Abstract
Genomes of RNA viruses encounter a continual threat from host cellular ribonucleases. Therefore, viruses have evolved mechanisms to protect the integrity of their genomes. To study the mechanism of 3′-end repair in dengue virus-2 in mammalian cells, a series of 3′-end deletions in the genome were evaluated for virus replication by detection of viral antigen NS1 and by sequence analysis. Limited deletions did not cause any delay in the detection of NS1 within 5 d. However, deletions of 7–10 nucleotides caused a delay of 9 d in the detection of NS1. Sequence analysis of RNAs from recovered viruses showed that at early times, virus progenies evolved through RNA molecules of heterogeneous lengths and nucleotide sequences at the 3′ end, suggesting a possible role for terminal nucleotidyl transferase activity of the viral polymerase (NS5). However, this diversity gradually diminished and consensus sequences emerged. Template activities of 3′-end mutants in the synthesis of negative-strand RNA in vitro by purified NS5 correlate well with the abilities of mutant RNAs to repair and produce virus progenies. Using the Mfold program for RNA structure prediction, we show that if the 3′ stem–loop (3′ SL) structure was abrogated by mutations, viruses eventually restored the 3′ SL structure. Taken together, these results favor a two-step repair process: non-template-based nucleotide addition followed by evolutionary selection of 3′-end sequences based on the best-fit RNA structure that can support viral replication.
Keywords: RNA virus, RNA replication, RNA-dependent RNA polymerase, quasi-species, RNA structure
INTRODUCTION
Dengue viruses (DENV), members of the flavivirus genus in the Flaviviridae family, have capped, ∼11-kb plus-strand RNA genomes lacking a poly(A) tail. The 5′- and 3′-untranslated regions (UTRs) are ∼100 and 450 nucleotides (nt) in length, respectively. The UTRs flank a single long ORF encoding a polyprotein that is cleaved to yield three structural and at least seven nonstructural (NS) proteins (Lindenbach and Rice 2003). Flavivirus 3′ UTRs contain stable stem–loop (SL) structures. The most stable SL structure is formed from the 3′-terminal ∼100 nt of the genome (3′ SL) (Brinton et al. 1986; Mohan and Padmanabhan 1991; for review, see Markoff 2003). The 3′ SL plays an essential role in RNA replication (Zeng et al. 1998; Yu and Markoff 2005) and contains potential binding sites for cellular and viral proteins, including NS5, the viral RNA-dependent RNA polymerase (RdRp) (Markoff 2003; Filomatori et al. 2006). The 3′ SL plays an essential role in negative-strand RNA synthesis. The DENV2 3′ SL could not be substituted by West Nile virus (WNV) 3′ SL (Zeng et al. 1998; Yu et al. 2008). Other evidence supports the role of 3′ SL in translation (Holden and Harris 2004; Chiu et al. 2005).
Specific RNA structural elements present in 5′- and 3′-terminal regions including two self-complementary cyclization sequences (CS) are important for viral replication (Khromykh et al. 2001; Lo et al. 2003) and RNA synthesis in vitro (You and Padmanabhan 1999; Ackermann and Padmanabhan 2001). Recently, Alvarez et al. identified a novel secondary structural element, the upstream AUG region, present within 5′- and 3′-terminal regions (5′-3′ UAR), that could potentially base pair to form two mutually exclusive structures, I and II (Alvarez et al. 2005). Mutations that abolish 5′-3′ UAR base-pairing in structure II affected viral replication, suggesting that the UAR in structure II plays an important role in this process.
RNA virus genomes are subject to 3′-end degradation due to activity of a variety of cellular ribonucleases (RNases) (Samuel 2001; Lu et al. 2005). Tomato bushy stunt virus recombination in a model host, yeast Saccharomyces cerevisiae, is enhanced 10- to 50-fold in host XRN1 5-3′ exoribonuclease-negative mutant cells (Serviene et al. 2006). Various housekeeping RNases that are abundantly distributed in intracellular compartments such as lysosomes, endoplastic reticulum, and ribosomes also could potentially act on the 3′ termini of viral RNAs (Irie 1999). Since replication is presumed to initiate from the 3′ end of viral RNAs, even loss of trivial numbers of 3′-end nucleotides may abrogate RNA synthesis. Thus, there is strong selection pressure to favor a mechanism for 3′-end protection and repair, and many RNA viruses have evolved such mechanism(s). For example, 5′- and 3′-terminal SL structures conserved in many positive-strand RNA viral genomes provide a passive defense against nucleases with single-strand specificities. In addition, enzymatic mechanisms for the potential repair of damaged 3′ ends have been suggested for various RNA viruses, including plant viruses such as brome mosaic virus and turnip crinkle virus (Rao et al. 1989; Nagy et al. 1997; Guan and Simon 2000), alphaviruses (Tomar et al. 2006), poliovirus (Andrews et al. 1985; Neufeld et al. 1994), and hepatitis C virus (HCV) (Ranjith-Kumar et al. 2001). However, except for studies in plant viruses, there have been no in vivo studies of 3′-end repair in RNA viruses, including flaviviruses.
In this study, we demonstrate that a 3′-end repair mechanism is functional for DENV2 in mammalian cells. 3′-end deletions introduced in DENV2 RNAs were repaired and viral progenies were produced depending on the length of the deletions. Analysis of 3′-terminal deletion mutants as templates for purified NS5 in in vitro RdRP assays (Ackermann and Padmanabhan 2001) revealed that progressive loss of template activities correlated well with lethal phenotypes of the mutant viral RNAs. Limited deletions that maintain functionality for negative-strand synthesis in vitro were able to restore the loss of nucleotides by 3′-end repair and produce progeny virions in cultured cells. Longer deletions caused a delay in the production of viral progenies, which eventually appeared with some heterogeneity in length and sequences at the 3′ end. This complexity was resolved and a consensus sequence emerged as the predominant species. Our results support a two-step model for 3′-end repair. The first step involves addition of nucleotides in a non-template-based manner by terminal nucleotidyl transferase (TNTase) activity of the viral polymerase. This is followed by selection of predominant species that are capable of viral replication. Moreover, we find that deletions that affect the potential to form structure I caused delayed virus growth, but eventually recovered genomes could form both structures I and II involving 5′-3′ CS and 5′-3′ UAR elements, suggesting that structure I also plays an important role in viral replication.
RESULTS
3′ ends in DENV2 deletion mutant RNAs
Dissection of functional domains of cis-acting elements in biologically active RNAs is made possible by synthesis of these RNA molecules by in vitro transcription of cDNAs linearized by restriction enzymes. Infectivity of viral RNAs produced by in vitro transcription followed by gene transfer into prokaryotic, eukaryotic, or plant cells has been demonstrated for numerous viruses. Both SP6 and T7 bacteriophage RNA polymerases were used for in vitro transcription from cDNAs encoding viral genomes (Mizutani and Colonno 1985; Tabler and Sanger 1985; Sarnow 1989). Often, unique restriction sites located at or in the vicinity of the authentic 3′ terminus of the desired RNA product from cDNA templates are used to linearize the cDNA templates for in vitro transcription.
