Abstract
Group II introns are mobile retroelements that invade their hosts. The Lactococcus lactis group II intron recruits cellular polymerases, nucleases, and DNA ligase to complete the retromobility process in Escherichia coli. Here we describe a genetic screen with a Tn5 transposon library to identify other E. coli functions involved in retromobility of the L. lactis LtrB intron. Thirteen disruptions that reproducibly resulted in increased or decreased retrohoming levels into the E. coli chromosome were isolated. These functions were classified as factors involved in RNA processing, DNA replication, energy metabolism, and global regulation. Here we characterize a novel mutant in the rne promoter region, which regulates RNase E expression. Retrohoming and retrotransposition levels are elevated in the rne∷Tn5 mutant. The stimulatory effect of the mutation on retromobility results from intron RNA accumulation in the RNase E mutant. These results suggest that RNase E, which is the central component of the RNA degradosome, could regulate retrohoming levels in response to cellular physiology.
Keywords: mobile retroelement, transposon screen, group II intron, regulation by RNase E
INTRODUCTION
Group II introns, found in archaeal, bacterial, and organellar genomes (Michel and Dujon 1983; Ferat and Michel 1993; Dai et al. 2003; Rest and Mindell 2003), are ribozymes that catalyze the splicing of their flanking exons (Michel and Ferat 1995; Bonen and Vogel 2001). Some group II introns are also mobile retroelements that site-specifically integrate into a homing site, through a process known as retrohoming; they can also integrate nonspecifically into ectopic sites through retrotransposition (Belfort et al. 2002; Lehmann and Schmidt 2003; Lambowitz and Zimmerly 2004; Lambowitz and Pyle 2006). The efficient retrohoming reaction and the less efficient retrotransposition reaction are both catalyzed by a ribonucleoprotein (RNP) complex that consists of the intron encoded protein (IEP) and the spliced intron lariat RNA (Belfort et al. 2002; Lambowitz and Zimmerly 2004; Lambowitz and Pyle 2006).
The group II intron Ll.LtrB is found in the conjugative plasmid pRS01 from the Gram-positive bacterium Lactococcus lactis (Mills et al. 1996; Shearman et al. 1996); it functions in Escherichia coli and encodes a protein, LtrA, that has reverse transcriptase, RNA maturase, and DNA endonuclease activities (Matsuura et al. 1997). In retrohoming, the RNP complex of the Ll.LtrB intron recognizes the retrohoming site through protein–DNA interactions between LtrA and the DNA target, along with base pairings between the intron's exon binding sequence (EBS/δ) and the target site's intron binding sequence (IBS/δ′) (Guo et al. 1997, 2000; Mohr et al. 2000; Singh and Lambowitz 2001). Next, the intron reverse-splices into the top strand of the DNA, and the bottom strand is cleaved nine nucleotides downstream of the integration site, using the endonuclease activity of the LtrA protein. The reverse transcriptase domain of the LtrA protein then synthesizes the first-strand cDNA copy of the intron RNA initiated from the 3′-hydroxyl present at the DNA cleavage site, through a process known as target-primed reverse transcription (TPRT) (Belfort et al. 2002; Lambowitz and Zimmerly 2004; Lambowitz and Pyle 2006). Following TPRT in E. coli, host-encoded DNA exonucleases (RecJ and MutD) and RNases (RNase H and Pol I) resect DNA and remove the intron RNA (Smith et al. 2005). Then, repair (Pol II, Pol IV, and Pol V) and replicative (Pol III) polymerases complete the process, including second-strand synthesis of the cDNA template. Finally, DNA ligase seals the nicks, completing the retrohoming process. This interplay between the intron's molecular machinary and the cellular replication/repair proteins reveals the complex nature of the group II intron-host relationship.
To extend the investigation of host functions involved in retrohoming of the Ll.LtrB intron, we screened a transposon library of mutant strains that contain the natural homing site inserted into the E. coli chromosome. A chromosomal homing site eliminates the need for the recipient plasmid while simplifying the genetic screen. We first measured retrohoming frequencies in a series of strains that contain the homing site placed at ∼10-min intervals within the chromosome. We then screened a mutant library in a strain that contains the homing site within the dedE gene on the E. coli genome, and identified a variety of host factors that either up- or down-regulate retrohoming of the Ll.LtrB intron. We chose mutants that have a similar effect with the homing site at a second chromosomal locus, the ycjV gene. Our findings indicate that retrohoming is perturbed by disturbances in nucleic acid and energy metabolism, as well as imbalances in global gene expression of the host. In this work, we characterize a novel disruption in the 5′-untranslated region (5′-UTR) of the RNase E gene, which elevates retrohoming levels, and suggests that this ribonuclease could silence group II introns in accord with the physiological status of the cell.
RESULTS
Retrohoming of the group II intron Ll.LtrB into the E. coli genome
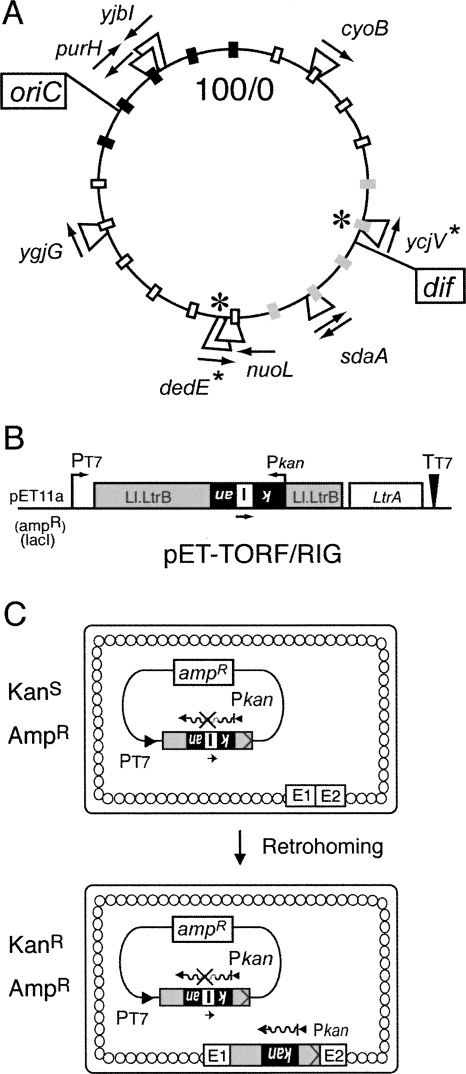
To eliminate the requirement for a recipient plasmid, we created a series of strains, each of which contains the Ll.LtrB intron homing site located at a different position in the chromosome (Fig. 1A). The homing site used was 45 base pairs (bp) in length, comprising residues −30 to +15, where the intron integrates between −1 and +1. This sequence was shown to contain all the elements required for efficient retrohoming (Cousineau et al. 1998; Guo et al. 2000; Mohr et al. 2000). The 45-bp homing site was inserted into eight different nonessential genes within the E. coli genome (Fig. 1A). Two of these genes (purH and sdaA) contained the homing site in opposite orientations. The retrohoming frequency in each of these strains was determined by inducing the Ll.LtrB intron from the pET-TORF/RIG donor plasmid (Fig. 1B). This plasmid contains the retromobility indicator gene (RIG) in Ll.LtrB and the LtrA protein coding sequence downstream of the intron (Coros et al. 2005). The RIG cassette consists of the kanamycin resistance (kanR) gene expressed in the opposite orientation relative to the Ll.LtrB intron, containing the group I td self-splicing intron in the same orientation as Ll.LtrB. The kanR gene is only expressed when Ll.LtrB integrates into a DNA target site through an RNA intermediate (Fig. 1C).
