genes. Mutations in either the β-catenin gene itself or its partner protein APC are responsible for the oncogenic activation of this pathway in colorectal tumors. Here we report the splicing factor SRp20 as a novel target gene of β-catenin/TCF4 signaling. Transfection of activated β-catenin mutants into colorectal cells increased expression of endogenous SRp20 transcript and protein and also stimulated a luciferase reporter construct containing the SRp20 gene promoter. In contrast, inhibition of endogenous β-catenin signaling by a dominant-negative TCF4 construct down-regulated both luciferase reporter and SRp20 expression. We further demonstrate that the β-catenin/TCF4-mediated increase in SRp20 protein levels is sufficient to modulate alternative splicing decisions in the cells. In particular, we observed a change in the alternative splicing pattern in a control minigene reporter as well as in the endogenous SRp20-regulated CD44 cell adhesion protein. These results demonstrate that the β-catenin/TCF4 pathway not only stimulates gene transcription, but also promotes the generation of transcript variants through alternative splicing. Our data support the recent notion that transcription and alternative splicing represent two different layers of gene expression and that signaling pathways act upon a coordinated network of transcripts in each layer.
Keywords: alternative splicing, β-catenin, CD44, colorectal cells, SR proteins, Wnt signaling
INTRODUCTION
The genetic information encoded in genomes is expressed in a differentiated and dynamic manner in order to generate specialized cellular phenotypes or the coordinated succession of developmental stages. Such gene expression programs are controlled by various mechanisms, including the activation of specific transcription factors in response to extracellular signals. Examples of important and evolutionarily conserved extracellular signals are the secreted Wnt glycoproteins. They play a fundamental role during embryonic development for the formation of germ layers and body axis (Cadigan and Nusse 1997; Yamaguchi 2001; Marikawa 2006), but also regulate stem cell renewal in colon crypts, hair follicles, and bone marrow (Reya and Clevers 2005). Some target genes have been identified, including the cell cycle–promoting cyclin D1 and c-myc genes as well as the tissue remodeling metalloproteinase matrilysin (Clevers 2006). When activated inappropriately, this pathway promotes malignant transformation, especially in colorectal cancer.
The Wnt1 class glycoproteins stimulate a canonical Wnt signaling pathway (for review, see Bienz and Clevers 2000; Clevers 2006), which employs the transcriptional coactivator β-catenin as a central effector. In unstimulated cells, free cytoplasmic β-catenin is constitutively marked for ubiquitination and subsequently degraded by the proteasome. The molecular mark recognized by the SCF complex is provided through phosphorylation of β-catenin at critical N-terminal serine and threonine residues by glycogen synthase kinase 3β (GSK3) and controlled by a multiprotein complex containing Axin and the APC tumor suppressor protein. Upon Wnt-ligand binding to its plasma membrane receptor, GSK3 is inhibited, and, in consequence, unphosphorylated β-catenin accumulates in the cell. Improper activation of this pathway can be due to mutations in the β-catenin gene that inactivate the GSK3 phosphorylation sites, as observed in various sporadic tumors of, for instance, colon (Ilyas et al. 1997; Morin et al. 1997), ovary (Sagae et al. 1999; Wright et al. 1999), liver (Koch et al. 1999; Legoix et al. 1999), and skin (Rubinfeld et al. 1997). In addition, loss of the adenomatous polyposis coli (APC) tumor suppressor gene activates the pathway and is a hallmark of hereditary and sporadic colorectal cancer (Fodde et al. 2001). Free cytoplasmic β-catenin translocates into the nucleus, where it binds to transcription factors of the TCF/Lef family and acts as a coactivator to promote the transcription of Wnt target genes.
Besides the activation of target gene transcription, recent genome-wide studies have revealed that tissues also differ in their gene expression profiles at the post-transcriptional level of alternative splicing. Current bioinformatics studies estimate that >70% of human genes can produce alternatively spliced variants (Ben-Dov et al. 2008), and numerous examples were characterized demonstrating a functional diversity between proteins encoded as splice variants from the same gene (for review, see Venables 2004; Srebrow and Kornblihtt 2006). In addition, splice variants can differ with respect to transcript stability or translation efficiency. Alternative splice variants thus expand transcriptome diversity and provide an important contribution to the functional diversity in gene expression programs. Another recent breakthrough from genome-wide gene expression analyses is that the majority of genes regulated by a given stimulus at the transcriptional level differ from those regulated at the alternative splicing level (Le et al. 2004; Pan et al. 2004; Blencowe 2006). This indicates that activation of signaling pathways can affect parallel layers of gene expression. It has been proposed that layers of alternative splicing are composed of specific groups of alternative spliced exons that are regulated together and thus constitute a coordinated physiological program in which specific splicing variants are generated to fulfill specific cellular functions (Blencowe 2006).
How are different splice variants turned on and off? Similar to the control of transcription through transcription factors that bind promoter, enhancer, or silencer elements, splicing decisions involve recognition of transcript-specific sequence elements by splice-regulating proteins. A cell-type-specific splicing pattern is the results of a complex combination of constitutive splicing components, of modulating splicing factors, and of their cooperative assembly on exon-specific regulatory sequence elements. For example, the highly conserved SR protein family of RNA-binding proteins (Cáceres et al. 1997) is essential for constitutive splicing and also influences the selection of alternative splice sites. Their activity is antagonized by members of the hnRNP family of proteins, and alterations in the expression ratio of these antagonistic factors cause drastic changes in splice-site selection. These factors establish RNA–protein and protein–protein interactions that define splice sites for the spliceosome. For each transcript, a specific combination of these factors regulates the recognition of alternative exons (Smith and Valcárcel 2000; Singh and Valcárcel 2005) and can involve their tissue-specific expression profiles or their phosphorylation and intracellular localization. Thus, the regulation of alternative splicing, much like that of transcription, is based on a combinatorial mode and responds to extracellular signals. Here, we describe that the β-catenin/TCF signaling pathway not only affects gene transcription, but also modulates alternative splicing decisions. We show that this pathway can directly activate transcription of the SFSR3 gene, encoding splicing factor SRp20. The subsequent increase in SRp20 protein levels propagates the signal to another layer of regulation and results in the generation of alternative splice variants on a subset of SRp20-target transcripts.
RESULTS
SRp20 protein levels correlate in colorectal cell lines with the extent of endogenous β-catenin/TCF4 signaling
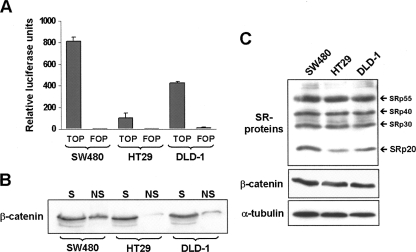
Colorectal cell lines are widely used to study β-catenin/TCF-4-driven gene transcription, but this activity can differ significantly between cell lines depending on which genetic alteration occurred in the pathway. In order to characterize pathway activation in three colorectal cell lines—SW480, HT29, and DLD-1—cells were transiently transfected with either the TOPglow luciferase reporter gene, which reflects the activity of β-catenin/TCF-mediated transcription, or the corresponding mutant reporter FOPglow (Korinek et al. 1997). A constitutively expressed Renilla luciferase reporter was also co-transfected in order to correct for the differences in transfection efficiencies between the cell lines. Analysis of luciferase activity in the three transfected cell types revealed that they varied up to fivefold in their endogenous β-catenin-driven transcriptional activity, which was highest in SW480 and lowest in HT29 cells (Fig. 1A). As further experimental support for the observed differences, we compared the amount of nuclear β-catenin in these cells by using a detergent-based cell fractionation methodology (see Materials and Methods) in which chromatin-associated β-catenin remains in the insoluble fraction. Figure 1B shows that the amount of insoluble nuclear β-catenin correlated well with the corresponding TOPglow activity in each cell line.
FIGURE 1.