In this study, we investigated the effects of 3′-end deletions on the ability of mutant RNAs to yield infectious virus. The cDNAs encoding full-length DENV2 were linearized using SacI, EheI, or BcgI restriction enzymes prior to in vitro transcription. The expected 3′ end of each of the in vitro transcripts, the parental DENV2 RNA and the deletion mutant RNAs, differed depending on the site chosen for linearization. For example, SacI-linearized cDNA would produce RNA with an extra G at the 3′ end, whereas EheI-cut cDNA would leave an extra GGC at the 3′ end. The nomenclature we used to describe the input RNAs used in our transfection experiments is described in Materials and Methods. The sequence analysis of in vitro synthesized input RNA shows that there is some additional nucleotide added to the 3′ end (Δ4+GCN, where N is any nucleotide) (see Table 2 below) at day 0, perhaps due to SP6 RNA polymerase, which is known to add nontemplated 3′ nucleotides.
TABLE 2.
Sequence analysis of individual input RNA transcripts and recovered viral genomes
Derivation of infectious virus from deletion-mutant RNAs
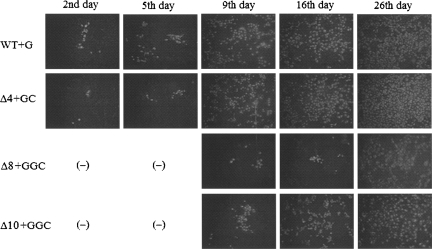
RNA transcripts were transfected into monkey kidney (LLC-MK2) cells. Transfected cells were split at days 2, 7, 14, 21, 28, and so on. An indirect immunofluorescence assay (IFA) specific for DENV2 NS1 was performed on cells at various times post-transfection to monitor virus replication (Fig. 1; Tables 1, 2). Deletion-mutant RNAs could be classified into three groups, based on the time required to detect progeny virions: (1) RNAs produced virus up to day 5 were described as fast-growing phenotype that replicated with kinetics similar to parental RNA by the criterion of IFA, (2) RNAs that produced virus after 7 d or later were grouped as slow-growing, and (3) nonviable RNAs that failed to produce progeny virus even after 58 d of observation. Input RNAs from Δ1+GGC, Δ3+GGC, Δ4+GC, and Δ4+C mutants belong to group 1; and those from Δ7+GGC, Δ8+GGC, and Δ10+GGC mutants to group 2; and finally, those from Δ14+GGC, Δ17+GC, and Δ22+GC mutants to group 3. Group 2 mostly failed to produce viruses, although they occasionally did yield progeny. Therefore, it was difficult to define the exact number of nucleotides that needed to be deleted to distinguish between slow-growing (group 2) and nonviable, or lethal (group 3), phenotypes.
FIGURE 1.
Immunofluorescence assay (IFA) to monitor virus replication. In vitro transcribed RNAs were used to transfect LLC-MK2 cells by electroporation. Cells were stained with a monoclonal antibody specific for DENV2 NS1 protein.
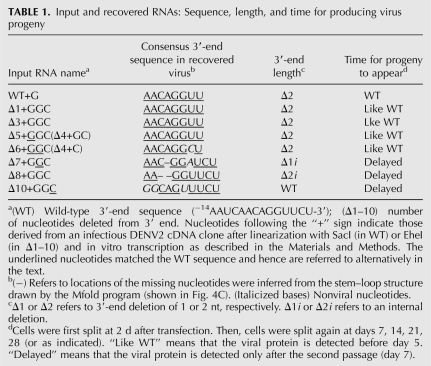
TABLE 1.
Input and recovered RNAs: Sequence, length, and time for producing virus progeny
Direct sequence analysis of the PCR products derived from the 5′ and 3′ ends of input and recovered viral RNAs
Next, we sought to sequence the input RNAs as well as the recovered genome RNAs. For recovered viral RNA, the supernatants were collected periodically (at least twice a week) after RNA transfection. RNAs were decapped, circularized, and then subjected to RT-PCR to amplify a 477-base-pair (bp) linear DNA representing the 5′- to 3′-end junctions (Supplemental Fig. 2A). For wild-type (WT) or fast-growing mutant viruses (group 1), viral RNAs were first obtained for amplification 9–14 d after transfection. For slow-growing (group 2) mutant viruses, RNAs were first obtained 16–40 d post-transfection. The consensus sequences of PCR products in the 3′-end to 5′-end direction consistently became unreadable or yielded mixed sequences at or upstream of the known 3′ end of the genome (see Supplemental Fig. 2B for a representative sequence from input of the Δ4+GC RNA-derived viral genome; data not shown for other mutants and will be provided upon request). However, consensus sequences of PCR products in the 5′ → 3′-end direction were consistently readable in the 5′-terminal region and then became unreadable due to mixing at the 3′ end (Supplemental Fig. 2C). These results indicated that the 5′ end of the DENV2 genome, including the flavivirus-conserved 5′ dinucleotide, AG, was present in all molecules, and the extra 5′ G present in the input RNAs was not observed. The mixed sequences were due to heterogeneity in the nucleotide sequences of 3′ ends in the recovered viral RNAs. Further, this heterogeneity was reduced over time during the course of viral replication, and genomes were gradually restored toward WT in length (Supplemental Fig. 2B).
The input RNAs with intended deletions were also heterogeneous as shown by unreadable sequences in the vicinity of the 3′ end. Sequence profiles of the PCR products from the 5′ end toward the 3′ end were clear until the exact 5′ end containing the extra G, and then became mixed at the 3′ end (data not shown). These results suggested that the SP6 polymerase prematurely terminated and produced RNA transcripts with 3′ ends of variable lengths.
The predominant species of recovered RNAs from the input WT+G, Δ1+GGC, Δ3+GGC, and Δ4+GC RNAs were missing 2 nt at the 3′ end but otherwise retained the consensus WT sequence (Table 1). Those derived from deletions ≥6 nt never reverted completely to WT sequence (Table 1).
5′- and 3′-terminal sequence analysis of cloned PCR products of recovered viruses from the input RNAs of Δ4+GC and Δ10+GGC mutants
Next, we cloned the PCR products to characterize the 5′- and 3′-terminal sequences of heterogeneous populations of RNAs from the recovered viruses. We sequenced more than 20 independent clones from the recovered viruses collected at each time point after transfection of input Δ4+GC and Δ10+GGC RNAs. As controls, the cloned PCR products of input RNAs used for transfection (Table 2, day 0) were also sequenced. For the Δ4+GC mutant, 50% of the input RNAs sequenced contained Δ6 or Δ7 and 25% contained Δ8 or Δ10 compared with the WT RNA sequence (Table 2). Here, it should be noted that all 3′-end sequences were readable because these were derived from cloned PCR products of different populations of RNAs (Supplemental Fig. 2D; data not shown for other mutants).
