Abstract
Mycoreovirus 1 (MyRV1), a member of the Reoviridae family possessing a genome consisting of 11 dsRNA segments (S1–S11), and the prototype hypovirus (CHV1-EP713) of the Hypoviridae family, which is closely related to the monopartite picorna-like superfamily with a ssRNA genome, infect the chestnut blight fungus and cause virulence attenuation and distinct phenotypic alterations in the host. Here, we present evidence for reproducible induction of intragenic rearrangements of MyRV1 S6 and S10, mediated by the multifunctional protein p29 encoded by CHV1. S6 and S10 underwent an almost full-length ORF duplication (S6L) and an internal deletion of three-fourths of the ORF (S10ss). No significant influence on symptom induction in the fungal host was associated with the S6L rearrangement. In contrast, S10-encoded VP10, while nonessential for MyRV1 replication, was shown to contribute to virulence reduction and reduced growth of aerial mycelia. Furthermore, p29 was found to copurify with MyRV1 genomic RNA and bind to VP9 in vitro and in vivo, suggesting direct interactions of p29 with the MyRV1 replication machinery. This study provides the first example of a viral factor involved in RNA genome rearrangements of a different virus and shows its usefulness as a probe into the mechanism of replication and symptom expression of a heterologous virus.
Keywords: RNA rearrangement, RNA silencing suppressor, mycoreovirus, hypovirus
INTRODUCTION
Recombination of RNA virus genomes is one of driving forces for their diversity and evolution (Lai 1992; Taniguchi and Urasawa 1995; Desselberger 1996; Nagy and Simon 1997). RNA recombination is a well established, widespread phenomenon in single-stranded RNA viruses, generating inter- and intramolecular recombinants (Lai 1992; Chetverin 1999; Nagy and Simon 1997). In members of the family Reoviridae with nine to 12 double-stranded (ds) RNA genome segments, intragenic rearrangement events commonly occur under natural and laboratory conditions. Replicase complexes are considered to switch their templates during RNA synthesis with the aid of secondary structures, direct repeats, and inverted repeats on the templates, resulting in considerable changes of sequences, frequently duplication, deletion, their combination, and sometimes alterations of open reading frames (ORFs) (Nuss 1984; Taniguchi and Urasawa 1995; Desselberger 1996; Murao et al. 1996).
Cryphonectria parasitica, the causal pathogen of chestnut blight disease, is the fungal host of a number of mycoviruses and provides a versatile system for studying virus/host and virus/virus interactions. Transfection protocols are available for two viruses, Chryphonectria hypovirus 1 (CHV1) and Mycoreovirus 1 (MyRV1) (Nuss 2005). The prototype hypovirus CHV1-EP713 reduces the levels of virulence, sporulation, and pigmentation of its filamentous fungal host, the chestnut blight fungus. CHV1 is evolutionarily related to the picorna-like superfamily that includes members of the plant-infecting Potyviridae and the animal-infecting Picornaviridae (Koonin et al. 1991). MyRV1-Cp9B21, the type species of the recently established genus Mycoreovirus in the family Reoviridae (Suzuki et al. 2004), carries a genome of 11 segments of dsRNA (S1–S11), ranging from 4127 to 732 base pairs (bp) in size and a total size of 23,433 bp. MyRV1 reduces the virulence of its host fungus, but it has little effect on asexual sporulation and pigmentation (Hillman et al. 2004). In mixed infection of C. parasitica by MyRV1 and CHV1, a one-way synergism is found in which replication enhancement of MyRV1 is mediated by the papain-like protease p29 of CHV1 (Sun et al. 2006).
The papain-like protease p29 (Choi et al. 1991) is a multifunctional protein, derived from the N-terminal portion of the polyprotein p69 encoded by the 5′-proximal ORF of CHV1. Functional domains mapped on p29 include the N-terminal 24 codons essential for virus viability, the adjacent region (amino acids 25–74) involved in symptom induction and in the elevation of replication and transmission of heterologous and homologous viruses, and the C-terminal half (amino acids 135–248) responsible for the cotranslational self-cleavage from precursor protein p69 (Hillman and Suzuki 2004; Sun et al. 2006). Furthermore, p29 is capable of suppressing RNA silencing in plant and fungal cells that is mediated by small-interfering RNAs, acting as a sequence-specific RNA degradation system (Segers et al. 2006). Some of the activities of p29 are shared with the multifunctional protein HC-Pro, which is encoded by members of the family Potyviridae within the picorna-like superfamily and was the first to be identified as an RNA silencing suppressor (Anandalakshmi et al. 1998; Kasschau and Carrington 1998). Importantly, C. parasitica was shown to utilize RNA silencing as one of the host defense responses against infecting mycoviruses (Segers et al. 2007). It is plausible that the enhancement in trans of MyRV1 replication by CHV1 is associated with the suppressive activities of RNA silencing of p29 (Sun et al. 2006).
During the study of mixed infections, we recently discovered that the MyRV1 genome segments underwent rearrangements. Here, we present evidence that the multifunctional protein p29 of CHV1 reproducibly and frequently induces, independently of virus replication, intragenic rearrangements of MyRV1 genome segments S6 and S10, creating a duplicated form of S6 (S6L) and an internally deleted form of S10 (S10ss). These inducible rearrangements reveal an unrecognized activity of the RNA silencing suppressor p29 of the hypovirus and provide insights into the functional roles of S6 and S10 in MyRV1 replication.
RESULTS
MyRV1 genome rearrangement occurred in fungal colonies co-infected by CHV1
While investigating a number of subcultured isolates and single conidial isolates from mixed infections by CHV1 and MyRV1 (Sun et al. 2006), we found that a minor portion of isolates had lost MyRV1 S10 of the authentic size (970 bp) and gained smaller fragments (S10ss1) of ∼400 bp instead (Fig. 1A, lane 2).
FIGURE 1.
Polyacrylamide gel electrophoresis profile of genomic RNA segments of MyRV1 and its variants. (A) SDS-PAGE analysis of rearranged genomic RNA segments. Double-stranded RNA was isolated from mycelia of EP155 or p29 transformants (Twtp29) infected singly or doubly with different MyRV1 variants and CHV1. Extracted RNA was separated on SDS-polyacrylamide gels and stained with ethidium bromide. The positions of authentic RNA segments and of altered segments S6 (S6L) and S10 (S10ss1 and S10ss2) are indicated. Under the condition of electrophoresis shown, MyRV1 segments S5 and S6 comigrated, as they have almost same length (2056 and 2023 bp, respectively). (Lanes 1,4) MyRV1, (lane 2) MyRV1+CHV1, (lane 3) CHV1, (lane 5) MyRV1/S6L, (lane 6) MyRV1/S6L+S10ss2, (lane 7) MyRV1/S10ss2. (B) Northern blot analysis of rearranged segments. Genomic dsRNA segments separated in A were blotted onto a nylon membrane as described in the Materials and Methods and hybridized with a mixture of DIG-UTP labeled probes specific for S6 and S10. Migration positions of S6, S10, S6L, and S10ss are denoted at both sides of the blots.
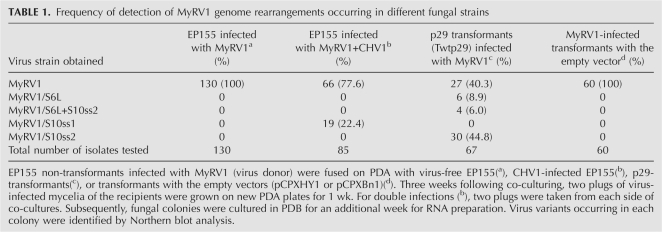
We first attempted to determine how frequently rearrangements occurred and prevailed in singly and doubly infected colonies. Protocols for purifying a mycovirus strain, like plaque purifications for animal or bacterial viruses and single-legion transfer for plant viruses, are unavailable. Therefore, the frequency of rearrangements was estimated as the ratio of virus-infected fungal colonies carrying rearranged segments to those with a normal dsRNA profile (Table 1, see footnote for experimental procedures). Of 85 colonies co-infected by CHV1 and MyRV1, 19 (22.4%) contained S10ss1 after culturing for 5 wk (Table 1, MyRV1+CHV1). Such an alteration was not observed in single infections of EP155 by MyRV1 when cultured in parallel (0/130) (Table 1). The genomic dsRNA prepared from single infections showed a normal electropherotype (Fig. 1A, lane 1).
TABLE 1.
Frequency of detection of MyRV1 genome rearrangements occurring in different fungal strains
Detection of two types of intragenic rearrangements of MyRV1 genome segments in p29 transformants
Based on previous observations that the multifunctional protein p29 encoded by CHV1 enhances the replication of MyRV1, irrespectively of whether it is expressed from the virus genome or host chromosomes (Sun et al. 2006), we hypothesized that p29 might also contribute to the MyRV1 rearrangements in the mixed infection. To test this possibility, fungal transformants with the coding domain of wild-type p29 (Twtp29) prepared previously (Suzuki et al. 2003; Sun et al. 2006) were fused as a recipient strain with MyRV1-infected EP155 (virus donor) by co-culturing for 3 wk (Table 1, see the footnote). A total of 67 cultures of recipients were examined, and three electropherotypes different from that of the wild-type MyRV1 were found (Fig. 1A, lanes 5–7; Table 1). A subpopulation of cultures contained a newly emerging large segment (S6L) (Fig. 1A, lane 5) that migrated between S1 and S2 in SDS-PAGE, and concomitantly the intensity of the comigrating S5–S6 bands had decreased. The second population lacked S10 at the expected position but carried smaller fragments (S10ss2) (Fig. 1A, lane 7). This is similar to the one found in the mixed infections. The last subpopulation contained both S6L and S10ss2 (Fig. 1A, lane 6). Although dsRNA preparations containing S6L, S6 and S10ss2, and S10ss2 may have consisted of mixed population with the wild-type and rearranged genomes, they were hereafter termed strains MyRV1/S6L, MyRV1/S6L+S10ss2, and MyRV1/S10ss2.