Activation of β-catenin/TCF signaling in colorectal cells lines. (A) Diagram showing the activity of TOPglow, a β-catenin/TCF-regulated transcriptional luciferase reporter construct (TOP) or a mutant control construct (FOP) after transfection into the three indicated colorectal cell lines. Results were normalized to the activity of a co-transfected, constitutively active Renilla luciferase construct. (B) Western blot showing the comparison of the nuclear chromatin-associated β-catenin pool in the three cell lines. A detergent-based cell fractionation methodology separated β-catenin into a soluble (S) and a nonsoluble, chromatin-bound (NS) pool. Note that the amount of non-soluble nuclear β-catenin correlated well with the corresponding TOPglow activity in each cell line. (C) Western blot analysis of the pattern of SR protein expression in the three cell lines. The pan-SR protein antibody 1H4 was used to detect the indicated endogenous SR proteins. Levels of β-catenin and α-tubulin were detected as loading controls.
Because preliminary experiments on alternative splicing in our laboratory (V. Gonçalves, P. Matos, and P. Jordan, unpubl.) indicated significant differences between these cell lines, we asked whether pathway activation would correlate with the expression levels of different SR proteins. Lysates from all cell lines were analyzed by Western blot using monoclonal antibody 1H4, which simultaneously detects a variety of endogenous SR proteins (Neugebauer and Roth 1997). As shown in Figure 1C, the immunoblot revealed that the three cell lines presented a similar band pattern, with most prominent expression of SRp55, SRp40, SRp30, and SRp20. Interestingly, the SRp20 levels varied between the cell lines: expression was lowest in HT29, increased in DLD-1, and highest in SW480 cells. Thus, SRp20 levels apparently correlated with the different activities of β-catenin-driven transcription in these cell lines.
β-catenin/TCF4 signaling modulates SRp20 expression
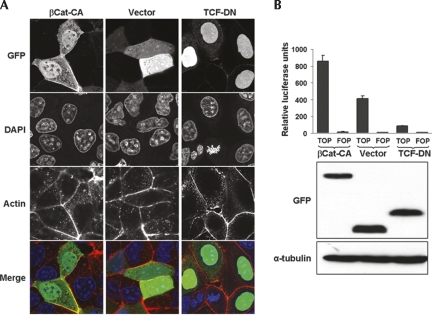
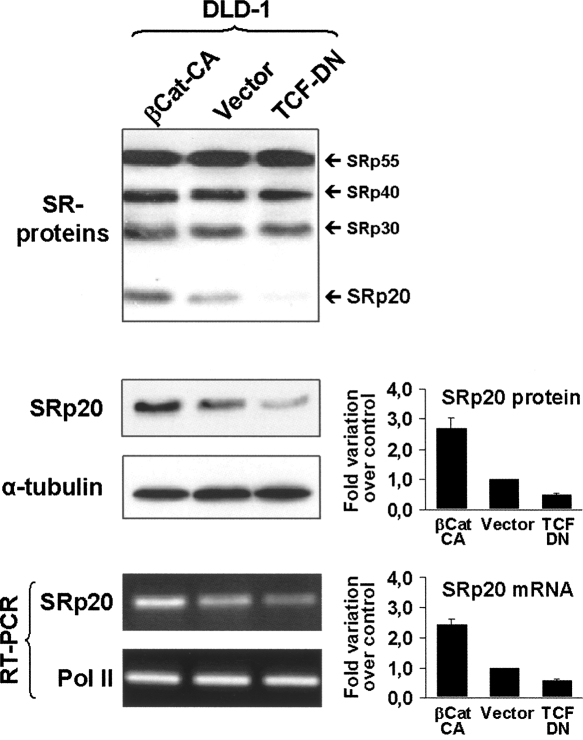
In order to study whether the differences in SRp20 expression levels occurred as a direct consequence of β-catenin signaling, we analyzed DLD-1 cells in more detail. These cells exhibited an intermediate level of endogenous β-catenin/TCF-4 activation and are therefore a suitable cellular system to induce either an increase or a decrease of pathway activity. For this, two previously reported mutant constructs were GFP-tagged and expressed in DLD-1 cells: a constitutively active mutant (βCat-CA) that increases β-catenin signaling is encoded by a nondegradable β-catenin lacking the first 47 amino acids (Kolligs et al. 1999), whereas a TCF-4 mutant encoding just the N-terminal β-catenin-binding domain of TCF-4 fused to a nuclear localization signal (TCF-DN), has a dominant-negative effect on β-catenin signaling (van de Wetering et al. 2002). Figure 2A shows experimental verification of the subcellular localization of these transfected mutants. In agreement with previously published data, βCat-CA localized to the nucleus as well as to the plasma membrane (Fig. 2A, left panels), while TCF-DN localized predominantly to the nucleus (Fig. 2A, right panels). When βCat-CA was coexpressed with the β-catenin/TCF-specific TOPglow reporter (Fig. 2B), an over twofold increase in luciferase activity was observed. In contrast, TCF-DN coexpression significantly inhibited the endogenous β-catenin/TCF-4 activity measured in DLD-1 cells. These data show that it was possible to increase or inhibit the β-catenin/TCF activity in DLD-1 cells, yielding levels comparable to those observed in SW480 or HT29 cells, respectively. Under these experimental conditions, the expression of SR proteins was again analyzed in transfected DLD-1 cells by Western blot (Fig. 3, upper panel). We found that the SRp20 band increased in the presence of βCat-CA but decreased in TCF-DN transfected cells, supporting the idea of a direct stimulation of SRp20 expression through β-catenin/TCF-4 activity. In order to confirm the results obtained with the pan-SR antiserum, a specific monoclonal antibody against SRp20 was used. Densitometric analysis of the Western blots, using α-tubulin as loading normalizer, revealed an over 2.5-fold increase in SRp20 protein levels upon βCat-CA expression in DLD-1 cells and a reduction to approximately one-half in the presence of TCF-DN (Fig. 3, middle panel).
FIGURE 2.
Characterization of GFP-tagged β-catenin and TCF-4 mutants and their effect on β-catenin/TCF transcriptional activity. (A) Subcellular localization of the mutants by confocal microscopy. Images show DLD-1 colorectal cells transfected with (left column) a non-degradable, constitutively active β-catenin mutant (βCat-CA), (middle column) the empty pEGFP control vector (vector), or (right column) a dominant-negative TCF-4 mutant (TCF-DN). Note the nuclear localization of βCat-CA and TCF-DN. (B) Variation in the endogenous β-catenin/TCF transcriptional activity of DLD-1 cells. (Upper panel) A diagram of the TOPglow and FOPglow reporter activities upon co-transfection with βCat-CA, empty pEGFP vector, or TCF-DN. (Middle and lower panels) Western blot analyses of the GFP-βCat-CA, pEGFP, and GFP-TCF-DN expression levels as well as α-tubulin as loading control. Note the increased luciferase activity in the presence of GFP-βCat-CA and the inhibition by TCF-DN.
FIGURE 3.
Effect of β-catenin/TCF transcriptional activity on the expression of SRp20 protein and transcript. DLD-1 cells were transfected with βCat-CA, empty pEGFP vector, or TCF-DN as indicated and then analyzed by Western blot for (upper panel) either SR protein levels or (middle panel) SRp20 protein level, and by RT-PCR for (lower panel) SRp20 transcript levels. Images from three different SRp20 Western blots as well as from RT-PCR experiments were digitalized, the band intensities were quantified, and the mean fold-changes are displayed graphically beside the corresponding panels.
The SFSR3 promoter responds to β-catenin/TCF4 signaling
We next asked whether the observed increase in SRp20 protein involved increased transcription as a response to β-catenin/TCF-4 activity. Indeed, the abundance of SRp20 mRNA increased when βCat-CA was expressed in DLD-1 cells, whereas the presence of TCF-DN led to reduced transcript levels (Fig. 3, lower panel). A semiquantitative RT-PCR methodology (see Materials and Methods) revealed that the observed changes mirrored the variations observed in SRp20 protein levels (Fig. 3, lower panel). These data indicated that β-catenin–TCF4 signaling might target the SRp20 promoter and regulate transcription of SRp20. We, therefore, cloned a 1982-base-pair (bp) genomic fragment containing the SRp20 gene promoter into a pGL2 luciferase reporter vector. This reporter construct, termed pRep5, was expressed in SW480, DLD-1, and HT29 cells and showed clear transcriptional activity when compared to the empty pGL2-vector (Supplemental Fig. S1B). In addition, the extent of pRep5 activation in these cell lines corresponded to their endogenous β-catenin-driven transcriptional activity, as determined in Figure 1.
