Abstract
Aptamers, an emerging class of therapeutics, are DNA or RNA molecules that are selected to bind molecular targets that range from small organic compounds to large proteins. All of the determined structures of aptamers in complex with small molecule targets show that aptamers cage such ligands. In structures of aptamers in complex with proteins that naturally bind nucleic acid, the aptamers occupy the nucleic acid binding site and often mimic the natural interactions. Here we present a crystal structure of an RNA aptamer bound to human thrombin, a protein that does not naturally bind nucleic acid, at 1.9 Å resolution. The aptamer, which adheres to thrombin at the binding site for heparin, presents an extended molecular surface that is complementary to the protein. Protein recognition involves the stacking of single-stranded adenine bases at the core of the tertiary fold with arginine side chains. These results exemplify how RNA aptamers can fold into intricate conformations that allow them to interact closely with extended surfaces on non-RNA binding proteins.
Keywords: aptamer, crystal structure, thrombin
INTRODUCTION
Aptamers are nucleic acids that adhere to a specific molecular target. Made by using iterative in vitro selection techniques (Ellington and Szostak 1990; Tuerk and Gold 1990), aptamers can be generated that bind essentially any protein or small molecule with high affinity and specificity (Nimjee et al. 2005). RNA and DNA aptamers have been generated to bind small molecule ligands such as arginine and adenosine monophosphate (Hermann and Patel 2000). They also have been selected for a range of clinically relevant targets including viral proteins (Sullenger et al. 1990; Kumar et al. 1997), growth factors (Green et al. 1996; Ruckman et al. 1998), transcription factors (Ishizaki et al. 1996; Lebruska and Maher 1999), and coagulation proteins (Bock et al. 1992; Kubik et al. 1994; Rusconi et al. 2002). Aptamers often bind their target proteins with affinities in the picomolar range, giving them binding affinities on par with the most avid monoclonal antibody/antigen interactions (Nimjee et al. 2005).
Several crystal structures of aptamers in complex with their target molecules have been determined. In the case of small molecule targets, the aptamers often form a cage that surrounds the ligand (Hermann and Patel 2000; Sussman et al. 2000; Tereshko et al. 2003). In structures of aptamers bound to nucleic acid binding proteins, the aptamers bind to the nucleic acid binding site and often mimic naturally occurring interactions (Convery et al. 1998; Jaeger et al. 1998; Cox et al. 2002; Huang et al. 2003; Ghosh et al. 2004). The crystal structure of a DNA aptamer bound to thrombin has been determined (Padmanabhan et al. 1993; Padmanabhan and Tulinsky 1996). This is the only crystal structure so far determined of an aptamer in complex with a protein that does not naturally bind nucleic acid. The DNA interacts with the surface of thrombin known as exosite-1 through van der Waals contacts and hydrogen bonds. The tertiary structure of the DNA is stabilized by a G-quadruplex (Macaya et al. 1993; Wang et al. 1993; Schultze et al. 1994; Padmanabhan and Tulinsky 1996).
Thrombin is a serine protease that plays a crucial role in blood coagulation. In addition to processing numerous proteins in the coagulation cascade, it has the unique ability to cleave soluble fibrinogen into fibrin, which polymerizes to help form a clot (for reviews, see Stubbs and Bode 1995; Hoffman and Monroe 2001; Di Cera 2003). Regions of the molecular surface of thrombin known as exosite-1 and exosite-2 influence interactions with numerous macromolecular substrates and are located on opposite sides of the molecule and away from the catalytic site (Stubbs and Bode 1995). Exosite-2 is the binding site for the oligosaccharide heparin, which is clinically used as an anticoagulant. Heparin binding leads to inhibition of thrombin by facilitating the interaction of thrombin with the endogenous thrombin inhibitor antithrombin (for review, see Rau et al. 2007).
Because of thrombin's essential role in blood coagulation, antithrombin therapeutics have been of great interest. Toggle-25t, an RNA aptamer that contains 2′fluoropyrimidine nucleotides, was selected to bind thrombin with high specificity and affinity (White et al. 2001). In contrast to the G-quadruplex architecture of the DNA aptamer, Toggle-25t has a predicted secondary structure of a stem–loop with an internal bulge. Biochemical analysis shows that Toggle-25t binds to exosite-2, not exosite-1, where the DNA aptamer binds (Jeter et al. 2004). In order to understand the interaction of an RNA aptamer with a protein that does not naturally bind nucleic acid, we determined the crystal structure of Toggle-25t in complex with human thrombin. We find that the simple secondary structure of the RNA forms an elaborate three-dimensional structure that presents an extended, undulatory surface that mates with the native surface of thrombin. Key RNA–protein contacts involve a succession of adenine–arginine stacking interactions, which we term the “A-Arg zipper.”