We tested whether this phenomenon specifically happened in p29-expressing transformant Twtp29, i.e., it was due to the action of p29, using different transformants. Twtp29 transformants harbored pCPXwtp29 with a hygromycin B phosphotransferase gene as the selectable marker. Using an independent transformant, strains with pCPXBn-wtp29 carrying a benomyl resistance gene were fused with EP155 infected with MyRV1 to transfer the virus. We observed that the MyRV1 genome alterations were also arising in these p29 transformants, whereas no rearrangement was seen in transformants with the empty vector carrying either drug resistance gene (Table 1, pCPXHY1 or pCPXBn1). These data strongly suggested that CHV1 p29 induced the genome rearrangements of MyRV1.
S6L and S10ss2 were derived from MyRV1 S6 and S10, respectively
To explore the origins of the segments S6L, S10ss1, and S10ss2, Northern analysis was performed using probes, each specific for one of the segments. As a result, the S6-specific probe hybridized to S6 and S6L, while the S10-specific probe allowed detection of S10, S10ss1, and S10ss2 (data not shown). Figure 1B shows a Northern profile obtained with a mixture of both probes specific for S6 and S10. Interestingly, S6L always occurred together with the authentic segment S6, although at varying relative levels. No intact S10 dsRNA was detectable in MyRV1/S10ss1, MyRV1/S10ss2, or MyRV1/S6L+S10ss2. From these results, the origins of S6L and S10ss were established as S6 and S10, respectively.
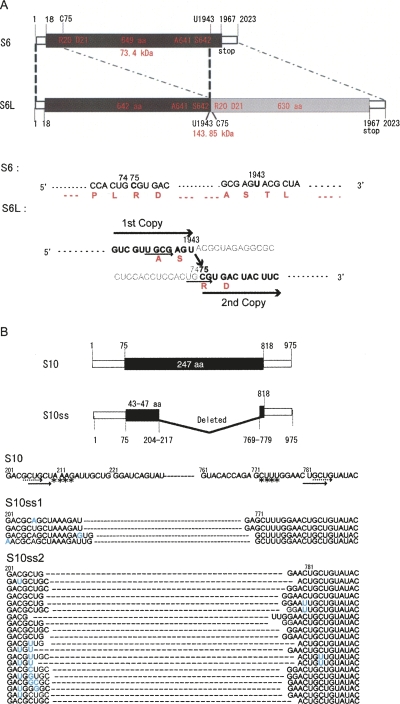
Assuming that the terminal conserved sequence domain is important for virus replication and possibly assembly (Anzola et al. 1987), primers complementary to the terminal sequences of MyRV1 S6 were used in RT-PCR to amplify cDNA of S6L. The primer set S6FL1 and S6FL2 (Supplemental Table S1) provided RT-PCR fragments of ∼4 kb. Sequencing of the 4-kb fragment showed that S6L was a head-to-tail, in-frame duplication form of the normal S6 ORF, although nucleotides 1944–2023 of the first segment and nucleotides 1–74 of the second segment, which corresponded to seven codons of the first ORF and the first 19 codons of the second ORF, were deleted (Fig. 2A). The resulting, extended ORF comprises 1273 codons that would encode a polypeptide of 144 kDa (Fig. 2A). The sequence of S6L was confirmed by sequencing other RT-PCR fragments obtained by different primer sets (Supplemental Table S1).
FIGURE 2.
Schematic representation of the organization of MyRV1 rearranged segments. (A) Diagram of the organization of MyRV1 RNA segment S6L. MyRV1 wild-type S6 is 2023 bp with a single ORF, which encodes 649 amino acids, starting at the AUG (map positions 18–20) and ending at 1965–1967 (Suzuki et al. 2004). Rearranged segment S6 (S6L) is 3892 bp with a duplication of almost the entire S6 segment. In S6L, two fragments spanning bases 1–1943 and bases 75–2023 are linked, resulting in an in-frame duplication of the ORF, which would code for a protein of 1272 amino acids. (Bottom panel) S6L sequence of the junction of two fragments in parallel with S6. (Black) Nucleotide numbers, (red) amino acid numbers. The light-colored nucleotide sequence is considered to be unused as a template during the rearrangement event. (Solid and dashed arrows) Direct repeats, (asterisks) inverted repeat in A and B. (B) Schematic representation of the organization of MyRV1-S10ss segments. Wild-type S10 comprises 975 bp possessing a single ORF extending from the AUG at nucleotides 75–77 to 818 and encoding 247 amino acids. S10ss RNAs arise from an internal deletion event that retains the 5′ terminal (nucleotide positions 1 to 204–217) and 3′ terminal portions (nucleotide positions 769–779 to 975). Sequences of 30 RT-PCR clones each from S10ss1 and S10ss2, grouped into four for S10ss1 and 20 for S10ss2, are shown in the diagram. (Blue) Nucleotide changes from S10 found in the clones. Numbers in the top and bottom panels refer to nucleotide positions.
RT-PCR using the primer set S10FL1 and S10FL2 (Supyani et al. 2007) led to amplification of fragments of ∼400 nucleotides (nt) in size, as expected from their migration position in SDS-PAGE (Fig. 1A). Sequence analysis of the fragments showed that both S10ss1 and S10ss2 contained internal deletions of different sizes at similar positions (Fig. 2B). The deletions extend from bases 216–218 to bases 768–770 for S10ss1, and bases 205–209 to bases 773–778 for S10ss2. The S10ss ORFs would comprise 51–61 codons (Fig. 2). Mismatches were found in many RT-PCR clones at or close to the deletion junctions (Fig. 2B), as described by Lai (1992) for aberrant homologous recombination.
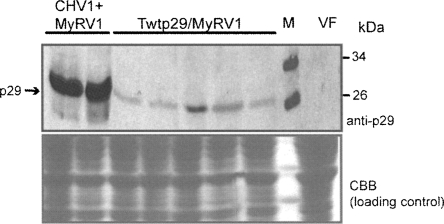
Taken together, MyRV1 rearrangements found in coinfected and p29-transformed cells were different in quality (S10ss1 versus S10ss2 and S6L) and in frequency (22.4% versus 59.7%). To gain some clues about the molecular mechanism underlying these differences, we compared expression levels of p29 in transformants with the p29 coding domain and CHV1-infected fungal colonies. Interestingly, p29 levels were different between the two strains; that is, p29 was expressed much more in mycelia coinfected with CHV1 and MyRV1 (Fig. 3, lanes CHV1+MyRV1) than in transformants (Fig. 3, lanes Twtp29). Although what makes the differences in MyRV1 rearrangements still remains largely unknown, it may be attributable to the different p29 expression levels between the two fungal strains. Alternatively, viral factors other than p29 may directly contribute to the MyRV1 rearrangement in coinfected fungal cells or lead to presumably different distributions of p29.
FIGURE 3.
Detection of p29 in mycelia infected with CHV1 or transformed with the p29 coding domain. Total proteins were extracted from mycelia infected with CHV1 alone or together with MyRV1 or transformed with the p29 coding sequence (Twtp29). Proteins were fractionated on an SDS-PAGE gel (12%) and visualized immunologically using anti-p29 antibodies after electroblotting to a nitrocellulose membrane (top panel) or by staining directly with Coomassie Brilliant Blue (bottom panel). Total protein fractions from a virus-free (VF) EP155 strain were treated in parallel.
MyRV1 S6L is expressed in fungal colonies infected with MyRV1, while S10ss2-encoded polypeptide failed to be detected
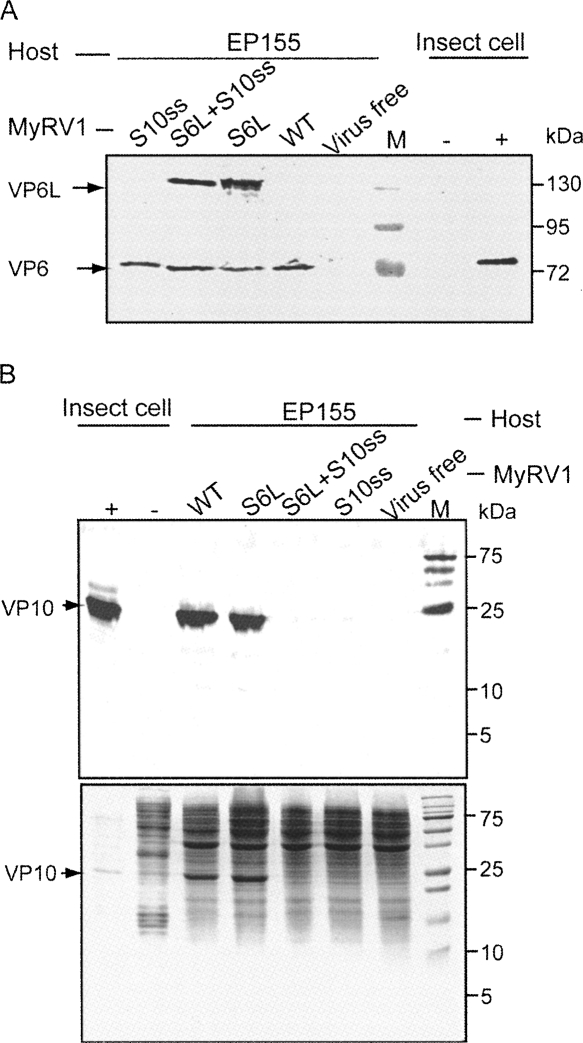
Sequence analysis of S6L and S10ss prompted us to identify proteins encoded by those segments in infected fungal cells. We introduced MyRV1/S6L, MyRV1/S6L+S10ss2, and MyRV1/S10ss2 from p29 transformants to EP155 nontransformants by anastomosis. Western blotting revealed that VP6 was detected in mycelia of MyRV1-infected EP155 specifically (Fig. 4A, lane WT), but not in mycelia of virus-free EP155 (Fig. 4, lane virus free). In fungal strains infected with MyRV1/S6L or MyRV1/S6L+S10ss2, the larger version of VP6 (VP6L), as expected from the sequence results shown in Figure 2, was detectable with anti-VP6 rabbit antibodies in addition to authentic VP6 (Fig. 4A).