FIGURE 1.
Homing-site insertions and chromosomal retrohoming assay. (A) Location of the genes that contain the −30/+15 homing site. The circular 100-min map of the E. coli genome is marked in 5-min intervals by small rectangles: black rectangles represent the Ori macrodomain, gray rectangles represent the Ter macrodomain, and unfilled rectangles represent the other macrodomains (Valens et al. 2004). The arrows indicate the direction of the homing site relative to the replication origin. The bidirectional replication origin (oriC) and the terminal recombination site (dif) are boxed. The location of the genes with the homing-site that were used to determine chromosomal retrohoming frequencies is marked with an asterisk. (B) The intron donor, pET-TORF/RIG. The ampR RIG donor plasmid carries a kanR gene (black) that is in the opposite transcriptional orientation to that of Ll.LtrB (gray), and is disrupted by the td intron (I) in the same orientation as Ll.LtrB. The kanR gene transcribed from Pkan is therefore not active, unless the Ll.LtrB transposes via an RNA intermediate and the td intron is spliced out. The coding sequence for LtrA has been deleted from the intron and is expressed downstream. PT7 is the T7 promoter, and TT7 is the T7 terminator. (C) Chromosomal retrohoming assay. With the RIG donor, only the Ll.LtrB intron that has lost the group I intron via retrohoming can confer KanR, thus allowing direct selection. The small arrow under the group I intron indicates transcription in the same orientation as the Ll.LtrB intron. Wavy lines depict kanR transcripts, and E1 and E2 represent the exon 1 and exon 2 of the −30/+15 homing site in the E. coli genome.
There was only a 2.2-fold difference in retrohoming frequency between the strain with the lowest frequency, dedE (1.7 × 10−6), and the strains with the highest frequency, sdaA and ycjV (3.7 × 10−6), at 37°C, and no significant difference when the homing site was present in different orientations relative to gene expression (Supplemental Table S1). These results are surprising given that endonuclease-dependent retrotransposition to ectopic chromosomal sites, which is ∼100-fold less efficient, displays a bipolar integration bias into the Ori and Ter macrodomains of the E. coli genome (Beauregard et al. 2006). These observations underscore differences in the localization of retrohoming to the wild-type target site (10−5–10−6 events per genome) and retrotransposition to ectopic sites (10−8–10−9 events per genome) (Coros et al. 2005). Despite the differences, these experiments set the stage for a mutant screen based on retrohoming into chromosomal sites.
Identification of transposon insertion mutants that affect Ll.LtrB retrohoming into the chromosome
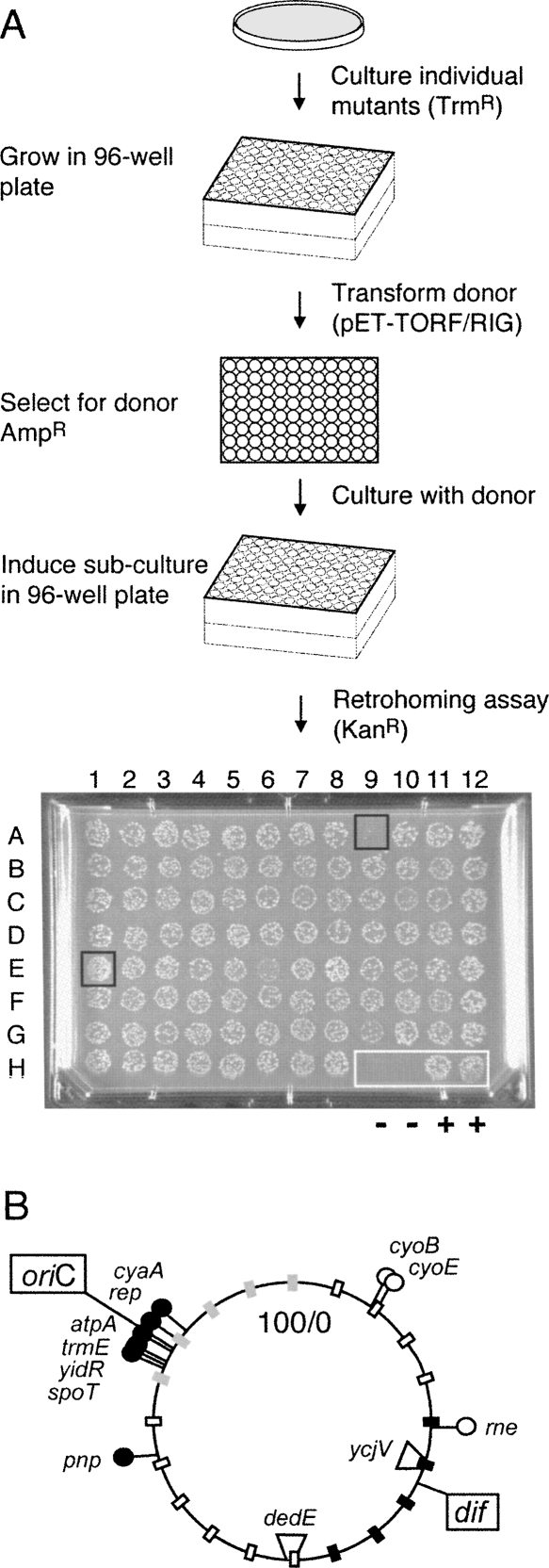
To identify viable mutants that affect retrohoming of the Ll.LtrB intron, a transposon insertion library was generated in the dedE homing-site strain using the Tn5 transpososome system (Goryshin et al. 2000). Tn5 insertion mutants generated by this method are randomly distributed throughout the E. coli chromosome and are stable due to the absence of the Tn5 transposase. We thus isolated 9936 independent insertion mutants, from three different transposon libraries. Subcultures were transformed by the pET-TORF/RIG-ampR donor plasmid. Mutants that affect retrohoming were identified by inducing the Ll.LtrB intron from the donor and spotting the cultures on kanamycin-containing plates (Fig. 2A).