As shown in Table 2, the cloned PCR products of input RNAs (0 d) or the recovered RNAs derived from the Δ4+GC viruses at days 9 and 13 possessed heterogeneous 3′ ends that included deletions of Δ6 to Δ8; these results are consistent with those obtained from direct sequencing (Supplemental Fig. 2). On the other hand, recovered genomes over time resolved to a predominant species containing Δ2 (Tables 1, 2, cf. day 20 and day 62). However, four out of 21 genomes were longer than the input RNAs at day 9. At later time points (days 20, 27, 34, 48, and 62), the fraction of genomes that were greater in length than the input Δ4+GC RNA gradually increased, and from day 27 onward genomes that were fully WT in sequence but lacking the 3′-terminal CU (Δ2) became the predominant species (Table 2). A variety of mutations that appeared at days 9, 13, and 20 were not observed at later time points (days 27–62) post-transfection. For example, on day 13, one clone had WT genome length but contained a mutation at −4 (U → G) (Table 2). Although genomes WT in length and sequence started appearing at day 20 and were present afterward, they remained a minor fraction of the total population (Table 2). This result was surprising in that the WT RNA that appeared at day 20 did not outcompete with other RNAs as the incubation was prolonged to day 62, and the Δ2 RNA remained as the predominant species. The preference for the appearance of the Δ2 species is not understood at present. However, the results shown in Table 2 also could not be explained by RNA ligase preferences, as the 3′ termini of recovered RNAs were diverse on days 9, 13, and 20. Moreover, we detected WT length (or 1 nt shorter than WT) with a 3′-terminal sequence of UUCU or UUC in a majority of clones derived from input Δ10+GGC RNA (Table 2). Therefore, it is unlikely that this distribution of 3′-end sequences is due to any bias that RNA ligase showed toward a specific terminal sequence or to secondary structure preferences in the circularization of viral RNAs (Romaniuk and Uhlenbeck 1983).
In contrast, the 3′-end sequences of the Δ10+GGC input RNA contained predominantly the intended deletion (Table 2, day 0). However, in a small fraction of molecules, there were deletions or extensions at the 3′ end of GGC, probably caused by SP6 polymerase. After transfection of this Δ10+GGC RNA into LLC-MK2 cells, the repair of deletions occurred with a delay. However, eventually, viral genomes containing WT length became predominant. Forty-one days after transfection into LLC-MK2 cells, 14 out of the 22 independent sequences derived from the 3′ termini of the viral genomes were of WT length or lacked only 1 nt.
By day 62 post-transfection, Δ10+GGC genomes had undergone 3′-end repair and contained shorter deletions; all genomes contained deletions of ≤6 nt but retained GGC (Table 2). The majority of recovered RNAs (17 out of 21 clones) contained UUCU or UUC at the 3′ end and all contained an adaptive mutation at −5 (G → U).
Titration of WT and mutant viruses by a plaque assay
The infectivity of viruses collected from supernatants of cells transfected with WT, Δ4+GC, or Δ10+GGC RNA at regular intervals post-transfection was quantified by a plaque assay using LLC-MK2 cells. The plaque titers (expressed as plaque forming units, pfu, per mL) of the WT virus and virus recovered from Δ4+GC mutant RNA at different time points were 8.0 × 103 pfu/mL for WT at day 20, and 1.2 × 104, 1.1 × 104, 7.5 × 104, and 1.5 × 104 pfu/mL for Δ4+GC RNA at days 13, 20, 27, and 48, respectively. The time periods for the appearance of viral antigen NS1 (Fig. 1) after transfection of WT and Δ4+GC RNAs were also similar. However, for viruses recovered from Δ10+GGC RNA, although these time periods were extended compared with the WT RNA, the virus titers reached 4.0 × 103 and 8.5 × 103 at day 48 and 62, respectively (Fig. 2). The plaque sizes of the Δ4+GC mutant virus (1.25 ± 0.44 mm) at day 13 were smaller in diameter than the WT (2.64 ± 1.06 mm). However, this difference was gradually reduced and the plaque sizes of the Δ4+GC mutant virus (1.25 ± 0.44 mm, 1.86 ± 0.57 mm, 2.41 ± 1.18 mm, and 2.67 ± 0.81 mm at days 13, 20, 27, and 48) or of mutant virus from Δ10+GGC RNA (1.52 ± 0.51 mm and 2.31 ± 0.83 mm at days 48 and 62, respectively) approached close to that of WT (Fig. 2).
FIGURE 2.
Plaque assays of recovered viruses from parental (WT+G) and 3′-deletion mutants (Δ4+GC and Δ10+GGC) input RNAs. Viruses were collected from supernatants on the indicated days and plaque titers were determined. The plaque assays were repeated three times and the plaque titers (pfu/mL) of a representative experiment are given. The average plaque sizes (mm) at indicated days are as follows. WT: day 20: 8.0 × 103, 2.64 ± 1.06 mm. Δ4+GC RNA: day 13: 1.2 × 104, 1.25 ± 0.44 mm; day 20: 1.1 × 104, 1.86 ± 0.57 mm; day 27: 7.5 × 104, 2.41 ± 1.18 mm; day 48: 1.5 × 104, 2.67 ± 0.81 mm. Δ10+GGC: day 48: 4.0 × 103, 1.52 ± 0.51 mm; day 62: 8.5 × 103, 2.31 ± 0.83 mm.
In vitro RdRp assays
According to the current model for viral replication (Westaway et al. 2003), the initiation of negative-strand RNA synthesis by the viral replicase begins at the 3′-terminal sequence of the flavivirus RNA genome. To assess the effects of deletion mutations on the activity of RNAs as templates for RdRp, we performed in vitro RdRp assays as described under in Materials and Methods using subgenomic RNA (sgRNA) templates comprised of the 5′-terminal 230 nt covalently linked to WT and mutant 3′ UTRs of the DENV2 genome and purified NS5 protein (Ackermann and Padmanabhan 2001). With the WT template, the in vitro assay previously was shown to yield two products (Fig. 3). The 1× product represents the de novo synthesized negative-strand RNA. The 2× product is a hairpin form of negative-strand RNA produced by self-priming at the 3′ end of the fold-back structure of template RNA, presumably at the site −8 from the 3′ end due to complementarity between the 3′-terminal UCU and an internal GGA sequence (−5, −6, −7 positions) (Ackermann and Padmanabhan 2001; Nomaguchi et al. 2003).
FIGURE 3.
In vitro template activity of sgRNAs containing WT or 3′-end deletions. sgRNAs containing WT or 3′-end deletions were analyzed for template activities in vitro using purified NS5. (A) sgRNAs containing Δ4+GC (lane 1), Δ8+GGC (lane 2), Δ10+GGC (lane 3), Δ14+GGC (lane 4), Δ17+GC (lane 5), and Δ22+GC (lane 6) were used as templates for negative-strand synthesis. (Lane 7) No RNA control. The products of the reaction were separated by electrophoresis and visualized by autoradiography. (1×) De novo product, (2×) double-stranded hairpin product. Ethidium bromide gel shows approximately equal amounts of sgRNAs used for RdRp assays. Experiments were repeated at least three times and the results of a representative experiment are shown. (B) Densitometric scan of gels were performed, and the sum of 1× and 2× RNAs was compared with WT. Error bars are based on three independent experiments. (C) Densitometric scan of 2× product only, compared with that of WT. (D) Densitometric scan of 1× RNA compared with the WT. (E) In vitro template activity of sgRNAs bearing adaptive mutations identified in recovered genomes. sgRNAs bearing 3′-end sequences, WT and 1–8, were used as templates in the in vitro assays. The RNA products were detected by autoradiography.