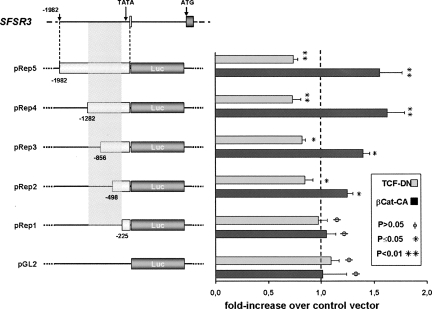
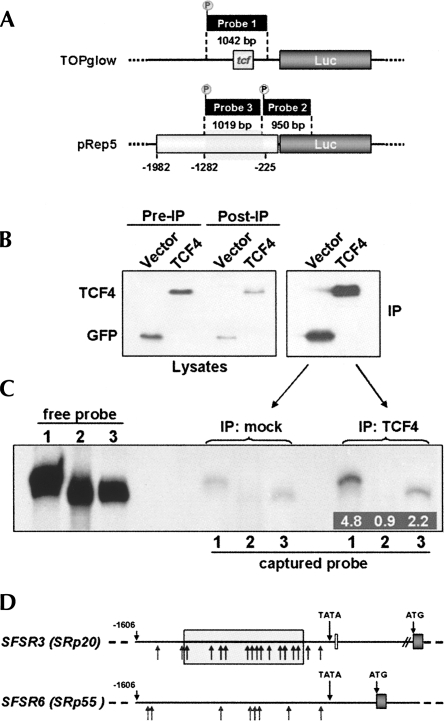
The pRep5 construct was then tested for its direct response to β-catenin–TCF4 signaling by transfecting the above-described β-catenin and TCF4 mutants into DLD-1 cells. The empty pEGFP control vector was transfected in parallel for normalization purposes so that background variations due to other transcription factors that may affect promoter activity could be eliminated. As shown in Figure 4, co-transfection of βCat-CA increased reporter activation, and the observed fold-increase was comparable to that obtained with the TOPglow reporter in the presence of βCat-CA (see Fig. 2). The pRep5 reporter also responded with inhibition to the presence of TCF-DN, while no such response was detected with the empty pGL2-vector (Fig. 4). Next, additional pGL2-based reporter constructs containing shorter fragments of the original 2-kb promoter region (Fig. 4, pRep1-4) were generated. When expressed in DLD-1 cells, these constructs allowed delimiting the responsive promoter region to the sequence between nucleotides −225 and −1282 (Fig. 4, gray shaded rectangle). Within this region, the transcriptional response increased progressively with promoter fragment size. Surprisingly, sequence analysis of this region using the TRANSFAC database (http://www.gene-regulation.com/pub/databases.html) revealed no high-score putative TCF/Lef family binding sites. We therefore tested the binding of TCF4 to the responsive promoter region by DNA/protein coimmunoprecipitation. GFP-TCF4 was expressed in DLD-1 cells, immunoprecipitated (Fig. 5B), and then incubated with different radio-labeled promoter fragments (Fig. 5A). As shown in Figure 5C, the positive control fragment with four consensus TCF binding sites prepared from the TOPglow reporter vector (Fig. 5A, probe 1) was clearly pulled down by GFP-TCF4, whereas the negative control fragment prepared from the pREP5 construct did not bind (Fig. 5A, probe 2). Under these conditions, the transcriptionally activated SFSR3 promoter fragment between nucleotides −225 and −1282 (Fig. 5A, probe 3) was also pulled down by TCF4, albeit less efficiently than the TOPglow fragment.
FIGURE 4.
Response of the SFSR3 gene promoter to β-catenin/TCF4 transcriptional activity. (Left side) Drawing showing the 1982-bp genomic fragment containing the SRp20 gene promoter that was subcloned to yield the luciferase reporter construct pRep5 and derived deletion mutants. (Gray shaded area) The responsive promoter region between nucleotides −225 and −1282. (Right side) The reporters' transcriptional activities in response to stimulation by βCat-CA or to inhibition by TCF-DN are also shown as diagrams for each individual construct and are given as fold-increase in luciferase activity compared to the control pEGFP vector.
FIGURE 5.
Binding of the SFSR3 gene promoter to TCF4. (A) Drawing showing the three promoter fragments used as radioactive probes in DNA/protein coimmunoprecipitation assays. Probes 1 and 2 served as positive or negative controls, respectively, whereas probe 3 corresponds to the transcriptionally responsive promoter region (gray shaded area between nucleotides −225 and −1282). (B) Anti-GFP-stained Western blot showing the expression of GFP (control) and GFP-Δ31TCF4 in cell lysates (Pre-IP) before and (Post-IP) after immunoprecipitation with monoclonal anti-GFP antibody ab1218, as well as the corresponding precipitated proteins (IP). (C) Autoradiograph of the polyacrylamide gel used to separate the radio-labeled DNA probes. (“free probe” lanes) Document comparable probe labeling efficiency; (“captured probe” lanes) show the amount of probe precipitated by either GFP (control) or GFP-Δ31TCF4. Inserted numbers indicate the band intensity quantification compared to the corresponding control bands. (D) Schematic comparison of the SFSR3 and SFSR6 gene promoters. (Arrows) Potential TCF4 binding sites. (The gray box) The SFSR3 promoter region identified in Figure 4 that was required to respond to β-catenin-TCF4 signaling.
Considering that the reporter transcriptional response increased progressively with promoter fragment size, these data suggest that the response of the SRp20 promoter to β-catenin/TCF signaling is regulated through several binding sites of lower affinity, spread across the responsive promoter region, rather than a highly conserved TCF recognition motif. Consistently, bioinformatics analysis of the cloned promoter sequence revealed 19 potential sites with considerable homology with the degenerated TCF/Lef transcription factor binding motif, 13 of which within the most responsive region between nucleotides −225 and −1282 (see sequence in Supplemental Fig. S2A). In contrast, considerably fewer potential binding sites were identified in the promoter of the SFSR6 gene encoding SRp55, which revealed no expression changes in the presence of βCat-CA (Fig. 5D; see also sequence in Supplemental Fig. S2B).
β-Catenin signaling modifies alternative splicing decisions
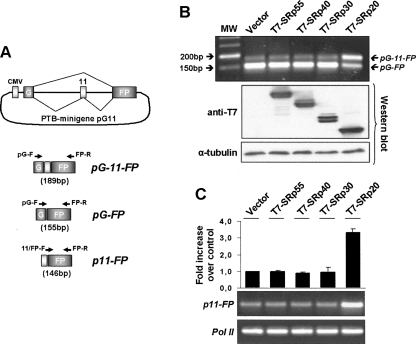
Alternative splicing is regulated by a combinatorial code that involves regulatory RNA sequence elements and changes in either the expression or the nuclear prevalence of competing splicing factors, including the SR proteins (Smith and Valcárcel 2000; Singh and Valcárcel 2005). In agreement with this notion, the observed increase in SRp20 could lead to different alternative splicing decisions in DLD-1 cells in a specific subset of genes. To test this, we employed minigene reporter constructs that would respond to increasing SRp20 levels and co-transfected T7-tagged SR proteins together with the different minigenes. As shown in Figure 6A, the PTB-derived minigene pG11, which contains the alternatively spliced exon 11 of PTB fused between GFP exons (Fig. 6A; Spellman et al. 2005), underwent alternative splicing in the presence of SRp20. The effect was SRp20-specific because co-transfected T7-tagged SRp30, SRp40, or SRp55 had no effect on the alternative splicing of this minigene in DLD-1 cells (Fig. 6B). In contrast, a FAS minigene (Förch et al. 2000) showed no response to co-transfected T7-tagged SRp20 (Supplemental Fig. S3).
FIGURE 6.