RESULTS AND DISCUSSION
Structure determination
Cocrystals of human thrombin in complex with the 25 nucleotide-long RNA aptamer, Toggle-25t (White et al. 2001), diffracted X-rays to 1.9 Å resolution. For crystallization, thrombin was covalently modified with the protease active site inhibitor D-Phe-Pro-Arg chloromethylketone (PPACK) in order to eliminate proteolysis. Electron density maps calculated from initial phases obtained by molecular replacement using the crystal structure of human thrombin (PDB ID number 1PPB) clearly indicated the phosphate backbone of the aptamer and allowed unambiguous model building of the RNA molecule. Table 1 contains a summary of X-ray data and crystallographic refinement statistics. The final model contains the entire 25 nucleotide aptamer, thrombin light-chain residues 1D through 14L and heavy-chain residues 16 through 246 (chymotrypsinogen numbering) (Bode et al. 1992), the PPACK inhibitor, 253 water molecules, and a single magnesium ion. The structure is refined to an R-free value of 0.25. Representative electron density is shown in Supplemental Figure 1.
TABLE 1.
Data collection and refinement statisticsa
RNA structure
Figure 1 shows the secondary structure of the aptamer. As predicted from sequence analysis (White et al. 2001), the aptamer has a stem–loop secondary structure with an internal bulge. The bulge is flanked by two short RNA duplex regions of three base pairs each. The crystal structure contains an additional base pair that was not predicted from sequence analysis, which is formed between C11 and G16 (Fig. 1A, dashed line). Of the 25 bases, 14 are in base pairs, and 11 are single-stranded. Extensive π-π stacking exists throughout the RNA molecule. In addition to the base-paired nucleotides, seven out of the 11 single-stranded bases participate in stacking interactions, either with other RNA bases or with arginine residues of the protein (see below). The stacking interactions are located at the core of the folded RNA, while unstacked bases are flipped-out, away from the core.
FIGURE 1.
Secondary structure of the aptamer and detailed interactions with the protein. (A) Secondary structure of the aptamer. Base pairs are indicated by filled circles or a dashed line. Mutations and their effect on affinity are indicated in gray (White et al. 2001). (B) Interactions with the protein. Interactions are colored according to type: red, ion pair; green, hydrogen bond; brown, van der Waals contact; blue, stacking interaction between protein and RNA. In the RNA diagram, rectangles represent bases, pentagons represent sugars, open circles represent the backbone phosphate position, and closed circles represent the 2′ position of the sugar ring.
A single divalent metal ion with no protein ligands coordinates nonbridging oxygen atoms of phosphates A8 and A9 of the RNA (Figs. 1B, 2A) and four water molecules. The metal ion was identified as magnesium on the basis of an average metal-to-oxygen distance of 2.13 Å, octahedral geometry, and a coordination number of 6 (Katz et al. 1996). This site was probably occupied by magnesium in the crystal because magnesium was present at 200 mM concentration during crystallization. Presumably this site could also be occupied by calcium because this ion was present during aptamer selection (White et al. 2001).
FIGURE 2.
Overall structure of the aptamer–thrombin complex. (A) Human thrombin (gray) complexed with the RNA aptamer (green), in stereo. Base-paired nucleotides are shown as yellow sticks, while single-stranded nucleotides are blue. Two protein arginine residues (brown) stack with single-stranded adenine bases. The protease inhibitor PPACK (dark red) is shown in the catalytic active site of thrombin. (B) The A-Arg zipper, shown as a stereo representation. Two protein arginines (yellow) intercalate with adenine RNA bases (green).
The aptamer contains 2′fluoro substitutions on uracil and cytosine nucleotides. In addition to conferring resistance to ribonuculeases that use the 2′OH group for cleavage of the adjacent phosphodiester bond (Pieken et al. 1991; Cummins et al. 1995), the 2′fluoro substitution dramatically increases the thermal stability of model duplexes because of an increase in the propensity of the sugar to adopt the C3′-endo conformation (Guschlbauer and Jankowski 1980; Cummins et al. 1995). In the structure, the sugars of all of the 2′fluoro nucleotides adopt the C3′-endo conformation, consistent with this propensity. The increased stability of the C3′-endo conformation for 2′fluoro nucleotides would likely serve to stabilize the tertiary fold of the aptamer because it reduces the flexibility of the phosphate backbone (Richardson et al. 2008). Of the purine nucleotides, which do not have a substitution at the 2′OH position, 10 adopt the C3′-endo conformation, and five adopt the C2′-endo conformation (A7, G13, A14, G16, A18). For these five, the phosphate backbone conformation prohibits C3′-endo sugar pucker (Richardson et al. 2008). This suggests that if these nucleotides were 2′fluoro modified, the resulting aptamer would have decreased binding affinity for thrombin because of destabilization of the RNA fold.
Interaction with the protein
The aptamer binds to the basic region on thrombin called exosite-2 (Figs. 2, 3A). This interaction buries a total of 1193 Å2 of solvent-accessible surface area. Most of the aptamer–protein contacts involve single-stranded nucleotides rather than base-paired ones. Figure 1B indicates all of the protein–RNA contacts. Interactions are colored according to type: red, ion pair; green, hydrogen bond; brown, van der Waals contact; blue, stacking interaction between protein and RNA. Five arginine and three lysine residues interact with the aptamer (Fig. 1B). Two of these residues (Arg 101 and Lys 236) form ion pairs with the phosphate backbone of the RNA. The others participate in hydrogen bond, van der Waals, or stacking interactions with the bases of the single-stranded regions (Fig. 1B). There is only one direct contact between a 2′fluoro group and the protein, which involves U17 and Arg 126 (Fig. 1B).
FIGURE 3.