FIGURE 4.
Western blotting analysis of protein products encoded by MyRV1 rearranged segments. (A) Detection of S6L-encoded proteins. Proteins were extracted from fungal mycelia of nontransformants (EP155) that had been infected with wild-type (WT) MyRV1 or its variants with MyRV1/S6L, MyRV1/S10ss2, or MyRV1/S6L+S10ss2 as shown above the lanes. Extracted proteins were electrophoresed in an SDS-polyacrylamide gel (10%), transferred to cellulose membrane, and probed with anti-MyRV1 VP6 antiserum. Proteins in virus-free EP155 and insect cells infected by baculovirus recombinants carrying (+) or not carrying (−) MyRV1 S6 cDNA were analyzed in parallel (Supyani et al. 2007). Insect cell lysates of the positive control were diluted 10 times more than those of the negative control. (M) Molecular mass standards (17–170 kDa; Fermentas, MD) (wt shown). (B) Detection of MyRV1 S10-encoded protein in mycelia infected by different virus strains. The same protein preparations as used in A were resolved in an SDS-PAGE gel (20%) and stained directly by Coomassie Brilliant Blue (bottom panel) or subjected to Western blotting. Anti-MyRV1 VP10 antiserum was used as the first antibody. Proteins in VP10-expressing (+) and non-expressing (−) insect cells (Supyani et al. 2007) were also used as positive and negative references. Molecular mass standards (5–250 kDa) were from Bio-Rad.
When stained by Coomassie Brilliant Blue, SDS-PAGE analysis revealed a polypeptide of ∼25 kDa readily detectable in total protein fractions from fungal colonies infected with MyRV1 and MyRV1-S6L, but not with MyRV1/S10ss2 or MyRV1/S6L+S10ss2 (Fig. 4B, bottom panel). As the expected size of VP10 was in the same range (Suzuki et al. 2004), these data led us to hypothesize the 25-kDa protein was MyRV1 VP10. To test this possibility, the proteins in mycelia were tested for reactivity with anti-MyRV1 VP10 antibodies. Single bands corresponding to 25 kDa were detected only in MyRV1- and MyRV1/S6L-infected mycelial cells (Fig. 4B, top panel). The 25-kDa protein showed the same migration position as VP10 expressed via the baculovirus vector system (Fig. 4B, top panel). Northern analysis showed that short transcripts were produced from S10ss1 (data not shown) and S10ss2 (Fig. 5), which could encode proteins of 51–61 amino acids. We used a gel with a high acrylamide concentration (20%) in an attempt to detect the potentially shorter version of VP10 by Western blotting. We failed to detect it, possibly because the shorter variant was not reactive to the antibody against authentic VP10 and/or it was unstable. Thus, it still remains unknown whether proteins encoded by S10ss1 and S10ss2 are expressed in infected fungal cells.
FIGURE 5.
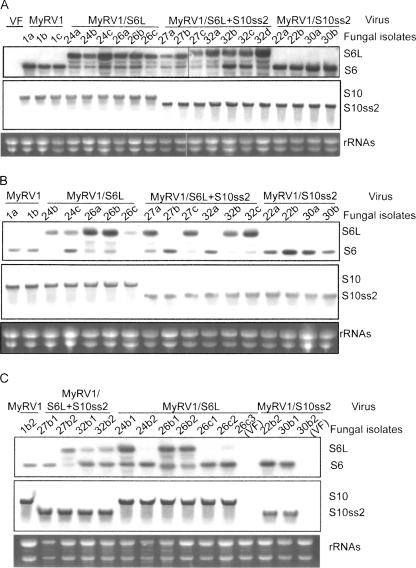
Northern blot analysis of ssRNAs isolated from fungal colonies obtained by serial subculturing or single conidial isolation. EP155 fungal colonies originally harboring MyRV1 (no. 1), MyRV1/S6L (nos. 24, 26), MyRV1/S6L+S10ss2 (nos. 27, 32), or MyRV1/S10ss2 (nos. 22, 30) (shown above the panels), were co-cultured with virus-free nontransformant EP155 for 2 wk. After lateral transfer of the viruses, a few plugs (a–c) obtained from different sites of the recipient EP155 side were cultured on PDA for 1 wk. This subculturing procedure was repeated serially a total of three times. Simultaneously, a few single conidial germlings carrying MyRV1 variants were obtained from colonies that had been subcultured once. Single-stranded RNAs (4 μg/lane), prepared from PDB cultures of fungal colonies subcultured once (A) or three times (B), and single conidial isolates (C), were electrophoresed under denaturing conditions and probed with S6- (top panels, A–C) or S10- (middle panels, A–C) specific probes. (Bottom panels) rRNAs stained with ethidium bromide as loading references. (VF) Virus-free. Same numbers mean same origins of strains. (a–c) Subcultures of single isolates designated A–C. For single conidial isolates originated from a subculture, additional numbers are placed at the right of the lower-case letters (C).
Lateral transfer of the rearranged segments in different host backgrounds in the absence of p29
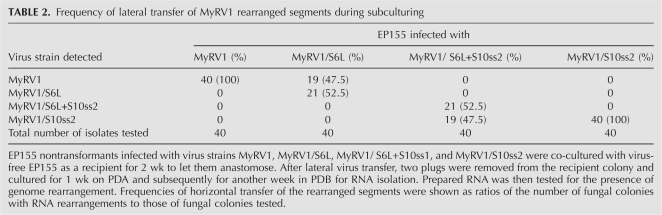
It was of interest to explore whether or not the transgenic expression of p29 is required for the maintenance of the rearranged segments. To this end, EP155 as a virus recipient was anastomosed with fungal strains (Twtp29) infected with MyRV1/S6L, MyRV1/S6L+S10ss2, or MyRV1/S10ss2 as a virus donor. After infection, the recipient EP155 was subcultured three times in PDA and examined for the presence of rearranged segments. Table 2 summarizes the frequencies of lateral transfer during subculturing, while Figure 5, A and B, shows the results of Northern analysis of representative colonies subcultured once and three times, respectively. EP155 infected with MyRV1/S6L and MyRV1/S6L+S10ss2 retained S6L well when subcultured once, but lost occasionally and gained wild-type S6 after being subcultured three times in the absence of a transgenic supply of p29 (Fig. 5B, e.g., 27b and 32a). Interestingly, subcultures even from single colonies showed different detection profiles of S6 and S6L when analyzed by Northern blotting (e.g., Fig. 5B, cf. 27a and 27c). The decrease in the ratio of S6L to S6 (“apparent reversion”) may be due to S6L being disadvantaged by its length over S6 as a template in transcription and replication steps.
TABLE 2.
Frequency of lateral transfer of MyRV1 rearranged segments during subculturing
By contrast, the S10ss2 in MyRV1/S10ss2 and MyRV1/S6L+S10ss2 was well maintained after serial subculturing in EP155 (Fig. 5; Table 2) or Twtp29 (data not shown).
Vertical transfer of the rearranged segments through conidia
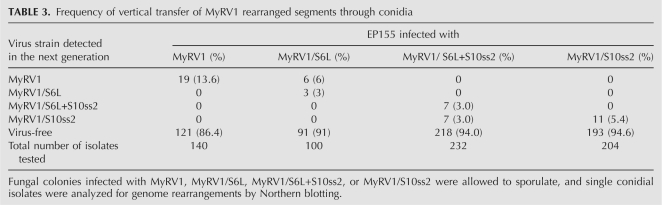
To determine the vertical transmissibility of rearranged segments, single conidial isolates were obtained from EP155 nontransformants infected with the virus variants and tested for rearrangements. For example, of 100 isolates from a colony infected with MyRV1/S6L, six harbored MyRV1, three contained MyRV1/S6L, and the rest were virus-free (Table 3). MyRV1/S6L segregated into MyRV1/S6L and MyRV1 (wild-type) in the next generations. MyRV1 S6 and S6L co-existing in single cells or maintained in different cells were possibly separated into independent asexual spores. Similarly, 232 isolates from a colony infected with MyRV1/S6L+S10ss2 were grouped into seven colonies infected with MyRV1/S6L+S10ss2, seven colonies infected with MyRV1/S10ss2, and 218 virus-free colonies (Table 3). The differences in ratios of S6L to S6 in MyRV1/S6L+S10ss and MyRV1/S6L as seen in Northern blots (Fig. 5C) are more complex. For example, S6L was predominantly (Fig. 5C, lane 24b1) or faintly detected over S6 (Fig. 5C, lane 24b2), even though isolates 24b1 and 24b2 were derived from the same parent 24b. Unlike S6L, no segregation of MyRV1/S10ss2 was observed during transmission through conidia. The transmission frequency of MyRV1/S10ss2 was 5.3%, which was slightly lower than that of the wild-type virus examined in parallel in the current study (13.6%) (Table 3) or previously reported (9.4%) (Sun et al. 2006).
TABLE 3.
Frequency of vertical transfer of MyRV1 rearranged segments through conidia
Phenotypic characteristics associated with S6L or S10ss
Examination of the possible phenotypic effects of rearranged segments alone was complicated by the fact that transgenic expression of p29 contributes to the phenotypic alteration in colonies uninfected or infected with MyRV1 or CHV1 (Craven et al. 1993; Suzuki et al. 2003; Sun et al. 2006). That is, p29 transformants infected with MyRV1 produced more aerial hyphae and induced greater lesions on apples than nontransformants infected with MyRV1. Therefore, taking advantage of the relative stability of the rearranged segments in EP155 nontransformants (Fig. 5; Table 2), we compared phenotypes of EP155 infected by wild-type MyRV1 and its variants with rearranged segments. When PDA media were inoculated by those fungal strains for phenotypic observation, PDB liquid media were simultaneously inoculated for dsRNA analysis to confirm the expected retention of the rearranged segments.
