Abstract
Feline immunodeficiency virus (FIV) is a lentivirus that infects cats and is related to human immunodeficiency virus (HIV). Although it is a common worldwide infection, and has potential uses as a human gene therapy vector and as a nonprimate model for HIV infection, little detail is known of the viral life cycle. Previous experiments have shown that its packaging signal includes two or more regions within the first 511 nucleotides of the genomic RNA. We have undertaken a secondary structural analysis of this RNA by minimal free-energy structural prediction, biochemical mapping, and phylogenetic analysis, and show that it contains five conserved stem–loops and a conserved long-range interaction between heptanucleotide sequences 5′-CCCUGUC-3′ in R/U5 and 5′-GACAGGG-3′ in gag. This long-range interaction is similar to that seen in primate lentiviruses where it is thought to be functionally important. Along with strains that infect domestic cats, this heptanucleotide interaction can also occur in species-specific FIV strains that infect pumas, lions, and Pallas' cats where the heptanucleotide sequences involved vary. We have analyzed spliced and genomic FIV RNAs and see little structural change or sequence conservation within single-stranded regions of the 5′ UTR that are important for viral packaging, suggesting that FIV may employ a cotranslational packaging mechanism.
Keywords: FIV, lentivirus, packaging signal, dimerization, RNA
INTRODUCTION
Feline immunodeficiency virus (FIV) is a lentivirus related to human immunodeficiency virus (HIV), the causative agent of AIDS (Olmsted et al. 1989; Talbott et al. 1989). Like HIV, FIV causes in its host, the domestic cat, a prolonged disease that is characterized by progressive depletion of CD4+ T cells, ultimately leading to a fatal immunodeficiency (Pedersen et al. 1987; Yamamoto et al. 1988; Siebelink et al. 1990). Many other parallels exist between the two viruses and these render FIV infection in cats a potentially useful small animal model for HIV (Olmsted et al. 1989; and reviewed in Elder et al. 2008). In addition to this, FIV is being developed as a human gene therapy vector as, like all lentiviruses, it is able to transduce nondividing cells while being associated with fewer safety concerns than primate lentiviral vectors (reviewed in Saenz and Poeschla 2004). FIV also poses a serious veterinary concern in its own right, as it infects domestic cats and big cats alike, with a worldwide distribution and a high seroprevalence (Troyer et al. 2005). A better understanding of the mechanisms of FIV replication is needed in order to utilize the virus to its full potential as an HIV model or as a gene therapy vector, and to seek ways to combat its spread in feline species. Examining the structure of functionally important regions of the FIV RNA will not only aid this, but may also enhance our understanding of the behavior of other lentiviral RNAs.
Full-length unspliced lentiviral RNA acts as both mRNA, coding for structural and enzymatic viral proteins, and as the viral genome, two copies of which are packaged into each virion. The virus is able to recognize and select this genomic RNA from among a pool of a vast excess of spliced viral and cellular RNAs. For FIV the nucleotide sequences responsible for this are partly mapped, but the specific structural signals and motifs that permit this recognition are not known. In some other retroviruses these RNA packaging signals, to which the viral Gag protein binds, have been elucidated (Rein 1994; Lever 2007). In some they are found in the region between the major splice donor (mSD) site and the gag start codon (Lever et al. 1989; D'Souza et al. 2001), thus ensuring that only full-length and unspliced RNAs are selected for packaging. In others they are found upstream of the mSD (Linial et al. 1978; McCann and Lever 1997) and other mechanisms such as cotranslational packaging are required to maintain specificity. Previous work suggests that the FIV genome encodes two major packaging determinants (Kemler et al. 2002; Browning et al. 2003a,b; Kemler et al. 2004; Mustafa et al. 2005; Ghazawi et al. 2006). Although some controversy exists as to their exact locations, one of these is located upstream of the mSD within the 5′ UTR and the other is near the 5′ end of the gag open reading frame. FIV is quite unique in its arrangement of packaging signals as the RNA between these two regions, including the mSD to the gag AUG, is dispensable for packaging in terms of both specific sequence and sequence length (Kemler et al. 2002; Browning et al. 2003a; Mustafa et al. 2005).
In addition to encapsidation, the 5′ end of the RNA genome of lentiviruses like HIV and FIV is involved in many functions during the viral life cycle, such as initiation of reverse transcription (Isel et al. 1993), polyadenylation (Weichs an der Glon et al. 1991) and genome dimerization (Darlix et al. 1990; and reviewed in Greatorex 2004). Both structural and sequence elements are thought to be important for these functions.
Here we present a structural analysis of the 5′ end of the FIV RNA genome and identify conserved structures. This RNA includes five stem–loops, one of which (SL2) is an extremely stable structure of ∼150 nucleotides (nt) that is present in both spliced and genomic RNAs and includes a region that has previously been shown to act as a packaging signal (Browning et al. 2003a). The first 105 nt are involved in extensive long-range interactions (LRIs) with those 3′ of the mSD, including the first 100 nt of gag. In particular, this structure appears to be anchored by complementary conserved heptanucleotide sequences in the R/U5 region and within the gag open reading frame. The structural model provides important functional correlates with published data on FIV packaging signals while extending the understanding of functional regions. In addition, the structure may offer an insight into the mode of RNA encapsidation and the possible role of RNA dimerization in this process. A very recently published paper (James and Sargueil 2008) suggests an alternative structure for the FIV leader RNA; however, the analysis did not identify the capacity for the RNA to form an LRI analogous to that seen in HIV-1 and HIV-2. For this and other reasons described in the Discussion we believe the model we present is correct and explains the known functional aspects of the 5′ leader region more cogently.
RESULTS
The 5′ end of the unspliced FIV RNA is predicted to fold into a stable structure containing either four or six medium or large stem–loops
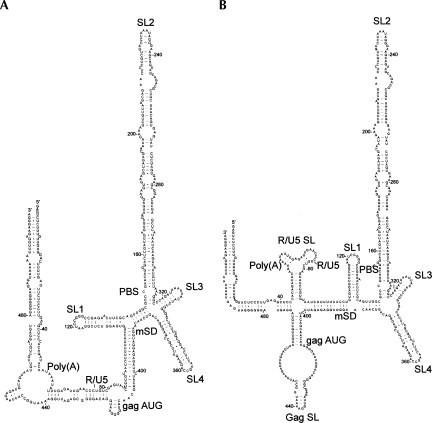
Minimal free-energy models of the 5′ 511 nt of genomic FIV RNA are shown in Figure 1. This RNA was chosen for analysis as it contains sequences necessary and sufficient for optimal packaging of the FIV genome (Browning et al. 2003b). Mfold prediction generated four possible structures, of which the two most stable are shown. In all four models a 171-base sequence (nucleotides 146–317) forms a single bulged stem–loop (SL2) with a predicted free energy of −69.50 kcal/mol. This stem–loop is a highly favored structure, as not only has it a low free energy, but it is also the only structure predicted to form when nucleotides 146–317 are folded in Mfold (data not shown). Stem–loops 1, 3, and 4 are also common to the two models shown in Figure 1. However, although the interactions between the extreme 5′ and 3′ ends of the RNA are similar in both, in Figure 1A an additional long-range interaction (LRI) is formed by exact complementarity between two heptanucleotide sequences 73–79 and 422–428. In Figure 1B, these sequences form two separate stem–loops; one within the R/U5 region and one at the start of the gag coding region. The two models have similar free energies of around −181 kcal/mol.