Effect of SRp20 on alternative splicing of the PTB-derived minigene pG11. (A) Drawing of the PTB-derived minigene pG11 showing the CMV promoter, the two constitutive GFP exons G and FP, and the alternative PTB exon 11. (Diagonal lines) Splicing patterns; amplified minigene-derived transcripts pG-11-FP, pG-FP, and p11-FP are depicted below. Positions of primers used for RT-PCR analysis are indicated. (B) RT-PCR analysis of exon 11 inclusion following overexpression of various SR proteins. Using forward primer pG-F in the first GFP exon and reverse primer FP-R in the second GFP exon, the inclusion of exon 11 can be distinguished as amplicons pG-FP of 155 bp and pG-11-FP of 189 bp, respectively. Note that in control DLD-1 cells, the pG11 minigene originates small amounts of the alternatively spliced pG-11-FP isoform, whereas overexpression of SRp20 strongly promotes pG-11-FP generation. The expression levels of the transfected SR proteins as well as of α-tubulin as loading control are shown as Western blots below the RT-PCR image. (C) Specific amplification of the PTB exon 11-containing transcripts. Primer 11/FP-F overlaps the splice junction between the PTB exon 11 and the second GPF exon, so that only exon 11-containing transcripts are amplified, allowing a more accurate semiquantitative determination of exon 11 inclusion. The diagram shows the fold-increase following transfection of DLD-1 cells with different SR proteins, and the two RT-PCR gel images show amplicons 11-FP as well as Pol II as control amplification.
We then wished to determine whether the increase in SRp20 that was observed in response to β-catenin/TCF4 signaling was sufficient to modulate alternative splicing of the PTB-derived minigene pG11. In order to allow a more accurate estimation of variations in pG11 alternative splicing, it was necessary to avoid competition with the abundant pG-FP amplicon (see Fig. 6C). Therefore, a semiquantitative RT-PCR assay was designed based on a primer that overlaps the junction between the PTB exon 11 and the second GPF exon (primer 11-FP-F), selectively amplifying the alternatively spliced form (amplicon 11-FP). Using this specific assay, we first validated the co-transfection of PTB-derived minigene pG11 with the different T7-tagged SR proteins and calculated that SRp20 overexpression increased inclusion of the alternative exon by more than threefold (Fig. 6C). Subsequently, we tested the alternative splicing response of the PTB-pG11 construct in cells coexpressing the GFP-fused mutants of β-catenin and TCF-4. Consistent with the described impact of β-catenin signaling on SRp20 expression (Fig. 3), the co-transfection of βCat-CA produced a roughly twofold increase in p11-FP mRNA levels, whereas TCF-DN reduced the inclusion of the alternative exon by ∼50% (Fig. 7).
FIGURE 7.
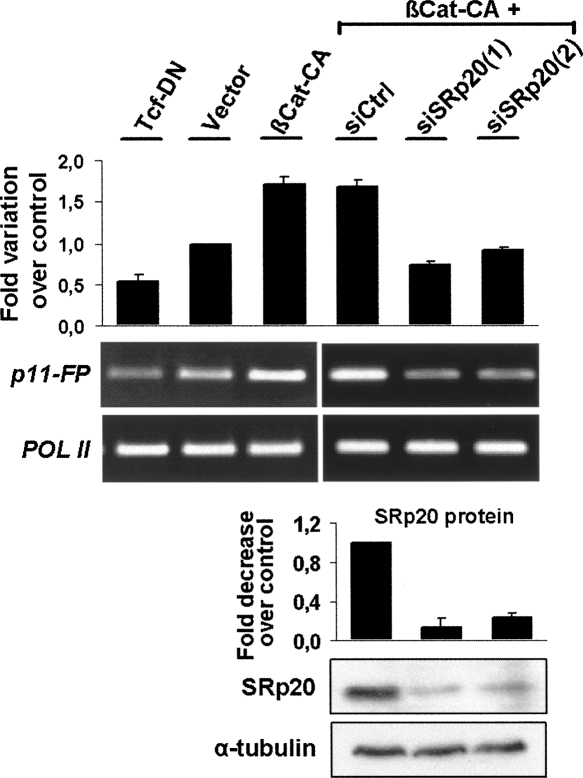
Effect of β-catenin/TCF4 transcriptional activity on alternative splicing of pG11. DLD-1 cells were co-transfected with the PTB-derived minigene pG11 and βCat-CA, empty pEGFP vector, or TCF-DN as indicated. The alternative exon 11-containing transcripts were specifically amplified by RT-PCR, and band intensities were quantified by densitometry using amplified Pol II levels as internal control. The stimulating effect of βCat-CA on exon 11 inclusion was further analyzed in cells previously transfected with either control or one of two SRp20-specific small interfering RNAs. Below the RT-PCR gel, a Western blot shows the degree of depletion of the endogenous SRp20 protein. Note that depletion of SRp20 prevented PTB exon 11 inclusion even in the presence of βCat-CA .
In order to directly demonstrate that these changes in alternative splicing were the result of increased SRp20 levels following β-catenin signaling, we used two specific siRNA oligos to knock down endogenous SRp20 expression in DLD-1 cells co-transfected with βCat-CA and the minigene. Figure 7 shows that two different SRp20 siRNAs reduced endogenous SRp20 protein levels to ∼20%, and this clearly prevented the βCat-CA-induced changes in pG11 alternative splicing.
β-Catenin signaling induces the tumor-associated splice variant CD44E
To investigate whether the observed effect of β-catenin/TCF-4 signaling on alternative splicing is physiologically relevant, we searched the literature for examples of reported SRp20 regulated genes. The most studied alternatively spliced gene in cancer is CD44, which is a transmembrane protein involved in cell–cell adhesion (for review, see Ponta et al. 2003). CD44 has over 20 experimentally observed isoforms due to variable incorporation of 10 alternative exons (v1–v10) in its proximal extracellular domain. Although the constitutionally expressed CD44 form (CD44H, which lacks all of the alternative exons) is always predominant, the increased expression of certain isoforms has been detected in several types of cancer (Naor et al. 2002). One such isoform, CD44E, contains the variable exons v8 to v10, and its splicing was reported to be stimulated by SRp20 due to the presence of an exonic splice enhancer (ESE) sequence within exon v9 (Galiana-Arnoux et al. 2003).
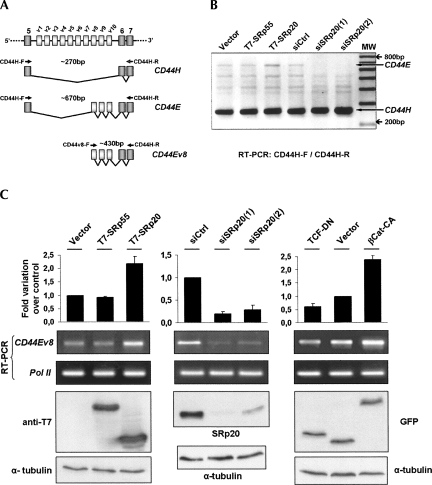
In order to determine whether β-catenin signaling promoted CD44E expression, we first amplified by RT-PCR all CD44 isoforms expressed in DLD-1 cells, including CD44H and E (Fig. 8A). We then compared the basal expression of these isoforms in DLD-1 cells following either overexpression of T7-SRp20 or depletion of endogenous SRp20 by RNA interference. As shown in Figure 8B, the expression level of the CD44H isoform (∼270 bp) was predominant, as reported, but amplification of CD44E (a 700-bp product, identity verified by direct sequencing) increased in the presence of T7-SRp20, but not of T7-SRp55, and completely disappeared upon depletion of endogenous SRp20 protein.
FIGURE 8.
Effect of β-catenin/TCF4 transcriptional activity and SRp20 protein levels on alternative splicing of endogenous CD44. (A) The drawing shows the variable exon region of the CD44 gene. (Diagonal lines) The two analyzed splicing patterns; the amplified minigene-derived transcripts CD44H and CD44E as well as the primers used for their amplification are depicted below. (B) RT-PCR analysis of endogenous CD44 transcripts with primers CD44H-F and CD44-R annealing to the constitutive exons that flank the variable exon region. CD44 transcripts were amplified from DLD-1 cells transfected with either T7-tagged SR proteins or siRNAs to deplete endogenous SRp20. Note the specific changes in CD44E, as indicated. (C) Quantitation of endogenous CD44E generation using an exon v8-specific forward primer. CD44E was amplified from DLD-1 cells transfected with (left panel) SRp55 or SRp20, (middle panel) with specific siRNAs to deplete endogenous SRp20, (right panel) or with βCat-CA and TCF-DN to modulate β-catenin/TCF4 transcriptional activity. Below the RT-PCR images, the corresponding Western blot images are shown to document the expression levels of overexpressed SR proteins, of depleted SRp20, or of βCat-CA and TCF-DN, respectively.