The results of in vitro RdRp assays show that Δ4+GC, Δ8+GGC, and Δ10+GGC sgRNAs were about as efficient as WT sgRNA as templates for total RNA synthesis (sum of 1× and 2× RNA products) (see Fig. 3A,B). However, the Δ4+GC, Δ8+GGC, and Δ10+GGC sgRNA templates were progressively less able to prime the synthesis of 2× products (Fig. 3C). This result was possibly because the deleted nucleotides in the mutant template sgRNAs are necessary for self-priming of negative-strand synthesis to produce the hairpin (2×) product. In contrast, these mutant sgRNA templates yielded progressively greater amounts of de novo (1×) RNA products; although the de novo product produced by Δ4+GC sgRNA was reduced by about 50%, the product produced by Δ8+GGC was either essentially at the same or a slightly reduced level compared with that of WT (Fig. 3D). sgRNAs bearing larger deletions (Δ14+GGC, Δ18+GGC, and Δ23+GGC) were markedly reduced in template activity, consistent with lethal phenotypes observed in LLC-MK2 cells transfected with viral RNAs.
In vitro template activity of sgRNAs bearing 3′-terminal adaptive mutations identified in recovered viral RNAs
From Δ10+GGC RNA-transfected LLC-MK2 cells, viruses were eventually recovered that contained WT-length genomes with mutations at the −10 and −9 (A → G) and −5 (G → U) positions (Fig. 3E; Sequence 1). The mutation at −10 (A → G) spontaneously reverted back to the WT (A) upon longer incubation of transfected LLC-MK2 cells, although the mutations at −9 and −5 were still retained (Fig. 3E, Sequence 2). When mosquito (C6/36) cells were infected with the recovered viruses containing all three mutations (at −10, −9, and −5), progeny viral RNAs showed reversions of −10 and/or −9 mutations to WT (Sequences 2, 3, 5) within a shorter time compared with LLC-MK2 cells (Supplemental Fig. 3B).
Next, we investigated the template activity of sgRNAs bearing these 3′-end mutations by in vitro RdRp assays (Fig. 3E). All of the sgRNAs bearing the 3′-end mutations shown in Sequences 1–3 and 5 (Fig. 3E) were active (Fig. 3E, lanes 1–3,5). None of the recovered genomes contained the −9 and −10 mutations in the absence of the −5 mutation (Fig. 3E, see Sequence 4). Therefore, we examined the effect of adaptive mutation at −5 by using sgRNAs containing only −9 and −10. As shown in Figure 3E, lane 4, a sgRNA containing only the −9 and −10 mutations was markedly reduced in template activity to ∼10% of the WT (Fig. 3E, lane 4). These in vitro results support the conclusion that the −5 mutation conferred fitness for replication of the genome derived from input Δ10+GGC RNA.
Genomes of the majority of Δ8+GGC-derived viruses contained an internal deletion of 2 nt (CA at positions −7 and −8; Δ2i) with respect to that of the WT genome and a substitution at the −13 position (A → G), (Fig. 3E, Sequence 7). The template activities of sgRNAs containing Δ2i with and without the −13 mutation as well as the sgRNA containing 3′-terminal dinucleotide deletions were tested in the in vitro RdRP assay. As shown in Figure 3E, the adaptive mutation at position −13 in the context of the internal deletion (Δ2i) had similar template efficiency as WT with regard to the amount of RNAs synthesized (Fig. 3E, cf. lane 7 and WT). However, it could potentially stabilize secondary structure at the 3′ SL in recovered genome RNA (see below).
Effects of altered 3′-end sequences on the secondary structure of the 3′ SL
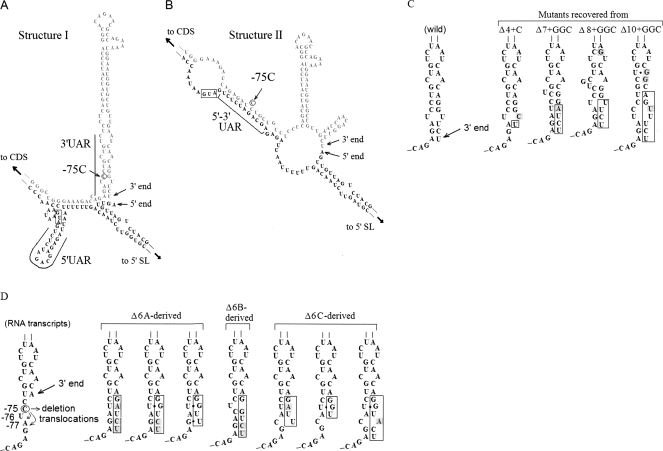
DENV RNA viral genomes have conserved secondary structural elements within the 3′- and 5′-terminal regions that play an important role in translation and replication as well as perhaps in assembly. When the 3′-end sequences were perturbed, mutant viruses introduced adaptive mutations and acquired improved virus growth. Therefore, we sought to examine whether perturbations of specific structural elements could explain the deleterious effects of these mutations on virus growth. To identify the RNA elements essential for replication, we used the Mfold program (Zuker et al. 1991; Alvarez et al. 2005). Alvarez et al. (2005) showed that a sgRNA containing the 5′-terminal region of the genome linked to the 3′-terminal 106 nt could assume two secondary structures, I and II, that differed in the base-paired region termed UAR, which is present in structure II (Fig. 4B) and not in structure I (Fig. 4A). By mutational analysis, Alvarez et al. (2005) further demonstrated that full-length genome RNAs containing mutations that would disrupt base-pairing in structure II of the 5′-3′ UAR were severely impaired for virus replication. Based on this observation, the investigators concluded that the ability to form structure II was a critical determinant of RNA replication competence.
FIGURE 4.
Analysis of secondary structures by Mfold. Structures I (A) and II (B) are similar to those described by Alvarez et al 2005. (C) Structure I representation of WT and consensus viral genomes recovered from Δ4+GC, Δ7+GGC, Δ8+GGC, Δ10+GGC viral RNAs, respectively, from left to right. (Shaded nucleotides) Substitution mutations, (boxes) recovered nucleotides. (D) Input RNA transcripts of Δ6, Δ6A, Δ6B, and Δ6C (see Table 3) are shown at left. The structure I representation of recovered genomes is shown on the right. Δ6A-derived viral genomes were classified as three types, based on the sequence alignments (see Table 3). Δ6B-derived genomes have a single sequence type, although they have variable lengths. Δ6C-derived genomes are classified as three sequence types. The secondary structures were drawn using the Mfold program (see Supplemental Table 3).
