Abstract
MicroRNAs (miRNAs) are short noncoding RNA molecules regulating the expression of mRNAs. Target identification of miRNAs is computationally difficult due to the relatively low homology between miRNAs and their targets. We present here an experimental approach to target identification where the cartilage-specific miR-140 was overexpressed and silenced in cells it is normally expressed in separate experiments. Expression of mRNAs was profiled in both experiments and the intersection of mRNAs repressed by miR-140 overexpression and derepressed by silencing of miR-140 was identified. The intersection contained only 49 genes, although both treatments affected the accumulation of hundreds of mRNAs. These 49 genes showed a very strong enrichment for the miR-140 seed sequence implying that the approach is efficient and specific. Twenty-one of these 49 genes were predicted to be direct targets based on the presence of the seed sequence. Interestingly, none of these were predicted by the published target prediction methods we used. One of the potential target mRNAs, Cxcl12, was experimentally validated by Northern blot analysis and a luciferase reporter assay.
Keywords: microRNA, target identification, cartilage development
INTRODUCTION
MicroRNAs (miRNAs) are short (21–22 nucleotides [nt]) noncoding RNA molecules that down-regulate the expression of protein-coding genes (Bartel 2004). miRNAs do not switch off their target genes completely but rather fine tune their expression, and it is now clear that they play an important role in apoptosis, proliferation, tumor formation, development, differentiation, metabolism, and disease development (Kloosterman and Plasterk 2006). miRNAs are incorporated into the RNA-induced silencing complex (RISC) and guide this complex to specific mRNAs that contain miRNA target sites, which can fall into three categories (Sethupathy et al. 2006). 5′-Dominant canonical target sites show perfect complementarity to the seed sequence of the miRNA (nucleotides 2–8) and extensive base pairing to the rest of the miRNA (Sethupathy et al. 2006). 5′-Dominant seed only target sites are also perfectly complementary to the seed sequence but have limited base pairing with the rest of the miRNA (Sethupathy et al. 2006). 3′-Compensatory target sites do not have a perfect match to the seed sequence but are compensated by extensive base pairing with the 3′-half of the miRNA (Brennecke et al. 2005). Target sites are usually in the 3′-untranslated region (UTR) of mRNAs but there are examples of target sites in the coding region as well (Kloosterman et al. 2005; Duursma et al. 2008), and their flanking regions can also influence interaction with miRNAs (Zhao et al. 2005; Didiano and Hobert 2006; Kertesz et al. 2007). Translation of mRNAs targeted by miRNAs is repressed and the steady-state level of some but not all mRNA targets is also reduced (Wightman et al. 1993; Bagga et al. 2005; Pillai et al. 2005; Petersen et al. 2006; Parker and Sheth 2007; Eulalio et al. 2008).
Identifying target mRNAs of miRNAs is an important step in elucidating the function of miRNAs, yet this step has proven computationally difficult due to the complexity of the miRNA–target interactions. Several target prediction programs have been developed, but the overlap between sets of predicted target genes for a given miRNA by different programs is surprisingly low (Sethupathy et al. 2006), suggesting that there are a number of false positive predictions. Genes that are targeted by miRNAs but not predicted by the programs are false negatives, and it is not known how many there could be. Nevertheless, these target prediction programs are very useful to define potential targets that can be validated experimentally, often using luciferase reporter assays (Lewis et al. 2003).
Another strategy for identifying targets is based on experimental data and usually involves the manipulation of miRNA activity. The first such study showed that overexpression of miR-1 and miR-124 in cells where these miRNAs are not normally expressed led to repression of many mRNAs (Lim et al. 2005). The opposite has also been demonstrated, in which silencing of miRNA activity resulted in derepression of many messages (Krützfeldt et al. 2005; Elmén et al. 2008a,b). Recently, two studies reported biochemical purification of RISC complexes using antibody against the Argonaute-2 protein. miRNA targets were identified by extracting mRNA from the purified complexes and identified by microarray hybridization or sequencing (Karginov et al. 2007; Beitzinger et al. 2007). However, introducing a miRNA into cells where it is normally not expressed and identifying mRNAs targeted by the miRNA has limited biological relevance.