Next, we used an exon v8-specific forward primer for selective amplification (Fig. 8A,C) and estimated more accurately the amount of CD44E transcript. SRp20 overexpression induced an over twofold increase in CD44E levels in these cells, while SRp20 depletion reduced exons v8–10 inclusion to below 25% of that found in control cells (Fig. 8C, left and middle panels, respectively). Finally, we expressed constitutively active β-catenin in DLD-1 cells and observed increased endogenous CD44E, whereas expression of dominant-negative TCF-4 mutants inhibited the endogenous level of exons v8–v10 inclusion (Fig. 8C, right panel).
DISCUSSION
The main conclusion from the present work is that the β-catenin/TCF4 signal transduction pathway stimulates gene transcription of the splicing factor SRp20, and this eventually generates a subset of transcript variants through alternative splicing.
Several lines of evidence support this conclusion. First, we show that the endogenous levels of SRp20 transcript and protein correlate with the activity of β-catenin-driven transcription in different colorectal cell lines. Furthermore, experimental stimulation or inhibition of β-catenin/TCF4 signaling affects SRp20 levels. Then we demonstrate that the isolated SRp20 gene promoter responds to stimulation and inhibition of β-catenin/TCF4 signaling. In addition, the β-catenin-stimulated increase of SRp20 protein levels is sufficient to promote alternative splicing decisions in colorectal cells, both in a minigene and in a selected endogenous gene, CD44. Finally, suppression of endogenous SRp20 by RNA interference prevents the β-catenin/TCF4-induced changes in alternative splicing.
Together, our data identify SFSR3 as a novel target gene for β-catenin/TCF4 signaling. Although microarray data with Wnt signal-dependent gene expression profiles of colorectal cells have been published and collected at the Wnt homepage (http://www.stanford.edu/∼rnusse/wntwindow.html; van de Wetering et al. 2002; Van der Flier et al. 2007; Vlad et al. 2008), SFSR3 has not been included in these gene lists. We observed an about twofold increase in SRp20 transcripts in our experiments, which is close to the threshold value set in most microarray experiments. Probably, SFSR3 values did not meet the stringent cut-off thresholds applied in order to filter significant changes in gene expression. However, the observed changes in SFSR3 expression have apparent physiological impact, because we detected clear changes in alternative splicing as a consequence of increased SRp20 levels. Thus, in case of splicing factors, subtle changes in gene expression may be sufficient to modulate alternative splicing decisions. Bearing in mind that the regulation of alternative splicing is based on a combinatorial mode, one can appreciate that relative changes in concentration between individual SR proteins and antagonizing hnRNP factors can affect splice-site choice in specific pre-mRNAs that contain the respective regulatory sequence elements (Smith and Valcárcel 2000).
Our analysis further revealed that the SFSR3 promoter contains 13 putative TCF binding sites within the β-catenin-responsive region and that TCF4 co-precipitates the corresponding promoter fragment. Consistently, the transcriptional activation of our reporter constructs to activated β-catenin increased proportionally with the length of the subcloned responsive promoter region, and therefore with the number of putative binding sites present (Fig. 4, cf. pRep2–pRep4). Others have also found up-regulated WNT target genes with a variable number of TCF/Lef sites in the promoter region between nucleotides −2000 and −100 (Schwartz et al. 2003; Ziegler et al. 2005). Although these data indicate that SFSR3 is a primary transcriptional TCF4 target, it cannot be excluded that other TCF-regulated transcription factors such as c-Myc (He et al. 1998), c-jun, or Fra-1 (Mann et al. 1999) also contribute to the regulation of SRp20 expression.
One endogenous target event in DLD-1 cells for the β-catenin-induced change in SRp20 levels is alternative splicing of CD44E. Suppression of endogenous SRp20 reduced the amount of generated CD44E, while ectopic expression of SRp20 protein or stimulation of its expression by activated β-catenin led to elevated CD44E levels. The family of CD44 cell-surface glycoproteins is involved in cell–matrix adhesion and growth factor presentation and was shown to influence cell growth, survival, and differentiation. Members of the CD44 family have been implicated in progression and metastasis of tumors (Ponta et al. 2003), including colorectal tumors (Wielenga et al. 1993, 1999; Herrlich et al. 1995; Gotley et al. 1996; Ropponen et al. 1998). In particular, CD44E was found significantly elevated in metastasic colorectal carcinomas (Takeuchi et al. 1995), and serum levels of CD44E were shown to double in blood from colorectal cancer patients with liver metastases, correlating with its immuno-histochemical expression (Yamaguchi et al. 1988, 1998). One possible explanation for a role for CD44E in metastatic cancers is that isoforms containing exon v10 provide a site for modification with chondritine-4-sulfate chains that promote anchoring of tumor cells to endothelial cells and may facilitate metastasis (Hayes et al. 2002). Our data therefore indicate that deregulated Wnt signaling in colorectal tumors may be one mechanism for promoting the expression of pro-invasive CD44 variants. Indeed, deregulated CD44 expression patterns have been reported in early lesions of colorectal neoplasia (Wielenga et al. 1999).
A connection of the Wnt pathway with pre-mRNA splicing has previously been observed for an E1A reporter transcript, the estrogen receptor variant Δ5-6, and splice variants WISP1v and FGFR3 AT-II (Sato et al. 2005; Lee et al. 2006, 2007). In these cases, the β-catenin/TCF4 complex apparently regulated the expression level of another splicing factor, SF1, and thus affected a different set of splicing events (Shitashige et al. 2007).
Altered expression of splicing factors has been frequently observed in various tumor types (Ghigna et al. 1998; Stickeler et al. 1999; Venables 2004), and human cancers express a large number of alternatively spliced transcripts (Wang et al. 2003; Venables 2004; Kalnina et al. 2005; Srebrow and Kornblihtt 2006). Our data on the regulation of SRp20 by β-catenin/TCF4 signaling provide a mechanistic insight into some of these observations.
In conclusion, these results demonstrate that the β-catenin/TCF pathway stimulates not only gene transcription, but also the generation of a subset of transcript variants through alternative splicing. Thus, two levels of a cellular gene expression program are affected by β-catenin/TCF signaling, first the transcriptional activation of the SFSR3 gene encoding splicing factor SRp20, and second the generation of alternative splice variants as a downstream consequence of increased SRp20 protein levels. These data further support the recent notion that transcription and alternative splicing represent two different layers of gene expression and that signaling pathways act upon a coordinated network of transcripts in each layer.
MATERIALS AND METHODS
Cell culture and transfection
DLD-1 and SW480 colorectal cells were maintained in DMEM supplemented with 10% (v/v) fetal calf serum (FCS) (Invitrogen), whereas HT29 cells were grown in RPMI. Cell lines were regularly checked for absence of mycoplasm infection. Cells were grown in 35-mm dishes to 60%–80% confluence, transfected using LipofectAMINE 2000 (Invitrogen) according to the manufacturer's instructions, and analyzed 16–20 h later. Total amounts of transfected DNA were 2 μg of DNA per dish for DLD-1 and SW480 cells or 4 μg for HT29 cells. For immunoprecipitation, DLD-1 cells were transfected with 4 μg of DNA per 60-mm dish. If required, the amount of DNA was adjusted with empty vector. Plasmid transfection efficiencies were judged microscopically by expression of 2 μg of GFP and reached 60%–80% in DLD-1 and SW480 cells or 30%–40% in HT29 cells. For RNA interference experiments, cells at 30%–40% confluence were transfected with 200 pmol of the indicated siRNAs using LipofectAMINE 2000 (Invitrogen) and analyzed after 48 h. The siRNA oligos were ordered from MWG Biotech with the following sequences:
siSRp20(1): 5′-GAGUGGAACUGUCGAAUGG;
siSRp20(2): 5′-GAGCUAGAUGGAAGAACAC; and
Control siGFP: 5′-GGCUACGUCCAGGAGCGCACCTT.
