Abstract
Pre-mRNA 3′ end formation is tightly linked to upstream and downstream events of eukaryotic mRNA synthesis. The two-step reaction involves endonucleolytic cleavage of the primary transcript followed by poly(A) addition to the upstream cleavage product. To further characterize the putative 3′ end processing endonuclease Ysh1p/Brr5p, we isolated and analyzed a number of new temperature- and cold-sensitive mutant alleles. We show that Ysh1p plays a crucial role in 3′ end formation and in RNA polymerase II (RNAP II) transcription termination on mRNA genes. In addition, we observed a range of additional functional deficiencies in ysh1 mutant strains, which were partially allele-specific. Interestingly, snoRNA 3′ end formation and RNAP II termination were defective on specific snoRNAs in the cold-sensitive ysh1-12 strain. Moreover, we observed the accumulation of several mRNAs including the NRD1 transcript in this mutant. We provide evidence that NRD1 autoregulation is associated with endonucleolytic cleavage and that this process may involve Ysh1p. In addition, the ysh1-12 strain displayed defects in RNA splicing indicating that a functional link may exist between intron removal and 3′ end formation in yeast. These observations suggest that Ysh1p has multiple roles in RNA synthesis and processing.
Keywords: 3′ end formation, 3′ endonuclease, RNA polymerase II transcription termination, snoRNA, Nrd1 autoregulation
INTRODUCTION
RNA polymerase II (RNAP II) transcribes protein-encoding genes and a subset of noncoding small nuclear RNAs (snRNA) and small nucleolar RNAs (snoRNA). In order to be biologically functional, all primary transcripts of RNAP II need to undergo extensive processing. For mRNAs, this includes capping at the 5′ end, removal of introns by splicing, and 3′ end processing.
Pre-mRNA 3′ end processing is initiated by endonucleolytic cleavage at the poly(A) site, followed by polyadenylation of the upstream cleavage product. In yeast, 3′ end processing is performed by a large complex of proteins, including cleavage and polyadenylation factor (CPF), cleavage factor IA (CF IA), and cleavage factor IB (CF IB) (Zhao et al. 1999). Ysh1p/Brr5p is associated with CPF and was first identified by sequence homology with mammalian CPSF73 (Chanfreau et al. 1996; Jenny et al. 1996). It was recognized early on that the protein carries a highly conserved β-lactamase fold commonly found in metal-dependent hydrolytic enzymes (Aravind 1999; Daiyasu et al. 2001; Callebaut et al. 2002). Although this suggested that catalytic activity was associated with Ysh1p, convincing structural and biochemical evidence for endonucleolytic activity has emerged only recently (Ryan et al. 2004; Wickens and Gonzalez 2004; Mandel et al. 2006).
Cleavage and polyadenylation factors are required for termination of RNAP II transcription (Connelly and Manley 1988; Proudfoot 2004; Buratowski 2005). Two general models have been proposed to explain the mechanism of termination (Proudfoot et al. 2002; Luo and Bentley 2004; Bentley 2005; Buratowski 2005). In the “anti-terminator” model, RNAP II complex undergoes conformational changes in response to the emerging terminator sequences on RNA either by recruiting termination factors and/or by displacing positive elongation factors (Logan et al. 1987; Orozco et al. 2002; Zhang et al. 2005; Zhang and Gilmour 2006). This renders the RNAP II complex termination competent and leads to the loss of processivity and gradual termination. The “torpedo” model proposes that RNAP II transcription termination is triggered by poly(A) site cleavage and subsequent degradation of the 3′ downstream RNA by Rat1p 5′-3′ exonuclease (Connelly and Manley 1988; Kim et al. 2004; West et al. 2004). Experimental support for either model has been provided in several eukaryotic experimental systems (Gilmour and Fan 2008), and a unified model of termination has been proposed that combines mechanistic predictions of both models (Luo et al. 2006).
All snRNAs and most snoRNAs in yeast are synthesized independently by RNAP II. The 3′ ends of snoRNA transcripts are produced by 3′-5′ exonucleolytic trimming that follows either endonucleolytic cleavage or RNAP II termination (Chanfreau et al. 1998; Allmang et al. 1999; van Hoof et al. 2000; Butler 2002). Numerous protein factors were reported to be essential for transcription termination on snoRNA genes, including the RNAP II subunits Rpb3p and Rpb11p, the sequence-specific RNA-binding proteins Nab3p and Nrd1p, the Sen1p helicase, the CTD kinase Ctk1p, and the RNAP II-associated Paf1 complex (Ursic et al. 1997; Conrad et al. 2000; Steinmetz et al. 2001, 2006a; Sheldon et al. 2005). Furthermore, several subunits of the 3′ end processing complexes CF IA (Pcf11p, Rna15p, and Rna14p) and CPF (Pta1p, Pti1p, Ref2p, Ssu72p, Swd2p, and Glc7p) were implicated in snoRNA termination (Fatica et al. 2000; Morlando et al. 2002; Dheur et al. 2003; Ganem et al. 2003; Nedea et al. 2003, 2008; Steinmetz and Brow 2003; Cheng et al. 2004; Dichtl et al. 2004; Kim et al. 2006).