FIGURE 2.
Mutant host screen and location of Tn5 insertions. (A) Schematic of retrohoming assay to screen E. coli mutant libraries. Strain MC1061 dedE∷HS contains the −30/+15 homing site in the dedE gene. Individual trimethoprim-resistant MC1061 dedE∷HS colonies, containing a randomly integrated EZ∷TN <DHFR> transposon, were recovered in selective medium and stored in 96-well plates. Each mutant was then transformed with the pET-TORF/RIG-ampR donor plasmid, and fresh transformants were grown overnight. Ll.LtrB intron expression was induced for 3 h in diluted log-phase cultures, which were spotted onto omnitray slabs containing kanamycin and incubated overnight. A sample slab is shown with two negative (−) control spots at H9 and H10 and with two positive (+) wild-type control spots at H11 and H12. Spot E1 represents a candidate for an “up” mutant and A9 for a “down” mutant. (B) Location on the E. coli genome of Tn5 insertions that affect retrohoming in dedE∷HS and ycjV∷HS strains. The filled circles indicate mutations that decrease retrohoming, and the open circles are mutations that increase retrohoming. The circular map is marked as in Figure 1A, with the location of the homing-site genes used to determine retrohoming frequencies indicated (dedE and ycjV).
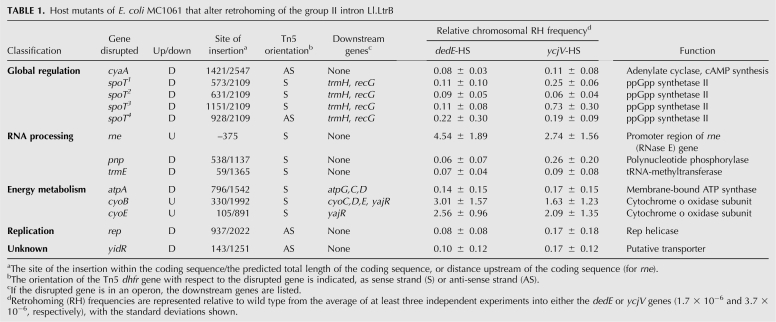
Based on visual inspection of KanR colonies, we identified 17 potential “up” mutants and 62 potential “down” mutants. The DNA flanking the Tn5 transposon was sequenced, and the insertion sites were located using BLAST searches against the E. coli genome. Of the 79 strains, 24 were shown to clearly reproduce the observed phenotype using a more quantitative, less dense plating format. To determine whether these mutants have a similar phenotype in another strain background, each of the 24 Tn5 transposon insertions were transduced into the ycjV homing-site strain and the retrohoming frequencies were determined. Strains with the homing site in the dedE and ycjV genes were selected because those gave the lowest and highest retrohoming levels, respectively (Supplemental Table S1), and they are well separated on the chromosome (Fig. 2B). Thirteen of the 24 mutants had similar effects in the two strain backgrounds (Table 1). Of these 13, three (cyoB, cyoE, and rne) increased the retrohoming frequency >2.5-fold and 10 (atpA, cyaA, pnp, rep, trmE, yidR, and four independent insertions in spoT) decreased the retrohoming frequency approximately fivefold in both strain backgrounds (Table 1). Interestingly, nine of the 10 down mutants listed in Table 1 map to within 5 min of oriC (Fig. 2B), although the significance of this observation is not known.
TABLE 1.
Host mutants of E. coli MC1061 that alter retrohoming of the group II intron Ll.LtrB
We classified the mutants according to their GO annotation on the EcoCyc website (http://ecocyc.org/) (Table 1). Five of the 13 independent mutations are located in two separate genes that affect global transcriptional regulation. One is in the cyaA gene, which encodes adenylate cyclase for cAMP synthesis, and the other four are in spoT, which plays a role in ppGpp synthesis. Three of the other insertions are in genes involved in RNA processing (rne, pnp, and trmE). Interestingly RNase E, the product of rne, and polynucleotide phosphorylase (PNPase), the product of pnp, have opposite effects on retrohoming (Table 1), although they are both components of the RNA degradosome (Carpousis 2007). Of the other 13 mutants, three affect energy metabolism (atpA, cyoB, and cyoE), one affects replication (rep), and one is defective in a putative transporter (yidR) (Table 1). Evidently, a variety of changes in the host influence the retrohoming efficiency, by either stimulating or inhibiting the process. To evaluate indirect effects, these mutants were screened for plasmid copy number, intron RNA expression, splicing efficiency, and LtrA protein expression (Supplemental Table S2). Modest fluctuations in these measurements could not account for the observed changes in retrohoming phenotypes, except for the trmE mutant, which had a 10-fold reduction in intron RNA. Here we characterize a mutant within the 5′-UTR of rne, which encodes RNase E.