DNA plasmids and constructs
The following published constructs were received as gifts: TCF-DN (encoding a dominant-negative TCF-4 containing just the β-catenin binding domain of TCF-4 fused to a nuclear localization signal [N-TCF-4]) (van de Wetering et al. 2002); β-Cat-CA (encoding a constitutively active nondegradable β-catenin mutant lacking the first 47 amino acids) (Kolligs et al. 1999); splicing reporter minigenes pG11 (Spellman et al. 2005) and FAS (Förch et al. 2000); and T7-tagged SRp20, SRp30, SRp40, and SRp55 (Cáceres et al. 1997). The TOPglow/FOPglow TCF Reporter Kit and the pCDNA3-HA-Δ31TCF4 construct (encoding a TCF-4 mutant with intact DNA binding but lacking the N-terminal β-catenin binding domain, thus avoiding interference with endogenous complexes in DNA/protein co-precipitation assays; see below) were purchased form Upstate (Millipore). The Δ31TCF4 mutant was subcloned into pEGFPc1 using EcoRI and KpnI restriction sites. The SFSR3 promoter-driven luciferase reporter constructs were amplified by PCR from genomic DNA and subcloning into pGL2-Enhancer Vector (Promega): pRep5 with primers pGL2SRp20_KpnF (5′-GGTACCTGACTGCTGTATCGTTTCCAA) and pGL2SRp20_MluR (5′-ACGCGTCCGATGAGTCTTCC), and pRep3 with primers pGL2SRp20_Kpn802 (5′-GGTACCTTGCGGCTCTGTCTTCGTAA) and pGL2SRp20_MluR. pRep4, pRep2, and pRep1 were derived from pRep5 by deleting restrictions fragments using SmaI, SmaI, and PvuII, or SmaI and StuI restriction enzymes, respectively. All constructs were confirmed by automated DNA sequencing.
Analysis of transcript expression and semiquantitative RT-PCR
Total RNA was extracted from cell lysates with the RNAeasy kit (QIAGEN), and 1 μg was reverse-transcribed using random primers (Invitrogen) and Ready-to-Go You-Prime Beads (GE Healthcare). Primers for specific amplification were as follows:
For SRp20: SRp20-F (5′-AACAAGACGGAATTGGAACG) and SRp20-R (5′-TGGGCCACGATTTCTACTTC);
For CD44: CD44H-F (5′-CATCTACCCCAGCAACCCTA) and CD44H-R (5′-GGTTGTGTTTGCTCCACCTT);
For CD44E: CD44v8-F (5′-TTTGGACAGGACAGGACCTC) and CD44H-R;
For pG11: minigene pG-F (5′-GGCAAAGAATTCGCCACCA) or 11/FP-F (5′-ATTCTTTTCGGAGCAAGGGC) and FP-R (5′-GAACTTCAGGGTCAGCTTGC);
For FAS minigene PT1 (5′-GTCGACGACACTTGCTCAAC) and PT2 (5′-AAGCTTGCATCGAATCAGTAG); and
For RNA polymerase II (Pol II) were Pol II-F (5′-GAGCGGGAATTTGAGCGGATGC) and Pol II-R (5′-GAAGGCGTGGGTTGATGTGGAAGA).
Amplification reactions were performed using AmpliTaq polymerase (Perkin-Elmer) at 60°C for 30 cycles for SRp20, pG11, FAS, and CD44E, at 58°C and 30 cycles for CD44, and at 66°C for 28 cycles for Pol II. All reactions included an initial denaturation of 5 min at 94°C and a final extension of 10 min at 72°C, and each cycle contained 30 sec of denaturation and extension. Semiquantitative analysis of transcript levels were previously established using serial dilutions of control cDNAs in order to determine the linear amplification phase for each PCR reaction. Products were separated on 2% agarose gels stained with ethidium bromide, images recorded digitally, and band intensity quantified using ImageJ software followed by normalization to Pol II expression levels. No amplification was obtained when RNA was mock reverse-transcribed without adding reverse transcriptase.
DNA/protein coimmunoprecipitation
For immunoprecipitation (IP), 2 × 106 DLD-1 cells were seeded in 60-mm dishes and transfected with either pEGFP-empty vector or pEGFP-Δ31TCF4. Following 16–20 h, cells were washed in cold PBS and lysed on ice in 250 μL of lysis buffer [50 mM Tris-HCl at pH 7.5, 0.5% (v/v) NP-40, 150 mM NaCl, 10% (v/v) glycerol, 2.5 mM MgCl2, and a protease inhibitor cocktail (Sigma)]. Lysates were prepared from four parallel dishes, then pooled, and cleared by centrifugation at 2500g for 5 min, and an aliquot of 0.1 vol was transferred to 2× Laemmli buffer (Pre-IP). The remaining supernatant from either GFP- or GFP-TCF4-containing lysates were split into four aliquots of 200 μL and each added to 50 μL of protein G-agarose beads (Roche) slurry (1:1 in lysis buffer), precoupled with 2 μg/mL−1 of mouse monoclonal anti-GFP antibody (ab1218; Abcam). After an incubation of 2 h at 4°C, beads were collected and an 0.1-vol aliquot of the supernatant (Post-IP) was added to 2× Laemmli buffer before the beads were washed six times with 1 mL of lysis buffer. Pre- and Post-IP aliquots as well as one of each precipitate were analyzed by Western blot to assess transgene expression levels and immunoprecipitation efficiency. The remaining three precipitates from each transfection were incubated with one of the following DNA probes: the TOPGLOW (TCF Reporter Plasmid; Upstate) digested with PvuI and XhoI restriction enzymes, to generate a 1042-bp fragment, containing the four canonical TCF binding sites (Probe 1, used as positive control); the pRep5 SRp20 promoter reporter construct digested with SalI (nucleotide −263) and EcoRI (nucleotide +590), to generate a 950-bp fragment with no transcriptional response (probe 2, used as negative control); or the pRep5 SRp20 promoter construct digested with SmaI (nucleotide −1282) and SalI (nucleotide −263) to generate a 1019-bp fragment containing all the putative TCF binding sites from the SRp20 promoter (probe 3, used as test probe). All probes were labeled by 5′-end labeling with [γ-32P]ATP using T4 polynucleotide kinase (kit, GE Healthcare) and unincorporated label was removed by gel-filtration chromatography on Sephadex G-25 spin columns (GE Healthcare). DNA/protein coimmunoprecipitation assays were adapted according to previously described methodologies (Aiyar and Sugden 1998; Kaldalu et al. 2000). Briefly, the DNA concentration of the probes was adjusted to 2 ng/μL, and 10 ng of each was added in parallel to the above-described GFP or GFP-Δ31TCF4 immunoprecipitates, in 40 μL of binding buffer [10 mM HEPES at pH 7.9, 20 mM MgCl2, 50 mM NaCl, 50 mM KCl, 10% (v/v) glycerol, 0.05% (v/v) Triton X-100] and incubated for 1 h at room temperature with gentle rotation. Beads were then washed six times with 1 mL of wash buffer [50 mM HEPES at pH 7.9, 100 mM KCl; 50 mM NaCl; 2.5 mM MgCl2; 0.1% (v/v) Triton X-100] and resuspended in 20 μL of 2× Laemmli buffer to separate bound probes from TCF4. Probes were denatured for 5 min at 65°C and resolved for 2 h at 100 V in a 5% polyacrylamide (29:1)-TBE gel, pre-run for 1 h at 100 V. The gel was then dried and exposed to X-ray film for 48–96 h. All results were confirmed in at least three independent experiments.
SDS-PAGE and Western blotting
Samples were prepared and detected as described (Matos and Jordan 2006). The antibodies used for Western blots were mouse anti-SR Proteins 1H4 and mouse anti-SRp20 (Neugebauer and Roth 1997) from Zymed-Invitrogen, mouse anti-β-catenin from BD Transduction Laboratories, monoclonal anti-α-tubulin clone B-5-1-2 (as loading control) from Sigma, rabbit anti-GFP ab290 from Abcam UK, and T7-Tag Antibody from Novagen (Merck). For densitometric analysis, films from at least three independent experiments were digitalized and analyzed using ImageJ software (NIH).