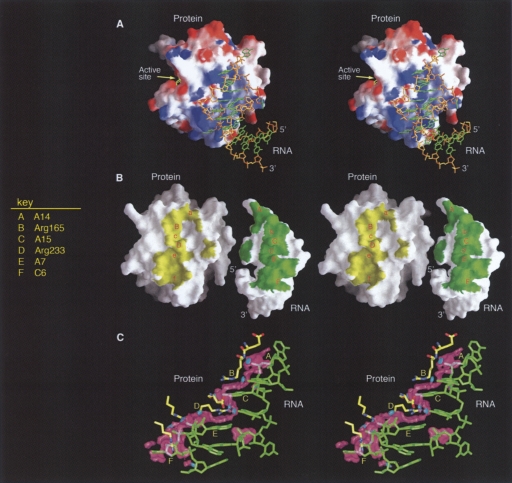
Surface and electrostatic complementarity at the protein–RNA interface. (A) Overall structure of the complex is shown with the molecular surface of thrombin colored according to surface electrostatic potential (red, negative; blue, positive) and the RNA aptamer as green and yellow sticks. (B) Complementary surfaces of protein and RNA. Here, the RNA molecule has been detached from the protein and rotated 180° to reveal the molecular surfaces buried in the complex (colored yellow and green). A series of protrusions and corresponding depressions are labeled with the letters A–F, with uppercase and lowercase type indicating protrusions and depressions, respectively. The primary residue responsible for each protrusion is indicated in the key. Buried surfaces were calculated using the SC algorithm (Lawrence and Colman 1993) with water molecules excluded and illustrated in GRASP (Nicholls 1992). (C) Side view of the interacting surfaces. The complex has been rotated 90° to the right from the orientation in A. The surface illustrated here has been drawn at positions where the molecular surface of the RNA (blue sticks) is <0.2 Å from the molecular surface of the protein (yellow sticks). The surface is colored magenta for van der Waals contacts and cyan for hydrogen bonds. The lettering scheme is the same as in B.
Five unpaired bases, all of which are adenine (A4, A5, A7, A18, A15), two arginine protein residues (Arg 165 and Arg 233), and the three base-paired nucleotides at the 5′ end of the RNA form a stack of π-π interactions (Fig. 1B). This stack is at the core of the RNA tertiary structure and extends ∼30 Å, almost the entire length of the aptamer (Fig. 2). The π-π interactions are nearly perfectly aligned along the entire length of the stack. The adenine base A15 is the only one that is displaced slightly (∼1 Å to the right in Fig. 2B) from a perfectly continuous stack. Both “halves” of the linear sequence of the aptamer contribute to this stack: A4, A5, and A7 from the first “half” and A15 and A18 from the second “half.” A cytosine base at position 6 is flipped out to allow the interdigitation of A18 into the stack. Near the end of the stack furthest away from the 5′ end of the RNA, Arg 165 and Arg 233 stack with adenine bases A15 and A7 in an alternating fashion (Fig. 2B). The stacking of an individual arginine residue with an RNA base is a common feature of protein–RNA interactions (Morozova et al. 2006). To our knowledge, however, this is the first example of an alternating arrangement of arginine residues and adenine bases in the same stack. We refer to this interaction as the A-Arg zipper.
Figure 3B shows the interacting molecular surfaces of thrombin and the aptamer. In this depiction, the aptamer has been detached from the protein to reveal the interacting surfaces: the surface of thrombin that contacts the aptamer is colored yellow, and the surface of the aptamer that contacts the protein is colored green. At the interface between the two molecules, there are six prominent complementary undulations. In Figure 3B, the protrusions are labeled with the uppercase letters “A” through “F,” and the complementary depressions are labeled with lowercase letters. The A-Arg zipper accounts for most of the protein–RNA interaction (protrusions labeled “B” through “E”). Two flipped-out nucleotides (C6 and A14) extend the interacting surfaces (Fig. 3B, labeled F and A, respectively). Another flipped-out nucleotide (U17) makes a second, smaller area of contact between the surfaces (Fig. 3B, isolated patches). Figure 3C shows a representation of the interacting surfaces that is colored according to the type of interaction. Magenta surfaces represent van der Waals contacts between protein residues (yellow sticks) and the RNA (green sticks), while hydrogen bonds are depicted by cyan disks.
The degree of shape complementarity (SC) for two interacting surfaces can be quantified with the SC statistic (Lawrence and Colman 1993). This statistic is independent of the buried surface area and has a maximum value of 1.0 for two perfectly complementary surfaces. The SC value for the thrombin–aptamer complex is 0.75. The SC values of a few naturally occurring protein–RNA complexes are presented as supplementary information and range from 0.60 to 0.81. The thrombin–aptamer complex has better shape complementarity than most antibody/antigen interactions (typical SC values of 0.64–0.68) (Lawrence and Colman 1993; Epa and Colman 2001). Its shape complementarity is as good as for subunit–subunit interfaces within oligomeric proteins, which are among the most highly complementary macromolecular surfaces in nature (typical SC values of 0.70–0.76).