FIGURE 1.
Minimal free-energy models of the FIV packaging signal RNA. Models were generated by the Mfold server using FIV Petaluma sequence RNA nt 1–511. The two most stable structures are shown. (A) δG = −180.83 kcal/mol; (B) δG = −181.60 kcal/mol.
Biochemical structure probing supports the existence of SL 1–SL 4 in vitro, as well as the R/U5-gag long-range interaction
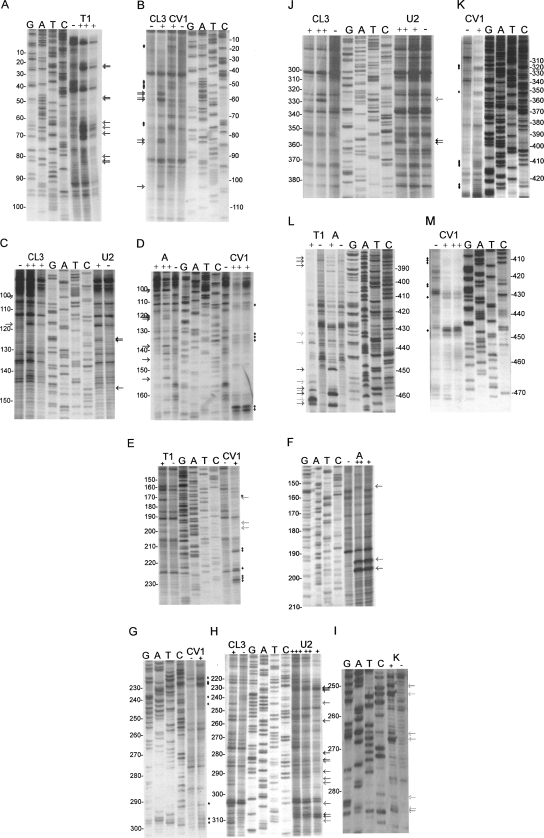
To further examine the structure of the FIV packaging signal, RNA corresponding to nucleotides 1–511 was transcribed in vitro. Transcripts were DNase-treated, purified, and digested with varying amounts of RNase (A, T1, CL3, U2, or CV1) or kethoxal. RNase A cleaves single-stranded (ss) Cytidine and Uracil bases, T1 and kethoxal cleave at ss Guanines, CL3 is specific for ss Cytidine, and in our hands, consistently but with lower avidity, ss Uracil. RNase U2 displays a preference for ss Adenine and to a lesser extent ss Guanine, and CV1 cleaves in, or just outside, helical, or stacked regions without base specificity. Cleavage sites were mapped by primer extension and incorporation of 33P-labeled dATP. 33P-labeled sequencing ladders were generated by cycle sequencing using both the DNA template from which RNAs were transcribed and the primer used in primer extension. Representative examples of ss and double-strand (ds)-specific nuclease digestions for each region of the RNA are shown in Figure 2. Negative control lanes contain cDNAs generated from the RNA without RNase/chemical treatment and show the reverse transcriptase pauses that occur when the enzyme/template interaction is destabilized by homopolymeric runs of nucleotides, or the formation of secondary structures ahead of or behind the catalytic site (Klarmann et al. 1993; Harrison et al. 1998; Klasens et al. 1999). Cleavage sites were defined as positions with more RT pausing product in the RNase/kethoxal-positive samples than in the negative control, and are shown in Figure 2 as arrows (ss cleavages) or diamonds (CV1). These data were then used to refine the secondary structural model of the region (Fig. 3), first as constraints in the Mfold algorithm, and then manually, to model the regions of the structure where both ss and ds cleavages were observed. It is possible that RNA molecules might adopt different conformations once they have been cleaved, and further cleavages would therefore not be indicative of the native structure. Hence, we used as structural constraints only cleavage sites that were observed with minimal concentrations of RNase, and were consistently seen in multiple independent experiments. The data used as constraints are shown in gray or yellow arrows (ss constraints) or black or yellow diamonds (ds constraints) in Figure 3. Orange arrows and diamonds in Figure 3 illustrate cleavages observed in Figure 2 but not always visible above background levels in repeat experiments. These are likely to represent sites of weak enzyme affinity or anomalous results and were not used as structural constraints for free energy minimization prediction.
FIGURE 2.
Biochemical analysis of the FIV packaging signal region RNA. FIV Petaluma strain nt 1–511 were transcribed in vitro and were digested with RNases or chemicals specific for ss (A, T1, CL3, U2, kethoxal) or ds (CV1) nucleotides. Primers were annealed to the RNA and cDNAs were made with AMV RT, incorporating 33P-dATP. Samples were separated on 10% polyacrylamide gels, alongside cycle sequencing ladders. Sequence is given in the genome sense for clarity. Different primers were used to examine different regions of the RNA. These regions were: A, B: R/U5; C, D: SL1 and PBS; E, F, G, H, I: SL2; J, K: SL3, SL4, gag; L, M: gag. Numbers represent FIV nucleotide numbers shown in Figure 3. Various concentrations of each RNase/chemical or incubation time were used; only lanes where RNAs were minimally cleaved are shown. These were: A, RNase T1 0.2/1 U; B, RNase CL3 1 U and RNase CV1 2 min; C, RNase CL3 0.1/1 U, RNase U2 0.1 U; D RNase A 0.2/2 pg, RNase CV1 30 sec/2 min; E, RNase T1 0.1 U, CV1 30 sec; F, RNase A 0.2/2 pg; G, RNase CV1 5 sec; H, RNase CL3 1 U, RNase U2 0.01/0.1/ 0.5 U; I, kethoxal 200 μg; J, RNase CL3 0.1/0.2 U, RNase U2 0.1/0.2 U; K, RNase CV1 2 min; L, RNase T1 0.1 U, RNase A 0.2 pg; M, RNase CV1 30 sec/2 min. RNase cleavage sites are indicated with arrows (ss nucleases, colored light to dark gray in the order T1, CL3, A, U2) or diamonds (CV1).