We assessed the predicted effects of 3′-end deletions on the putative secondary structures of sgRNAs using the Mfold program. The analysis indicated that RNAs that conferred a slow-growth phenotype (derived from Δ7+GGC, Δ8+GGC, and Δ10+GGC RNAs) (group 2) still exhibited a propensity to form structure II with a stability similar to that of WT RNA (ΔG≥ −89.3 kcal mol−1); however, the potential to form structure I was abrogated by 3′-end deletions. The abrogation in the probability of RNAs to form structure I was obviously due to a direct result of deleting nucleotides from the 3′ end; this deletion would be predicted to reduce the thermal stability of the long stem in the 3′ SL that is present in structure I but not in structure II (Fig. 4A,B). In contrast, the 3′-terminal sequences of a majority of the recovered genomes were predicted to form both structures I and II with similar thermal stability (ΔG≥ −88.5) (Supplemental Table 3). The Δ8+GGC-derived mutation at the −13 position (A → G) might stabilize structure I due to the formation of a C–G base pair (Fig. 4C). Although a previous study (Alvarez et al. 2005) indicated the importance of structure II in viral replication, our results indicate that propensity to form structure II alone is not sufficient. This conclusion is supported by the phenotypes of mutants, Δ7+GGC, Δ8+GGC, and Δ10+GGC input RNAs, that retain their ability to form structure II but are inefficient in replication. However, the recovered RNAs regained the propensity to form structure I and still retained their potential to form structure II and replicated efficiently upon passage (Supplemental Table 3). These results suggest that both structures I and II are required in a multi-step process involved in RNA replication.
Mechanism of 3′-end repair
Based on our results, we hypothesized that the 3′-end repair of sequential deletions is mediated by an activity of NS5 that adds nucleotides to the 3′ end of deletion mutants in the 3′ SL (in structure I). To test this hypothesis, we introduced mutations between nucleotides −79 and −74 (5′-AGAUCC-3′) (Fig. 4D). We used input RNAs from cDNAs linearized at the Bcg I restriction site (Supplemental Fig. 1). If 3′-end repair of Δ6 RNA containing point mutations within −79 to −74 involved the addition of nucleotides complementary to −79 to −74, any mutation introduced within this region would be revealed as a consequence of repair in the Δ6 genome. We sought to ensure that such mutations do not affect secondary structure II (see Fig. 4B). A single non-base-paired C at position 75 (C-75) in structure II is deleted (Δ6A), or either translocated with U at position −76 (−78GAUC → −78GACU; Δ6B RNA) or after A at −77 (−78GAUC → −78GCAU; Δ6C) RNAs (see Fig. 4D; Table 3).
TABLE 3.
Initial RNA transcripts and sequences of the recovered viral genomes
Mutant RNAs (Δ6, Δ6A, Δ6B, and Δ6C; see Table 3) were transfected into LLC-MK2 cells. Viruses were recovered from the transfected cells at day 20 post-transfection, and the 3′ ends of genomes in replicating viruses were cloned and sequenced as previously described (Table 3). There were no spontaneous reversions occurring from mutations introduced into the −75 to −77 region in Δ6A-C RNAs to WT during replication (Fig. 4D). Out of seven clones sequenced from the input Δ6 RNA, one contained a WT sequence and length and the others contained WT sequences but were shorter in length.
On the other hand, recovered RNA species from transfected mutant Δ6A-C RNAs were heterogeneous in their 3′-end sequences. This mixture of RNAs included WT and mutant sequences in each of the transfected Δ6A-C RNAs; each gave rise to distinct proportions of 3′-end sequences as the predominant species of RNAs (Table 3). In Δ6A-derived viral RNAs, the sequence complementary to −75 to −79, 5′-GAUCU-3′, was observed in four out of 15 clones. In the case of mutant Δ6B and Δ6C RNA-transfected cells, the exact complementary sequence was not observed (Fig. 4D; Table 3). However, as shown in Figure 4D, the recovered genomes sustained the bottom of the 3′ SL in structure I. These results suggested that the 3′ end is repaired by the addition of nucleotides relatively at random, but the requirement to form secondary structure I plays a role in the selection of replication-competent sequences.
DISCUSSION
The current model for flavivirus replication suggests that translation of the viral RNA, processing of the polyprotein, assembly of the viral RNA replicase, and recruitment of viral RNA into a specific compartment are sequential steps before RNA synthesis begins (Uchil and Satchidanandam 2003; Westaway et al. 2003). It is possible that viral genomes are susceptible to the loss of terminal nucleotides to 3′ → 5′ exoribonuclease(s) during any of these steps. To protect their genomes, viruses have evolved mechanisms to repair the ends by one or more pathways (Nagy et al. 1997; Guan and Simon 2000; Panavas et al. 2005).
Sequence analysis of progeny viral genomes revealed that repair of 3′-terminal nucleotides occurred depending on the extent of deletions of the input RNA. Deletions of 1–5 nt but ending with GGC are repaired and the GGC nucleotides are removed. The progeny viruses grew without any noticeable delay compared with parental RNA, and the viral RNAs contained WT sequences with the 3′ end ending with GGUU but missing the conserved 3′ CU (Table 1, rows 1–4). Interestingly, when 6 nt were removed and nonviral GGC were added to the 3′ end, although the time for the progeny to appear was the same as WT, the 3′ end of recovered viral RNA ended with GGCU but still lacked the conserved 3′ CU (Table 1, row 5). In contrast, if the deletions were longer (e.g., Δ7+GGC), the nonviral GGC nucleotides were retained in the repaired region and the genome length became close to the WT. Adaptive mutation(s) at the 3′ end were also generated, although the preferred 3′-terminal sequences were UU, CU, UCU, or UUCU (Table 1, rows 6–8). This is a surprising and novel finding that genomes lacking 3′-terminal nucleotides, CU, became the predominant species among the recovered viral RNAs after 3′-end repair. This observation can be rationalized in two possible ways. One is that even if the terminal dinucleotide CU is missing in the majority of viral RNAs, these RNAs may be functional in replication. This possibility is consistent with the results of a recent study using an in vitro RdRP assay containing a recombinant Japanese encephalitis virus RdRP. It was shown that initiation of RNA synthesis occurred at the −3 position, skipping the 3′-terminal CU (U81) (Kim et al. 2007).
Another possible explanation is that although the predominant species in evolving virion RNAs contain 3′ UU, missing the 3′ CU in the repaired region, they are present as a heterogeneous mixture or quasi-species (e.g., Table 2, day 62). These results suggest that the presence of viral RNAs as quasi-species may confer a selective advantage for virus survival as shown for poliovirus (Vignuzzi et al. 2006). These results suggest that Δ2 is potentially adapted in a replication-competent environment in a cell culture system.