Structure–activity relationship
Figure 1A shows the effect that nucleotide substitutions within the aptamer have on its binding affinity for thrombin (White et al. 2001). Consistent with stabilization of the fold by the base-paired regions, elimination of base pair interactions results a dramatic loss in affinity. Substitution of U17 with adenine has essentially no effect on affinity. U17, one of the flipped-out nucleotides, only contacts the protein through a hydrogen bond between its sugar and Arg 126 that would presumably be formed with a substituted base (Fig. 1B). Substitution of U12 with adenine results in a nearly three-orders-of-magnitude reduction in binding affinity. In the structure, the imino nitrogen of U12 forms a single hydrogen bond with the N3 nitrogen of A15 in a non-Watson–Crick base pair. This hydrogen bond stabilizes the orientation of A15, which is one of the bases involved in the A-Arg zipper.
The nucleotide sequences resulting from the RNA aptamer selection against thrombin contained a conserved motif (White et al. 2001). This sequence conservation was used to make the trimmed-down Toggle-25t aptamer. In the motif, the adenine residues that contribute to the A-Arg zipper are absolutely conserved. This conservation suggests that π-π stacking with arginine residues is an important feature for thrombin binding and that adenine bases are particularly well suited for this interaction. As the A-Arg zipper accounts for most of the interacting surface area, the importance of this interaction is consistent with the structure. Why adenine bases are preferred in the motif is not readily apparent from the structure because adenine, uracil and cytosine bases generally stack with arginine residues with approximately the same frequency in RNA–protein structures (Morozova et al. 2006). Analysis of the selected RNA sequences also showed that for the three nucleotides that are flipped-out in the structure, C6 is conserved in the motif, while A14 and U17 are not. In the structure, C6 makes extensive van der Waals contact with the protein, while A14 and U17 have minimal interactions with the protein (Fig. 1B).
Amino acid substitutions of thrombin and their effect on Toggle-25t interaction have been studied by measuring the extent of inhibition of thrombin by antithrombin in the presence and absence of the aptamer (Jeter et al. 2004). Mutation of either Arg 101 or Arg 233 eliminates the effect of the aptamer, while mutation of Gln 239 has no effect. (These amino acids are numbered Arg 98, Arg 245, and Gln 251, respectively, in Jeter et al. 2004.) These results are consistent with interactions observed in the structure. In the structure, Arg 233 is part of the A-Arg zipper, Arg 101 forms a bidentate ion pair with the RNA phosphate backbone of A8, and Gln 251 has no direct contacts with the aptamer (Fig. 1B).
The aptamer recognizes the native surface of thrombin
We sought to assess the extent of conformational changes in the protein upon aptamer binding by comparing the aptamer complex with other crystal structures of thrombin. Thrombin is an excellent protein for this analysis because there are >100 crystal structures of human thrombin present in the PDB database. Because many of these represent similar crystal packing arrangements, we selected the highest resolution crystal structure from each of 16 different crystal forms for the comparison. The structures were also chosen based on the criteria that they must not have a ligand bound at exosite-2 and must be determined to 2.4 Å resolution or better. From this ensemble of structures, a mean structure was calculated. The average root mean square (RMS) deviation of the ensemble to the mean structure is 0.4 Å for backbone atoms. The RMS deviation of thrombin in the aptamer complex from the mean structure is 0.2 Å. This extent of deviation is within the limit of precision with which the crystallographic data define the atomic positions at 1.9 Å resolution. At the level of the backbone, therefore, thrombin is not altered from its native conformation upon RNA binding.
We also examined the side-chain residues at the aptamer–thrombin interface. Because none of the structures within the ensemble has a ligand bound at exosite-2, the structures represent the unliganded states of these residues and give an estimate of the conformational space available to each side chain. Some residues, particularly lysine and arginine, adopt a wide range of conformations, while others have very discrete conformations (Supplemental Fig. 2). Most of the residues that interact with the RNA adopt conformations in the aptamer complex that are also observed in the ensemble of structures. The only exceptions are Asp 178 and Arg 233, which adopt side-chain rotamer conformations (Lovell et al. 2000) that do not occur within the ensemble (Supplemental Fig. 2). These conformations, which are shifted by 1–2 Å from the unliganded states, are necessary to accommodate the A-Arg zipper. The conformation of Arg 165, which is the other arginine that is part of the A-Arg zipper, does not change upon aptamer binding. Arg 165 is stabilized by hydrogen bonds to the backbone carbonyl oxygens of Thr 177 and Met 180. Therefore, aside from the minor rotamer changes of Asp 178 and Arg 233, the aptamer recognizes the same conformation of thrombin that is observed in the absence of a ligand at exosite-2. This suggests that the aptamer achieves high affinity, in part, by minimizing conformational changes in the protein from its native state. An open question is how much the RNA must rearrange to bind the protein. This cannot be answered definitively without structural information for the aptamer in the absence of thrombin.
Comparisons with other complexes
The oligosaccharide heparin binds thrombin through a nonspecific electrostatic association (Olson et al. 1991). Crystal structures of thrombin with heparin (Carter et al. 2005) or heparin mimetics (Dementiev et al. 2004; Li et al. 2004) show that negatively charged sulfate and carboxylate moieties on heparin interact with arginine and lysine residues of exosite-2. Consistent with a nonspecific electrostatic association, the conformation of the oligosaccharides varies among the cocrystal structures. While the aptamer also contacts all of the residues that interact with heparin in the structures, the nature of the interaction with the aptamer is quite different. The extensive van der Waals contacts, the high degree of shape complementarity, and the π-π stacking interactions with arginine residues are features not present in the binding of heparin to thrombin. Lack of these features may partially account for the difference in binding affinity: heparin binds to thrombin with a K d of ∼6 μM (Olson et al. 1991), while the aptamer binds with a K d of 0.5 nM (White et al. 2001). Comparisons of the aptamer and heparin complexes with thrombin are shown in Supplemental Figure 3.