The rne mutation alters expression of RNase E, which regulates retrohoming
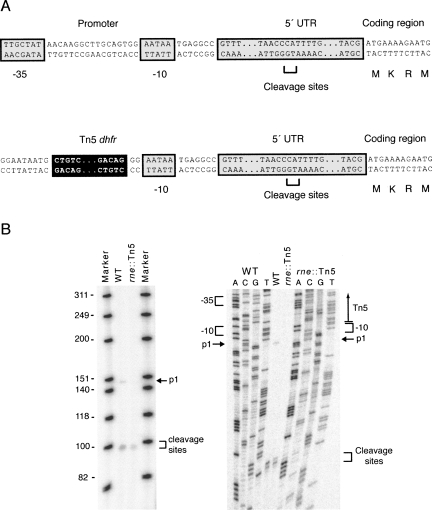
The rne gene contains an unusually long 5′-UTR (Fig. 3A; Claverie-Martin et al. 1991), which folds into a secondary structure and is cleaved by RNase E, resulting in the autoregulation of the rne transcript (Jain and Belasco 1995). The rne transcript levels in E. coli are also regulated by the presence of multiple promoters that may be influenced by the presence of gene b1085, which is transcribed on the noncoding strand within the 5′-UTR of rne (Ow et al. 2002). In a complementation assay with the b1085 gene, we showed that the b1085 gene is not directly affecting retrohoming (data not shown). Therefore, to determine whether the rne mutation in the 5′-UTR affects expression of the rne gene directly, we performed primer extension assays in the wild type and in the rne∷Tn5 mutant using an rne-specific primer. The primary transcription start site (p1) of the rne gene was undetectable in the 5′-UTR mutant (Fig. 3B, left and right), but the rne gene was still being expressed to some extent and controlled, as evidenced by the RNase E–specific autoregulated cleavage products present in the mutant. Low-level expression could account for viability of the rne∷Tn5 mutant.
FIGURE 3.
The 5′-UTR rne mutant regulates RNase E. (A) Schematic of the wild-type rne promoter region. The P1 promoter -35 and -10 sites are shown relative to the unusually long 5′ UTR upstream of the coding sequence. The P1 promoter is disrupted by the presence of the Tn5 insertion, depicted below. (B) Primer extension assay of the rne promoter region of the wild type and rne∷Tn5 5′-UTR mutant. The P1 transcript is not apparent in rne∷Tn5, but the cleavage product is.
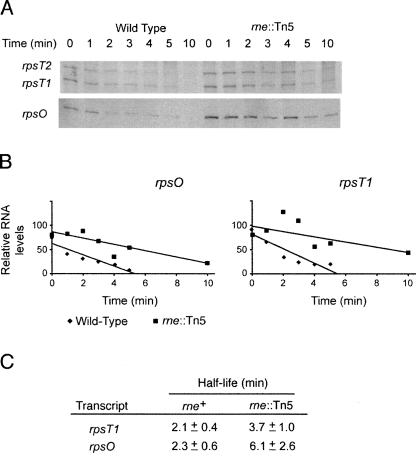
To further explore RNase E expression in the rne∷Tn5 mutant, we performed stability assays on the rne-regulated transcripts, rpsO and rpsT. RNA was extracted at the times indicated, after addition of rifamycin to a log-phase culture to inhibit transcription, and the RNAs were identified and quantified by Northern blot. The rpsT gene has two transcripts of equivalent stabilities, initiating at two promoters (Mackie and Parsons 1983). The rpsO and rpsT transcript levels and half-lives were found to be increased 1.8- to 2.7-fold in the mutant strain (Fig. 4), indicating that the Tn5 insertion in the 5′-UTR of rne is affecting the expression of RNase E–regulated transcripts. This increase in half-life is consistent with the 1.4- to 2.2-fold increase in expression when separate rne promoters are deleted (Ow et al. 2002).
FIGURE 4.
RNase E–regulated transcripts are stabilized in the 5′-UTR rne∷Tn5 mutant. (A) Northern blots of E. coli transcripts. Blots were probed with rpsT- and rpsO-specific probes following the addition of rifampicin in the wild-type and rne∷Tn5 mutant strains. Time after the addition of rifampicin is shown above the blot. Probes for the Northern blots are described in Supplemental Table S4. (B) Decay of transcripts. Scans of the Northern blots were quantitated. All numbers in the graph have been normalized to a blot of 16S ribosomal RNA. (C) Half-lives of RNase E–regulated transcripts. Half-lives were derived from decay curves (panel B) from three independent experiments.
We had previously shown that RNase E inhibits retrohoming into a plasmid (Smith et al. 2005), in accord with enhanced retrohoming into the chromosome in the rne∷Tn5 mutant. This result indicates that enhanced retrohoming in RNase E mutants is independent of the replicon that contains the homing site, suggesting that RNase E acts at the level of the RNP, rather than at the level of the target DNA. Furthermore, we measured mobility to ectopic sties in E. coli lacking the homing site and showed that retrotransposition in the rne∷Tn5 mutant was ∼100-fold elevated (C.L. Piazza, unpubl.).
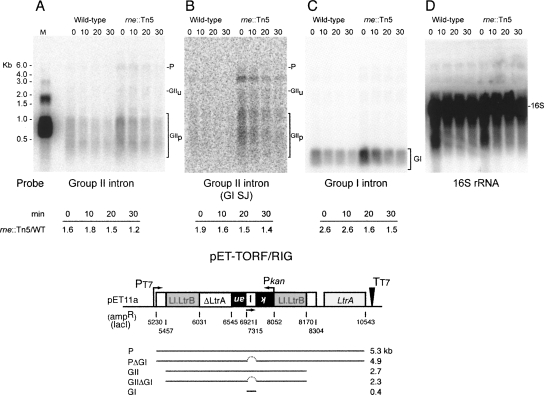
To determine whether the effect of the Tn5 insertion is direct, via stabilization of intron RNA, or indirect, via expression of a factor involved in the retromobility pathway, Northern blots were performed. Intron expression was induced for 1.5 h with isopropyl β-D-1-thiogalactopyranoside (IPTG) from pET-TORF/RIG-ampR, the cells were washed to remove the IPTG, and RNA was extracted 0–30 min thereafter. The Northern blot indicated 1.2- to 1.8-fold greater stability of intron RNA when probed with a group II intron–specific probe (Fig. 5A). A second probe within the group II intron, at the group I td intron splice junction in the kanR gene, revealed a similar degree of group II intron RNA stabilization (1.4- to 1.9-fold) (Fig. 5B). Likewise, the group I intron RNA was 1.5- to 2.6-fold more stable in the rne mutant, as observed with the group I td intron probe (Fig. 5C). In contrast, 16S rRNA was equivalently stable in the parental strain and the rne∷Tn mutant (Fig. 5D). This work demonstrates that RNase E has a direct effect on intron RNA stability.