Cell fractionation
Nuclear proteins were separated into a soluble pool not retained in the nucleus and into a chromatin-bound insoluble pool according to modification of previously described procedures (Solan et al. 2002). Briefly, cells washed in cold PBS were scraped and lysed on ice for 10 min in 200 μL of fractionation buffer [50 mM Tris at pH 7.9, 0.1% (v/v) NP-40; 1.5 mM MgCl2, 10 mM KCl, and a protease inhibitor cocktail (Sigma)]. The soluble fraction was collected by centrifuging the lysate at 3500g for 5 min and adding the supernatant to 50 μL of 5× Laemmli SDS sample buffer. The pellet containing the insoluble nuclear fraction was washed once in fractionation buffer and then resuspended in 250 μL of 1× Laemmli sample buffer supplemented with 5 mM MgCl2 and 50 U of endonuclease (Benzonase; Sigma) for cleavage of nucleic acids. Equal volumes of both fractions were analyzed side-by-side on Western Blots. Results were confirmed in at least three independent experiments.
Confocal immunofluorescence microscopy
Cells were grown on 10mm × 10mm glass coverslips, transfected, and incubated as indicated above, then washed twice in PBS, immediately fixed with 4% (v/v) formaldehyde in PBS for 30 min at room temperature, and subsequently permeabilized with 0.2% (v/v) Triton X-100 in PBS for 10 min at room temperature. Cells were then labeled for 30 min with a 1:200 dilution of phalloidin-TRITC (Sigma) washed three times in PBS, briefly stained with 0.5 ng/mL DAPI (Sigma), washed again, and the coverslips were mounted in VectaShield (Vector Laboratories) and sealed with nail polish. Images were recorded with the 405-nm, 488-nm, and 532-nm laser lines of a Leica TCS confocal microscope and processed with Adobe Photoshop software.
Luciferase reporter assay
Approximately 5 × 105 DLD-1 cells were seeded in 35-mm dishes, transfected with 100 ng of the pRL-TK luciferase reporter (for constitutive expression of Renilla luciferase as internal control; Promega), 1 μg of each specific luciferase reporter (pReps or TOPglow), and 1 μg of βCat-CA, Tcf-DN, or pEGFP control vector. After 16–20 h, transfected cells were lysed, assayed with the Dual Luciferase Reporter Assay (Promega) following the manufacturer's instructions, and measured in an Anthos Lucy-2 Luminometer. Lysates were assayed in duplicates, and additional aliquots were analyzed by Western blot to document protein expression levels. All Firefly luciferase values were normalized by the control values obtained for Renilla luciferase and, when indicated, plotted as fold-increase over the value of vector control. Displayed values correspond to at least three independent transfection assays.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank H. Clevers, J. Collard, J. Cáceres, J. Valcárcel, and C. Smith for providing plasmid vectors used in this study. This work was supported by the Fundação para a Ciência e Tecnologia, Portugal (grant POCTI 47546/02), the Programa de Financiamento Plurianual do CIGMH, and fellowships BD 18262/2004 to V.G. and BPD 20531/2004 to P.M.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1253408.
REFERENCES
- Aiyar A., Sugden B. Fusions between Epstein-Barr viral nuclear antigen-1 of Epstein-Barr virus and the large T-antigen of simian virus 40 replicate their cognate origins. J. Biol. Chem. 1998;273:33073–33081. doi: 10.1074/jbc.273.49.33073. [DOI] [PubMed] [Google Scholar]
- Ben-Dov C., Hartmann B., Lundgren J., Valcárcel J. Genome-wide analysis of alternative pre-mRNA splicing. J. Biol. Chem. 2008;283:1229–1233. doi: 10.1074/jbc.R700033200. [DOI] [PubMed] [Google Scholar]
- Bienz M., Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- Blencowe B.J. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Cáceres J.F., Misteli T., Screaton G.R., Spector D.L., Krainer A.R. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan K.M., Nusse R. Wnt signaling: A common theme in animal development. Genes & Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Fodde R., Smits R., Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer. 2001;1:55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- Förch P., Puig O., Kedersha N., Martínez C., Granneman S., Séraphin B., Anderson P., Valcárcel J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Galiana-Arnoux D., Lejeune F., Gesnel M.C., Stevenin J., Breathnach R., Del Gatto-Konczak F. The CD44 alternative v9 exon contains a splicing enhancer responsive to the SR proteins 9G8, ASF/SF2, and SRp20. J. Biol. Chem. 2003;278:32943–32953. doi: 10.1074/jbc.M301090200. [DOI] [PubMed] [Google Scholar]
- Ghigna C., Moroni M., Porta C., Riva S., Biamonti G. Altered expression of heterogenous nuclear ribonucleoproteins and SR factors in human colon adenocarcinomas. Cancer Res. 1998;58:5818–5824. [PubMed] [Google Scholar]
- Gotley D.C., Fawcett J., Walsh M.D., Reeder J.A., Simmons D.L., Antalis T.M. Alternatively spliced variants of the cell adhesion molecule CD44 and tumour progression in colorectal cancer. Br. J. Cancer. 1996;74:342–351. doi: 10.1038/bjc.1996.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes G.M., Chiu R., Carpenito C., Dougherty S.T., Dougherty G.J. Identification of sequence motifs responsible for the adhesive interaction between exon v10-containing CD44 isoforms. J. Biol. Chem. 2002;277:50529–50534. doi: 10.1074/jbc.M206719200. [DOI] [PubMed] [Google Scholar]
- He T.C., Sparks A.B., Rago C., Hermeking H., Zawel L., da Costa L.T., Morin P.J., Vogelstein B., Kinzler K.W. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Herrlich P., Pals S., Ponta H. CD44 in colon cancer. Eur. J. Cancer. 1995;31A:1110–1112. doi: 10.1016/0959-8049(95)00252-e. [DOI] [PubMed] [Google Scholar]
- Ilyas M., Tomlinson I.P.M., Rowan A., Pignatelli M., Bodmer W.F. β-Catenin mutations in cell lines established from human colorectal cancers. Proc. Natl. Acad. Sci. 1997;94:10330–10334. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldalu N., Toots U., de Lorenzo V., Ustav M. Functional domains of the TOL plasmid transcription factor XylS. J. Bacteriol. 2000;182:1118–1126. doi: 10.1128/jb.182.4.1118-1126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalnina Z., Zayakin P., Silina K., Linē A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342–357. doi: 10.1002/gcc.20156. [DOI] [PubMed] [Google Scholar]
- Koch A., Denkhaus D., Albrecht S., Leuschner I., von Schweinitz D., Pietsch T. Childhood hepatoblastomas frequently carry a mutated degradation targeting box of the β-catenin gene. Cancer Res. 1999;59:269–273. [PubMed] [Google Scholar]
- Kolligs F.T., Hu G., Dang C.V., Fearon E.R. Neoplastic transformation of RK3E by mutant β-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol. Cell. Biol. 1999;19:5696–5706. doi: 10.1128/mcb.19.8.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V., Barker N., Morin P.J., van Wichen D., de Weger R., Kinzler K.W., Vogelstein B., Clevers H. Constitutive transcriptional activation by a β-catenin–TCF complex in APC-/colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Le K., Mitsouras K., Roy M., Wang Q., Xu Q., Nelson S.F., Lee C. Detecting tissue-specific regulation of alternative splicing as a qualitative change in microarray data. Nucleic Acids Res. 2004;32:e180. doi: 10.1093/nar/gnh173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Choi Y.S., Park Y.A., Jeong S. Modulation of oncogenic transcription and alternative splicing by β-catenin and an RNA aptamer in colon cancer cells. Cancer Res. 2006;66:10560–10566. doi: 10.1158/0008-5472.CAN-06-2526. [DOI] [PubMed] [Google Scholar]
- Lee H.K., Kwak H.Y., Hur J., Kim I.A., Yang J.S., Park M.W., Yu J., Jeong S. β-Catenin regulates multiple steps of RNA metabolism as revealed by the RNA aptamer in colon cancer cells. Cancer Res. 2007;67:9315–9321. doi: 10.1158/0008-5472.CAN-07-1128. [DOI] [PubMed] [Google Scholar]