A DNA aptamer has been selected that also binds to human thrombin (Bock et al. 1992). The crystal structure of this aptamer in complex with thrombin revealed that it binds to thrombin at exosite-1, which is on the opposite side of thrombin from exosite-2 (Fig. 2A; Padmanabhan et al. 1993; Padmanabhan and Tulinsky 1996). Although there is ambiguity in the polarity of the DNA in the crystal structure that makes a detailed analysis difficult (Padmanabhan and Tulinsky 1996), protein recognition involves both hydrogen bonds and van der Waals contacts. Using the crystal structure (PDB ID 1HAO) that has the same DNA polarity as was determined for the free DNA aptamer by NMR (Macaya et al. 1993; Wang et al. 1993; Schultze et al. 1994), 677 Å2 of solvent-accessible surface area is buried by the binding of the DNA aptamer to thrombin. This area is approximately half of the amount buried by the RNA aptamer (1193 Å2). The SC value of the DNA aptamer for thrombin is 0.64 (using PDB ID 1HAO), compared with 0.75 for the RNA aptamer.
In addition to mediating binding to heparin, crystal structure analyses from two different groups demonstrated that exosite-2 on thrombin also interacts with the platelet surface glycoprotein Ib-alpha (GPIbα) (Celikel et al. 2003; Dumas et al. 2003). Consistent with this observation and our crystal structure, we previously showed that the Toggle-25t RNA aptamer is a potent inhibitor of thrombin-mediated activation of platelets (White et al. 2001). Based upon their structural studies, Celikel and colleagues (2003) hypothesized that the binding of GPIba to exosite-2 is important for exocite-1 modulation of fibrinogen binding and thrombin-mediated clotting. In an accompanying manuscript by Nimjee et al. (2008) we provide data that supports this hypothesis. We demonstrate that the DNA aptamer and the RNA aptamer Toggle-25t, which bind to exosite-1 and exosite-2, respectively, synergistically inhibit coagulation.
Vascular endothelial growth factor (VEGF) is an important regulator of both normal and pathological angiogenesis (Ferrara 2004). The binding of heparin to certain isoforms of VEGF (such as VEGF165) that have a C-terminal heparin-binding domain modulates angiogenic activity (Ferrara 2004). An RNA aptamer-based therapeutic, Macugen (also known as pegaptanib sodium), which targets VEGF165, is an FDA-approved treatment for the wet form of age-related macular degeneration (Ruckman et al. 1998; Bell et al. 1999; Ng and Adamis 2005). Although the three-dimensional structure of this aptamer is not known, similarities exist between it and the Toggle-25t aptamer and its interaction with thrombin. The aptamers are about the same length and have the same general secondary structure: a stem–loop with and internal bulge that is flanked by two short duplex regions (Bell et al. 1999; Lee et al. 2005). Single-stranded adenine bases of Toggle-25t play an important role in thrombin binding. Among the 15 bases that are predicted to be single stranded in Macugen, seven are adenine. By using NMR to measure backbone amide chemical shifts in VEGF upon aptamer binding, it has been shown that, like the thrombin aptamer, Macugen targets the heparin binding site (Stauffer et al. 2002; Lee et al. 2005). Three arginine residues (Arg 124, Arg 145, and Arg 156) are among those that undergo significant backbone chemical shifts upon aptamer binding to VEGF, suggesting that, as is the case for the thrombin–RNA interaction, arginine residues play an important role in aptamer binding to VEGF. These similarities suggest that recognition of the VEGF heparin binding site by Macugen may involve features present in the thrombin–aptamer interaction such as extensive stacking of single-stranded bases at the core of the fold, π-π interactions of arginine residues with bases, and possibly an A-Arg zipper.
In summary, an RNA aptamer can fold in such a way as to present a surface that is extremely complementary to the unliganded state of the target protein. The molecular contortions and conformational space available permit the aptamer to mate with the native surface of its protein target, indeed rivaling the complementarity observed within the most specific naturally occurring macromolecular complexes. The intricacies of the RNA/protein interface presented here help explain how RNA can be selected to bind protein targets with such high affinity and specificity.
MATERIALS AND METHODS
Crystallization
We used PPACK (D-Phe-Pro-Arg chloromethylketone)-inhibited human thrombin to limit proteolysis during crystallization (Haemtech). The Toggle-25t RNA aptamer (White et al. 2001) was chemically synthesized by Dharmacon Research. In it, all pyrimidine nucleotides contain 2′-fluoro groups. The protein:RNA complex was formed prior to crystallization by incubating protein at 3 mg/mL with the aptamer in 150 mM NaCl, 2 mM CaCl2, 20 mM HEPES pH 7.3 for 10 min at 37°C and then on ice. The molar ratio for protein:RNA used was 1:1.5. Crystals were grown at 17°C from hanging drops containing 2 μL of protein:RNA complex mixed with 2 μL of reservoir solution (14% [w/v] PEG-8000, 200 mM magnesium acetate, 100 mM sodium cacodylate pH 6.5) and equilibrated against the reservoir solution using vapor diffusion. Crystals appeared within 2 wk and grew to 100 × 70 × 150 150 μm3 in size. The crystals were transferred stepwise into a cryoprotectant solution containing the buffer components of an equilibrated hanging drop and 34% (w/v) PEG-8000, 15% (w/v) PEG-400. They were then flash cooled by plunging into liquid nitrogen.