FIGURE 5.
Intron RNA accumulates in the rne∷Tn5 mutant. (A) Northern blot probed with a group II intron probe. P indicates precursor; GIIu, unprocessed group II intron; and GIIp, processed group II intron. M is the marker lane in which a group II intron of 903 nucleotides was separated along with size markers. This and other probe sequences are listed in Supplemental Table S4. (B) A second group II intron probe. The same blot was probed with a group I intron splice junction probe, which hybridizes to the kanR gene in the group II intron. The band below the weak precursor band is likely to represent the precursor lacking the group I intron, but this has not been confirmed. (C) A group I intron probe also reveals enhanced stability. (D) 16S rRNA probe. A schematic of the pET-TORF/RIG transcript below the blots indicates the theoretical length of the precursor (P), the precursor lacking the group I intron (PΔGI), the group II intron (GII), the group II intron lacking the group I intron (GIIΔGI), and the group I intron (GI).
DISCUSSION
Our findings highlight the complex nature of the group II intron–host relationship. Whereas group II introns are known to rely on host factors to complete the mobility process, including DNA polymerases, ribo- and deoxyribonucleases, and DNA ligase (Smith et al. 2005), little is known about the introns' responsiveness to other factors involved in cellular physiology. Here we have developed a mutant screen to address this matter. We thus showed that group II introns appear to respond to changes in global regulation (cyaA and spoT), RNA processing (rne, pnp, and trmE), energy metabolism (atpA, cyoE, and cyoB), DNA replication (rep), and possibly transport functions (putative; yidR), but in some of these cases, effects of insertions on downstream gene expression remain to be addressed.
It is noteworthy that a diverse set of host factors also influences the mobility of DNA elements (Twiss et al. 2005). In particular, a series of mutants that affect DNA and intermediary metabolism were identified as factors that regulate transposition of the DNA-based elements IS903, Tn10, and Tn552, suggesting that these elements are also stimulated by nutritional stress. With respect to retrotransposition, particularly in Saccharomyces cerevisiae, a plethora of host mutants was identified in genes that affect Ty1 (Scholes et al. 2001; Griffith et al. 2003) and Ty3 elements (Aye et al. 2004). Thus factors that affect transcriptional regulation, RNA processing, and metabolism were implicated. Although these various screens identified some similar host functions, very few overlapping genes were discovered in any of these studies, suggesting that individual elements draw on a different repertoire of host proteins to contribute to and/or regulate mobility (for review, see Beauregard et al. 2008).
Retrohoming and RNase E
RNA processing genes have been shown to have dramatic effects on the mobility of Ll.LtrB (Smith et al. 2005). In particular, RNase H has been implicated in the removal of the intron RNA from the RNA–DNA hybrid intermediate, after first-strand cDNA synthesis. In contrast to the facilitatory role of RNase H, it was demonstrated that RNase I and RNase E inhibit retrohoming. The inhibition occurs either directly, through degradation of the intron, either prior to reverse splicing or cDNA synthesis (Smith et al. 2005), or indirectly, through control of a function that regulates retromobility. Since RNase E is an essential protein, the previous study used a temperature-sensitive mutation, rne-1, with retrohoming into a plasmid target being ∼10-fold elevated. The mutation that we have identified here in the 5′-UTR of rne has a similar phenotype, with chromosomal retrohoming being three- to fivefold elevated. Given the increase in retrohoming into plasmid and chromosomal targets in the rne-1 and 5′-UTR rne strains (Table 1; Smith et al. 2005; data not shown), it seemed likely that the intron is more stable when the rne gene is compromised. In accord with this prediction, the rne∷Tn5 mutation increased the RNA levels of Ll.LtrB in the cell (Fig. 5). These results argue against an RNase E–regulated factor causing increases in retrohoming, and in favor of RNase E being directly involved in the degradation of the intron.
Retromobility and functions of the degradosome
RNase E, which down-regulates retrohoming, is the organizing component of the degradosome, a complex that in E. coli also includes PNPase, RhlB, and enolase (for review, see Carpousis 2007). RNase E is the only essential enzyme in the RNA decay complex, cleaving in intercistronic regions of polycistronic messages. PNPase acts as a 3′-5′ exoribonuclease, whereas RhlB is a DEAD-box helicase. Enolase, a glycolytic enzyme, is postulated to act as a sensor, linking cellular energetics to mRNA degradation.
An intriguing observation is that two components of the degradosome have opposing effects on retrohoming. Although the rne∷Tn5 mutant had elevated retrohoming levels, the pnp∷Tn5 mutant suffered a more than 10-fold reduction in retrohoming (Table 1). This result implies that RNase E and PNPase act in different pathways, and that PNPase has a stimulatory role in retromobility. The concept of multiple pathways is consistent with the RNA degradosome being a dynamic structure that can consist of different RNase E–based multiprotein complexes (Morita et al. 2005; Carpousis 2007).
PNPase can act independently of RNase E in several cellular processes. These include modulation of polyadenylation (Mohanty and Kushner 2000) and selective RNA degradation (Yamanaka and Inouye 2001). Also, PNPase can bind DNA and has been hypothesized to inhibit DNA replication (Bermudez-Cruz et al. 2002). Furthermore, the absence of PNPase affects expression of almost one-half of the mRNA in the cell (Mohanty and Kushner 2003). Given these diverse roles, it is difficult to ascribe the stimulation of retrohoming by PNPase to any particular function, but we have two favored hypotheses, based on both RNA levels and splicing being maintained in the mutant (Supplemental Table S2). First, a stimulatory role for PNPase could relate to the need for degradation of intron RNA in the RNA–DNA hybrid, after RNA-templated cDNA synthesis, and before second-strand cDNA synthesis can proceed. We have previously shown that the endonucleolytic activity of RNase H1 and the 5′-3′ exoribonucleolytic activity of Pol I are likely involved in this function (Smith et al. 2005). Given that the directionality of the PNPase exonuclease is 3′-5′, it would not be surprising if a third enzyme were involved in removal of the RNA before second-strand cDNA synthesis. Second, the Ll.LtrB group II intron can integrate at replication forks (Ichiyanagi et al. 2002; Zhong and Lambowitz 2003). If indeed PNPase inhibits replication (Bermudez-Cruz et al. 2002), causing stalled forks, PNPase may promote retrohoming indirectly. These two hypotheses are not mutually exclusive. Given the differences in RNA degradation pathways in Gram-positive organisms (Condon 2007), it will be of particular interest to determine the roles of ribonucleases in L. lactis.