Thus, to identify biologically relevant targets we both overexpressed and silenced the activity of miR-140 separately in the same cell line that normally expresses miR-140. Here we show that the intersection of mRNAs that are repressed or derepressed upon miRNA overexpression and silencing, respectively, is relatively small in this cell line. However, the identified mRNAs are enriched for miR-140 target sites, suggesting that our method is feasible for experimental identification of direct miRNA targets. Interestingly, none of the identified 21 targets were predicted by any of the available target prediction programs, indicating that target predictions have a significant number of false negatives. We also validated one of the predicted targets, Cxcl12, that is involved in cartilage development.
RESULTS AND DISCUSSION
Overexpression and antagonism of miR-140
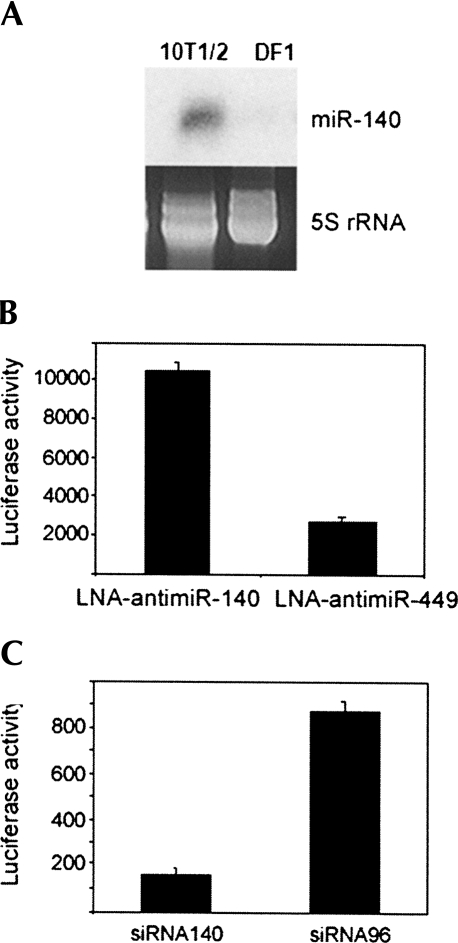
It has been shown that miRNA expression can be mimicked by transfecting cells with siRNAs with identical sequences to miRNAs that are not expressed in the cells (Lim et al. 2005). Transcripts that are repressed and contain miRNA seed sites are predicted to be direct miRNA targets, but the biological relevance of target messages identified by this approach is not clear. We hypothesized that transcripts showing repression upon miRNA overexpression should also be derepressed when the miRNA is silenced, and this control could be useful in experimental target identification. We used miR-140 as a model miRNA to test this hypothesis and searched for a cell line that expresses miR-140 in moderate levels, which would make it feasible to both antagonize it or increase its expression level. Since miR-140 is specifically expressed in the cartilage tissue of mouse and zebrafish embryos (Wienholds et al. 2005; Tuddenham et al. 2006) we tested the murine C3H10T1/2 fibroblast cell line that can be differentiated into chondrocytes, adipocytes, or muscle cells. Figure 1A shows that undifferentiated C3H10T1/2 cells express miR-140 and, hence, provided an appropriate cell line to test our hypothesis.
FIGURE 1.
Overexpression and antagonism of miR-140. (A) Expression of miR-140 in C3H10T1/2 cells was confirmed by Northern blot analysis. DF1 chicken fibroblast cells were used as negative control. Equal loading is shown by ethidium bromide stained 5S rRNA. (B) Luciferase activity was increased when pLuc-140 (a construct containing a perfect match to miR-140 downstream from the luciferase gene) was cotransfected with LNA-antimiR-140 compared with pLuc-140 cotransfected with LNA-antimiR-449 (negative control). Units are arbitrary. (C) Luciferase activity was decreased when pLuc-140 was cotransfected with siRNA-140 compared with pLuc-140 cotransfected with siRNA-96 (negative control). Units are arbitrary.
The efficiency of miRNA overexpression and antagonism was monitored by luciferase reporter assays. A construct was generated that contained a 21-nucleotide sequence with perfect complementarity to miR-140 (pLuc-140), in the 3′-UTR region of the luciferase gene, which was expected to be efficiently targeted by miR-140. Luc-140 was cotransfected into C3H10T1/2 cells with either siR-140 (an siRNA with the sequence of miR-140) or LNA-antimiR-140 (locked nucleic acid modified DNA oligonucleotide complementary to miR-140) using an siRNA mimicking miR-96 and an LNA-antimiR specific to miR-449 as negative controls. The miR-140 sensor construct showed strong derepression in the presence of LNA-antimiR-140 (Fig. 1B) and strong repression when it was cotransfected with siR-140 (Fig. 1C). Having shown that the activity of miR-140 can be increased or antagonized by siRNA or LNA-antimiR treatments, respectively, we used this approach on a genomic scale. Total RNA was extracted from C3H10T1/2 cells transfected with siRNA-140, siRNA-96, LNA-antimiR-140 or LNA-antimiR-449 and the mRNA expression profiles were determined using Affymetrix microarrays.