Previous studies indicated that mutations at 3′-terminal nucleotides of WNV or Kunjin replicon RNAs significantly affected replication efficiency (Khromykh et al. 2003; Tilgner and Shi 2004). The conclusions of those studies are consistent with our results showing that the recovered viruses evolved to having the same or nearly close to WT 3′ termini such as UU, CU, UCU, or UUCU. Our previous results obtained by in vitro assays using DENV2 RdRP, WT, and mutant sgRNAs containing a 3′-terminal CU → UU mutation indicated that the activity of the mutant template UU was not affected. Moreover, at the 3′ end, U is preferred over A–G, or C (Nomaguchi et al. 2003).
We compared the 3′-end repair of viral RNAs (generated from BcgI-linearized cDNAs) containing the 3′ deletions without any nonviral nucleotides with the repair of those containing GGC nucleotides. The 3′ deletions up to Δ3 were fast-growing (group 1), like WT, whereas deletions from Δ4 to Δ7 were slow-growing (group 2), and Δ8 and larger deletion mutants were not recovered at all (group 3 mutants). Sequence analysis showed that the major species of recovered RNAs again contained Δ2 at the 3′ end (data not shown). These results were in contrast to those shown in Table 2, that in longer deletions the presence of GGC at the 3′ ends of Δ8 or Δ10 RNAs produced progeny virus. Moreover, part of these nonviral GGC nucleotides were retained in the recovered genomes together with additional mutations upstream of UU, CU, UCU, and UUCU at the 3′ end. It is noteworthy that these nonviral sequences in the recovered RNAs maintained structure I of the 3′ SL (Fig. 4C).
One possible mechanism for the 3′-end repair of DENV2 is non-template-based addition by terminal nucleotidyl transferase (TNTase), which is an intrinsic activity of viral RNA polymerase, NS5. This mechanism was postulated in studies of plant viruses (Rao et al. 1989; Guan and Simon 2000). However, this mechanism has not been shown for RNA viruses that replicate in mammalian cells, and for the first time, this study describes the results of 3′-end repair utilized by DENV2 polymerase. This intrinsic property of RdRPs has been suggested in in vitro experiments involving viral polymerases of DENV2 (Ackermann and Padmanabhan 2001), HCV (Ranjith-Kumar et al. 2001), poliovirus (Arnold et al. 1999), and Sindbis virus (Tomar et al. 2006). In most cases, the nature of the nucleotides added by RdRPs was not random and seemed to be influenced by the 3′-end nucleotides. Alternatively, a host 3′ → 5′ exoribonuclease activity may be involved in trimming the 3′ end prior to the TNTase activity of the viral RdRP (or the host enzyme). Such exonuclease activity was recently identified in the replication of tomato bushy stunt virus, a positive-strand plant virus, in a model host (yeast) genome-wide screen (Panavas et al. 2005).
Results of the in vitro RdRP assays show that sgRNA mutants containing deletions of Δ5, Δ8, and Δ10 but having nonviral GGC at the 3′ end had good template activities. In contrast, deletions of 14 nt, or more, essentially abolished template activities in vitro (Fig. 3). These results are consistent with the ability of viral RNAs to repair their genomes in cultured cells. However, we also observed that the ratios of de novo to 2× hairpin sgRNA products were altered. This observation suggests the possibility that the deleted nucleotides are important for self-priming of RNA synthesis to yield the 2× product.
Mosquito-borne flavivirus genomes have conserved sequences at the 5′- and 3′-terminal regions (5′- and 3′-CS1). These motifs are complementary and were postulated to be involved in circularization of the genome (Hahn et al. 1987). Physical and functional interaction between 5′- and 3′-CS1 were required for RNA synthesis in vitro (You and Padmanabhan 1999; Ackermann and Padmanabhan 2001; You et al. 2001). RNA–RNA interaction resulting in circularization of the genome was elegantly established by atomic force microscopy (Alvarez et al. 2005). These investigators also identified novel motifs capable of forming base-paired interactions termed 5′- and 3′-UAR. Using the Mfold program (Zuker et al. 1991), it was demonstrated that the 5′- and 3′-terminal regions of DENV2 RNA could form two structures, I and II, with similar free energies (Fig. 4). Although both structures show identical 5′- and 3′-CS1 interaction, only structure II shows base-pair interactions between UARs. Mutagenesis of nucleotides involved in the formation of structure II reduced viral replication of DENV2 in cultured cells (Alvarez et al. 2005). The relationship between these two structures and their requirements for viral replication had not been explored.
We investigated this relationship in detail in the input and progeny virion RNAs recovered from transfected cells. Our results indicate that if the input RNAs containing deletions retain the ability to form both structures I and II, then the recovered viruses grow at the same rate as WT. However, if the input RNAs lost the ability to form structure I but are capable of forming structure II, then the recovered viruses regain the ability to form both structures I and II (Supplemental Table 3). Our results are consistent with those of Tilgner and Shi (2004); mutants of the WNV replicon that maintained base-pairing within the bottom part of the 3′ SL (synonymous with structure I, [Alvarez et al. 2005]) are able to replicate with minimum efficiency. The importance of structure I in viral replication is further exemplified by our result that 3′-end SL mutations introduced in the −75 to −77 region impinged on the selection of optimal nucleotides in 3′-end repair replication. Based on our results, we propose a two-step model for 3′-end repair in DENV2. The first step involves the non-template-based addition of nucleotides by viral replicase, followed by evolutionary selection of nucleotides dictated by structural constraints for optimal replication of the viral genome.
MATERIALS AND METHODS
Construction of DENV2 infectious cDNAs containing deletions/mutations in the 3′ termini
A full-length cDNA of DENV2 (New Guinea C strain) cloned into yeast shuttle vector pRS424, named pRS424FLD2, was previously described (Polo et al. 1997). Deletions and/or mutations of the 3′ termini were generated by homologous recombination essentially as described (Zeng et al. 1998). Briefly, PCR products were made that included 30 nt of vector sequence and ∼300 nt of DENV2 3′-end sequence while incorporating the desired 3′-end mutations and either a EheI or BcgI restriction site (for a list of primers used, see Supplemental Table 1). Plasmid pRS424FLD2 was digested by SacI, which cleaves at the 3′ terminus of DENV2. Yeast YPH857 cells were cotransfected with the linearized pRS424FLD2 and each PCR product, and yeasts harboring recombinant plasmids were selected on solid media lacking tryptophan. Extracted DNA from yeast colonies was used to transform Escherichia coli STBL2 (Invitrogen). Plasmid DNAs were isolated from the bacterial colonies, and the DNA sequences were verified using an ABI 377 automated DNA sequencer and Big Dye terminator chemistry (Applied Biosystems).