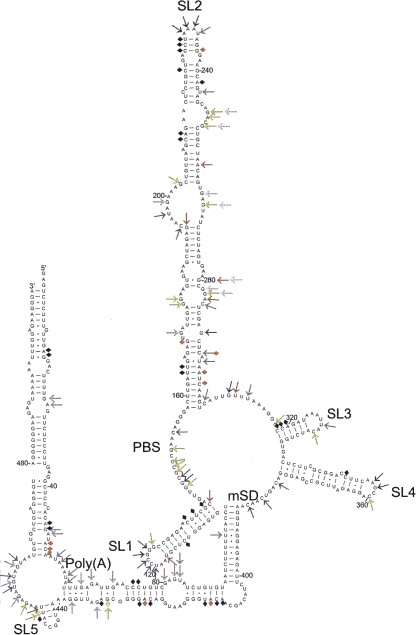
FIGURE 3.
Secondary structural model of the FIV packaging signal RNA. Model generated based on minimal free energy as determined by Mfold, and biochemical mapping constraints. Arrows represent cleavage sites for ss RNases A, T1, CL3, or U2 or kethoxal. Diamonds represent CV1 cleavage sites. White, gray, and black indicate cleavage sites shown in Figure 2 and observed in multiple independent experiments (shades of gray represent different RNases). Yellow represents cleavage sites not shown in Figure 2 but observed in multiple independent experiments. Orange shows cleavage sites shown in Figure 2 but not always observed in repeat experiments.
As shown in Figure 3, the biochemical data strongly support the RNA base pairing of the free-energy minimized model displayed in Figure 1A including long-range interactions between the R/U5 region and the start of gag, and also the stem–loops 1–5. The base of SL2 is not paired, and this leaves much of the primer binding site (nucleotides 142–159) accessible for annealing of the tRNA primer in vivo. Apart from this region at the base of SL2, biochemical probing supports the Mfold prediction of SL 1–SL 4 shown in Figure 1, with the exception of a paired region between two internal loops of SL2 which instead forms a large, purine-rich internal loop (nucleotides 194–203 and 263–269). The biochemical data, in particular CV1 cleavage sites at nucleotides 74 and 75 and 424–426 (Fig. 2B,K), suggest that the R/U5 and gag regions interact in a way similar to that predicted in Figure 1A. A palindromic stem–loop (SL5) forms in the gag coding region instead of the two small stem–loops shown in Figure 1A. The CV1 cleavages in this region might result from dynamic dimerization of a small proportion of the RNA at this stem–loop. Native agarose gel analysis revealed a very minor amount of RNA of a size consistent with a dimer (not shown), but the exact nature of this species was not analyzed separately due to its small quantity.
Although the RNase digestions suggest that most areas of the structure exist in an exclusively single-stranded or paired form, there are two areas of the structure in which the data are consistent with conformational flexibility. Nucleotides 61–72 and 429–439 are cleaved by both ss and ds nucleases, and U100 is cleaved by ss nucleases, but is in the middle of a paired region. The presence of a number of G–U pairs within both of these regions would also be consistent with helices of lower stability.
Further chemical probing experiments (kethoxal and CMCT) covering much of the remaining 511 nt are in agreement with this model (data not shown).
The secondary structural model is supported by sequence conservation and base-pair covariation between different FIV isolates
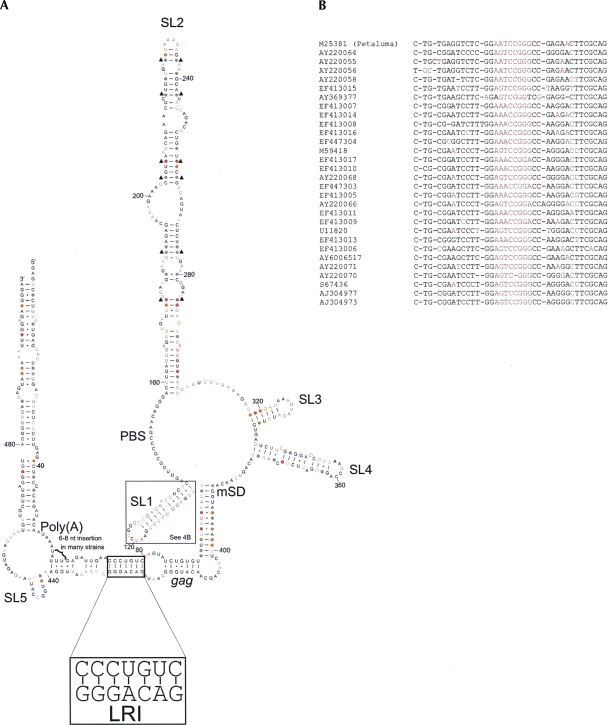
In order to examine sequence conservation within the FIV packaging signal region, a sequence alignment of 77 FIV isolates from domestic cats was produced by discontinuous megablast of FIV Petaluma nucleotides 1–511 (Supplemental Fig. 1). Sequence conservation relative to the Petaluma strain was calculated at each nucleotide position and is shown in color in Figure 4. In general, there is a low degree of sequence conservation within the loops of SL1–SL 4, and within internal loops of SL2. Where isolates vary in sequence within paired regions, the structure is often conserved by substitution of a G–C or A–U pair for a G–U pair or vice versa. This is illustrated in Figure 4 by dots in place of nucleotides. In some instances, both nucleotides vary within the same isolate, leading to maintenance of canonical base pairing and maintaining bond strength. Where this occurs in most or all mutants, it suggests that there is strong selective pressure on the virus to maintain structural integrity at that location. Nucleotide pairs for which this occurs are indicated with black triangles. Structural conservation is evident particularly in the stem regions of SL2, at the base of SL3, and within the paired region between nucleotides 92–105 and 388–401. In many instances where a base pair is not conserved a nearby internal loop or bulge is closed, maintaining structural stability. One example of this is shown on Figure 4A (SL4, open circles), although this also occurs in other locations, notably at the A–A bulge at the base of SL2 (see Supplemental Fig. 1 for details). Nucleotide insertions are not shown on Figure 4; where these occur they are generally confined to a small number of strains. An exception is found around nucleotides 60–70, where many isolates have a 6–8 nt insertion. Although sequence conservation of SL1 is low, structural conservation is high, as all isolates are able to form a stem of ∼12 base pairs with one or two internal loops or bulges, as shown in Figure 4B.