Structure determination
Diffraction data were measured at 98 K with an R-axis IV image plate system mounted on a Rigaku RU-H3R generator (Molecular Structure Corporation). Data reduction and scaling were done using the DENZO and SCALEPACK programs (Otwinowski and Minor 1997). Initial phases were determined by molecular replacement using coordinates for human thrombin (PDB ID number 1PPB) as a search model in CNS (Brunger et al. 1998). The crystals belong to space group P21212 and contain one complex per asymmetric unit. The starting R factor was 45% for the resolution range 15–4 Å. At this point, difference electron density adjacent to the thrombin molecule clearly indicated the location of the RNA phosphate backbone. The model was built using the molecular visualization program O (Jones et al. 1991) and refined in CNS (Brunger et al. 1998) using a bulk solvent correction. Table 1 contains a summary of diffraction data and refinement statistics; electron density is shown in Supplemental Figure 1.
Structure comparison and SC analysis
Crystal structures of thrombin (protein data bank IDs: 1ABJ, 1AD8, 1B7X, 1BTH, 1DE7, 1DOJ, 1DX5, 1H8D, 1JMO, 1JOU, 1NY2, 1OOK, 1PPB, 1THP, 1TMT, and 4HTC; www.pdb.org) were superimposed to the mean calculated structure using X-PLOR (Brünger 1992). The regions of thrombin included in the superposition were those present in all 16 coordinate files (light-chain residues 1B through 14J, and heavy-chain residues 16–146 and 150–242; chymotrypsinogen numbering) (Bode et al. 1992). The aptamer complex was aligned to the mean structure and compared with the ensemble using X-PLOR (Brünger 1992) and the visualization program O (Jones et al. 1991). The SC statistic was calculated using the CCP4 suite of programs (Lawrence and Colman 1993; Collaborative Computational Project N 1994). Atomic radii for nucleic acid atoms were set according to values for chemically similar protein atoms. For the SC calculations, all parameters were set to their default values. Buried surface areas were calculated using the program GRASP (Nicholls 1992). Figures were made using Ribbons (Carson 1997), GRASP (Nicholls 1992), probe (Word et al. 1999), and pymol (www.pymol.org).
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank the Duke University Structural Biology Resource for the use of their facility, George Pitoc for affinity binding measurements, Frank Church, Don Sullenger, and Daniel Gewirth for useful discussions, and Dinshaw Patel and Nikola Pavletich for comments on the manuscript. This work was funded by a grant from the NIH (B.A.S.). The coordinates and structure factors have been deposited in the protein data bank (www.pdb.org, ID 3DD2).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1239308.
REFERENCES
- Bell C., Lynam E., Landfair D.J., Janjic N., Wiles M.E. Oligonucleotide NX1838 inhibits VEGF165-mediated cellular responses in vitro. In Vitro Cell. Dev. Biol. Anim. 1999;35:533–542. doi: 10.1007/s11626-999-0064-y. [DOI] [PubMed] [Google Scholar]
- Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- Bode W., Turk D., Karshikov A. The refined 1.9-Å X-ray crystal structure of D-Phe-Pro-Arg chloromethylketone-inhibited human α-thrombin: Structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure–function relationships. Protein Sci. 1992;1:426–471. doi: 10.1002/pro.5560010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. X-PLOR version 3.1: A system for X-ray crystallography and NMR. Yale University Press; New Haven, CT: 1992. [Google Scholar]
- Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Carson M. Ribbons. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- Carter W.J., Cama E., Huntington J.A. Crystal structure of thrombin bound to heparin. J. Biol. Chem. 2005;280:2745–2749. doi: 10.1074/jbc.M411606200. [DOI] [PubMed] [Google Scholar]
- Celikel R., McClintock R.A., Roberts J.R., Mendolicchio G.L., Ware J., Varughese K.I., Ruggeri Z.M. Modulation of α-thrombin function by distinct interactions with platelet glycoprotein Ibalpha. Science. 2003;301:218–221. doi: 10.1126/science.1084183. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project N. The CCP4 suite: Programs for X-ray crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Convery M.A., Rowsell S., Stonehouse N.J., Ellington A.D., Hirao I., Murray J.B., Peabody D.S., Phillips S.E., Stockley P.G. Crystal structure of an RNA aptamer–protein complex at 2.8 Å resolution. Nat. Struct. Biol. 1998;5:133–139. doi: 10.1038/nsb0298-133. [DOI] [PubMed] [Google Scholar]