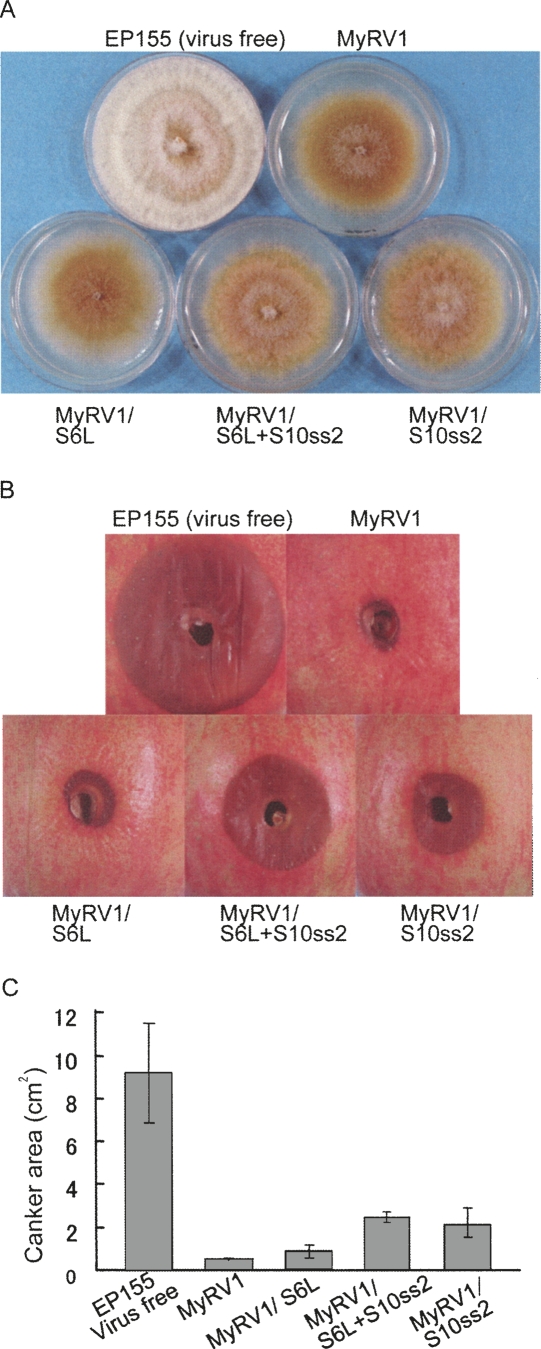
As shown in Figure 6A, virus-free EP155 grown in parallel produced orange pigments and conidia-containing pustules, and grew faster. Fungal colonies infected with MyRV1/S6L were indistinguishable from EP155 infected with wild-type MyRV1 that showed increased brown pigmentation and reduced growth of aerial mycelia and vegetative growth rate relative to uninfected EP155. Infection by MyRV1/S10ss2 and MyRV1/S6L+S10ss2 enhanced slightly the growth of aerial hyphae relative to that by MyRV1 or MyRV1/S6L (Fig. 6A, see white mycelia at the central portions of colonies infected by MyRV1/S10ss2 or MyRV1/S6L+S10ss2).
FIGURE 6.
Phenotype of fungal colonies infected with MyRV1 and its variants with rearranged segments. (A) Colony morphology of Cryphonectria parasitica strain EP155 infected with MyRV1 variants. MyRV1 variants containing S6L, S10ss2, and S6L+S10ss2 were introduced into EP155 by anastomosis. The fungal strains were then cultured on 60-mm PDA for 8 d on the bench top. EP155 transfected with purified particles of MyRV1 and virus-free EP155 was grown in parallel. (B) Representative lesions induced by each strain on apples. Fresh cultures of each strain either uninfected (EP155) or infected with MyRV1, MyRV1/S6L, MyRV1/S10ss2, MyRV1/S6L+S10ss2, or both viral strains were used to inoculate apples. Lesions produced 10 d post-inoculation are shown. (C) Levels of virulence of fungal strains. Averages and standard errors of sizes of lesions induced on apples 10 d after inoculation were calculated from five measurements for virulence of each strain.
The virulence of fungal colonies infected with the viral strains with the rearrangements was determined with an apple assay. As reported earlier (Hillman et al. 2004), EP155 infected with MyRV1 induced extremely small lesions on apples, in contrast to virus-free EP155 that formed much greater lesions of ∼9 cm2 in size (Fig. 6B,C). MyRV1/S6L and MyRV1 were similar in their ability to induce lesions. Lesions induced by fungal colonies infected with MyRV1/S10ss2 or MyRV1/S6L+S10ss2 were significantly greater (∼2–3 cm2), albeit much smaller than EP155 (∼9 cm2), than MyRV1 or MyRV1/S6L-infected EP155 (∼1 cm2) (Fig. 6B,C).
These combined results suggested that S10-coded VP10 contributed directly or indirectly to the suppression of the production of aerial hyphae and lesion formation on apples caused by MyRV1.
p29 copurifies with MyRV1 genomic RNA and interacts with MyRV1 VP9 in vitro and in vivo
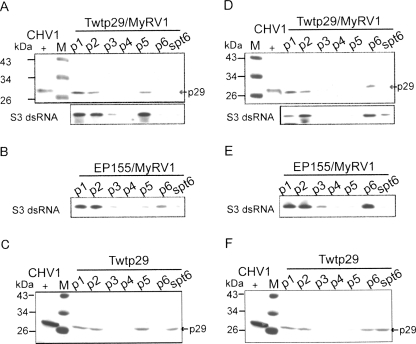
Based on the assumption that p29 might interact with proteins encoded by MyRV1 and/or its RNA to induce the rearrangements, we conducted three experiments: fractionation of cellular components, far-Western analysis, and immunoprecipitation of recombinant proteins of MyRV1 expressed in insect cells. Extracts of protoplasts prepared from mycelia that expressed p29 transgenically and were infected with MyRV1(Twtp29/MyRV1), virus-free Twtp29, and MyRV1-infected non-transformants (EP155/MyRV1) were fractionated in the absence (Fig. 7A–C) or presence (Fig. 7D–F) of nonionic detergent NP-40, and were analyzed by Western and Northern blotting. As shown in Figure 7A, p29 and MyRV1 dsRNA were concomitantly detected in fractions p1, p2, and p5 that were rich in cell debris, cell debris, and plasma membrane/vesicles (Jacob-Wilk et al. 2006) when fractionated without NP-40. However, MyRV1 dsRNA was predominantly observed in p1, p2, and p6 in the absence of p29 (Fig. 7B), while p29 was fractionated in the p1, p2, p5, and spt6 fractions without MyRV1 infection (Fig. 7C). Importantly, fractionation profiles of MyRV1 dsRNA were different between the two host backgrounds, EP155 and Twtp29 (Fig. 7, cf. A and B). When NP-40 was used in fractionation, p29 and genomic RNA were also cofractionated in p1, p2, and p6 (Fig. 7B,E). The same fractions and spt6, obtained from virus-free Ttp29, contained p29 (Fig. 7F). The observation that p29 altered a fractionation profile of MyRV1 genomic RNA only in the absence of NP-40 suggests an association of p29 and the MyRV1 replication machinery (genomic RNA) that is disrupted by the detergent.
FIGURE 7.
Fractionation analysis of extracts from p29 transformants infected with MyRV1. Spheroplasts were prepared from p29-expressing mycelia (Suzuki et al. 2003) that had been infected (Twtp29/MyRV1, A,D) or uninfected (Twtp29, C,F) with MyRV1. MyRV1-infected nontransformant strain EP155 (EP155/MyRV1, B,E) was also utilized in parallel. Protoplasts (1 × 107) were then lysed in 1 mL of 50 mM Tris-HCl at pH 8.0, 75 mM NaCl, in the absence (A–C) or presence (D–F) of 1% NP-40 by a freeze–thaw method. The lysates were centrifuged at 4°C serially at 100g for 10 min, at 1000g for 10 min, at 5000g for 30 min, at 10,000g for 30 min, at 20,000g for 30 min, and at 100,000g for 180 min. Resulting pellets at each step (p1–p6) were suspended in 1 mL of lysis buffer and directly used for Western blotting with anti-p29 antibodies and for Northern blotting with a DIG-labeled probe specific for MyRV1 S3 after extraction by phenol/chroloform and ethanol precipitation. Total protein fractions of CHV1-infected mycelia (CHV1+) were included in Western blotting. (M) Positions of molecular mass standards (43 kDa, 34 kDa, 26 kDa, and 17 kDa; Fermentas, MD).