RNA transfection and recovery of viruses
Recombinant plasmid DNA (containing WT or 3′-end deletion/mutations) was linearized at the 3′ end of DENV2 by digestion with SacI, EheI, or BcgI. The linearized DNA was used as the template for in vitro transcription catalyzed by SP6 RNA polymerase (New England Biolabs) in the presence of the m7GpppG cap structure analog. We used the following nomenclature to name the RNAs used for transfections based on their 3′ ends. The in vitro transcription of the SacI-linearized plasmid would give rise to parental RNA containing an extra G at the 3′ end (WT-G) (see Supplemental Fig. 1). The BcgI-linearized plasmid would yield an RNA with no additional nucleotides at the 3′ end of WT or different deletion mutants (Δ1, Δ2, Δ3, etc). On the other hand, the EheI-linearized plasmid with different 3′-end deletions would yield mutant RNAs containing nonviral GGC nucleotides added to the 3′ end of deletion mutants Δ5+GGC (or Δ4+GC, as addition of GGC to the 3′ end of the mutant RNAs replaced the deleted G at the −5 position), Δ8+GGC, Δ10+GGC, etc. In addition, SP6 polymerase prefers a G at a +1 start site (Jobling et al. 1988), and hence all transcribed RNAs would contain an extra G at the 5′ end. RNA transcripts (3 μg) were electroporated (Bio-Rad) into LLC-MK2 cells (Polo et al. 1997). After electroporation, cells were incubated in a T-12.5 flask, and on days 2 and day 9, cells were trypsinized and transferred into a T-25 and T-75 flask, respectively. This procedure was repeated using one-third of the trypsinized cells and a T-75 flask every 7 d.
Immunofluorescence
Indirect immunofluorescence staining was performed on cells that had been seeded to a 1-cm2 chamber slide (LabTek). Acetone-fixed cells were incubated with a 1:200 dilution of a monoclonal antibody against DENV2 NS1 antigen (7E11). FITC-labeled goat anti-mouse immunoglobulin G (Kirkegaard & Perry Laboratories, Inc.) was used (1:100) as detector antibodies. Photomicrographs (200× magnification) were acquired with a Leitz Diaplan microscope coupled to a Leica/Wild MPS48 automated photographic system.
Extraction of viral RNA and sequence analysis
After clarification of the culture media at 13,000 rpm for 3 min at 4°C, viral RNA was extracted from the supernatant using a Mini-RNA kit (Qiagen). RNA was treated with Tobacco acid pyrophosphatase (Epicenter) to remove the cap structure, and then 3′ and 5′ ends of the RNA were ligated with T4 RNA ligase (Roche Molecular Biochemicals) as described previously (Chen et al. 1994). After phenol–chloroform extraction and ethanol precipitation, the pellet was resuspended in 10 μL of diethylpyrocarbonate-treated H2O; 2.5 μL of this solution was used for RT-PCR with primers designed to amplify the 5′–3′ joined ligation product between nucleotides 10,420 and 177 (Supplemental Table 1). The RT-PCR products were directly sequenced to determine the consensus sequence. To investigate the distribution of individual sequences within the population, each PCR product was subcloned into the pGEM-T Easy plasmid (Promega). At least 20 independent clones were sequenced for each PCR product.
Plaque assay
Titers of WT and mutant DENV2 viruses were determined by plaque assay on LLC-MK2 cells. Serially diluted supernatants recovered from the culture medium were incubated with cells in paired wells of six-well plates for 2 h. Cells were overlaid with media containing 0.5% Sea-Kem GTG agarose (FMC Bioproducts) and were incubated for 9 d at 37°C and visualized by staining with neutral red (Zeng et al. 1998).
In vitro RdRp assay
The cDNA plasmid encoding DENV2 sgRNA is as described previously (You et al. 2001). Sequential 3′-end deletions were introduced into the cDNA by PCR using the 5′ primer from a region upstream of the T7 RNA polymerase promoter and various 3′ primers containing the desired mutations (Supplemental Table 2). sgRNAs were obtained by in vitro transcription of PCR-amplified DNA templates with T7 RNA polymerase. The standard RdRp assay mixture (50 μL) contained 50 mM Tris-HCl at pH 8.0, 50 mM NaCl, 5 mM MgCl2, template RNA (1 μg), 500 μM each of ATP, GTP, and UTP, 10 μM CTP, and 10 μCi of [α-32P] CTP along with 0.5 μg of purified NS5. The reaction mixture was incubated for 1 h at 30°C and terminated by acid phenol/chloroform extraction, followed by ethanol precipitation. The RNA pellet was collected by centrifugation (12,000g for 5 min), dried, and resuspended in 50 μL of nuclease-free H2O. Labeled samples were passed through a Bio-Rad P-30 gel filtration column to remove unincorporated rNTPs; the flow-through fraction was precipitated with ethanol. The dried RNAs were analyzed by formaldehyde–agarose gel electrophoresis and visualized by autoradiography. Band intensities were measured with a PhosphorImager (Molecular Dynamics).
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Mike Klutch for excellent technical support in sequence analysis. The work was supported by grants from NIAID, AI54776, and AI 070791 and an ORISE grant.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1051208.
REFERENCES
- Ackermann M., Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 2001;276:39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- Alvarez D.E., Lodeiro M.F., Luduena S.J., Pietrasanta L.I., Gamarnik A.V. Long-range RNA–RNA interactions circularize the dengue virus genome. J. Virol. 2005;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews N.C., Levin D., Baltimore D. Poliovirus replicase stimulation by terminal uridylyl transferase. J. Biol. Chem. 1985;260:7628–7635. [PubMed] [Google Scholar]
- Arnold J.J., Ghosh S.K., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): Divalent cation modulation of primer, template, and nucleotide selection. J. Biol. Chem. 1999;274:37060–37069. doi: 10.1074/jbc.274.52.37060. [DOI] [PubMed] [Google Scholar]
- Brinton M.A., Fernandez A.V., Dispoto J.H. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology. 1986;153:113–121. doi: 10.1016/0042-6822(86)90012-7. [DOI] [PubMed] [Google Scholar]
- Chen Z., Faaberg K.S., Plagemann P.G. Determination of the 5′ end of the lactate dehydrogenase-elevating virus genome by two independent approaches. J. Gen. Virol. 1994;75:925–930. doi: 10.1099/0022-1317-75-4-925. [DOI] [PubMed] [Google Scholar]
- Chiu W.W., Kinney R.M., Dreher T.W. Control of translation by the 5′- and 3′-terminal regions of the dengue virus genome. J. Virol. 2005;79:8303–8315. doi: 10.1128/JVI.79.13.8303-8315.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomatori C.V., Lodeiro M.F., Alvarez D.E., Samsa M.M., Pietrasanta L., Gamarnik A.V. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes & Dev. 2006;20:2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H., Simon A.E. Polymerization of nontemplate bases before transcription initiation at the 3′ ends of templates by an RNA-dependent RNA polymerase: An activity involved in 3′-end repair of viral RNAs. Proc. Natl. Acad. Sci. 2000;97:12451–12456. doi: 10.1073/pnas.97.23.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C.S., Hahn Y.S., Rice C.M., Lee E., Dalgarno L., Strauss E.G., Strauss J.H. Conserved elements in the 3′-untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 1987;198:33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- Holden K.L., Harris E. Enhancement of dengue virus translation: Role of the 3′-untranslated region and the terminal 3′ stem–loop domain. Virology. 2004;329:119–133. doi: 10.1016/j.virol.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Irie M. Structure–function relationships of acid ribonucleases: Lysosomal, vacuolar, and periplasmic enzymes. Pharmacol. Ther. 1999;81:77–89. doi: 10.1016/s0163-7258(98)00035-7. [DOI] [PubMed] [Google Scholar]