- Cox J.C., Hayhurst A., Hesselberth J., Bayer T.S., Georgiou G., Ellington A.D. Automated selection of aptamers against protein targets translated in vitro: From gene to aptamer. Nucleic Acids Res. 2002;30:e108. doi: 10.1093/nar/gnf107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins L.L., Owens S.R., Risen L.M., Lesnik E.A., Freier S.M., McGee D., Guinosso C.J., Cook P.D. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995;23:2019–2024. doi: 10.1093/nar/23.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dementiev A., Petitou M., Herbert J.M., Gettins P.G. The ternary complex of antithrombin–anhydrothrombin–heparin reveals the basis of inhibitor specificity. Nat. Struct. Mol. Biol. 2004;11:863–867. doi: 10.1038/nsmb810. [DOI] [PubMed] [Google Scholar]
- Di Cera E. Thrombin interactions. Chest. 2003;124:11S–17S. doi: 10.1378/chest.124.3_suppl.11s. [DOI] [PubMed] [Google Scholar]
- Dumas J.J., Kumar R., Seehra J., Somers W.S., Mosyak L. Crystal structure of the GpIbα–thrombin complex essential for platelet aggregation. Science. 2003;301:222–226. doi: 10.1126/science.1083917. [DOI] [PubMed] [Google Scholar]
- Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Epa V.C., Colman P.M. Shape and electrostatic complementarity at viral antigen–antibody complexes. Curr. Top. Microbiol. Immunol. 2001;260:45–53. doi: 10.1007/978-3-662-05783-4_3. [DOI] [PubMed] [Google Scholar]
- Ferrara N. Vascular endothelial growth factor: Basic science and clinical progress. Endocr. Rev. 2004;25:581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- Ghosh G., Huang D.B., Huxford T. Molecular mimicry of the NF-κB DNA target site by a selected RNA aptamer. Curr. Opin. Struct. Biol. 2004;14:21–27. doi: 10.1016/j.sbi.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Green L.S., Jellinek D., Jenison R., Ostman A., Heldin C.H., Janjic N. Inhibitory DNA ligands to platelet-derived growth factor B-chain. Biochemistry. 1996;35:14413–14424. doi: 10.1021/bi961544+. [DOI] [PubMed] [Google Scholar]
- Guschlbauer W., Jankowski K. Nucleoside conformation is determined by the electronegativity of the sugar substituent. Nucleic Acids Res. 1980;8:1421–1433. doi: 10.1093/nar/8.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann T., Patel D.J. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- Hoffman M., Monroe D.M., 3rd A cell-based model of hemostasis. Thromb. Haemost. 2001;85:958–965. [PubMed] [Google Scholar]
- Huang D.B., Vu D., Cassiday L.A., Zimmerman J.M., Maher L.J., 3rd, Ghosh G. Crystal structure of NF-κB (p50)2 complexed to a high-affinity RNA aptamer. Proc. Natl. Acad. Sci. 2003;100:9268–9273. doi: 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki J., Nevins J.R., Sullenger B.A. Inhibition of cell proliferation by an RNA ligand that selectively blocks E2F function. Nat. Med. 1996;2:1386–1389. doi: 10.1038/nm1296-1386. [DOI] [PubMed] [Google Scholar]
- Jaeger J., Restle T., Steitz T.A. The structure of HIV-1 reverse transcriptase complexed with an RNA pseudoknot inhibitor. EMBO J. 1998;17:4535–4542. doi: 10.1093/emboj/17.15.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeter M.L., Ly L.V., Fortenberry Y.M., Whinna H.C., White R.R., Rusconi C.P., Sullenger B.A., Church F.C. RNA aptamer to thrombin binds anion-binding exosite-2 and alters protease inhibition by heparin-binding serpins. FEBS Lett. 2004;568:10–14. doi: 10.1016/j.febslet.2004.04.087. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Katz A.K., Glusker J.P., Beebe S.A., Bock C.W. Calcium ion coordination: A comparison with that of beryllium, magnesium, and zinc. J. Am. Chem. Soc. 1996;118:5752–5763. [Google Scholar]
- Kubik M.F., Stephens A.W., Schneider D., Marlar R.A., Tasset D. High-affinity RNA ligands to human α-thrombin. Nucleic Acids Res. 1994;22:2619–2626. doi: 10.1093/nar/22.13.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P.K., Machida K., Urvil P.T., Kakiuchi N., Vishnuvardhan D., Shimotohno K., Taira K., Nishikawa S. Isolation of RNA aptamers specific to the NS3 protein of hepatitis C virus from a pool of completely random RNA. Virology. 1997;237:270–282. doi: 10.1006/viro.1997.8773. [DOI] [PubMed] [Google Scholar]
- Lawrence M.C., Colman P.M. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Lebruska L.L., Maher L.J., 3rd Selection and characterization of an RNA decoy for transcription factor NF-κ B. Biochemistry. 1999;38:3168–3174. doi: 10.1021/bi982515x. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Canny M.D., De Erkenez A., Krilleke D., Ng Y.S., Shima D.T., Pardi A., Jucker F. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. Proc. Natl. Acad. Sci. 2005;102:18902–18907. doi: 10.1073/pnas.0509069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Johnson D.J., Esmon C.T., Huntington J.A. Structure of the antithrombin–thrombin–heparin ternary complex reveals the antithrombotic mechanism of heparin. Nat. Struct. Mol. Biol. 2004;11:857–862. doi: 10.1038/nsmb811. [DOI] [PubMed] [Google Scholar]
- Lovell S.C., Word J.M., Richardson J.S., Richardson D.C. The penultimate rotamer library. Proteins. 2000;40:389–408. [PubMed] [Google Scholar]