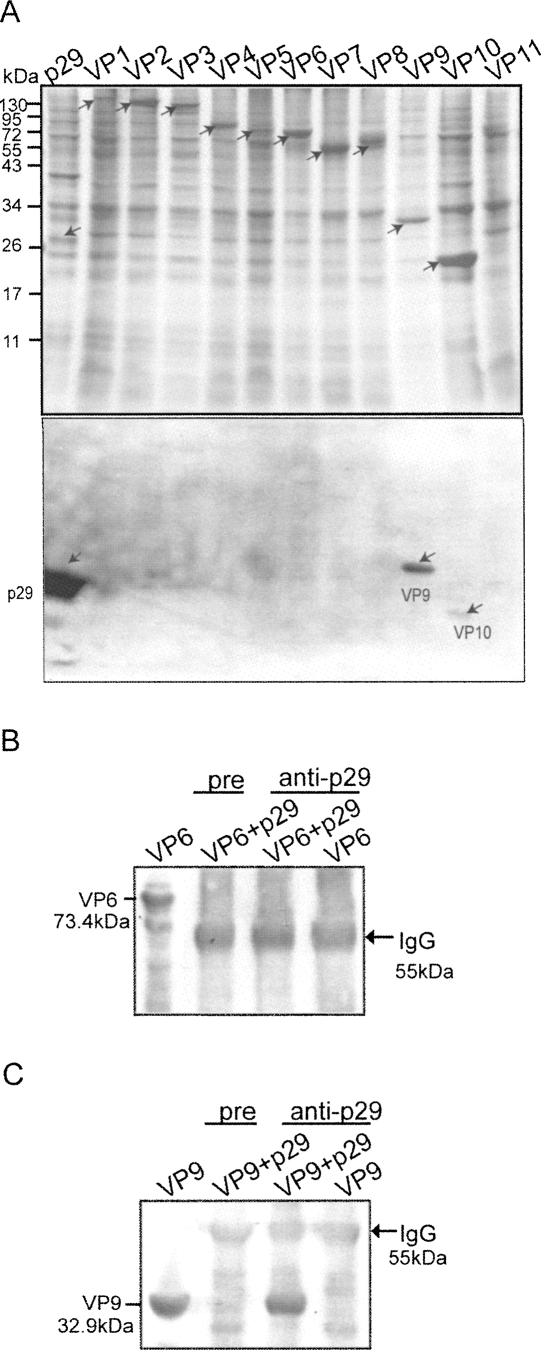
Taking advantage of the baculovirus expression system, we have previously expressed all MyRV1-encoded proteins (Fig. 8A; Supyani et al. 2007). The recombinant proteins were subjected to far-Western blotting. Consequently, p29 was found to bind VP9 strongly and VP10 faintly by this assay (Fig. 8A, bottom panel). We performed an inverse far-Western analysis in which p29 was overlaid with MyRV1 VP9 and probed by anti-VP9. This assay also showed the ability of p29 to bind to VP9 (data not shown). To further confirm these interactions, an immunoprecipitation assay was performed. Multiple proteins, p29 and VP6, and p29 and VP9 were co-expressed in Spodoptera frugiperda cells and immunoprecipitated by anti-p29 antibodies. Sedimented immunocomplex was probed by the respective antibodies. MyRV1 VP9 was co-precipitated only when p29 was expressed simultaneously and anti-p29 antibodies were used for immunoprecipitation (Fig. 8C). However, MyRV1 VP9 was not detected when it was expressed alone or when a preimmune serum was used for immunocomplex precipitation. Binding of p29 with coexpressed VP6 was not observed with either anti-p29 antiserum or pre-immune serum.
FIGURE 8.
Far Western and immunoprecipitation assays for detection of MyRV1 viral proteins binding to p29. (A) Far-Western blotting of proteins encoded by the MyRV1 genome segments expressed in S. frugiperda cells. MyRV1 proteins VP1–VP11 (arrows) were resolved in an SDS-polyacrylamide gel (12.5%) and stained directly with Coomassie Brilliant Blue (top panel), or were transferred to a PVDF membrane. The membrane was treated with p29 expressed in insect cells and was probed with anti-p29 antibodies (bottom panel). (Arrows) Strong and weak binding reactions. (B,C) Immunoprecipitation of MyRV1 proteins with co-expressed p29. MyRV1 VP6 or VP9 was co-expressed with p29 in insect cells via a baculovirus vector system. Immunoprecipitates with anti-p29 antibodies (anti-p29) or pre-immune serum (pre) along with IgG sorb were subjected to Western blotting in which blots were probed with anti-VP6 (B) and anti-VP9 antibodies (C). One of the major bands commonly found is the heavy chain subunit of IgG (arrow). Molecular mass standards (17–130 kDa) were from Fermentas (MD).
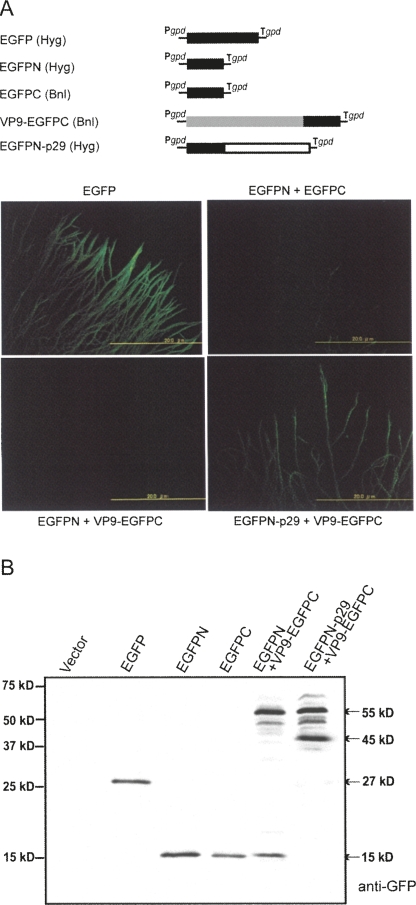
One may argue that the in vitro binding between the two proteins shown by far-Western and coimmunoprecipitation analyses would not reflect their in vivo binding nature. To demonstrate their association in vivo, we used bimolecular fluorescence complementation (BiFC) or a split GFP assay. Transformants expressing EGFP fluoresced brightly (Fig. 9A, EGFP), while fungal strains transformed with the N- or C-terminal halves of EGFP alone (data not shown) or together (Fig. 9A, EGFPN+EGFPC) were not green fluorescent. Transformants coexpressing EGFPN and VP9 fused with EGFPC failed to show green fluorescence (Fig. 9A, VP9/EGFPN+EGFPC). Only fungal strains expressing both EGFPN fused with p29 and VP9 fused with EGFPC regained the ability to fluoresce (Fig. 9A, EGFPN/p29+VP9/EGFPC). Western blotting confirmed an expected expression profile of protein products in the transformed colonies used in BiFC (Fig. 9B). That is, while no protein was detected in mycelia transformed with the empty vector (Vector), a 15-kDa protein, expected to be EGFPN or EGFPN, was observed in transformants with EGFPN and EGFPC. Cotransformation of the fungal strain EP155 with EGFPN+VP9/EGFPC or p29/EGFPN+VP9/EGFPC resulted in expression of two major proteins of ∼55 kDa and 15 kDa, and 55 kDa and 45 kDa, respectively. The proteins of 55 kDa and 45 kDa were considered to be the fusion products VP9/EGFPC and EGFPN/p29.
FIGURE 9.
In vivo interactions of CHV1 p29 and MyRV1 VP9. (A) Schematic representation of constructs used in the bimolecular fluorescence complementation assay (BiFC). The entire (EGFP, amino acids 1–239), N-terminal half (EGFPN, amino acids 1–140), or C-terminal half (EGFPC, amino acids 120–229) coding domains of EGFP were prepared. Fusion versions designed to express a p29/Ntertminal EGFP product (p29/EGFPN) and a VP9/Cterminal EGFP product (VP9/EGFPC) were also generated. These were cloned into the fungal expression vector pCPXHY1 or pCPXBN1 that carried the hygromycin (Hyg) or benomyl resistance gene (Bnl). BiFC assay of fungal strains transformed with the constructs shown in A alone or in combination. Mycelia grown under cover glass on PDA were observed with fluorescence microscopy under a UV lamp (Suzuki et al. 2000; Parsley et al. 2002). Fungal strains observed were: transformants with EGFP, EGFPN+EGFPC, EGFPN+VP9/EGFPC, and p29/EGFPN+EGFPC. (B) Western blot analysis of fungal transformants. Total protein fractions were obtained from transformants with empty vector (Vector), EGFP, EGFPN, EGFPC, EGFPN+VP9/EGFPC, and p29/EGFPN+EGFPC, as shown on the top of the panel. Proteins were fractionated on a 12.5% PAGE gel in the presence of SDS, and probed with anti-GFP antiserum (Invitrogen).
All these combined results suggested that p29 copurified with the MyRV1 genomic RNA and interacted with VP9 in vitro and in vivo.
DISCUSSION
In contrast to many well-studied examples of mixed infections in plants and animals, only a few reports are available on virus/virus interactions in fungal hosts. As an example in fungi, Sun et al. (2006) previously reported one-way synergistic interactions between CHV1 and MyRV1 mediated by the RNA silencing suppressor p29 encoded by CHV1. The current study showed another example of interplay between the two viruses in which p29 as a major player, reproducibly induced two types of alterations in the MyRV1 genome segments: an intragenic duplication of almost full-length ORF of S6 (S6L) and a 75% internal deletion of the S10 ORF (S10ss1 and S10ss2). To our knowledge this is the first case of rearrangement of reovirus segments induced by a protein encoded by a distinct virus, CHV1, which is closely associated with the picorna-like superfamily (Koonin et al. 1991).
It is known that members of the family Reoviridae infecting animals undergo a variety of rearrangements, including intragenic and intergenic extension and deletion of dsRNA segments, in naturally infected host individuals or in cultured host cells inoculated with reoviruses, most frequently with high multiplicity of infection (Taniguchi and Urasawa 1995; Desselberger 1996; Taniguchi et al. 1996; Cao et al. 2008). In plant reoviruses, artificial (non-natural) maintenance exclusively in one of their hosts, either in plants by vegetative propagation of infected plants or in insect vector cell cultures by subculturing, leads to the appearance of mutated segments often together with their cognate segments (Nuss 1984; Maoka et al. 1993; Tomaru et al. 1997). The rearrangement phenomenon reported in this study is different from those of other reports in that MyRV1 rearrangements can be induced and reproduced within 4–5 wk by a heterologous virus or protein (Fig. 1; Table 1).