The phylogenetic plasticity of the RNase E–based degradosome has been proposed to be an evolutionary adaptation to prevailing environmental conditions of the bacterial cell (Marcaida et al. 2006) and may regulate group II introns accordingly. Indeed, elevation of the group II intron RNP when RNase E levels are depressed has important evolutionary consequences. Target-based stimulation confines group II intron movement to within a cell, supposedly enhancing genetic diversity. In addition to such intracellular mobility, as is the case for some of the mutants described in Table 1 (C.J. Coros, C.L. Piazza, V.R. Chalamcharla, D. Smith, and M. Belfort, in prep.), RNP-based stimulation, as when RNase E is down-regulated, could conceivably promote extracellular horizontal intron transfer.
It has recently been shown that components of human L1 elements, which like group II introns are target-primed retrotransposons, localize to stress granules, which also contain an RNA silencing complex (Goodier et al. 2007). It was therefore suggested that this localization provides a means whereby the cell mitigates the potential mutagenic effects of retrotransposition by targeting the L1 RNPs for degradation. It is tempting to speculate that like the human stress granules, the bacterial degradosome is a metabolic sensor, possibly acting via enolase, to gauge the physiological status of the cell and accordingly silence group II intron retromobility.
MATERIALS AND METHODS
Bacterial strains and media
Retrohoming assays were performed in TBYE (1% Bactone tryptone, 0.5% NaCl, 0.1% Bacto yeast extract) using λDE3 lysogens, which express T7 RNA polymerase from a prophage, upon induction with IPTG (Studier et al. 1990). The E. coli strains used for the retrohoming assays were derived from MC1061(λDE3) (Supplemental Table S3A).
The λRed system was used to insert the homing site into eight different locations in the E. coli genome, according to published procedures (Yu et al. 2000). Basically, the homing-site−FRT−kan−FRT cassette, amplified from plasmid pKD4∷HS, was targeted individually to the cyoB, ycjV, nuoL, ygjG, dedE, sdaA, purH, and yjb1 genes by homologous recombination in the λRed-expressing strain DY329. The marked homing site was next transferred to MC1061(λDE3) by generalized P1 transduction (Sambrook and Russell 2001). The kanR gene was then excised by expressing the FLP recombinase protein from the temperature-sensitive plasmid pCP20 (Datsenko and Wanner 2000). PCR was used to confirm the loss of the marker and the presence of the insertion.
Plasmids
Plasmids are listed in Supplemental Table S3B. Construction of the donor plasmid pET-TORF/RIG-ampR, which has the LtrA protein transcribed downstream of the Ll.LtrB intron was previously described (Coros et al. 2005). The pKD4∷HS template plasmid was created by inserting the −30/+15 homing site upstream of the FRT−kan−FRT cassette in the plasmid pKD4 (Datsenko and Wanner 2000), by inverse-PCR with primers IDT0183 and IDT0184. All oligonucleotides are listed in Supplemental Table S4. The plasmids pGEM-5′-UTR and pGEM-5′-UTR-rne were created using the pGEM-T vector system (Promega) with the primer set IDT0953-IDT0954, which amplified sequences from MC1061 and MC1061-5′-UTR-rne genomic DNA, respectively.
Mutant library screen and plasmid-to-chromosome retrohoming assay
Electro-competent MC1061(λDE3) dedE∷HS cells were electroporated with the EZ∷TN <DHFR> transpososome (Epicentre) and were plated on medium containing 10 μg/mL trimethoprim. Individual trimethoprim-resistant colonies, carrying the dihydrofolate reductase gene, were recovered in selective medium and stored in 96-well plates at −80°C. Approximately 10,000 individual cultures in 96-well format were then transformed with the pET-TORF/RIG-ampR donor plasmid, and fresh transformants were grown overnight. Cultures were diluted 1 in 100 into 1 mL of selective medium and were grown to an OD600 of ∼0.2 in 96-deep-well plates at 37°C, with shaking at 300 RPM. Ll.LtrB intron expression was induced with 100 μM IPTG. After 3 h of induction, the 1 mL culture was spun down, and 7.5-μL aliquots were spotted onto omnitray slabs containing medium with 40 μg/mL of kanamycin and incubated overnight at 37°C. Levels of mobility were observed for each “spot,” and the mutants were identified as having an increased, decreased, or no effect on retromobility (Fig. 2). Retromobility frequencies were then determined for mutant strains that appeared to increase or decrease retrohoming using a less dense plating format that allowed for the quantification of KanR colonies versus total cells.
Mapping of Tn5 insertion sites
Transposon (EZ∷TN <DHFR>) integration sites for mutant strains that increased or decreased retrohoming were determined by inverse PCR sequencing (Ochman et al. 1988). Genomic DNA was isolated using the QIAGEN DNeasy Tissue Kit, according to manufacturer's protocol. Chromosomal DNA was digested with Sau3AI or MseI, recircularized by ligation, and amplified with primers IDT0326 and IDT0327 or with primers IDT0330 and IDT0331 for Sau3AI or MseI products, respectively. The first PCR product was then amplified with nested primers IDT0328 and IDT0333 or with nested primers IDT0332 and IDT0329 for the Sau3AI or MseI products, respectively, and sequenced. NCBI BLAST searches were performed to determine the integration sites within the E. coli genome using the website www.ncbi.nlm.nih.gov and gene functions were assigned using the EcoCyc http://ecocyc.org/ website.
Northern blot analysis
Analysis of rpsT and rpsO was performed in strain MC1061(λDE3) dedE∷HS with or without rne∷Tn5. Cells were diluted 1 in 100 in 25 mL TBYE and grown to an OD600 0.6–0.8, at which time 150 μg/mL rifamycin was added. One-milliliter samples were taken at 0–10 min and were added to 500 μL of frozen 50 mM NaN3. RNA was extracted using the Invitrogen TRIzol Max Bacterial RNA Isolation Kit and was separated on a 6% polyacrylamide denaturing gel. The RNA was transferred to GE Healthcare Amersham Hybond-N+ membrane, UV-crosslinked, and hybridized according to the manufacturer's protocol for oligonucleotide probes. Membranes were probed for rpsT and rpsO transcripts with 32P-labeled oligonucleotides IDT1085 and IDT1083, respectively.