Identifying potential miR-140 targets
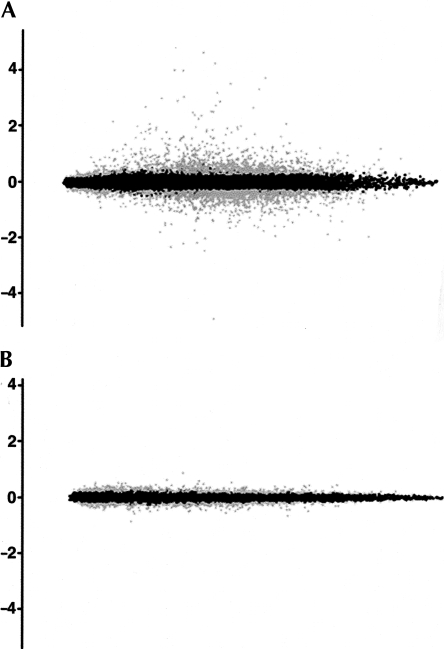
Recent studies that characterized miRNA target sites in mammals made two main conclusions: first, most miRNA target sites are located in the 3′-UTR of the mRNA; and second, the most important factor in the recognition of the target site appears to be the sequence to which the first eight bases of the miRNA will pair, usually called the “seed” sequence (Grimson et al. 2007; Lewis et al. 2005). Microarray data were obtained for the two samples in the miRNA overexpression experiment (cells transfected with either siRNA-140 or siRNA-96) and the two samples in the miRNA silencing experiment (cells transfected with either LNA-antimiR-140 or LNA-antimiR-449). Using the criteria detailed in the methods section we found 1236 and 466 probe sets differentially expressed between the two compared samples in the overexpression and silencing experiments, respectively (Fig. 2).
FIGURE 2.
Differentially expressed mRNAs (MvA Plots). Each dot in the plot represents a probe set. The coordinates of each probe set represent the logarithm of the average (x axis) and the logarithm of the ratio (y axis) of the expression values of the probe set in the two samples being compared. Probe sets fulfilling the differential expression criteria are colored in gray, the remainder in black. (A) Comparison of siRNA-96 and siRNA-140 transfected samples. Targets of miR-140 are expected to have lower expression value for the siRNA-140 sample, corresponding to a positive log-ratio of expression (upper half of the graph). (B) Comparison of LNA-antimiR-449 and LNA-antimiR-140 transfected samples. Targets of miR-140 should have a higher expression value for the LNA-antimiR-140 treated sample, corresponding to a negative log-ratio of expressions (lower half of the graph).
To evaluate the global effect of the siRNA and antimiR transfections on the transcriptome we plotted the distribution of normalized differential expression values for all the probe sets and for the probe sets which map to mRNAs containing at least one microRNA seed site in its 3′-UTR (see Supplemental Figs. 1,2). The empirical distribution curve corresponding to genes containing miR-140 seed sites is shifted to the right in the plot obtained from the overexpression experiment. The explanation is that comparing the seed site-containing mRNAs to the set of all mRNAs generally present higher z-values, which means they are expressed at lower levels in the cells transfected with siRNA-140 than in the cells transfected with siRNA-96 control. The opposite effect is observed for the mRNAs containing a miR-96 seed site. This result suggests that a significant fraction of these mRNAs are repressed by siRNA-140 or siRNA-96. In the silencing experiment similar results are obtained for miR-140 and miR-449. The effect observed for mRNAs containing a miR-449 seed might be the result of antagonizing miR-449 or a result of nonspecific binding of LNA-antimiR-449 to miR-34, which shares the seed sequence of miR-449. Additionally, the longer the seed sequence, the more marked this effect is, supporting the hypothesis that additional base-pairing in the seed region strengthens the efficacy of miRNA targeting (Nielsen et al. 2007). In the silencing experiment the same effect is observed, although with lower significance levels for mRNAs with a miR-140 seed site. The P-values obtained using the Wilcoxon test are presented in Supplemental Tables 1 and 2.