Reverse genetics systems based on engineered cDNA of limited segments or a complete set of cDNAs of the genome segments have been established recently for the genera Rotavirus (Komoto et al. 2006) and Orthoreovirus (mammalian reovirus, MRV; Kobayashi et al. 2007) of the Reoviridae family, but not for other reovirus members. Therefore, it is still difficult to assign a function to each genome segment of a reovirus other than rotaviruses and MRV. As an alternative, genetics of reoviruses with reassorted or rearranged segments has been used toward determining the functional assignments of reoviral segments (Taniguchi et al. 1994, 1996; Ramig 1997; Gault et al. 2001; Matthijnssens et al. 2006a). In the current study, three strains of MyRV1 containing S6L, S10ss, and both S6L and S10ss2 provided insights into the functional roles of those segments. S6L and S10ss2 are maintained, while S6L occasionally decreases in amount (“apparent reversion”), even after serial subculturing of infected fungal colonies or conidiation (Fig. 5; Table 2). The reduction of S6L may be due to less efficient transcription and replication relative to S6. Further, S6L and S10ss2 mRNAs were detected in infected mycelia (Fig. 5). Taken together, these data suggest that the altered segments retain the necessary information for proper packaging, transcription, and replication, assuming that reovirus transcription occurs in core particles. The failure to detect VP10 or its shorter version (Fig. 4) of the expected size of 6 kDa in colonies infected with MyRV1/S6L+S10ss2 or MyRV1/S10ss2 suggests that VP10 may be dispensable for virus viability. Moreover, infection of EP155 with MyRV1/S10ss or MyRV1/S6L+S10ss results in enhanced growth of aerial hyphae and increased virulence on apples, compared with infection with MyRV1 or MyRV1/S6L (Fig. 5), suggesting an involvement of VP10 in the repression of aerial hyphae caused by MyRV1 and supporting the proposal made previously by Sun et al. (2006) that growth of aerial mycelia is correlated with virulence on apples.
MyRV1 VP6 possesses the NTP binding motifs that are conserved in the counterparts of other members of the turreted group within the family Reoviridae (Suzuki et al. 2004). The MRV counterpart μ2, best characterized among them, is known to be a minor structural protein and part of the replication complex with the dsRNA genome segments and other protein components within the core particles (Schiff et al. 2007). Thus, it was anticipated that extension of the ORF in S6L might have effects on replication and/or symptom induction. However, in contrast to S10ss viruses, no significant effect of S6L was observed on morphological alterations induced by MyRV1 or its replication (Fig. 6). The predominant amount of S6L mRNA in some single conidial isolates without reducing the accumulation of other segments may suggest that VP6L is functional in viral replication (Figs. 1, 5). Nevertheless, interpretation of these results is complicated by the concomitance of S6 and S6L and their protein products VP6 and VP6L (Fig. 4), allowing the possibility of transcomplementation of VP6L by VP6 when both forms of the products coexist in single infected cells.
Preliminary results of Northern blot analysis suggest that in addition to S6L and S10ss RNAs, partially duplicated or deleted forms of S1, S2, S4, and S8, as well as the intact segments, are observed in Twtp29 originally infected with MyRV1 (data not shown). S6L is maintained relatively stably through conidiation and subculturing in the absence of p29 (Fig. 5; Tables 2, 3). These observations suggest that p29 may act promiscuously on MyRV1 segments to rearrange, rather than to select, recombinants preexisting in a minor amount, and only modifications (S10ss and S6L) tolerable for and/or advantageous to virus viability can be detected. In rotaviruses and phytoreoviruses, certain segments are inclined to be rearranged or altered at higher rates than other segments, suggesting that modifications of those segments are better tolerated (Nuss 1984; Taniguchi and Urasawa 1995; Desselberger 1996).
A few hypotheses were proposed for rearrangements of rotavirus occurring during plus-sense synthesis (Kojima et al. 1996) or minus-sense strand synthesis (Gault et al. 2001), based upon primary and secondary sequence structures of genome segments. Recently a “loop model” was newly proposed by integrating information on the 3D structure of the λ3 RNA-dependent RNA polymerase (RdRp) of MRV to explain duplication rearrangmenets, in which a template switch happens between 5′ and 3′ portions of the negative strand template in the center of RdRp (catalytic core) (Matthijnssens et al. 2006b). In any event, viral RdRp complexes disengage at a certain site of the template and re-engage at a different site of the same template molecule, in which certain RNA template structures like direct repeats and inverted repeats facilitate backward (extension) and forward (deletion) transfer of the polymerase. Fundamentally, this is similar to the RdRp-driven template switch model proposed by Nagy and Simon (1997) for the recombination event of ss(+) RNA viruses. In this regard, it is noteworthy that direct and inverted repeats of 4–5 nt are found near the recombination site of S6L and S10ss (Fig. 2A,B). Due to the lack of biochemical evidence for the RdRp-driven template switch, we cannot rule out the possibility of a nonreplicative, breaking and joining mechanism to explain the MyRV1 rearrangements, which is established in ssRNA virus recombination (Chetverin 1999).
In reoviruses, plus-strand mRNA is synthesized in core particles using virion-associated RdRp and genomic dsRNA as a template in a conservative manner, while minus-sense RNA synthesis occurs on plus-sense strands assorted into core particles. These events occur in cytoplasmic inclusion bodies, termed viroplasms (Fields 1996). MyRV1, like other reoviruses, may form viroplasms that serve as a virus factory in which virus assembly and RNA replication occur. On the other hand, hypovirus is believed to be replicated in Golgi-derived membraneous vesicles induced by infection to which p29 is assumed to be recruited along with other viral proteins (Jacob-Wilk et al. 2006). The CHV1 p29 protein was recently shown to be an RNA silencing suppressor (Segers et al. 2006). Like other RNA silencing suppressors, p29 is multifunctional in the replication cycle, being involved in viral protein processing, symptom determination, and replication enhancement. The mechanism of how p29 induces the genome rearrangement of a heterologous virus when expressed from the virus genome or transgenically and where its activity domain resides on p29 remain unknown. However, the data of Figure 7 revealed cofractionation of p29 with MyRV1 genomic RNA. This is consistent with the observation that many RNA silencing suppressors have dsRNA binding ability (Merai et al. 2006). Figures 8 and 9 also showed specific in vitro and in vivo interactions of p29 with MyRV1 VP9. It is an interesting speculation that p29 interacts with parts of the MyRV1 replication machinery in which MyRV1 VP9 plays an as-yet-unraveled role, thereby leading to genome rearrangements. The functional analysis of VP9 is underway. The future establishment of an in vitro rearrangement assay, like the Qβ replicase system (Chetverin 1999), using purified virus intact particles or core particle should help address those questions.
MATERIALS AND METHODS
Fungal strains, culturing, and virulence assay
C. parasitica strain EP155 and its transformants expressing wild-type p29 (Twtp29) were prepared previously (Suzuki et al. 2003; Sun et al. 2006). EP155 and p29 transformants infected with MyRV1 were described earlier by us (Hillman et al. 2004; Sun et al. 2006). EP155 infected with CHV1-EP713 was prepared earlier (Sun et al. 2006).
Fungal colonies were grown for 5–10 d under bench-top conditions at 24°–26°C on potato dextrose agar (PDA, Difco) for morphological observation, and in potato dextrose broth (PDB, Difco) when mycelia were used for RNA extraction. For maintenance, strains were cultured on regeneration plates (Churchill et al. 1990) and stored at 4°C until use.
Virulence of fungal colonies was estimated by an assay with apple fruits described by Fulbright (1984) and Hillman et al. (2004). Apples were inoculated with plugs of freshly grown mycelia. Inoculated sites were sealed with parafilm and left for 7 d at 25°C, and were further incubated for an additional week at 25°C without parafilm. Areas of necrotic lesions were measured at day 10 post-inoculation.
Transformation of C. parasitica spheroplasts
Spheroplasts were prepared by digesting cell walls of mycelia of C. parasitica virus-free strain EP155 cultured in PDB by the methods of Churchill et al. (1990). Transformation with the coding domain of wild-type p29 was performed according to the method of Choi and Nuss (1992) in which two vectors, pCPXHY1 (Craven et al. 1993) and pCPXBn1 (Deng et al. 2007), carrying hygromycin and benomyl resistance genes as selectable marker genes, were used.
Sequence determination of rearranged segments
The dsRNA was fractionated in SDS-PAGE gels and eluted from gel slices as described by Suzuki et al. (1990). The purified dsRNA was denatured in DMSO for 15 min at 65°C (Asamizu et al. 1985), reverse-transcribed in a reaction of 20 μL (50 mM Tris-HCl at pH 8.6, 40 mM KCl, 1 mM MnSO4, 1 mM DTT, 0.5 mM of each dNTP, 200 units of MMLV reverse transcriptase). DNAs complementary to different portions of rearranged segments were amplified by RT-PCR with different combinations of primer sets and cloned into pGEM T-easy (Promega). When necessary, the amplified fragments were digested with appropriate restriction enzymes before cloning. Plasmid clones were sent to Macrogen, Inc. for sequencing on an ABI PRISM 3700 DNA analyzer.
RNA preparation and Northern blot analysis
Total RNA was prepared from C. parasitica mycelia cultured in 50 mL of PDB as described by Suzuki and Nuss (2002). To eliminate fungal chromosomal DNA, extracted nucleic acids were treated twice with RNase-free RQ1 DNase I (Promega), followed by phenol, phenol/chloroform, and chloroform extractions and ethanol precipitation. Single-stranded RNA fractions were obtained from total RNA by LiCl (2 M) precipitation. Double-stranded RNA was isolated from total RNA fractions using CC41 (Whatman) cellulose column chromatography (Isogai et al. 1998).
