Abstract
One mechanism for the origin of new plant microRNAs (miRNAs) is from inverted duplications of transcribed genes. However, even though many young MIRNA genes have recently been identified in Arabidopsis thaliana, only a subset shows evidence for having evolved by this route. We propose that the hundreds of thousands of partially self-complementary foldback sequences found in a typical plant genome provide an alternative path for miRNA evolution. Our genome-wide analyses of young MIRNA genes suggest that some arose from DNA that either has self-complementarity by chance or that represents a highly eroded inverted duplication. These observations are compatible with the idea that, following capture of transcriptional regulatory sequences, random foldbacks can occasionally spawn new miRNAs. Subsequent stabilization through coevolution with initially fortuitous targets may lead to fixation of a small subset of these proto-miRNA genes.
Keywords: Arabidopsis thaliana, microRNAs, evolution
INTRODUCTION
Similar to their animal counterparts, plant miRNAs are produced from endogenous transcripts that contain self-complementary foldbacks. These precursors are processed by DICER-LIKE1 (DCL1), generating the mature miRNAs that are incorporated into RISC, a protein complex that uses miRNAs as specificity components to regulate target genes (for reviews, see Jones-Rhoades et al. 2006; Chapman and Carrington 2007).
While the biogenesis and the mechanisms of action of miRNAs are increasingly well understood, less is known about the evolutionary origins of individual MIRNA genes. Allen and colleagues (2004) showed that in plants, miRNAs genes could arise from inverted duplication of what will then become a target of the miRNA. More elaborate scenarios for an inverted duplication origin have been described (Rajagopalan et al. 2006; Fahlgren et al. 2007), but common to all of them is that the origin of the new MIRNA is dependent on duplication and inversion events.
However, these scenarios do not seem to account for the appearance of all new miRNAs. Recently, ultradeep sequencing of Arabidopsis thaliana small RNA (sRNA) populations (Rajagopalan et al. 2006; Fahlgren et al. 2007) showed that several recently evolved miRNAs could not be explained by the inverted duplication hypothesis. Searching for MIRNA gene candidates, Jones-Rhoades and Bartel (2004) had previously found 138,864 imperfect inverted repeats in the genome of A. thaliana. We speculated that such genomic regions with the potential to generate hairpin-like RNAs could be the source of new miRNAs, as proposed recently also by Axtell (2008). We report that analysis of miRNAs that are unique to A. thaliana (i.e., not found in A. lyrata, poplar, or rice) suggests that some of these miRNAs arose from sequences that either have self-complementarity by chance or that represent highly degenerate inverted duplications. We propose that miRNAs can evolve spontaneously from foldback sequences after these have come under the control of transcriptional regulatory sequences.
RECENTLY EVOLVED MIRNA GENES IN A. THALIANA
One of the premises for studying the evolutionary origin of individual miRNAs is the identification of young MIRNA genes, i.e., ones that are species specific, and hence more likely to have evolved recently. These young MIRNA genes are expected to retain some sequence similarity to the region from which they have originated, making it possible to track their evolutionary history. On the other hand, miRNAs deeply conserved across species must have originated a long time ago, and the accumulated mutations will obscure their origin. In A. thaliana, several recently evolved MIRNA genes have high similarity to their locus of origin, indicating that MIRNAs can arise by inverted duplication of such sequences (Allen et al. 2004; Rajagopalan et al. 2006; Fahlgren et al. 2007).
Recently, the results for several exhaustive small RNA sequencing efforts have been reported for A. thaliana (Lu et al. 2006; Rajagopalan et al. 2006; Fahlgren et al. 2007). Among the miRNAs newly discovered in these studies, several were not found in the monocot species rice, Oryza sativa, or even in the more closely related poplar, Populus trichocarpa. These miRNAs include four new miRNA candidates that we had identified before the results of deep sequencing efforts had been published, using a newly developed functional assay (see Supplemental Figs. 1,2; Supplemental Tables 1–4). We used this set of miRNAs with limited conservation in subsequent analyses.
EVOLUTIONARY ORIGIN OF MIRNA GENES
According to the inverted duplication hypothesis (Allen et al. 2004), a recently evolved MIRNA gene should have long stretches of sequence similarity to the gene that gave origin to it, allowing the identification of the founder gene. The same is true for new MIRNA genes that originated by related mechanisms involving duplication (Rajagopalan et al. 2006).