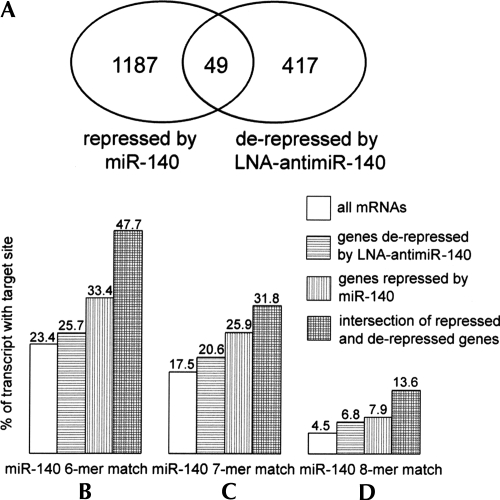
Another piece of statistical evidence that many of the miR-140 targets were affected by the manipulation of miR-140 concentration can be obtained by examining the percentage of mRNAs that contain seed sites (see Fig. 3). Regardless of the length of the seed sequence (six, seven, or eight nucleotides) the same pattern can be observed: (1) the percentage of mRNAs with seed sites is higher in the sets of mRNAs which are repressed during overexpression or derepressed upon antagonism of miR-140 than in the set of all messages; and (2) the percentage of mRNAs with seed sites is higher in the set of mRNAs for which at least one of the corresponding probe sets is both repressed in the overexpression and derepressed in the silencing experiments than the percentages obtained for either of the two experiments.
FIGURE 3.
Number of differentially expressed genes and percentage of miR-140 seed matches. Panel A shows the number of genes differentially expressed upon siRNA-140 and LNA-antimiR-140 treatment and the intersection of these genes. Panels B, C, and D show the percentage of miR-140 seed matches in all mRNAs (open bar), genes derepressed after treatment with LNA-antimiR-140 (horizontal lines), gene repressed by siRNA-140 (vertical lines), and genes derepressed after treatment with LNA-antimiR-140 and repressed by siRNA-140 (horizontal and vertical lines).
The first result confirms the hypothesis that a significant proportion of the observed differential expression can be directly attributed to the different concentrations of miR-140 in the compared samples. The second result suggests that taking the intersection of the sets of mRNAs obtained in the two experiments is an effective way to filter the relevant miRNA targets. We note that no such pattern is observed in the set of mRNAs derepressed by siRNA-140 or the set of mRNAs repressed by LNA-antimiR-140 treatment. Upon repeating the same analysis for miR-96 we observed an increased percentage of seed sites for genes that have lower expression values in the sample transfected with siRNA-96 than in the sample transfected with siRNA-140 (two- to threefold increase depending on the length of seed sequence). This indicates that a significant number of genes were targeted by siRNA-96. However, taking the intersection with the mRNAs derepressed by LNA-antimiR-449 treatment does not yield an increase in the percentage of mRNAs containing a miR-96 seed site, which is consistent with the fact that the seed sites of miR-96 and miR-449 are different. This implies that the enrichment observed for miR-140 seed sites in the intersection of differentially expressed mRNA sets is specific.
Next, we analyzed the intersection of mRNAs that were repressed by siRNA-140 and derepressed by LNA-antimiR-140. This list consisted of only 49 messages (Supplemental Table 3), although siRNA-140 repressed 1236 mRNAs and LNA-antimir-140 derepressed 466 mRNAs. These 49 mRNAs can be either direct or indirect miR-140 targets and, although the priority usually is to identify direct targets of miRNAs, information about indirect targets can be also useful. Indirect targets are regulated by direct targets; therefore, these genes can provide information about the downstream effects of the miRNA.