Total RNA was electrophoresed in a nondenatured agarose gel (1.4%) in 1× TAE buffer (40 mM Tris-acetate–1 mM EDTA at pH 7.8). For separation of ssRNA fractions, a 1% agarose gel was used with the 1× TAE buffer system to distinguish viral mRNA from genomic dsRNA that might contaminate the ssRNA fraction. Fractionated RNA in an agarose gel was capillary-transferred onto a Hybond-N+ nylon membrane (Amersham Biosciences) and denatured in 50 mM NaOH, 10 mM NaCl for 5 min. Viral genomic dsRNA was subjected to SDS-polyacrylamide gel (12%) electrophoresis (SDS-PAGE) in Laemmli's buffer system (Sambrook and Russell 2001). After washing in 1× TBE (89 mM Tris-borate, 89 mM boric acid, 2.5 mM EDTA at pH 8.3), the separated viral dsRNAs was electrotransferred from the SDS-PAGE gel to a Hybond-N+ nylon membrane in a submarine electroblot apparatus (Marysol Model KS8452) at a constant current of 400 mA for 5 h, and denatured as stated above.
After washing twice in 2× SSC (0.3 M NaCl, 0.03 M NaOAc at pH 7.0), and twice in 0.1× SSC (15 mM NaCl, 1.5 mM NaOAc at pH 7.0), the membrane was baked for 2 h at 80°C regardless of electrophoresis types. RNA was then probed with dioxigenin (DIG)-11-dUTP-labeled DNA or RNA fragments prepared according to the method recommended by the manufacturer (Roche Diagnostics). Prehybridizations and hybridizations were carried out as described by Suzuki et al. (2003). Chemiluminescent signals were visualized on a film.
Baculovirus expression of MyRV1 genome segments
Recombinant baculoviruses carrying genome segments of MyRV1 were prepared by using the Bac-to-Bac Baculovirus Expression System (Invitrogen) as described by Supyani et al. (2007). Culturing of S. frugiperda cells and subsequent protein analysis were performed according to Matsuura et al. (1987) and Suzuki et al. (1994).
Western and far-Western blot analyses
Polyclonal antibodies to CHV1 p29 and MyRV1 VP6, VP9, and VP10 were prepared as described by Suzuki et al. (1994). Those proteins were expressed as GST-fusion products in Escherichia coli using expression vectors pGEX-2T (p29) and pGEX-6P-1 (VP6, VP9, VP10). The recombinant proteins (1 mg/injection) purified according to the manufacturer's protocols (Promega) were used to immunize Japanese white rabbits by intradermal (first and second) and intramuscular (third through fifth) injections. Antibodies specific for p29 were purified using a sepharose affinity column (NHS-activated sepharose 4 Fast Flow, Amersham Pharmacia Biotech) to which p29, expressed and purified by the maltose binding protein expression system (pMAL-c2X, New England Biolabs), was bound, following the manufacturer's instructions.
Mycelia were homogenized in a mortar and pestle in liquid nitrogen. The homogenates were mixed with the lysis buffer (50 mM Tris-HCl at pH 8.0, 75 mM NaCl, 1% Nonidet P-40 [NP-40]), and centrifuged at 14,000g for 5 min. Proteins in the supernatant were denatured by heating for 10 min at 100°C in 2× SDS-PAGE sample buffer (Sambrook and Russell 2001). Proteins separated in the gel were electrotransferred to a Trans-Blot nitrocellulose membrane (Bio-Rad) in a semi-dry fashion. Membranes were treated with blocking solution, TBS (20 mM Tris-HCl at pH 7.5, 0.5 M NaCl) containing 1% BSA. Specific antibody–antigen reactions were detected by the method of Suzuki et al. (1994). For far-Western blotting, insect cells were lysed in 40 mM Tris-HCl at pH 6.8, 0.25 mM DTT, 0.05% glycerol. After blocking with TBS containing 2% BSA and 0.2% polyvinylpyrolidon (PVP), the membranes were incubated overnight with lysates of insect cells infected with Acwtp29 and expressing p29 (Sun et al. 2006). The concentration of p29 in insect cell lysates was adjusted to 100–200 ng/mL, which was estimated based on the band intensity of an SDS-PAGE gel stained by Coomassie Brilliant Blue. The blots were treated with antibodies to p29 and the second antibody, followed by detection of specific reactions as described above for Western blotting.
Immunoprecipitation
S. frugiperda cells were inoculated with baculovirus recombinant Acwtp29 (Sun et al. 2006) alone or in combination with recombinants Ac9BS6 or Ac9BS9 (Supyani et al. 2007). Acwtp29, Ac9BS6, and Ac9BS9 allow high-level expression of CHV1 p29, MyRV1 VP6, and MyRV1 VP9, respectively. After washing three times with phosphate buffered saline, infected insect cells were lysed in 50 mM Tris-HCl, 75 mM NaCl, 0.5% NP-40, and 1% (v/v) protease inhibitor cocktail (Nacalai Tesque) and centrifuged at 15,000g for 45 min. Proteins interacting with CHV1 p29 in the supernatant were indirectly bound to anti-p29 antibodies and subsequently to protein A-sepharose beads (IgG sorb, Tago, Inc.). Immunocomplexes were washed in the lysis buffer four to five times and suspended in SDS-PAGE sample buffer. Dissociated proteins were probed with anti-MyRV1 VP6 or anti-MyRV1 VP9. Specific antibody–antigen reactions were visualized with a chemiluminescent technique (Roche Diagnostics).
Fractionation of cellular extracts by centrifugation
Protoplasts (1 × 107) were lysed in 1 mL of 50 mM Tris-HCl at pH 8.0, 75 mM NaCl, with or without 1% Nonidet P-40 (NP-40), by freezing in liquid nitrogen and thawing at 37°C. Extracts in isolated cells were fractionated by serial centrifugation (100g for 10 min ∼100,000g for 180 min), and obtained as pellets. The supernatant after the final centrifugation was also recovered. Each fraction was analyzed by Western and Northern blotting.
Bimolecular fluorescence complementation assay
Several derivatives from EGFP (Invitrogen) constructs were prepared using PCR techniques. The N- and C-terminal regions (140 and 120 amino acids) were amplified by high-fidelity KOD DNA polymerase (Toyobo) using primer sets NEGFP5F and NEGFP5R, and CGFP5F and CGFP3R, and then cloned into either pCPXHY1 or pCPXBn1 to obtain EGFPN and EGFPC. EGFP fusion constructs, with CHV1 p29 (EGFPN/p29) or MyRV1 VP9 (VP9/EGFPC), were designed as described by Hu et al. (2002). The p29 and VP9 coding sequences were amplified using the primer sets p29F and p29R, and VP9-5F and VP9-3R and cloned into pGEM-T-easy (Promega). Amplified DNAs were linked with the coding fragments of EGFPN and EGFPC using appropriate restriction enzymes added during PCR. Resulting fragments were cloned into pCPXHY1 or pCPXBn1 for preparation of VP9/EGFPC and EGFPN/p29. These transforming constructs were introduced into fungal chromosomes as described above. Sequences of the primers used in PCR and overlap PCR are listed in Supplemental Table S1.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported in part by the Okayama University COE program “Establishment of Plant Health Science” to N.S., financial support from Yomogi, Inc. to N.S., and the Sasakawa Scientific Research Grant from the Japan Science Society to L.S. We thank Dr. Ulrich Desselberger for critical reading of the manuscript, and Drs. Donald L. Nuss, William MacDonald, and Bradley I. Hillman for generous gifts of fungal strains 9B21 and EP155 and vector constructs.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1125408.