- Jobling S.A., Cuthbert C.M., Rogers S.G., Fraley R.T., Gehrke L. In vitro transcription and translational efficiency of chimeric SP6 messenger RNAs devoid of 5′ vector nucleotides. Nucleic Acids Res. 1988;16:4483–4498. doi: 10.1093/nar/16.10.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh A.A., Meka H., Guyatt K.J., Westaway E.G. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001;75:6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh A.A., Kondratieva N., Sgro J.Y., Palmenberg A., Westaway E.G. Significance in replication of the terminal nucleotides of the flavivirus genome. J. Virol. 2003;77:10623–10629. doi: 10.1128/JVI.77.19.10623-10629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.G., Yoo J.S., Kim J.H., Kim C.M., Oh J.W. Biochemical characterization of a recombinant Japanese encephalitis virus RNA-dependent RNA polymerase. BMC Mol. Biol. 2007;8:59. doi: 10.1186/1471-2199-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B.D., Rice C.M. Molecular biology of flaviviruses. Adv. Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- Lo M.K., Tilgner M., Bernard K.A., Shi P.Y. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′-untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 2003;77:10004–10014. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Maduro M., Li F., Li H.W., Broitman-Maduro G., Li W.X., Ding S.W. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans . Nature. 2005;436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markoff L. 5′- and 3′-noncoding regions in flavivirus RNA. Adv. Virus Res. 2003;59:177–228. doi: 10.1016/S0065-3527(03)59006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani S., Colonno R.J. In vitro synthesis of an infectious RNA from cDNA clones of human rhinovirus type 14. J. Virol. 1985;56:628–632. doi: 10.1128/jvi.56.2.628-632.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan P.M., Padmanabhan R. Detection of stable secondary structure at the 3′ terminus of dengue virus type 2 RNA. Gene. 1991;108:185–191. doi: 10.1016/0378-1119(91)90433-c. [DOI] [PubMed] [Google Scholar]
- Nagy P.D., Carpenter C.D., Simon A.E. A novel 3′-end repair mechanism in an RNA virus. Proc. Natl. Acad. Sci. 1997;94:1113–1118. doi: 10.1073/pnas.94.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld K.L., Galarza J.M., Richards O.C., Summers D.F., Ehrenfeld E. Identification of terminal adenylyl transferase activity of the poliovirus polymerase 3Dpol. J. Virol. 1994;68:5811–5818. doi: 10.1128/jvi.68.9.5811-5818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomaguchi M., Ackermann M., Yon C., You S., Padmanbhan R. De novo synthesis of negative-strand RNA by dengue virus RNA-dependent RNA polymerase in vitro: Nucleotide, primer, and template parameters. J. Virol. 2003;77:8831–8842. doi: 10.1128/JVI.77.16.8831-8842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas T., Serviene E., Brasher J., Nagy P.D. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc. Natl. Acad. Sci. 2005;102:7326–7331. doi: 10.1073/pnas.0502604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S., Ketner G., Levis R., Falgout B. Infectious RNA transcripts from full-length dengue virus type 2 cDNA clones made in yeast. J. Virol. 1997;71:5366–5374. doi: 10.1128/jvi.71.7.5366-5374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar C.T., Gajewski J., Gutshall L., Maley D., Sarisky R.T., Kao C.C. Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: Implication for viral RNA synthesis. J. Virol. 2001;75:8615–8623. doi: 10.1128/JVI.75.18.8615-8623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A.L., Dreher T.W., Marsh L.E., Hall T.C. Telomeric function of the tRNA-like structure of brome mosaic virus RNA. Proc. Natl. Acad. Sci. 1989;86:5335–5339. doi: 10.1073/pnas.86.14.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk P.J., Uhlenbeck O.C. Joining of RNA molecules with RNA ligase. Methods Enzymol. 1983;100:52–59. doi: 10.1016/0076-6879(83)00045-2. [DOI] [PubMed] [Google Scholar]
- Samuel C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow P. Role of 3′-end sequences in infectivity of poliovirus transcripts made in vitro. J. Virol. 1989;63:467–470. doi: 10.1128/jvi.63.1.467-470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serviene E., Jiang Y., Cheng C.P., Baker J., Nagy P.D. Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J. Virol. 2006;80:1231–1241. doi: 10.1128/JVI.80.3.1231-1241.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabler M., Sanger H.L. Infectivity studies on different potato spindle tuber viroid (PSTV) RNAs synthesized in vitro with the SP6 transcription system. EMBO J. 1985;4:2191–2199. doi: 10.1002/j.1460-2075.1985.tb03914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner M., Shi P.Y. Structure and function of the 3′-terminal six nucleotides of the West Nile virus genome in viral replication. J. Virol. 2004;78:8159–8171. doi: 10.1128/JVI.78.15.8159-8171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar S., Hardy R.W., Smith J.L., Kuhn R.J. Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J. Virol. 2006;80:9962–9969. doi: 10.1128/JVI.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchil P.D., Satchidanandam V. Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 2003;278:24388–24398. doi: 10.1074/jbc.M301717200. [DOI] [PubMed] [Google Scholar]
- Vignuzzi M., Stone J.K., Arnold J.J., Cameron C.E., Andino R. Quasi-species diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westaway E.G., Mackenzie J.M., Khromykh A.A. Kunjin RNA replication and applications of Kunjin replicons. Adv. Virus Res. 2003;59:99–140. doi: 10.1016/s0065-3527(03)59004-2. [DOI] [PubMed] [Google Scholar]
- You S., Padmanabhan R. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′ end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 1999;274:33714–33722. doi: 10.1074/jbc.274.47.33714. [DOI] [PubMed] [Google Scholar]
- You S., Falgout B., Markoff L., Padmanabhan R. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 2001;276:15581–15591. doi: 10.1074/jbc.M010923200. [DOI] [PubMed] [Google Scholar]
- Yu L., Markoff L. The topology of bulges in the long stem of the flavivirus 3′ stem–loop is a major determinant of RNA replication competence. J. Virol. 2005;79:2309–2324. doi: 10.1128/JVI.79.4.2309-2324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Nomaguchi M., Padmanabhan R., Markoff L. Specific requirements for elements of the 5′- and 3′-terminal regions in flavivirus RNA synthesis and viral replication. Virology. 2008;374:170–185. doi: 10.1016/j.virol.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Falgout B., Markoff L. Identification of specific nucleotide sequences within the conserved 3′-SL in the dengue type 2 virus genome required for replication. J. Virol. 1998;72:7510–7522. doi: 10.1128/jvi.72.9.7510-7522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Jaeger J.A., Turner D.H. A comparison of optimal and suboptimal RNA secondary structures predicted by free energy minimization with structures determined by phylogenetic comparison. Nucleic Acids Res. 1991;19:2707–2714. doi: 10.1093/nar/19.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]