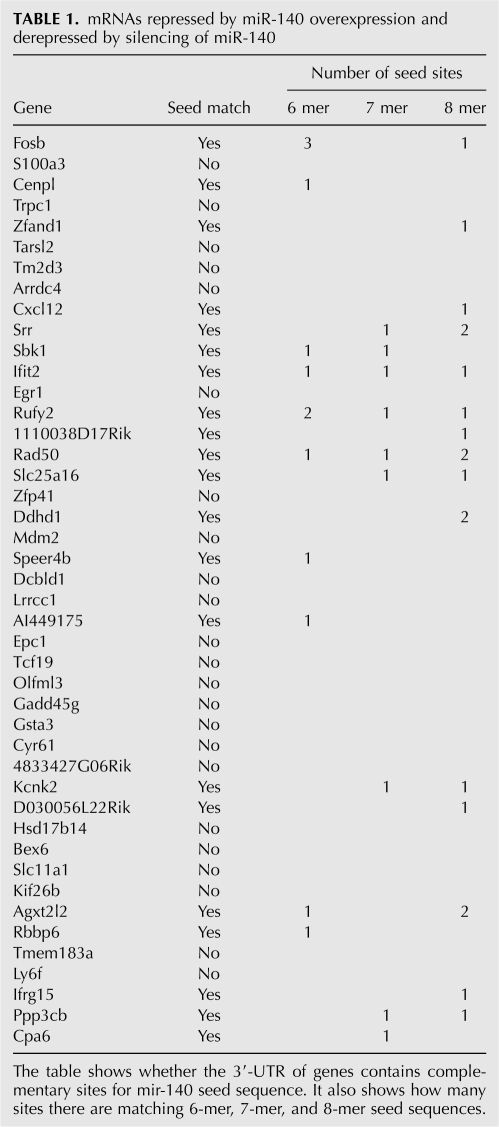
To identify potential direct targets we searched for seed sequences in the 3′-UTR sequences of the mRNAs. Twenty-one out of the 49 mRNAs contained at least one 6-mer seed sequence, and these were considered as potential direct miR-140 targets. However, most target genes (17 out of 21) contained 7- or 8-mer target sites. The list of target genes and the distribution of target sites are shown on Table 1. We compared the list of 21 candidate mRNAs with miR-140 targets predicted by TargetScan, MiRBase, and PicTar. Surprisingly, none of the 21 mRNAs were predicted by any of the programs, suggesting that these programs generate a significant number of false negatives.
TABLE 1.
mRNAs repressed by miR-140 overexpression and derepressed by silencing of miR-140
The 28 genes without a potential target site are considered as indirect targets, although we cannot rule out that some of them are direct targets. Orom et al. (2008) demonstrated that genes encoding for ribosomal proteins are regulated by miR-10 in spite of the absence of sequence motifs complementary to the seed sequence of miR-10. There are no bioinformatics tools at the moment to identify this kind of direct targets; therefore, the 28 genes need to be analyzed in the future to establish whether they are directly targeted by miR-140.
Validation of Cxcl12 as a miR-140 target
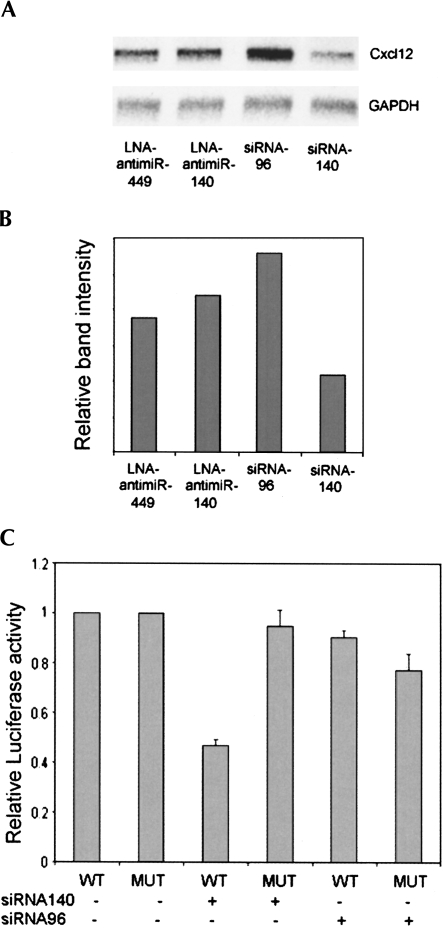
We chose one potential target mRNA identified from the list of candidate messages for experimental validation based on its reported involvement in osteogenesis (Zhu et al. 2007). First, we validated that the level of the CXC group of chemokine ligand 12 (Cxcl12, also known as Stromal-derived factor 1; SDF-1) mRNA changes when miR-140 activity is manipulated. Northern blot data confirmed the slight derepression of Cxcl12 as result of LNA-antimiR-140 treatment and the strong repression by siR-140 (Fig. 4A). Next, we tested whether Cxcl12 is a direct target of miR-140. 3′-UTR sequence of Cxcl12 was inserted downstream from the luciferase gene (luc-Cxcl12-wt) to assess the effect of miR-140 on luciferase activity. A mutant version of Cxcl12 3′-UTR, carrying point mutations in the miR-140 target site, was also cloned downstream from the luciferase gene (luc-Cxcl12-mut). The wild-type and mutant 3′-UTR containing luciferase plasmids were cotransfected with or without siRNA-140 or with siR-96 as negative control. Luciferase activity was significantly lower in cells cotransfected with luc-Cxcl12-wt and siR-140 than in cells cotransfected with luc-Cxcl12-wt and siR-96, luc-Cxcl12-mut and siR-140 or luc-Cxcl12 without siRNA (Fig. 4B). The Northern blot analysis and luciferase assay together confirmed that Cxcl12 is directly targeted by miR-140.