FIGURE 4.
Sequence conservation and base-pair covariation in FIV leader RNA. (A) Secondary structural model of the 5′ end of the FIV Petaluma strain RNA showing sequence conservation between 77 isolates (see Supplemental Fig. 1 for details). Nucleotides in black are 100% conserved, those in gray are >95% conserved, those in orange are 75%–95% conserved, and those in pink are <75% conserved between isolates. Filled circles indicate nucleotides which vary in sequence between isolates but form a Watson-Crick or G–U pair in >95% of mutated isolates, indicating structural conservation. Open circles on SL4 indicate structural conservation so that one base-pair and one internal loop are always present. Black triangles indicate base pairs where both nucleotides are substituted in the same isolate and canonical base pairing is maintained. Structural conservation in SL1 is not shown in A. (B) Sequence alignment of the FIV SL1 sequence. The Petaluma SL1 sequence is shown at the top, and sequences of SL1 variant isolates are shown below alongside the accession numbers. Pink represents unpaired nucleotides. Black indicates nucleotides able to form a base pair between the left-hand and the right-hand sequence.
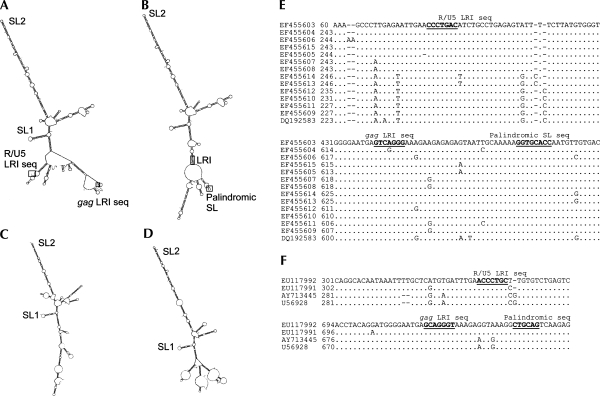
The role of SL1 and SL2 in the viral lifecycle is not understood, although their presence in the predicted structure of the 5′ UTR of related viruses, FIVPco (Cougar lentivirus, accession number EF455603) (Fig. 5A,B), FIVPleo (Lion lentivirus, accession number EU117992) (Fig. 5C), and FIVOma (Pallas' cat lentivirus, accession number U56928) (Fig. 5D) suggests that they are of vital importance to strains of FIV that infect both large and small cats. The ability to form a long-range interaction between sequences in R/U5 and gag is also conserved in FIV strains that infect these other feline species, where the heptanucleotide sequences vary slightly within strains, but also between FIVPetaluma and FIVPco and FIVPleo/Oma, always maintaining complementarity (Fig. 5E,F). Mfold models suggest that when this LRI occurs in FIVPco, a small palindromic stem–loop forms within gag (Fig. 5B,E). A palindromic sequence is found in a similar location in FIVOma and FIVPleo gag sequences (Fig. 5F).
FIGURE 5.
SL1, SL2, and the LRI sequence is conserved among FIV strains that infect other feline species. (A) Mfold structural prediction of the 5′ end of the Cougar lentivirus genome (accession number EF455603). (B) Mfold structural prediction of the 5′ end of the Cougar lentivirus genome with LRI paired. (C) Mfold structural prediction of the 5′ end of the FIVPleo genome. (D) Mfold structural prediction of the 5′ end of the FIVOma genome. (E) Sequence alignment of the 5′ end of FIV strains that infect cougars, showing conservation of LRI and palindromic regions. (F) Sequence alignment of the 5′ end of FIV strains that infect Pallas' cats (AY713445 and U56928) and lions (EU117991 and EU117992), showing conservation of LRI and palindromic regions.
SL1 and SL2 are present in spliced as well as genomic RNA
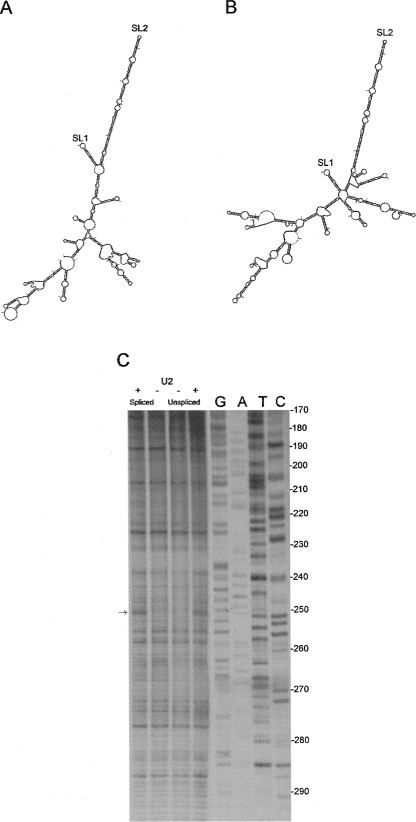
To examine which areas of the structure might distinguish between spliced and genomic RNA, and could therefore contribute to packaging signal specificity, spliced RNAs were examined in silico. Mfold structural predictions were used to visualize the FIV Petaluma 5′ UTR nucleotides 1–385 spliced either to orf A (Fig. 6A), vif (Fig. 6B), or rev (data not shown) ORFs. SL2 is predicted to be present in all three cases. SL1 is predicted to form in orf A and vif RNA, but not rev RNA, which may be of significance. As SL2 has previously been postulated to act as a packaging signal (Browning et al. 2003a), we undertook a biochemical analysis of FIV leader RNA that terminated at the mSD (nucleotides 1–381) or that included the mSD and the start of the gag ORF (nucleotides 1–484). Figure 6C shows an RNase U2 digestion of these two RNAs. An identical nuclease cleavage site is visible in both at A250. Crucially, the RT pausing pattern, which is dependent on the secondary structure of the RNA, is identical in spliced and genomic RNA, suggesting that SL2 is present in both.
FIGURE 6.
SLs 1 and 2 are present within spliced and unspliced RNA. Mfold structural predictions of FIV Petaluma 5′ UTR spliced to orf A (A) and vif (B). (C) Biochemical analysis of the SL2 region of “spliced” (nucleotides 1–381) versus “unspliced” (nucleotides 1–511) RNA. In vitro transcribed RNAs were digested with 0 U (−) or 0.2 U (+) RNase U2 and primer extension was performed, incorporating 33P-dATP and using a primer that annealed 3′of SL2. cDNAs were separated on a 10% polyacrylamide gel alongside a sequencing ladder that was generated using the same primer. DNA sequence is given in the genome sense for clarity.