In this study, we isolated a number of conditional ysh1 mutant alleles and characterized them for defects in mRNA and snoRNA synthesis. We found that all analyzed ysh1 mutants were deficient in pre-mRNA 3′ end formation and RNAP II transcription termination on mRNA genes. Moreover, a cold-sensitive ysh1 mutant strain displayed distinct defects in snoRNA 3′ end formation, termination on snoRNA genes, and RNA splicing. We also provide evidence that implies endonucleolytic cleavage and functional Ysh1p in the process of regulated premature termination at the NRD1 locus. Altogether, this study underscores the central role of the 3′ end processing endonuclease Ysh1p in cellular RNA metabolism.
RESULTS
Ysh1p is required for 3′ end cleavage and polyadenylation of pre-mRNA in vitro
To address the cellular role of the putative 3′ endonuclease Ysh1p, we initially produced point mutations within the β-lactamase consensus signature motif H68X69H70X71D72H73, which is located at the amino-terminal end of the protein and which contributes to formation of the catalytic core of the protein (Fig. 1C; Aravind 1999; Mandel et al. 2006). We found that alanine substitutions of the highly conserved histidines H68 and H70 or of aspartate D72 resulted in lethality (Fig. 1A). This observation underscored the functional significance of this motif but hampered further functional analyses. Therefore, we isolated temperature-sensitive (ts) and cold-sensitive (cs) alleles of the YSH1/BRR5 gene using random mutagenesis (Fig. 1B; see Materials and Methods). Of the mutant strains chosen for further analysis, three were ts, with ysh1-32 being lethal at 33°C, and ysh1-13 and ysh1-15 being lethal at 37°C; in contrast, the cs ysh1-12 strain ceased growth at 15°C (Fig. 1B). Amino acid changes within mutant Ysh1p proteins were checked by DNA sequencing and found to localize predominantly to the nonconserved C-terminus (Fig. 1C); this part of the protein is involved in both catalytic steps of 3′ end processing and mediates the interaction with Pta1p (Zhelkovsky et al. 2006).
FIGURE 1.
Isolation of conditional ysh1 mutants. (A) The conserved β-lactamase signature H68X69H70X71D72H73 is essential for cell viability. Plasmid shuffling was used to test the requirement of H68, H70, and D72 residues of the β-lactamase consensus motif for cell viability. The LM109 strain with a disrupted chromosomal YSH1 gene and carrying YSH1-URA3 plasmid was transformed with the ADE2-plasmid bearing either the wild-type YSH1 gene or its mutant versions H68A, H70A, and D72A, followed by counterselection on 5′-FOA plates. (B) Tenfold serial dilutions of cultures of wild-type and ysh1 mutant strains were spotted onto YPAD medium and incubated for 3 d at 23°–37°C or 5 d at 15°C. (C) Schematic representation of ysh1 mutant sequences underlying the respective temperature- or cold-sensitive phenotypes. (Shaded boxes) The β-lactamase, β-CASP, and C-terminal domains and the conserved H68F69H70L71D72H73 signature are marked approximately.
Initially, we tested the ysh1 mutant strains for cleavage and polyadenylation in vitro (Fig. 2A,B). Total cell extracts were produced from wild-type and mutant strains (ysh1-12, ysh1-13, ysh1-15, ysh1-32) and assayed on a synthetic CYC1 substrate for cleavage and on pre-cleaved CYC1-pre RNA for polyadenylation. Comparing the effect of the mutations to wild-type, we observed that both cleavage and polyadenylation were reduced in all the ysh1 mutant strains, with the defects being more pronounced at the restrictive temperatures (Fig. 2A,B).
FIGURE 2.
Ysh1p is required for cleavage and polyadenylaion of pre-mRNA in vitro. (A, upper panel) In vitro cleavage and (lower panel) polyadenylation assays with protein extracts prepared from wild-type and ysh1 temperature-sensitive strains as indicated. Input shows a control where no protein was added. Positions of substrate RNAs, 5′ and 3′ end cleavage products and polyadenylation products bands are shown. HpaII-digested pBR322 fragments were 5′ end labeled and served as markers. Internally [32P]-labeled substrate RNAs, CYC1 for the cleavage assay and CYC1-pre for specific polyadenylation, were used. Reactions were performed either at 30°C (lanes 1–6) or at 37°C (lanes 7–12). Extracts were preincubated at restrictive temperatures for 10 min prior to assaying. (B) As in A, except reactions were performed either at the permissive temperature 30°C (lanes 1–3) or at the non-permissive temperatures 17°C (lanes 4–6) for cleavage, and at 15°C for polyadenylation. (C,D) Reconstitution of specific cleavage and polyadenylation activities of the ysh1-32 extract in vitro. (C) Cleavage and (D) polyadenylation assays were performed essentially as in A, with protein extracts prepared from wild-type and the ysh1-32 strain as indicated. The ysh1-32 extract was combined with purified CPF to restore the specific 3′end processing activity. Twice more CPF was used for the reconstitution of polyadenylation than for the reconstitution of cleavage.