FIGURE 4.
Validation of Cxcl12 as a miR-140 target. (A) Northern blot analysis of Cxcl12 expression following transfection of cells with LNA-antimiR-449, LNA-antimiR-140, siRNA-96, or siRNA-140. (B) Quantification of the bands shown on panel A. The analysis confirmed the array result: slight derepression of Cxcl12 after treatment with LNA-antimiR-140 and strong repression by siRNA-140. (C) Luciferase assays were carried out to confirm that Cxcl12 is directly targeted by miR-140. The wild-type 3′-UTR sequence of Cxcl12 was cloned downstream from the luciferase gene, and this plasmid (WT) was transfected with or without siRNAs mimicking miR-140 or miR-96 (negative control). A mutant (MUT) construct was also used that contained mutations in the predicted target site of miR-140. Luciferase activities were normalized to transfections without siRNAs.
In addition to Cxcl12 we also validated the mRNA level of Lfrg15 and Rad50 by Northern blot (data not shown). mRNAs of another two genes (FosB and ZFand1) were below detection level by Northern blot, and therefore were not possible to validate (data not shown).
We present here an experimental approach to identify miRNA targets that relies on separate overexpression and silencing of a miRNA in the same cell line. The intersection of mRNAs that are repressed by miRNA overexpression and derepressed by miRNA silencing showed a specific and strong enrichment for a sequence signature that was complementary to the seed sequence of the manipulated miRNA. This approach revealed a relatively small number of target genes for miR-140, but it does not necessarily mean that this miRNA targets only these genes. This approach, similarly to other approaches, can identify targets that are expressed in the cell line used in the experiment. We expect that other targets can be found for miR-140 in other cell lines using this protocol. We envisage that this approach is feasible for other miRNAs and, thus, represents an attractive alternative to other target identification approaches. The advantage of this approach is that it uses a combination of routine techniques such as overexpression and silencing of miRNAs and mRNA array analysis. The weakness of the method is that it can only identify target genes that show altered accumulation at the mRNA level. This is demonstrated by the fact that one of the previously validated miR-140 targets, HDAC4 (Tuddenham et al. 2006), was missed by this analysis because its mRNA level does not change upon manipulation of miR-140 level (data not shown). However, the mRNA array analysis can be replaced with a proteomics profiling to maximise the potentials of the approach.
MATERIALS AND METHODS
Analysis of microarray data
Affymetrix GeneGhip Mouse Genome 430 2 Arrays were used for mRNA profiling. Data were imported to R (http://www.r-project.org/) using the Bioconductor (http://www.bioconductor.org/) package affy. For each of the four samples a microarray experiment was performed in triplicate. Probe set expression values were obtained from probe intensities by using the AffyProbeMiner software (Liu et al. 2007) including annotation for chip probes and grouping into probe sets. Expression values were normalized using the Robust Multichip Average (RMA) methodology (Irizarry et al. 2003).
To decide which probe sets should be considered as differentially expressed we used two approaches. In the first approach Bioconductor package limma (Smyth 2004) was used for each probe set to build a linear model and perform a t-test. Because siRNA transfections caused a much stronger effect on expression profile than LNA-antimiR transfections we used different cutoffs for the two experiments. A cutoff of 5% with a P-value adjusted for multiple testing was used for the siRNA experiments and a cutoff of 1% using the nonadjusted P-value was used for the LNA-antimiR experiments. In the second approach probe sets were ordered by average expression and for each of them a locally normalized ratio of expressions or z-value was calculated (Yang et al. 2002). We considered a cutoff of 1.96, corresponding to a probability of 5%.