DISCUSSION
We have investigated the structure of the 5′ UTR of FIV by means of minimal free energy, biochemical, and phylogenetic analyses, and shown that the first 511 nt of the genomic transcript fold into a complex secondary structure, containing five stem–loops stabilized by long-range interactions.
Mfold analyses suggest that the 5′ UTR of the FIV RNA can fold into two different structures with similar free energies. However we have shown by in vitro biochemical assays and phylogenetic analyses that the 5′ UTR forms a structure similar to that shown in Figure 1A, with four stem–loops, and an additional small stem–loop toward the start of gag. Much of the 5′ and 3′ region of this RNA is involved in LRIs, rather than forming R/U5 and Gag SLs as seen in Figure 1B.
The biochemical evidence for this structure, obtained by RNase mapping experiments using ss-specific RNases T1, A, CL3, and U2 and CV1 to map paired regions, strongly supports the structure in Figure 1A. Almost all of the cleavage sites observed map within expected regions, as shown in Figure 3. Nearly all ss-specific cleavage sites are within ss regions, or are at the closing pairs of loops or bulges. The only exceptions are within G–U-rich regions, which may exhibit conformational flexibility, and within the gag SL, where CV1 cleavages are also found. However, as discussed below, the findings would be compatible with conformational mutability involved in initiation of RNA dimerization.
CV1 cleavages map within base-paired regions, and mainly to helices of 4 or more pairs, as expected. CV1 has been postulated to recognize the conformation of the sugar-phosphate backbone, and hence, is also able to cleave unpaired nucleotides, either when the bases are stacked, or when two or more short helices are joined by small internal loops or bulges (Lowman and Draper 1986). This may be the case for the cleavages seen at nucleotidess 51–53, 170, 293, 227–229, and 236.
An important feature of the FIV 5′ RNA secondary structure is the conserved heptanucleotide LRI between the R/U5 junction and the start of gag. We postulate that this interaction occurs on the basis of the biochemical, phylogenetic, and in silico modeling analysis presented. A similar heptanucleotide LRI sequence has also been observed in HIV-1, HIV-2, and SIV (Paillart et al. 2002). Without this interaction in FIV, the structural characteristics of both R/U5 and gag would change and would be likely to form stem–loops as in Figure 1B. Interestingly, deletions within R and/or U5, as well as at the start of gag have previously been shown to lower encapsidation efficiency of FIV (Kemler et al. 2002). Specifically, deleting R alone, U5 alone, or R and U5 together led to similar relative encapsidation efficiency (REE) of around 0.25 compared with a native FIV (Kemler et al. 2002). As the R/U5 junction is at nucleotides 75–76, which is in the middle of this LRI, deletion of either R or U5 would destabilize it and would be expected to have an impact upon the surrounding RNA structure. In support of this, deletions of as little as 13 nt at the 5′ of gag, which includes 3 nt of the LRI, gave a threefold reduction in REE (Kemler et al. 2002). This situation may be similar to that seen in HIV-1, where a LRI between the R/U5 region and the start of gag stabilizes a secondary structure of the intervening region that is important for virus dimerization and packaging (Abbink and Berkhout 2003; Song et al. 2008).
RNA encapsidation is initiated by binding of the viral Gag polyprotein to single-stranded regions of the packaging signal structure. The 5′ end of the FIV RNA involves many long-range interactions but contains few single-stranded regions that are conserved. This gives us important clues as to the viral life cycle and the packaging process. Previously, it has been shown that several regions of the RNA contain important encapsidation determinants (Kemler et al. 2002; Browning et al. 2003b; Kemler et al. 2004; Ghazawi et al. 2006). In addition to R/U5 and the beginning of gag, these include SL1 and most of SL2. However, there are a limited number of conserved single-stranded regions in the first 100 nt, and the presence of SL1 and SL2 in spliced as well as genomic RNAs would seem to preclude them from acting alone as specific packaging determinants. The strong implication is that, like HIV-2 (Griffin et al. 2001), FIV employs a cotranslational packaging mechanism. The close proximity of the packaging signal to the gag initiation codon raises the problem encountered by all lentiviruses of competition between Gag binding leading to packaging and ribosome scanning for translational initiation, and this has been clearly shown for HIV-1 (Anderson and Lever 2006). Interestingly, in studies on FIV packaging there is a suggestion that leaving a longer region of the gag sequence intact may decrease packaging from an optimum level, suggesting that a similar competitive effect may be occurring and further supporting the cotranslational model (Kemler et al. 2002). We favor this idea, as the regions containing SL1 and SL2, and in particular the tip region of SL2, were previously shown to aid encapsidation (Browning et al. 2003a; Mustafa et al. 2005), and are highly conserved structures, as seen in other packaging signals. The lack of conserved sequences or structures confined to genomic but not spliced RNAs lends further support to a cotranslational packaging model for FIV. This is clearly an important issue if FIV is to be used efficiently and safely as a gene therapy vector.
From examination of the secondary structure, as has been suggested for some other retroviruses, FIV may select a dimeric genome for packaging. A dimerization initiation site has not previously been postulated in the FIV leader region. Such sites tend to be palindromic sequences or GACG tetraloops, neither of which are conserved within the 5′ UTR. However our proposed secondary structure of the FIV 5′ RNA does contain a stem–loop within the first 100 nt of the gag ORF (SL5), which is a palindromic sequence (AAUGGCCAUU), the central 6 nt of which are virtually 100% conserved (one nucleotide substitution in 63 published sequences, possibly a sequencing error). A palindrome of different sequence but similar length is also found in strains of FIV that infect other feline species such as pumas and Pallas' cats. If FIV dimerization is initiated by a site within gag and packaging is dependent on genome dimerization, this would confer high specificity packaging limited exclusively to the unspliced viral RNA. Previously it has been shown that the FIV genome requires nucleocapsid to chaperone dimerization (Moscardini et al. 2002). However, these experiments were performed using only nucleotides 1–358, and none of gag. If the dimerization initiation site lies within gag, then the requirement for NC may be less stringent.
Although the sequence of SL 1–SL 4 varies between FIV isolates, their structures remain conserved, as shown in Figure 4. This is particularly evident for SL1 and SL2. The sequence of SL1 is variable between isolates, but consistently forms a stem–loop of 12–14 base pairs with one or more bulged nucleotides and a 6–8 nt loop with consensus sequence AG/AT/ACC/ΔGGG/A. The tip of SL1 may act as a protein docking site, with the bulged nucleotide(s) contributing to the metastability of the stem, unwinding it and enabling further protein–RNA interactions. Although the sequence of SL2 is variable, the overall structure is maintained between isolates, as shown by the base-pair covariation observed. The approximate sizes and positions of the internal loops and bulges are also maintained, although much of their sequence is not. This suggests that it is the structure of SL2 rather than its sequence that is functionally important. Such a large stem–loop structure would be a formidable energetic barrier for proteins that scan or unwind the RNA. In addition to acting as a potential packaging signal, it could also function as an IRES, such as that recently proposed within the FIV 5′ UTR (Camerini et al. 2008), or it may act to stabilize the transcripts. Alternatively, it might act as a cellular transport signal, as there is a precedent for large RNA stem–loops to do so (Pasquinelli et al. 1997).