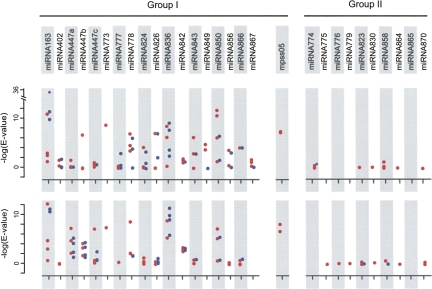
To test the additional hypothesis that random foldbacks could lead to new miRNAs, we selected 29 A. thaliana specific miRNAs, which were not detectable in a preliminary assembly of the A. lyrata genome using microHARVESTER (Supplemental Table 5; Dezulian et al. 2006). We first divided the MIRNA foldbacks into miRNA and miRNA* containing arms and aligned the arms to the set of all annotated cDNAs (from now on called “transcriptome”) and the reference genome sequence of A. thaliana. Based on these results, two groups of MIRNA genes were distinguished (Fig. 1).
FIGURE 1.
Detection of MIRNA related sequences in the A. thaliana transcriptome (blue) and genome (red). MIRNA foldbacks of A. thaliana specific miRNAs were divided into miRNA containing arm (top), and miRNA* containing arm (bottom). Each arm was aligned using FASTA, and the best four hits are reported. Group I contains MIRNAs with significant similarity to some other genomic/transcriptomic region (E value ≤ 0.05). MIRNA genes for which no significant similarity could be found are indicated in Group II.
The first group contains MIRNA foldbacks with at least one arm that has significant similarity to some other genomic region (E VALUE ≤ 0.05). This group includes MIRNA genes that apparently arose through an inverted duplication (miR163, miR447, miR778, miR824, miR842, miR843, miR856, and miR866) (Fahlgren et al. 2007), and one of our candidates that has not yet been confirmed by other studies, mpss05 (see Supplemental Materials). Among these, the best alignment of miR842 was between the miRNA* arm and At1g52130, a gene encoding a jacalin lectin and belonging to the same family as two validated targets (Supplemental Fig. 2, At5g38550 and At1g60130). These results suggest that the origin of miR842 is likely through duplication from a gene related to its target. Both arms of the mpss05 candidate had high similarity to two separate regions of the A. thaliana genome (chromosome 3: 16,815,951–16,816,018, and chromosome 4: 6009,736–6,009,804). In silico folding of the chromosome 3 region indicates a self-complementary structure that is related to the MIRNA foldback (Supplemental Fig. 3). Thus, mpss05 could have originated by direct duplication/transposition of a genomic region that contained a foldback structure by chance.
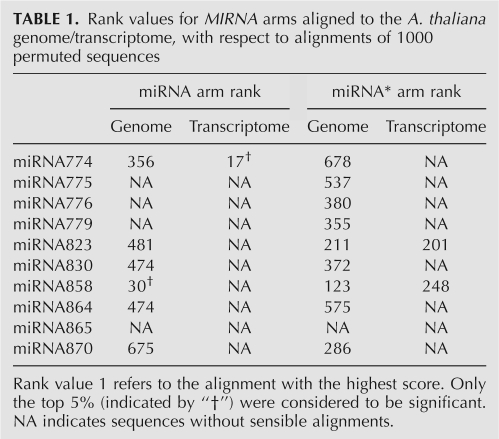
The second group of MIRNA genes included those for which no statistically significant alignment with another region of the genome could be found. To evaluate alignments with scores above the significance threshold, we randomly shuffled the sequence of both arms 1000 times and again aligned against the transcriptome and genome. We define rank as the number of alignments of permuted sequences that had higher alignment scores than the original sequence. Scores with low rank indicate that the original alignment, while highly degenerate, was statistically significant (Table 1). This exercise showed that the similarity between MIR858 and a genomic region on chromosome 4 (10,406,453–10,406,508), as well as between MIR774a and At3g19890, a validated target (Supplemental Fig. 2; Lu et al. 2006), is significant. For the other MIRNA genes, any similarity to other regions of the genome is apparently fortuitous.
TABLE 1.