Sequence data
miRNA sequences were retrieved from the miRBase database (http://miRNA.sanger.ac.uk/sequences/). 3′-UTR sequences were used from two sources to maximize the number of genes with retrieved 3′-UTRs. Both UCSC (http://genome.ucsc.edu/) and Ensembl (http://www.ensembl.org/) genome browsers were consulted. RefSeq IDs were used to map probe sets to UCSC database and Emsembl Gene IDs were used to map probesets to Ensembl database. When different 3′-UTR sequences were reported by the two databases both sequences were used.
To predict direct miRNA targets we considered three types of seed matches, based on Grimson et al. (2007). The first type of seed match was six nucleotides in length and was complementary to nucleotides 2 to 7 in the miRNA. The second group of seed matches was 7 nt, and was complementary to nucleotides 1–7 in the miRNA or nucleotides 2–7 in the miRNA with “A” at the first position. The third class of seed matches was 8 nt, and matched nucleotides 1–8 in the miRNA or nucleotides 2–8 in the miRNA with an “A” at the first position.
DNA constructs
A modified pGL3 control vector (Tuddenham et al. 2006) was used for cloning the Cxcl12 3′-UTR. For the wild-type construct, 796 base pairs (bp) of the Cxcl12 3′-UTR were PCR amplified from mouse genomic DNA using a pair of oligonucleotides (5′-AGCCCAATTCGAAGCCAGTCTCTAG-3′ and 5′-TGGCGCCCAAGGGAATTCTGGCAG-3′) and cloned into pGemT-Easy (Promega). The 3′-UTR was then PCR amplified from pGemT-Easy to incorporate SacI and NheI restriction sites to the 5′ and 3′ ends, respectively, and inserted into the modified pGL3. For the mutant pGL3 construct, the seed sequence of the predicted target site was replaced with a HindIII site introducing eight point mutations. The perfect match (pLuc-140) plasmid was obtained by cloning the mir-140 complementary sequence into the modified pGL3 vector.
Transfections and luciferase reporter assays
3T3, C3H10T1/2, and DF1 cell lines were cultured in Dulbecco's modified Eagle medium (DMEM) containing 2 mM L-glutamine and 10% fetal bovine serum (FBS) (Gibco, Invitrogen). Cells (3 × 104 cells/well) were transfected with Lipofectamine 2000 (Invitrogen) with either pLuc-140, wild-type, or mutant constructs (200 ng), with and without siRNA-140 or siRNA-96 (30 nM) (Sigma), or LNA-antimiR-140: 5′-[FAM]-TAgGgTAaAamCmCamCT-3′ or LNA-antimiR-449: 5′-[FAM]-TaAmCaATacAmCTgmCmC-3′ (complete phosphorothioate backbone; LNA uppercase; DNA lowercase; mC denotes LNA methylcytosine; 5 nM). SiRNA-140 consisted of 5′-CAGUGGUUUUACCCUAUGGUAG-3′ and 5′-ACCAUAGGGUAAAACCACUGAG-3′ oligonucleotides and siRNA-96 was made up of 5′-UUUGGCACUAGCACAUUUUUGCUUG-3′ and 5′-AGCAAUCAUGUGUAGUGCCAAUAU-3′ oligonucleotides.
As a positive control, the modified pGL3 control vector was used without a 3′-UTR insert. Lipofectamine-only treated cells served as negative controls. Transfections were carried out six times in triplicate using two independent plasmid preparations. Luciferase activity was measured 48 h later using a multilabel counter (Victor2, Perkin-Elmer). Relative reporter activity for siRNA-140 treated cells was obtained by normalization to non-siRNA-140 treated wild-type or mutant constructs, respectively.
Northern blot analysis
Total RNA was extracted from cells using Trizol reagent, following the recommendations of the supplier (Invitrogen). To detect Cxcl12 mRNA, 20 μg of total RNA from each sample was separated on 1% denaturing agarose gel, blotted to membrane, and hybridized to radioactively labeled probes overnight at 37°C in ULTRAhyb hybridization buffer (Ambion). Probes were labeled with [α-32P]dCTP using Ready-to-Go DNA labeling beads (Amersham Pharmacia Biotech), following the instructions of the supplier. The Cxcl12 probe was the same 796-bp fragment used in the luciferase assays. The loading control Gapdh probe is a 298-bp fragment PCR amplified using the oligonucleotide pair 5′-ATTTGGCCGTATTGGGCGCCTGGTCACCA-3′ and 5′-AAGACACCAGTAGACTCCACGACATAC-3′. To detect miR-140 expression, 30 micrograms of each total RNA sample was resolved on a 15% denaturing polyacrylamide gel and transferred to Zeta-probe membrane (Bio-Rad) using a semidry electroblotting apparatus. Membranes were hybridized overnight at 37°C in ULTRAhyb-Oligo hybridization buffer (Ambion) with γ-ATP labeled oligonucleotides complementary to miR-140. Membranes were exposed to Kodak Phosphor Screen SD230 for quantification. After exposure, the screen was scanned on a Molecular Imager FX reader (Bio-Rad).