Although there is some phylogenetic evidence for the structural conservation of SL 3 and 4, previous packaging assays have shown that the region is not important for encapsidation (Browning et al. 2003a; Mustafa et al. 2005), and hence, they are likely to play another, as yet undetermined, role in the viral life cycle. The LRI between nucleotides 92–105 and 388–401 is also structurally conserved, although deletions between the mSD and the gag AUG, which would delete most of the right-hand side of this interaction, have minimal effects on encapsidation (Kemler et al. 2002; Browning et al. 2003a), again suggesting an alternative role for this region.
During the preparation of this manuscript an alternative structure for the FIV 5′ RNA was published (James and Sargueil 2008). As this study differed from ours in the length of transcript used and its method of preparation (denaturing and refolding rather than directly from transcription), it is possible that differences in secondary structure do exist due to kinetic folding traps or global energetic phenomena. However, their phylogenetic analysis did not identify the conserved complementary heptanucleotide sequences (Fig. 4) and, partly because of this, did not recognize the potential LRI in the RNA. The majority of their biochemical probing is completely consistent with the structure we present here, and indeed, they find CV1 cleavages consistent with the heptanucleotide duplex which stabilizes the LRI. In addition, all of the cleavage data presented for nucleotides 158–303 maps accurately onto our proposed SL2 with the exception of 3 single-strand cleavage sites at the base. Some aspects of their structure coincide with our own, including SL1; however, we disagree with much of their interpretation of the available data. Among other concerns we note that in our phylogenetic analysis we used only validated individual isolates, whereas James and Sargueil (2008) have analyzed a data set containing identical clones of several isolates which will skew any consensus toward the overrepresented strains. They identify a palindromic sequence “GAGCTC” in the UTR which is labeled upon their model as a dimer intitation site (DIS), yet it is <90% conserved and there is no mention of the much more highly conserved palindrome “UGGCCA” ∼30 nt downstream of the gag AUG, which is presented on a stem–loop in the LRI model. The fact that they do not find dimeric RNA when using a gag sequence alone but do when the 5′ UTR and gag are expressed is consistent with the palindrome we have identified only being presented for a kissing loop interaction when the full-length UTR + gag RNA folds into the LRI structure.
Despite a low sequence identity between FIV and other lentiviruses, the model that we propose forms a highly ordered structure that is stabilized by long-range interactions between R/U5 and gag, similar to that observed in primate lentiviruses. This is an example of how the study of FIV can identify RNA sequences and structures that are common to all lentiviruses and are, hence, important potential drug targets. FIV contains fewer accessory genes than HIV and SIV, and the relative simplicity of its genome might aid our understanding of individual lentiviral processes, and the minimal requirements for efficient viral replication. Thus, it presents a very valuable model to study lentiviral biology.
MATERIALS AND METHODS
In silico analyses of FIV RNA
Secondary structural analysis of FIV RNAs was performed using the Mfold server (Mathews et al. 1999; Zuker 2003) and standard constraints, unless specified. Phylogenetic comparisons were made using discontinuous megablast (http://blast.ncbi.nlm.nih.gov/Blast.cgi) of part or all of FIV Petaluma strain nucleotides 216–727 (Talbott et al. 1989), FIVPco nucleotides 184–751 (Bruen and Poss 2007), or FIVPleo accession number EU117992 nucleotides 262–802 (Pecon-Slattery et al. 2008). A consensus sequence was taken for molecular clones that originated from the same viral isolate.
Preparation of in vitro transcribed RNAs
DNA templates containing the T7 RNA polymerase promoter at the 5′ end were prepared by PCR using 1× Biomix Red (Bioline), 2 μM each primer, and 0.5 μg plasmid TR394 (Browning et al. 2001). Primers used were 1 (5′-GACTTTAATACGACTCACTATAGGAGTCTCTTTGTTGAGGAC-3′) 484 (5′-CCCCTACTCCTACAGCAAC-3′) 381 (5′CTGTCCCTCGGCGAATCTC-3′) 511 (5′-TCCCTTCTCCAAATTTTTTAC-3′). Numbers correspond to the FIV nucleotide to which the 5′ end of the primer anneals (with the exception of 1, which has a T7 RNA polymerase promoter at the 5′ end). FIV nucleotides are numbered according to the transcription start site (1 = nucleotide 216 from Talbott et al. 1989). The size and purity of PCR products was assessed by electrophoresis on 1% agarose gels. The remainder of each sample was purified using a Qiagen PCR purification kit in accordance with the manufacturer's instructions. In vitro transcriptions were performed in 20 μL and contained 7.5 mM dNTPs, 1× buffer (Megashortscript buffer, Ambion), 2 μg template DNA, and 2 μL T7 RNA polymerase (Megashortscript enzyme mix, Ambion) for 4 h at 37°C. DNA templates were degraded with 4 U DNase (turboDNase, Ambion) for 20 min at 37°C. The size and integrity of RNA transcripts was verified by electrophoresis on 1% agarose gels. RNA was purified on filter cartridges (MegaClear, Ambion). RNAs were subjected to enzymatic cleavage immediately.
Enzymatic and chemical cleavage of RNAs
Two micrograms in vitro transcribed RNA were digested with RNase CV1 (0 or 0.1 U), T1 (0–1 U), A (0–50 pg; all from Ambion), or CL3 (0–0.2 U; Industrial Research Laboratories [IRL]) in 10 μL 1× structure buffer (Ambion) or with RNase U2 (0–0.2 U, IRL) in 20 mM sodium acetate, pH 4.8, 2 mM MgCl2, 100 mM KCl, or structure buffer (Ambion), as above. Digestions contained 0.1 μg/μL yeast tRNA (Ambion). RNase T1, CL3, U2, and A digestions took place at 37°C for 10–15 min and CV1 digestions were incubated at 37°C for 5 sec to 8 min. Reactions were terminated with 20 μL inactivation/precipitation buffer (Ambion) followed by overnight incubation at −20°C. Kethoxal digests contained 2 μg of RNA and 200−400 μg kethoxal in 70 mM HEPES–KOH pH 7.8, 10 mM MgCl2, 270 mM KCl, and 2 mM DTT. Tubes were incubated at 30°C for 10 min and reactions were terminated with 10 μg yeast tRNA, 0.4 M sodium acetate pH 6, 40 mM boric acid, and 3 volumes of ethanol and storage at −20°C overnight. Chemically or enzymatically cleaved RNAs were then washed in 70% ethanol, air dried, and resuspended in 6 μL H2O.