Rank values for MIRNA arms aligned to the A. thaliana genome/transcriptome, with respect to alignments of 1000 permuted sequences
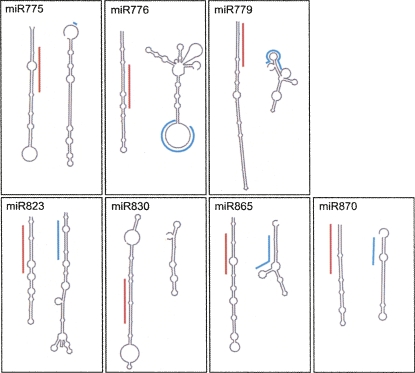
Finally, for each of the A. thaliana MIRNA genes without significant alignment scores, we examined their orthologous regions in the genome of A. lyrata, which diverged from A. thaliana about 5 million years ago (Koch et al. 2000). First, we identified orthologs for the protein-coding genes flanking each of the new MIRNA genes. In seven cases the syntenic relationships of the orthologous genes were conserved in A. lyrata, allowing the comparison of the MIRNA-containing regions between the protein coding genes with their respective counterparts in A. lyrata. In none of the cases was the entire foldback including the miRNA substantially conserved, confirming the microHARVESTER results, which had indicated that no homologs were present in A. lyrata (Fig. 2). The exception is miR823, which seems to be conserved in A. lyrata. Both, miRNA and foldback can be easily recognized in the homologous region of A. lyrata, but the fragment that can be aligned to the foldback contains two insertions. This causes a drastic change of the predicted secondary structure, although this alternative structure could still be subject to DCL1-dependent processing (Fig. 3). In four other cases, there was partial sequence conservation with the possibility of a foldback (Fig. 3), but the miRNA and miRNA* sequences themselves were not conserved. In the remaining three cases, the flanking genes were on different contigs in the A. lyrata genome sequence or the MIRNA foldback could not be meaningfully aligned to the A. lyrata intergenic region.
FIGURE 2.
Alignments of MIRNA-foldback regions and surrounding sequences from A. thaliana with their orthologous counterparts in A. lyrata. Nucleotides involved in the MIRNA foldback are represented in green and the mature miRNA in red. Numbers next to the alignments indicate the position within the respective intergenic region.
FIGURE 3.
Secondary structure of A. thaliana miRNA foldbacks (left) compared to predicted secondary structure of the orthologous sequences from A. lyrata (right). The red line indicates the mature A. thaliana miRNA sequence, while the blue line refers to the corresponding A. lyrata sequence.
In addition, we examined in detail the genomes of Carica papaya and P. trichocarpa, the two closest Arabidopsis relatives for which advanced drafts of genome sequences are available (Tuskan et al. 2006; Ming et al. 2008). The synteny-based strategy applied to A. lyrata failed, because we could not detect homologs of the MIRNA flanking genes in these two species. However, this does not exclude the possibility that MIRNA homologous sequences are located in different regions of the genome. For this reason, we also performed a whole-genome search against P. trichocarpa and C. papaya using Blast and blat (Altschul et al. 1990; Kent 2002). None of the MIRNAs had significant conserved counterparts in the other two genomes. These observations corroborate the idea of new miRNAs being spawned by random sequences that have appeared only recently in evolution.
CONCLUSIONS
The only hypotheses that have so far explicitly been advanced for the origin of A. thaliana miRNAs rely on the duplication of genic regions that subsequently will become the target of the new miRNA (Allen et al. 2004; Rajagopalan et al. 2006; Fahlgren et al. 2007). In some cases, such a newly evolved miRNA could also target another gene that is unrelated to the founder locus (Fahlgren et al. 2007). Alternatively, as suggested by Rajagopalan and colleagues (2006), a new MIRNA gene could arise from the duplication/transposition of a gene that has been the subject of a prior duplication event. Finally, Axtell (2008) has speculated that spurious transcription of random foldbacks could be a first step in the evolution of new miRNAs in plants.