- Macaya R.F., Schultze P., Smith F.W., Roe J.A., Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova N., Allers J., Myers J., Shamoo Y. Protein–RNA interactions: Exploring binding patterns with a three-dimensional superposition analysis of high-resolution structures. Bioinformatics. 2006;22:2746–2752. doi: 10.1093/bioinformatics/btl470. [DOI] [PubMed] [Google Scholar]
- Ng E.W., Adamis A.P. Targeting angiogenesis, the underlying disorder in neovascular age-related macular degeneration. Can. J. Ophthalmol. 2005;40:352–368. doi: 10.1016/S0008-4182(05)80078-X. [DOI] [PubMed] [Google Scholar]
- Nicholls A. GRASP: Graphical representation and analysis of surface properties. Columbia University Press; New York, NY: 1992. [Google Scholar]
- Nimjee S.M., Rusconi C.P., Sullenger B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- Nimjee S.M., Oney S., Volovyk Z., Long S.B., Hoffman M., Sullenger B. Synergistic effect of oligonucleotide ligands that inhibit exosites 1 and 2 on thrombin. RNA. 2008 doi: 10.1261/rna.1240109. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson S.T., Halvorson H.R., Bjork I. Quantitative characterization of the thrombin–heparin interaction. Discrimination between specific and nonspecific binding models. J. Biol. Chem. 1991;266:6342–6352. [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276A:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Padmanabhan K., Tulinsky A. An ambiguous structure of a DNA 15-mer thrombin complex. Acta Crystallogr. D Biol. Crystallogr. 1996;52:272–282. doi: 10.1107/S0907444995013977. [DOI] [PubMed] [Google Scholar]
- Padmanabhan K., Padmanabhan K.P., Ferrara J.D., Sadler J.E., Tulinsky A. The structure of α-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- Pieken W.A., Olsen D.B., Benseler F., Aurup H., Eckstein F. Kinetic characterization of ribonuclease-resistant 2′-modified hammerhead ribozymes. Science. 1991;253:314–317. doi: 10.1126/science.1857967. [DOI] [PubMed] [Google Scholar]
- Rau J.C., Beaulieu L.M., Huntington J.A., Church F.C. Serpins in thrombosis, hemostasis and fibrinolysis. J. Thromb. Haemost. 2007;5(Suppl. 1):102–115. doi: 10.1111/j.1538-7836.2007.02516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J.S., Schneider B., Murray L.W., Kapral G.J., Immormino R.M., Headd J.J., Richardson D.C., Ham D., Hershkovits E., Williams L.D., et al. RNA backbone: Consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution) RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckman J., Green L.S., Beeson J., Waugh S., Gillette W.L., Henninger D.D., Claesson-Welsh L., Janjic N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998;273:20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- Rusconi C.P., Scardino E., Layzer J., Pitoc G.A., Ortel T.L., Monroe D., Sullenger B.A. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. 2002;419:90–94. doi: 10.1038/nature00963. [DOI] [PubMed] [Google Scholar]
- Schultze P., Macaya R.F., Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- Stauffer M.E., Skelton N.J., Fairbrothe W.J. Refinement of the solution structure of the heparin-binding domain of vascular endothelial growth factor using residual dipolar couplings. J. Biol. NMR. 2002;23:57–61. doi: 10.1023/a:1015346504499. [DOI] [PubMed] [Google Scholar]
- Stubbs M.T., Bode W. The clot thickens: Clues provided by thrombin structure. Trends Biochem. Sci. 1995;20:23–28. doi: 10.1016/s0968-0004(00)88945-8. [DOI] [PubMed] [Google Scholar]
- Sullenger B.A., Gallardo H.F., Ungers G.E., Gilboa E. Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell. 1990;63:601–608. doi: 10.1016/0092-8674(90)90455-n. [DOI] [PubMed] [Google Scholar]
- Sussman D., Nix J.C., Wilson C. The structural basis for molecular recognition by the vitamin B-12 RNA aptamer. Nat. Struct. Biol. 2000;7:53–57. doi: 10.1038/71253. [DOI] [PubMed] [Google Scholar]
- Tereshko V., Skripkin E., Patel D.J. Encapsulating streptomycin within a small 40-mer RNA. Chem. Biol. 2003;10:175–187. doi: 10.1016/s1074-5521(03)00024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Wang K.Y., McCurdy S., Shea R.G., Swaminathan S., Bolton P.H. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry. 1993;32:1899–1904. doi: 10.1021/bi00059a003. [DOI] [PubMed] [Google Scholar]
- White R., Rusconi C., Scardino E., Wolberg A., Lawson J., Hoffman M., Sullenger B. Generation of species cross-reactive aptamers using “toggle” SELEX. Mol. Ther. 2001;4:567–573. doi: 10.1006/mthe.2001.0495. [DOI] [PubMed] [Google Scholar]
- Word J.M., Lovell S.C., LaBean T.H., Taylor H.C., Zalis M.E., Presley B.K., Richardson J.S., Richardson D.C. Visualizing and quantifying molecular goodness-of-fit: Small-probe contact dots with explicit hydrogen atoms. J. Mol. Biol. 1999;285:1711–1733. doi: 10.1006/jmbi.1998.2400. [DOI] [PubMed] [Google Scholar]