- Legoix P., Bluteau O., Bayer J., Perret C., Balabaud C., Belghiti J., Franco D., Thomas G., Laurent-Puig P., Zucman-Rossi J. β-Catenin mutations in hepatocellular carcinoma correlate with a low rate of loss of heterozygosity. Oncogene. 1999;18:4044–4046. doi: 10.1038/sj.onc.1202800. [DOI] [PubMed] [Google Scholar]
- Mann B., Gelos M., Siedow A., Hanski M.L., Gratchev A., Ilyas M., Bodmer W.F., Moyer M.P., Riecken E.O., Buhr H.J., et al. Target genes of β-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc. Natl. Acad. Sci. 1999;96:1603–1608. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marikawa Y. Wnt/β-catenin signaling and body plan formation in mouse embryos. Semin. Cell Dev. Biol. 2006;17:175–184. doi: 10.1016/j.semcdb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Matos P., Jordan P. RAC1, but not RAC1B, stimulates RELB-mediated gene transcription in colorectal cancer cells. J. Biol. Chem. 2006;281:13724–13732. doi: 10.1074/jbc.M513243200. [DOI] [PubMed] [Google Scholar]
- Morin P.J., Sparks A.B., Korinek V., Barker N., Clevers H., Vogelstein B., Kinzler K.W. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Naor D., Nedvetzki S., Golan I., Melnik L., Faitelson Y. CD44 in cancer. Crit. Rev. Clin. Lab. Sci. 2002;39:527–579. doi: 10.1080/10408360290795574. [DOI] [PubMed] [Google Scholar]
- Neugebauer K.M., Roth M.B. Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes & Dev. 1997;11:1148–1159. doi: 10.1101/gad.11.9.1148. [DOI] [PubMed] [Google Scholar]
- Pan Q., Shai O., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Ponta H., Sherman L., Herrlich P.A. CD44: From adhesion molecules to signaling regulators. Nat. Rev. Mol. Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- Reya T., Clevers H. Wnt signaling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Ropponen K.M., Eskelinen M.J., Lipponen P.K., Alhava E., Kosma V.M. Expression of CD44 and variant proteins in human colorectal cancer and its relevance for prognosis. Scand. J. Gastroenterol. 1998;33:301–309. doi: 10.1080/00365529850170900. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B., Robbins P., El-Gamil M., Albert I., Porfiri E., Polakis P. Stabilization of β-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- Sagae S., Kobayashi K., Nishioka Y., Sugimura M., Ishioka S., Nagata M., Terasawa K., Tokino T., Kudo R. Mutational analysis of β-catenin gene in Japanese ovarian carcinomas: Frequent mutations in endometrioid carcinomas. Jpn. J. Cancer Res. 1999;90:510–515. doi: 10.1111/j.1349-7006.1999.tb00777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Idogawa M., Honda K., Fujii G., Kawashima H., Takekuma K., Hoshika A., Hirohashi S., Yamada T. β-Catenin interacts with the FUS proto-oncogene product and regulates pre-mRNA splicing. Gastroenterology. 2005;129:1225–1236. doi: 10.1053/j.gastro.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Schwartz D.R., Wu R., Kardia S.L.R., Levin A.M., Huang C.-C., Shedden K.A., Kuick R., Misek D.E., Hanash S.M., Taylor J.M.G., et al. Novel candidate β-catenin/T-cell factor signaling identified expression profiling of ovarian endometrioid adenocarcinomas. Cancer Res. 2003;63:2913–2922. [PubMed] [Google Scholar]
- Shitashige M., Naishiro Y., Idogawa M., Honda K., Ono M., Hirohashi S., Yamada T. Involvement of splicing factor-1 in β-catenin/T-cell factor-4-mediated gene transactivation and pre-mRNA splicing. Gastroenterology. 2007;132:1039–1054. doi: 10.1053/j.gastro.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Singh R., Valcárcel J. Building specificity with nonspecific RNA-binding proteins. Nat. Struct. Mol. Biol. 2005;12:645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- Smith C.W., Valcárcel J. Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- Solan N.J., Miyoshi H., Carmona E.M., Bren G.D., Paya C.V. RelB cellular regulation and transcriptional activity are regulated by p100. J. Biol. Chem. 2002;277:1405–1418. doi: 10.1074/jbc.M109619200. [DOI] [PubMed] [Google Scholar]
- Spellman R., Rideau A., Matlin A., Gooding C., Robinson F., McGlincy N., Grellscheid S.N., Southby J., Wollerton M., Smith C.W. Regulation of alternative splicing by PTB and associated factors. Biochem. Soc. Trans. 2005;33:457–460. doi: 10.1042/BST0330457. [DOI] [PubMed] [Google Scholar]
- Srebrow A., Kornblihtt A.R. The connection between splicing and cancer. J. Cell Sci. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- Stickeler E., Kittrell F., Medina D., Berget S.M. Stage-specific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene. 1999;18:3574–3582. doi: 10.1038/sj.onc.1202671. [DOI] [PubMed] [Google Scholar]
- Takeuchi K., Yamaguchi A., Urano T., Goi T., Nakagawara G., Shiku H. Expression of CD44 variant exons 8–10 in colorectal cancer and its relationship to metastasis. Jpn. J. Cancer Res. 1995;86:292–297. doi: 10.1111/j.1349-7006.1995.tb03053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Flier L.G., Sabates-Bellver J., Oving I., Haegebarth A., De Palo M., Anti M., Van Gijn M.E., Suijkerbuijk S., Van de Wetering M., Marra G., et al. The intestinal Wnt/TCF signature. Gastroenterology. 2007;132:628–632. doi: 10.1053/j.gastro.2006.08.039. [DOI] [PubMed] [Google Scholar]
- van de Wetering M., Sancho E., Verweij C., de Lau W., Oving I., Hurlstone A., van der Horn K., Batlle E., Coudreuse D., Armáis A.P., et al. The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Venables J.P. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- Vlad A., Röhrs S., Klein-Hitpass L., Müller O. The first five years of the Wnt targetome. Cell. Signal. 2008;20:795–802. doi: 10.1016/j.cellsig.2007.10.031. [DOI] [PubMed] [Google Scholar]
- Wang Z., Lo H.S., Yang H., Gere S., Hu Y., Buetow K.H., Lee M.P. Computational analysis and experimental validation of tumor-associated alternative RNA splicing in human cancer. Cancer Res. 2003;63:655–657. [PubMed] [Google Scholar]
- Wielenga V.J., Heider K.H., Offerhaus G.J., Adolf G.R., van den Berg F.M., Ponta H., Herrlich P., Pals S.T. Expression of CD44 variant proteins in human colorectal cancer is related to tumor progression. Cancer Res. 1993;53:4754–4756. [PubMed] [Google Scholar]
- Wielenga V.J., Smits R., Korinek V., Smit L., Kielman M., Fodde R., Clevers H., Pals S.T. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am. J. Pathol. 1999;154:515–523. doi: 10.1016/S0002-9440(10)65297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright K., Wilson P., Morland S., Campbell I., Walsh M., Hurst T., Ward B., Cummings M., Chenevix-Trench G. β-Catenin mutation and expression analysis in ovarian cancer: Exon 3 mutations and nuclear translocation in 16% of endometrioid tumours. Int. J. Cancer. 1999;82:625–629. doi: 10.1002/(sici)1097-0215(19990827)82:5<625::aid-ijc1>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T. Heads or tails: Wnts and anterior–posterior patterning. Curr. Biol. 2001;11:R713–R724. doi: 10.1016/s0960-9822(01)00417-1. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A., Goi T., Taguchi S., Ohtaki N., Seki K., Hirose K., Nakagawara G., Urano T., Furukawa K. Clinical significance of serum levels of CD44 variant exons 8–10 protein in colorectal cancer. J. Gastroenterol. 1988;33:349–353. doi: 10.1007/s005350050095. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A., Goi T., Seki K., Ohtaki N., Maehara M., Kobayashi T., Niimoto S., Katayama K., Hirose K., Nakagawara G., et al. Clinical significance of combined immunohistochemical detection of CD44v and sialyl LeX expression for colorectal cancer patients undergoing curative resection. Oncology. 1998;55:400–403. doi: 10.1159/000011885. [DOI] [PubMed] [Google Scholar]
- Ziegler S., Röhrs S., Tickenbrock L., Möröy T., Klein-Hitpass L., Vetter I.R., Müller O. Novel target genes of the Wnt pathway and statistical insights into Wnt target promoter regulation. FEBS J. 2005;272:1600–1615. doi: 10.1111/j.1742-4658.2005.04581.x. [DOI] [PubMed] [Google Scholar]