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported by the European Commission (FP6 Integrated Project SIROCCO LSHG-CT-2006-037900) to T.D. and by grants from the Danish National Advanced Technology Foundation and Danish Medical Research Council to S.K. The Wilhelm Johannsen Centre for Functional Genome Research was established by the Danish National Research Foundation, Copenhagen, Denmark (www.dg.dk). H.P. is a student of Instituto Gulbenkian de Ciência's Ph.D. Program in Computational Biology (sponsored by Fundação Para a Ciência e a Tecnologia [FCT], Fundação Calouste Gulbenkian, Siemens SA Portugal) and was supported by FCT fellowship SFRH/BD/33204/2007.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1221108.
REFERENCES
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Beitzinger M., Peters L., Zhu J.Y., Kremmer E., Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNAtarget recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiano D., Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA–target interactions. Nat. Struct. Mol. Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- Duursma A.M., Kedde M., Schrier M., le Sage C., Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J., Lindow M., Silahtaroglu A., Bak M., Christensen M., Lind-Thomsen A., Hedtjärn M., Hansen J.B., Hansen H.F., Straarup E.M., et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008a;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J., Lindow M., Schütz S., Lawrence M., Petri A., Obad S., Lindholm M., Hedtjärn M., Hansen H.F., Berger U., et al. LNA-mediated microRNA silencing in nonhuman primates. Nature. 2008b;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K.K., Garrett-Engele P., Lim L.P., Bartel D.P. miRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of affymetrix genechip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov F.V., Conaco C., Xuan Z., Schmidt B.H., Parker J.S., Mandel G., Hannon G.J. A biochemical approach to identifying microRNA targets. Proc. Natl. Acad. Sci. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. The role of site accessibility in microrna target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Plasterk R.H. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Wienholds E., Ketting R.F., Plasterk R.H. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2005;32:6284–6291. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with “antagomirs.”. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are miRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu H., Zeeberg B.R., Qu G., Koru A.G., Ferrucci A., Kahn A., Ryan M.C., Nuhanovic A., Munson P.J., Reinhold W.C., et al. Affyprobeminer: A web resource for computing or retrieving accurately redefined affymetrix probe sets. Bioinformatics. 2007;23:2385–2390. doi: 10.1093/bioinformatics/btm360. [DOI] [PubMed] [Google Scholar]
- Nielsen C.B., Shomron N., Sandberg R., Hornstein E., Kitzman J., Burge C.B. Determinants of targeting by endogenous and exogenous miRNAs and sirnas. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom U.A., Nielsen F.C., Lund A.H. MicroRNA-10a binds the 5′-UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Petersen C.P., Bordeleau M.E., Pelletier J., Sharp P.A. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Sethupathy P., Megraw M., Hatzigeorgiou A.G. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat. Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- Tuddenham L., Wheeler G., Ntounia-Fousara S., Waters J., Hajihosseini M.K., Clark I., Dalmay T. The cartilage specific microRNA-140 targets Histone Deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. Post-transcriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans . Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Yang I., Chen E., Hasseman J., Liang W., Frank B., Wang S., Sharov V., Saeed A., White J., Li J., Lee N., Yeatman T., Quackenbush J. Within the fold: Assessing differential expression measures and reproducibility in microarray assays. Genome Biol. 2002;3:research0062.1–research0062.12. doi: 10.1186/gb-2002-3-11-research0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Samal E., Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- Zhu W., Boachie-Adjei O., Rawlins B.A., Frenkel B., Boskey A.L., Ivashkiv L.B., Blobel C.P. A novel regulatory role for stromal-derived factor-1 signaling in bone morphogenic protein-2 osteogenic differentiation of mesenchymal C2C12 cells. J. Biol. Chem. 2007;282:18676–18685. doi: 10.1074/jbc.M610232200. [DOI] [PubMed] [Google Scholar]