In support of the hypothesis of a random origin of some A. thaliana MIRNA genes, we have found that some evolutionarily young A. thaliana MIRNA genes have no similarity to other regions of the A. thaliana genome, which suggests that they have evolved directly from a sequence that fortuitously contained certain features of MIRNA genes, such as the ability to produce an RNA with a hairpin-like structure. Indeed, in silico folding of the A. thaliana reference genome has shown that it has the potential to form hundreds of thousands of imperfect foldbacks (Jones-Rhoades and Bartel 2004). It is conceivable that acquisition of promoters could lead to transcription of such foldbacks, which in turn could become substrates for DCL1 processing. Svoboda and Di Cara (2006) had speculated that animal miRNAs could originate from random sequences, emphasizing that a random match between miRNA and target would be much more likely in animals, because of the much lower sequence complementarity required for animal miRNA targeting. Based on a comparison of three Drosophila species, a random origin, accompanied by high birth and death rates, has been proposed for the majority of miRNAs in this genus (Lu et al. 2008). Among the evolutionarily young MIRNA genes, none appeared to have formed by inverted duplication, and only a few shared a common origin with other MIRNA loci. Therefore, Lu and colleagues (2008) suggested that such MIRNAs originated from non-miRNA sequences after accumulation of mutations.
Our analysis of orthologous regions between A. lyrata and A. thaliana revealed limited sequence conservation for several A. thaliana MIRNA genes. Although we cannot exclude that the MIRNA genes have degenerated in A. lyrata, the fact that these MIRNA genes are also not conserved in C. papaya and P. trichocarpa (nor in the more distantly related O. sativa) indicates that they all arose after the split between A. thaliana and its nearest relative 5 million years ago. This observation suggests that these regions were not under strong selective pressure and therefore available for mutations that eventually led to the origin of new MIRNA genes. If in any of these cases a newly evolved miRNA fortuitously guides cleavage of an mRNA, this interaction could become the subject of either negative selection (if the interaction is deleterious for the organism) or positive selection (if the interaction is advantageous). This potential route of miRNA/target coevolution would be similar to what has been suggested for transcription factor binding sites, which are often surprisingly transient, with considerable turnover rates (Dermitzakis and Clark 2002).
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Jim Carrington and his group for discussion and for releasing a large small RNA set at the ASRP website before publication, and Jeremy Schmutz, Pedro Pattyn, Yves van de Peer, and Dan Rokshar's group at DOE-JGI for access to an unannotated draft assembly of the A. lyrata MN47 genome sequence. This research was supported by a DAAD fellowship to F.F.F., ERA-PG (DFG) project ARelatives, European Community FP6 IP SIROCCO (contract LSHG-CT-2006-037900), and a Gottfried Wilhelm Leibniz Award to D.W., and the Max Planck Society.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1149408.
REFERENCES
- Allen E., Xie Z., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana . Nat. Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Axtell M.J. Evolution of microRNAs and their targets: Are all microRNAs biologically relevant? Biochim. Biophys. Acta. 2008 doi: 10.1016/j.bbagrm.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Chapman E.J., Carrington J.C. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- Dermitzakis E.T., Clark A.G. Evolution of transcription factor binding sites in mammalian gene regulatory regions: Conservation and turnover. Mol. Biol. Evol. 2002;19:1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- Dezulian T., Remmert M., Palatnik J.F., Weigel D., Huson D.H. Identification of plant microRNA homologs. Bioinformatics. 2006;22:359–360. doi: 10.1093/bioinformatics/bti802. [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Howell M.D., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., et al. High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS One. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Kent W.J. BLAT: The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M.A., Haubold B., Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol. Biol. Evol. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- Lu C., Kulkarni K., Souret F.F., MuthuValliappan R., Tej S.S., Poethig R.S., Henderson I.R., Jacobsen S.E., Wang W., Green P.J., et al. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Shen Y., Wu Q., Kumar S., He B., Shi S., Carthew R.W., Wang S.M., Wu C.I. The birth and death of microRNA genes in Drosophila . Nat. Genet. 2008;40:351–355. doi: 10.1038/ng.73. [DOI] [PubMed] [Google Scholar]
- Ming R., Hou S., Feng Y., Yu Q., Dionne-Laporte A., Saw J.H., Senin P., Wang W., Ly B.V., Lewis K.L., et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana . Genes & Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P., Di Cara A. Hairpin RNA: A secondary structure of primary importance. Cell. Mol. Life Sci. 2006;63:901–908. doi: 10.1007/s00018-005-5558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G.A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]