Reverse transcription and cycle sequencing assays
Primer (50 mM) was added to 0.67 μg RNA in 15 mM Tris pH 7.5, 25 mM KCl, and was annealed to the RNA by heating to 72°C and cooling slowly to 42°C. The reaction volume was 4 μL and the primers used were 511 (as above), 484 (as above), 381 (as above), 283 (5′-CCGCTTCACTAGAGATACTCA-3′), 252 (5′-CGTCTGCTACTGCTTCCCT-3′), 199 (5′-CTATTGCTCTAGCTTCACTTCC-3′), 146 (5′-CGCCAACTGCGAAGTTCTCGG-3′), or 125 (5′-CCCGGATTCCGAGACCTCAC-3′). Reverse transcription was performed at 42°C for 30 min by the addition of 6 μL of 25 mM Tris pH 8.3, 50 mM KCl, 12.5 mM MgCl2, 10 mM DTT, 0.5 mM dTTP, 0.5 mM dGTP, 0.5 mM dCTP, 5 nM dATP, 0.4 μCi/μL α-33PdATP (Perkin-Elmer), 1.6 U/μL RNasin (Promega), 1 U/μL AMV RT (Promega). Sequencing ladders were prepared with the fmol cycle sequencing system (Promega) using the same DNA templates as the in vitro transcriptions and the same primers as the reverse transcriptions, and 5 μCi α-33P dATP per 10 μL reaction. Ten microliters of formamide-containing loading dye (gel-loading buffer II, Ambion) were added to all samples before heating to 80°C for 5 min and separating by electrophoresis on 10% acrylamide, 7.5 M urea gels (Sequagel, National Diagnostics) in 1× TBE (89 mM Tris base, 89 mM boric acid, 2 mM EDTA). Gels were dried on Whatmann 3MM paper and were visualized by autoradiography for 6 h to 14 d at −70°C.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Chi Ngai Chan for help in setting up the project. This work was funded by grants 078007/Z/05/Z and 080984/Z/06/Z from the Wellcome Trust, the Charles and Elsie Sykes Trust, and the Biomedical Research Centre.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1284908.
REFERENCES
- Abbink T.E., Berkhout B. A novel long-distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003;278:11601–11611. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- Anderson E.C., Lever A.M. Human immunodeficiency virus type 1 Gag polyprotein modulates its own translation. J. Virol. 2006;80:10478–10486. doi: 10.1128/JVI.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning M.T., Schmidt R.D., Lew K.A., Rizvi T.A. Primate and feline lentivirus vector RNA packaging and propagation by heterologous lentivirus virions. J. Virol. 2001;75:5129–5140. doi: 10.1128/JVI.75.11.5129-5140.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning M.T., Mustafa F., Schmidt R.D., Lew K.A., Rizvi T.A. Delineation of sequences important for efficient packaging of feline immunodeficiency virus RNA. J. Gen. Virol. 2003a;84:621–627. doi: 10.1099/vir.0.18886-0. [DOI] [PubMed] [Google Scholar]
- Browning M.T., Mustafa F., Schmidt R.D., Lew K.A., Rizvi T.A. Sequences within the gag gene of feline immunodeficiency virus (FIV) are important for efficient RNA encapsidation. Virus Res. 2003b;93:199–209. doi: 10.1016/s0168-1702(03)00098-4. [DOI] [PubMed] [Google Scholar]
- Bruen T.C., Poss M. Recombination in feline immunodeficiency virus genomes from naturally infected cougars. Virology. 2007;364:362–370. doi: 10.1016/j.virol.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini V., Decimo D., Balvay L., Pistello M., Bendinelli M., Darlix J.L., Ohlmann T. A dormant internal ribosome entry site controls translation of feline immunodeficiency virus. J. Virol. 2008;82:3574–3583. doi: 10.1128/JVI.02038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix J.L., Gabus C., Nugeyre M.T., Clavel F., Barre-Sinoussi F. cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Mol. Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- D'Souza V., Melamed J., Habib D., Pullen K., Wallace K., Summers M.F. Identification of a high affinity nucleocapsid protein binding element within the Moloney murine leukemia virus Psi-RNA packaging signal: Implications for genome recognition. J. Mol. Biol. 2001;314:217–232. doi: 10.1006/jmbi.2001.5139. [DOI] [PubMed] [Google Scholar]
- Elder J.H., Sundstrom M., de Rozieres S., de Parseval A., Grant C.K., Lin Y.C. Molecular mechanisms of FIV infection. Vet. Immunol. Immunopathol. 2008;123:3–13. doi: 10.1016/j.vetimm.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazawi A., Mustafa F., Phillip P.S., Jayanth P., Ali J., Rizvi T.A. Both the 5′ and 3′ LTRs of FIV contain minor RNA encapsidation determinants compared to the two core packaging determinants within the 5′-untranslated region and gag. Microbes Infect. 2006;8:767–778. doi: 10.1016/j.micinf.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Greatorex J. The retroviral RNA dimer linkage: Different structures may reflect different roles. Retrovirology. 2004;1:22. doi: 10.1186/1742-4690-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin S.D., Allen J.F., Lever A.M. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: Cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 2001;75:12058–12069. doi: 10.1128/JVI.75.24.12058-12069.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison G.P., Mayo M.S., Hunter E., Lever A.M. Pausing of reverse transcriptase on retroviral RNA templates is influenced by secondary structures both 5′ and 3′ of the catalytic site. Nucleic Acids Res. 1998;26:3433–3442. doi: 10.1093/nar/26.14.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isel C., Marquet R., Keith G., Ehresmann C., Ehresmann B. Modified nucleotides of tRNA3 Lys modulate primer/template loop–loop interaction in the initiation complex of HIV-1 reverse transcription. J. Biol. Chem. 1993;268:25269–25272. [PubMed] [Google Scholar]