REFERENCES
- Anandalakshmi R., Pruss G.J., Ge X., Marathe R., Mallory A.C., Smith T.H., Vance V.B. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. 1998;95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzola J.V., Xu Z.K., Asamizu T., Nuss D.L. Segment-specific inverted repeats found adjacent to conserved terminal sequences in wound tumor virus genome and defective interfering RNAs. Proc. Natl. Acad. Sci. 1987;84:8301–8305. doi: 10.1073/pnas.84.23.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu T., Summers D., Motika M.B., Anzola J.V., Nuss D.L. Molecular cloning and characterization of the genome of wound tumor virus: A tumor-inducing plant reovirus. Virology. 1985;144:398–409. doi: 10.1016/0042-6822(85)90281-8. [DOI] [PubMed] [Google Scholar]
- Cao D., Barro M., Hoshino Y. Porcine rotavirus bearing an aberrant gene stemming from an intergenic recombination of the NSP2 and NSP5 genes is defective and interfering. J. Virol. 2008;82:6073–6077. doi: 10.1128/JVI.00121-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverin A.B. The puzzle of RNA recombination. FEBS Lett. 1999;460:1–5. doi: 10.1016/S0014-5793(99)01282-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G.H., Nuss D.L. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science. 1992;257:800–803. doi: 10.1126/science.1496400. [DOI] [PubMed] [Google Scholar]
- Choi G.H., Pawlyk D.M., Nuss D.L. The autocatalytic protease p29 encoded by a hypovirulence-associated virus of the chestnut blight fungus resembles the potyvirus-encoded protease HC-Pro. Virology. 1991;183:747–752. doi: 10.1016/0042-6822(91)91004-z. [DOI] [PubMed] [Google Scholar]
- Churchill A.C.L., Ciufetti L.M., Hansen D.R., Van Etten H.D., Van Alfen N.K. Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 1990;17:25–31. [Google Scholar]
- Craven M.G., Pawlyk D.M., Choi G.H., Nuss D.L. Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J. Virol. 1993;67:6513–6521. doi: 10.1128/jvi.67.11.6513-6521.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng F., Allen T.D., Hillman B.I., Nuss D.L. Comparative analysis of alterations in host phenotype and transcript accumulation following hypovirus and mycoreovirus infections of the chestnut blight fungus Cryphonectria parasitica . Eukaryot. Cell. 2007;6:1286–1298. doi: 10.1128/EC.00166-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desselberger U. Genome rearrangements of rotaviruses. Adv. Virus Res. 1996;46:69–95. doi: 10.1016/s0065-3527(08)60070-6. [DOI] [PubMed] [Google Scholar]
- Fields B. Reoviridae. In: Fields B.N., editor. Fields virology. 3rd ed. Lippincott-Raven; Philadelphia: 1996. pp. 1553–1555. [Google Scholar]
- Fulbright D.W. Effect of eliminating dsRNA in hypovirulent Endothia parasitica . Phytopathology. 1984;74:722–724. [Google Scholar]
- Gault E., Schnepf N., Poncet D., Chenon A.G. A human rotavirus with rearranged genes 7 and 11 encodes a modified NSP3 protein and suggests an additional mechanism for gene rearrangement. J. Virol. 2001;75:7305–7314. doi: 10.1128/JVI.75.16.7305-7314.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman B.I., Suzuki N. Viruses in the chestnut blight fungus. Adv. Virus Res. 2004;63:423–472. doi: 10.1016/S0065-3527(04)63007-7. [DOI] [PubMed] [Google Scholar]
- Hillman B.I., Supyani S., Kondo H., Suzuki N. A reovirus of the fungus Cryphonectria parasitica that is infectious as particles and related to the Coltivirus genus of animal pathogens. J. Virol. 2004;78:892–898. doi: 10.1128/JVI.78.2.892-898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.D., Chienenov Y., Keppola T.K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;4:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Isogai M., Uyeda I., Lindsten K. Taxonomic characteristics of fijiviruses based on nucleotide sequences of the oat sterile dwarf virus genome. J. Gen. Virol. 1998;79:1479–1485. doi: 10.1099/0022-1317-79-6-1479. [DOI] [PubMed] [Google Scholar]
- Jacob-Wilk D., Turina M., Van Alfen N.K. Mycovirus Cryphonectria hypovirus 1 elements cofractionate with trans-Golgi network membranes of the fungal host Cryphonectria parasitica . J. Virol. 2006;80:6588–6596. doi: 10.1128/JVI.02519-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau K.D., Carrington J.C. A counter-defensive strategy of plant viruses: Suppression of post-transcriptional gene silencing. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Antar A.A.R., Boehme K.W., Danthi P., Eby E.A., Guglielmi K.M., Holm G.H., Johnson E.M., Maginnis M.S., Naik S., et al. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe. 2007;1:147–157. doi: 10.1016/j.chom.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K., Taniguchi K., Urasawa T., Urasawa S. Sequence analysis of normal and rearranged NSP5 genes from human rotavirus strains isolated in nature: Implications for the occurrence of the rearrangement at the step of plus strand synthesis. Virology. 1996;224:446–452. doi: 10.1006/viro.1996.0551. [DOI] [PubMed] [Google Scholar]
- Komoto S., Sasaki J., Taniguchi K. Reverse genetics system for introduction of site-specific mutations into the double-stranded RNA genome of infectious rotavirus. Proc. Natl. Acad. Sci. 2006;103:4646–4651. doi: 10.1073/pnas.0509385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Choi G.H., Nuss D.L., Shapira R., Carrington J.C. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc. Natl. Acad. Sci. 1991;88:10647–10651. doi: 10.1073/pnas.88.23.10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M. RNA recombination in animal and plant viruses. Microbiol. Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maoka T., Omura T., Harjosudarmo J., Tsuchizaki T. Loss of vector-transmissibility by maintaining rice ragged stunt virus in rice plants without vector transmission. Ann. Phytopathol. Soc. Japan. 1993;59:185–187. [Google Scholar]
- Matthijnssens J., Rahman M., Martella V., Xuelei Y., De Vos S., De Leener K., Ciarlet M., Buonavoglia C., Van Ranst M. Full genomic analysis of human rotavirus strain B4106 and lapine rotavirus strain 30/96 provides evidence for interspecies transmission. J. Virol. 2006a;80:3801–3810. doi: 10.1128/JVI.80.8.3801-3810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J., Rahman M., Van Ranst M. Loop model: Mechanism to explain partial gene duplications in segmented dsRNA viruses. Biochem. Biophys. Res. Commun. 2006b;340:140–144. doi: 10.1016/j.bbrc.2005.11.165. [DOI] [PubMed] [Google Scholar]
- Matsuura Y., Possee R.D., Overton H.A., Bishop D.H.L. Baculovirus expression vectors: The requirements for high level expression of proteins, including glycoproteins. J. Gen. Virol. 1987;68:1233–1250. doi: 10.1099/0022-1317-68-5-1233. [DOI] [PubMed] [Google Scholar]
- Merai Z., Kerenyi Z., Kertesz S., Magna M., Lakatos L., Silhavy D. Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J. Virol. 2006;80:5747–5756. doi: 10.1128/JVI.01963-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murao K., Uyeda I., Ando Y., Koganezawa H. Genomic rearrangement in genome segment 12 of rice dwarf phytoreovirus. Virology. 1996;216:238–240. doi: 10.1006/viro.1996.0054. [DOI] [PubMed] [Google Scholar]
- Nagy P.D., Simon A.E. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- Nuss D.L. Molecular biology of wound tumor virus. Adv. Virus Res. 1984;29:57–93. doi: 10.1016/s0065-3527(08)60405-4. [DOI] [PubMed] [Google Scholar]
- Nuss D.L. Hypovirulence: Mycoviruses at the fungal–plant interface. Nat. Rev. Microbiol. 2005;3:632–642. doi: 10.1038/nrmicro1206. [DOI] [PubMed] [Google Scholar]
- Parsley T.B., Chen B., Geletka L.M., Nuss D.L. Differential modulation of cellular signaling pathways by mild and severe hypovirus strains. Eukaryot. Cell. 2002;1:401–413. doi: 10.1128/EC.1.3.401-413.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramig R.F. Genetics of the rotaviruses. Annu. Rev. Microbiol. 1997;51:225–255. doi: 10.1146/annurev.micro.51.1.225. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. Molecular cloning: A laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Schiff L.A., Nibert M., Tyler K.L. Orthoreoviruses and their replication. In: Knipe D.M., et al., editors. Fields virology. 5th ed. Wolters Kluwer/Lippincott-Raven; Philadelphia: 2007. pp. 1853–1915. [Google Scholar]
- Segers G.C., van Wezel R., Zhang X., Hong Y., Nuss D.L. Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot. Cell. 2006;5:896–904. doi: 10.1128/EC.00373-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers G.C., Zhang X., Deng F., Sun Q., Nuss D.L. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. 2007;104:12902–12906. doi: 10.1073/pnas.0702500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.-Y., Nuss D.L., Suzuki N. Synergism between a mycoreovirus and a hypovirus mediated by the papain-like protease p29 of the prototypic hypovirus CHV1-EP713. J. Gen. Virol. 2006;87:3703–3714. doi: 10.1099/vir.0.82213-0. [DOI] [PubMed] [Google Scholar]
- Supyani S., Hillman B.I., Suzuki N. Baculovirus expression of the 11 mycoreovirus-1 genome segments and identification of the guanylyltransferase-encoding segment. J. Gen. Virol. 2007;88:342–350. doi: 10.1099/vir.0.82318-0. [DOI] [PubMed] [Google Scholar]
- Suzuki N., Nuss D.L. The contribution of p40 to hypovirus-mediated modulation of fungal host phenotype and viral RNA accumulation. J. Virol. 2002;76:7747–7759. doi: 10.1128/JVI.76.15.7747-7759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Watanabe Y., Kusano T., Kitagawa Y. Sequence analysis of rice dwarf phytoreovirus genome segments S4, S5 and S6: Comparison with the equivalent wound tumor virus segments. Virology. 1990;179:446–454. doi: 10.1016/0042-6822(90)90313-g. [DOI] [PubMed] [Google Scholar]
- Suzuki N., Sugawara M., Kusano T., Mori H., Matsuura Y. Immunodetection of rice dwarf phytoreoviral proteins in both insect and plant hosts. Virology. 1994;202:41–48. doi: 10.1006/viro.1994.1320. [DOI] [PubMed] [Google Scholar]
- Suzuki N., Geletka L.M., Nuss D.L. Essential and dispensable virus-encoded replication elements revealed by efforts to develop hypoviruses as gene expression vectors. J. Virol. 2000;74:7568–7577. doi: 10.1128/jvi.74.16.7568-7577.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Maruyama K., Moriyama M., Nuss D.L. Hypovirus papain-like protease p29 is an enhancer of viral dsRNA accumulation and vertical transmission. J. Virol. 2003;77:11697–11707. doi: 10.1128/JVI.77.21.11697-11707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Supyani S., Maruyama K., Hillman B.I. Complete genome sequence of Mycoreovirus 1/Cp9B21, a member of a new genus within the family Reoviridae, from the chestnut blight fungus Cryphonectria parasitica . J. Gen. Virol. 2004;85:3437–3448. doi: 10.1099/vir.0.80293-0. [DOI] [PubMed] [Google Scholar]
- Taniguchi K., Urasawa S. Diversity in rotavirus genomes. Semin. Virol. 1995;6:123–131. [Google Scholar]
- Taniguchi K., Nishikawa K., Kobayashi N., Urasawa S. Differences in plaque size and VP4 sequence found in SA11 virus clones having simian authentic VP4. Virology. 1994;198:325–330. doi: 10.1006/viro.1994.1035. [DOI] [PubMed] [Google Scholar]
- Taniguchi K., Kojima K., Urasawa S. Nondefective rotavirus mutants with an NSP1 gene which has a deletion of 500 nucleotides, including a cysteine-rich zinc finger motif-encoding region (nucleotides 156 to 248), or which has a nonsense codon at nucleotides 153–155. J. Virol. 1996;70:4125–4130. doi: 10.1128/jvi.70.6.4125-4130.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaru M., Maruyama W., Kikuchi A., Yan J., Zhu Y., Suzuki N., Isogai M., Oguma Y., Kimura I., Omura T. The loss of outer capsid protein P2 results in nontransmissibility by the insect vector of rice dwarf phytoreovirus. J. Virol. 1997;71:8019–8023. doi: 10.1128/jvi.71.10.8019-8023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]