Ll.LtrB intron expression was induced with 100 μM IPTG in strain MC1061(λDE3) dedE∷HS rne∷Tn5 containing the pET-TORF/RIG-ampr donor plasmid. After 1.5 h of induction, samples were centrifuged and resuspended in fresh media without IPTG. Aliquots were taken at 0–30 min and 1 mL culture was added to 500 μL of frozen 50 mM NaN3. RNA was extracted using the QIAGEN RNeasy Kit and separated on a 1.2% agarose/formaldehyde gel. The RNA was transferred and hybridized as above. Membranes were probed for group II Ll.LtrB intron, group I td intron, the group I td intron splice junction in the kan gene of the pET-TORF/RIG-ampR construct, and 16S rRNA with 32P-labeled oligonucleotides IDT0653, MB14, W1609, and IDT0861, respectively. For both Northern blots, membranes were exposed on phosphor screens and scanned with the Amersham Bioscience Typhoon 9400. Bands were quantified using the GE Healthcare ImageQuant 5.2 software and normalized to 16S rRNA bands.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Sydney Kushner, Bill Reznikoff, and Harry Taber for useful discussions and advice and Maryellen Carl and John Dansereau for their help in preparing the manuscript and figures, respectively. We acknowledge the use of the Wadsworth Center DNA Sequencing Core. This work was supported by NIH grants GM39422 and GM44844.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1247608.
REFERENCES
- Aye M., Irwin B., Beliakova-Bethell N., Chen E., Garrus J., Sandmeyer S. Host factors that affect Ty3 retrotransposition in Saccharomyces cerevisiae . Genetics. 2004;168:1159–1176. doi: 10.1534/genetics.104.028126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauregard A., Chalamcharla V.R., Piazza C.L., Belfort M., Coros C.J. Bipolar localization of the group II intron Ll.LtrB is maintained in Escherichia coli deficient in nucleoid condensation, chromosome partitioning and DNA replication. Mol. Microbiol. 2006;62:709–722. doi: 10.1111/j.1365-2958.2006.05419.x. [DOI] [PubMed] [Google Scholar]
- Beauregard A., Curcio M.J., Belfort M. The take and give between retrotransposable elements and their hosts. Annu. Rev. Genet. 2008;42 doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort M., Derbyshire V., Parker M.M., Cousineau B., Lambowitz A.M. Mobile introns: Pathways and proteins. In: Craig N., et al., editors. Mobile DNA II. ASM Press; Herndon, VA: 2002. pp. 761–783. [Google Scholar]
- Bermudez-Cruz R.M., Garcia-Mena J., Montanez C. Polynucleotide phosphorylase binds to ssRNA with same affinity as to ssDNA. Biochimie. 2002;84:321–328. doi: 10.1016/s0300-9084(02)01385-8. [DOI] [PubMed] [Google Scholar]
- Bonen L., Vogel J. The ins and outs of group II introns. Trends Genet. 2001;17:322–331. doi: 10.1016/s0168-9525(01)02324-1. [DOI] [PubMed] [Google Scholar]
- Carpousis A.J. The RNA degradosome of Escherichia coli: An mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- Claverie-Martin F., Diaz-Torres M.R., Yancey S.D., Kushner S.R. Analysis of the altered mRNA stability (ams) gene from Escherichia coli. Nucleotide sequence, transcriptional analysis, and homology of its product to MRP3, a mitochondrial ribosomal protein from Neurospora crassa . J. Biol. Chem. 1991;266:2843–2851. [PubMed] [Google Scholar]
- Condon C. Maturation and degradation of RNA in bacteria. Curr. Opin. Microbiol. 2007;10:271–278. doi: 10.1016/j.mib.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Coros C.J., Landthaler M., Piazza C.L., Beauregard A., Esposito D., Perutka J., Lambowitz A.M., Belfort M. Retrotransposition strategies of the Lactococcus lactis Ll.LtrB group II intron are dictated by host identity and cellular environment. Mol. Microbiol. 2005;56:509–524. doi: 10.1111/j.1365-2958.2005.04554.x. [DOI] [PubMed] [Google Scholar]
- Cousineau B., Smith D., Lawrence-Cavanagh S., Mueller J.E., Yang J., Mills D., Manias D., Dunny G., Lambowitz A.M., Belfort M. Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell. 1998;94:451–462. doi: 10.1016/s0092-8674(00)81586-x. [DOI] [PubMed] [Google Scholar]
- Dai L., Toor N., Olson R., Keeping A., Zimmerly S. Database for mobile group II introns. Nucleic Acids Res. 2003;31:424–426. doi: 10.1093/nar/gkg049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferat J.-L., Michel F. Group II self-splicing introns in bacteria. Nature. 1993;364:358–361. doi: 10.1038/364358a0. [DOI] [PubMed] [Google Scholar]
- Goodier J.L., Zhang L., Vetter M.R., Kazazian H.H., Jr LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryshin I.Y., Jendrisak J., Hoffman L.M., Meis R., Reznikoff W.S. Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat. Biotechnol. 2000;18:97–100. doi: 10.1038/72017. [DOI] [PubMed] [Google Scholar]
- Griffith J.L., Coleman L.E., Raymond A.S., Goodson S.G., Pittard W.S., Tsui C., Devine S.E. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae . Genetics. 2003;164:867–879. doi: 10.1093/genetics/164.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Zimmerly S., Perlman P.S., Lambowitz A.M. Group II intron endonucleases use both RNA and protein subunits for recognition of specific sequences in double-stranded DNA. EMBO J. 1997;16:6835–6848. doi: 10.1093/emboj/16.22.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Karberg M., Long M., Jones J.P., III, Sullenger B., Lambowitz A.M. Group II introns designated to insert into therapeutically-relevant DNA target sites in human cells. Science. 2000;289:452–457. doi: 10.1126/science.289.5478.452. [DOI] [PubMed] [Google Scholar]
- Ichiyanagi K., Beauregard A., Lawrence S., Smith D., Cousineau B., Belfort M. Retrotransposition of the Ll.LtrB group II intron proceeds predominantly via reverse splicing into DNA targets. Mol. Microbiol. 2002;46:1259–1272. doi: 10.1046/j.1365-2958.2002.03226.x. [DOI] [PubMed] [Google Scholar]