- James L., Sargueil B. RNA secondary structure of the feline immunodeficiency virus 5′ UTR and Gag coding region. Nucleic Acids Res. 2008;36:4653–4666. doi: 10.1093/nar/gkn447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemler I., Azmi I., Poeschla E.M. The critical role of proximal gag sequences in feline immunodeficiency virus genome encapsidation. Virology. 2004;327:111–120. doi: 10.1016/j.virol.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Kemler I., Barraza R., Poeschla E.M. Mapping the encapsidation determinants of feline immunodeficiency virus. J. Virol. 2002;76:11889–11903. doi: 10.1128/JVI.76.23.11889-11903.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarmann G.J., Schauber C.A., Preston B.D. Template-directed pausing of DNA synthesis by HIV-1 reverse transcriptase during polymerization of HIV-1 sequences in vitro. J. Biol. Chem. 1993;268:9793–9802. [PubMed] [Google Scholar]
- Klasens B.I., Huthoff H.T., Das A.T., Jeeninga R.E., Berkhout B. The effect of template RNA structure on elongation by HIV-1 reverse transcriptase. Biochim. Biophys. Acta. 1999;1444:355–370. doi: 10.1016/s0167-4781(99)00011-1. [DOI] [PubMed] [Google Scholar]
- Lever A.M. HIV-1 RNA packaging. Adv. Pharmacol. 2007;55:1–32. doi: 10.1016/S1054-3589(07)55001-5. [DOI] [PubMed] [Google Scholar]
- Lever A., Gottlinger H., Haseltine W., Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J. Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial M., Medeiros E., Hayward W.S. An avian oncovirus mutant (SE 21Q1b) deficient in genomic RNA: Biological and biochemical characterization. Cell. 1978;15:1371–1381. doi: 10.1016/0092-8674(78)90062-4. [DOI] [PubMed] [Google Scholar]
- Lowman H.B., Draper D.E. On the recognition of helical RNA by cobra venom V1 nuclease. J. Biol. Chem. 1986;261:5396–5403. [PubMed] [Google Scholar]
- Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- McCann E.M., Lever A.M. Location of cis-acting signals important for RNA encapsidation in the leader sequence of human immunodeficiency virus type 2. J. Virol. 1997;71:4133–4137. doi: 10.1128/jvi.71.5.4133-4137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscardini M., Pistello M., Bendinelli M., Ficheux D., Miller J.T., Gabus C., Le Grice S.F., Surewicz W.K., Darlix J.L. Functional interactions of nucleocapsid protein of feline immunodeficiency virus and cellular prion protein with the viral RNA. J. Mol. Biol. 2002;318:149–159. doi: 10.1016/S0022-2836(02)00092-X. [DOI] [PubMed] [Google Scholar]
- Mustafa F., Ghazawi A., Jayanth P., Phillip P.S., Ali J., Rizvi T.A. Sequences intervening between the core packaging determinants are dispensable for maintaining the packaging potential and propagation of feline immunodeficiency virus transfer vector RNAs. J. Virol. 2005;79:13817–13821. doi: 10.1128/JVI.79.21.13817-13821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted R.A., Hirsch V.M., Purcell R.H., Johnson P.R. Nucleotide sequence analysis of feline immunodeficiency virus: Genome organization and relationship to other lentiviruses. Proc. Natl. Acad. Sci. 1989;86:8088–8092. doi: 10.1073/pnas.86.20.8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillart J.C., Skripkin E., Ehresmann B., Ehresmann C., Marquet R. In vitro evidence for a long-range pseudoknot in the 5′-untranslated and matrix coding regions of HIV-1 genomic RNA. J. Biol. Chem. 2002;277:5995–6004. doi: 10.1074/jbc.M108972200. [DOI] [PubMed] [Google Scholar]
- Pasquinelli A.E., Ernst R.K., Lund E., Grimm C., Zapp M.L., Rekosh D., Hammarskjold M.L., Dahlberg J.E. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecon-Slattery J., McCracken C.L., Troyer J.L., VandeWoude S., Roelke M., Sondgeroth K., Winterbach C., Winterbach H., O'Brien S.J. Genomic organization, sequence divergence, and recombination of feline immunodeficiency virus from lions in the wild. BMC Genomics. 2008;9:66. doi: 10.1186/1471-2164-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C., Ho E.W., Brown M.L., Yamamoto J.K. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science. 1987;235:790–793. doi: 10.1126/science.3643650. [DOI] [PubMed] [Google Scholar]
- Rein A. Retroviral RNA packaging: A review. Arch. Virol. Suppl. 1994;9:513–522. doi: 10.1007/978-3-7091-9326-6_49. [DOI] [PubMed] [Google Scholar]
- Saenz D.T., Poeschla E.M. FIV: From lentivirus to lentivector. J. Gene Med. 2004;6(Suppl)(1):S95–S104. doi: 10.1002/jgm.500. [DOI] [PubMed] [Google Scholar]
- Siebelink K.H., Chu I.H., Rimmelzwaan G.F., Weijer K., van Herwijnen R., Knell P., Egberink H.F., Bosch M.L., Osterhaus A.D. Feline immunodeficiency virus (FIV) infection in the cat as a model for HIV infection in man: FIV-induced impairment of immune function. AIDS Res. Hum. Retroviruses. 1990;6:1373–1378. doi: 10.1089/aid.1990.6.1373. [DOI] [PubMed] [Google Scholar]
- Song R., Kafaie J., Laughrea M. Role of the 5′ TAR stem–loop and the U5-AUG duplex in dimerization of HIV-1 genomic RNA. Biochemistry. 2008;47:3283–3293. doi: 10.1021/bi7023173. [DOI] [PubMed] [Google Scholar]
- Talbott R.L., Sparger E.E., Lovelace K.M., Fitch W.M., Pedersen N.C., Luciw P.A., Elder J.H. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc. Natl. Acad. Sci. 1989;86:5743–5747. doi: 10.1073/pnas.86.15.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer J.L., Pecon-Slattery J., Roelke M.E., Johnson W., VandeWoude S., Vazquez-Salat N., Brown M., Frank L., Woodroffe R., Winterbach C., et al. Seroprevalence and genomic divergence of circulating strains of feline immunodeficiency virus among Felidae and Hyaenidae species. J. Virol. 2005;79:8282–8294. doi: 10.1128/JVI.79.13.8282-8294.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichs an der Glon C., Monks J., Proudfoot N.J. Occlusion of the HIV poly(A) site. Genes & Dev. 1991;5:244–253. doi: 10.1101/gad.5.2.244. [DOI] [PubMed] [Google Scholar]
- Yamamoto J.K., Sparger E., Ho E.W., Andersen P.R., O'Connor T.P., Mandell C.P., Lowenstine L., Munn R., Pedersen N.C. Pathogenesis of experimentally induced feline immunodeficiency virus infection in cats. Am. J. Vet. Res. 1988;49:1246–1258. [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]