- Jain C., Belasco J.G. RNase E autoregulates its synthesis by controlling the degradation rate of its own mRNA in Escherichia coli: Unusual sensitivity of the rne transcript to RNase E activity. Genes & Dev. 1995;9:84–96. doi: 10.1101/gad.9.1.84. [DOI] [PubMed] [Google Scholar]
- Lambowitz A.M., Zimmerly S. Mobile group II introns. Annu. Rev. Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- Lambowitz A.M., Pyle A.M. Group II introns: Ribosymes that splice RNA and invade DNA. In: Atkins J.F., editor. The RNA World. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 469–505. [Google Scholar]
- Lehmann K., Schmidt U. Group II introns: Structure and catalytic versatility of large natural ribozymes. Crit. Rev. Biochem. Mol. Biol. 2003;38:249–303. doi: 10.1080/713609236. [DOI] [PubMed] [Google Scholar]
- Mackie G.A., Parsons G.D. Tandem promoters in the gene for ribosomal protein S20. J. Biol. Chem. 1983;258:7840–7846. [PubMed] [Google Scholar]
- Marcaida M.J., DePristo M.A., Chandran V., Carpousis A.J., Luisi B.F. The RNA degradosome: Life in the fast lane of adaptive molecular evolution. Trends Biochem. Sci. 2006;31:359–365. doi: 10.1016/j.tibs.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Matsuura M., Saldanha R., Ma H., Wank H., Yang J., Mohr G., Cavanagh S., Dunny G.M., Belfort M., Lambowitz A.M. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: Biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes & Dev. 1997;11:2910–2924. doi: 10.1101/gad.11.21.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2:33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Ferat J.-L. Structure and activities of group II introns. Annu. Rev. Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- Mills D.A., McKay L.L., Dunny G.M. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J. Bacteriol. 1996;178:3531–3538. doi: 10.1128/jb.178.12.3531-3538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty B.K., Kushner S.R. Polynucleotide phosphorylase, RNase II and RNase E play different roles in the in vivo modulation of polyadenylation in Escherichia coli . Mol. Microbiol. 2000;36:982–994. doi: 10.1046/j.1365-2958.2000.01921.x. [DOI] [PubMed] [Google Scholar]
- Mohanty B.K., Kushner S.R. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol. Microbiol. 2003;50:645–658. doi: 10.1046/j.1365-2958.2003.03724.x. [DOI] [PubMed] [Google Scholar]
- Mohr G., Smith D., Belfort M., Lambowitz A.M. Rules for DNA target site recognition by a Lactococcal group II intron enable retargeting of the intron to specific DNA sequences. Genes & Dev. 2000;14:559–573. [PMC free article] [PubMed] [Google Scholar]
- Morita T., Maki K., Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes & Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Gerber A.S., Hartl D.L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow M.C., Liu Q., Mohanty B.K., Andrew M.E., Maples V.F., Kushner S.R. RNase E levels in Escherichia coli are controlled by a complex regulatory system that involves transcription of the rne gene from three promoters. Mol. Microbiol. 2002;43:159–171. doi: 10.1046/j.1365-2958.2002.02726.x. [DOI] [PubMed] [Google Scholar]
- Rest J.S., Mindell D.P. Retroids in archaea: phylogeny and lateral origins. Mol. Biol. Evol. 2003;20:1134–1142. doi: 10.1093/molbev/msg135. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. Molecular cloning: A laboratory manual, Vol. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. pp. 10.11–10.49. [Google Scholar]
- Scholes D.T., Banerjee M., Bowen B., Curcio M.J. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159:1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman C., Godon J.-J., Gasson M. Splicing of a group II intron in a functional transfer gene of Lactococcus lactis . Mol. Microbiol. 1996;21:45–53. doi: 10.1046/j.1365-2958.1996.00610.x. [DOI] [PubMed] [Google Scholar]
- Singh N.N., Lambowitz A.M. Interaction of a group II intron ribonucleoprotein endonuclease with its DNA target site investigated by DNA footprinting and modification interference. J. Mol. Biol. 2001;309:361–386. doi: 10.1006/jmbi.2001.4658. [DOI] [PubMed] [Google Scholar]
- Smith D., Zhong J., Matsuura M., Lambowitz A.M., Belfort M. Recruitment of host functions suggests a repair pathway for late steps in group II intron retrohoming. Genes & Dev. 2005;19:2477–2487. doi: 10.1101/gad.1345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W., Rosenberg A.H., Dunn J.J., Dubendorff J.W. Use of T7 RNA polymerase to direct expression of cloned genes. In: Goeddel D.V., editor. Methods in enzymology. Vol. 185. Academic Press; New York: 1990. pp. 60–89. [DOI] [PubMed] [Google Scholar]
- Twiss E., Coros A.M., Tavakoli N.P., Derbyshire K.M. Transposition is modulated by a diverse set of host factors in Escherichia coli and is stimulated by nutritional stress. Mol. Microbiol. 2005;57:1593–1607. doi: 10.1111/j.1365-2958.2005.04794.x. [DOI] [PubMed] [Google Scholar]
- Valens M., Penaud S., Rossignol M., Cornet F., Boccard F. Macrodomain organization of the Escherichia coli chromosome. EMBO J. 2004;23:4330–4341. doi: 10.1038/sj.emboj.7600434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K., Inouye M. Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli . J. Bacteriol. 2001;183:2808–2816. doi: 10.1128/JB.183.9.2808-2816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Ellis H.M., Lee E.C., Jenkins N.A., Copeland N.G., Court D.L. An efficient recombination system for chromosome engineering in Escherichia coli . Proc. Natl. Acad. Sci. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., Lambowitz A.M. Group II intron mobility using nascent strands at DNA replication forks to prime reverse transcription. EMBO J. 2003;22:4555–4565. doi: 10.1093/emboj/cdg433. [DOI] [PMC free article] [PubMed] [Google Scholar]