To confirm that the observed defects in pre-mRNA 3′ end formation were due to inactive CPF (the factor that contains Ysh1p), we aimed to reconstitute its activity by addition of purified wild-type CPF to the ysh1-32 extract (Fig. 2C,D). We found that CPF partially rescued the cleavage defect of the mutant extract at both permissive and restrictive temperatures (Fig. 2C, lanes 4,9). Likewise, specific polyadenylation activity of the ysh1-32 extract was restored by addition of CPF (Fig. 2D, lanes 4,9). CPF alone was not able to cleave the CYC1 RNA (Fig. 2C, lanes 5,10) and gave unspecific hyperadenylation of the CYC1-pre RNA substrate as previously observed (Fig. 2D, lanes 5,10; Preker et al. 1997). Taken together, the above results underscore an important role for Ysh1p in both catalytic steps of pre-mRNA 3′ end formation.
Distinct effects of mutations within Ysh1p on mRNA abundance
Next, we performed Northern blotting to analyze the steady-state levels of mRNAs in ysh1 mutant strains before and after shift to restrictive temperatures. We found that several mRNAs (ASC1, ASN1, PGK1, RPS16A) were significantly reduced in ysh1-32 strains following growth at 37°C (Fig. 3A, lanes 6–8); similar phenotypes were observed for ysh1-13 and ysh1-15 mutant strains (data not shown). The well-characterized rna15-1 strain was used as a control and showed the expected strong reduction of mRNAs after 1-h shift to 37°C (Fig. 3A, lane 4; Minvielle-Sebastia et al. 1994). Unexpectedly, the reverse effect was observed in ysh1-12 cells, where the levels of all tested mRNAs were relatively stable or even increased upon shift to non-permissive temperature (Fig. 3A, lanes 12–14); this included NAB2 mRNA, which increased up to 13-fold. Interestingly, Nab2p is believed to autoregulate the levels of its own mRNA in a process that requires the nuclear exosome component Rrp6p (Roth et al. 2005), and our results suggest a role for Ysh1p in this process (see Discussion).
FIGURE 3.
Ysh1p is required for normal mRNA accumulation and poly(A) site selection. (A) Northern blot analysis of mRNA steady-state levels in ysh1 strains. Wild-type, ysh1-32, and rna15-1 strains were grown in YPD at 25°C and shifted for 1, 3, and 6 h to 37°C. The cold-sensitive strain ysh1-12 was grown in YPD at 30°C and shifted for 1, 3 and 6 h to 15°C. Total RNA was extracted from wild-type and mutant ysh1 cells and separated on formaldehyde/1.2% agarose gels. Filters were developed with either random-primed labeled DNA probes or end-labeled oligonucleotides directed against RNA species indicated on the left. 18S rRNA and SCR1 served as loading controls. (B) Poly(A) tails shorten in the ysh1-32 strain after shift to restrictive temperature. 3′ End labeling of poly(A) tails with total RNA extracted from (left panel) wild-type and ysh1-32 or (right panel) wild-type and ysh1-12 mutant strains after growth at permissive temperatures (23°C or 30°C, respectively), and after shift for 1, 3, and 6 h to respective restrictive temperatures (37°C or 15°C). Poly(A) tail length (in nucleotides) is indicated on the left. (C) Western blot analysis of wild-type, ysh1-32, and ysh1-12 mutant extracts prepared from cells grown analogously to A. Equal amounts of total protein were loaded in each lane. Blots were probed with antibodies directed against the proteins indicated on the left. (D) Northern blot analysis of total RNAs extracted from wild-type and mutant ysh1 cells. The ysh1-13, ysh1-15, ysh1-32, and rna15-1 strains were grown in YPD at 23°C and shifted for 1, 3, and 6 h to 37°C. ysh1-12 was grown at 30°C and shifted for 1, 3, and 6 h to 15°C. The analysis was performed essentially as in A. 18S rRNA served as a loading control. The positions of ACT1 poly(A) sites I–V are labeled. The scheme below the panel represents the relative positions of the different ACT1 poly(A) sites. (E) Analysis of ACT1 poly(A) site usage in wild-type, ysh1-13, and ysh1-15 cells. Total RNAs extracted from strains grown as described in E were treated with RNase H and oligonucleotides ACT1-RNase H and oligo(dT), and analyzed by polyacrylamide Northern blotting with a probe specific for the 3′ end of ACT1 mRNA. Positions of the poly(A) sites I–V are indicated on the right. RNA levels were quantified by PhosphorImager scanning (Molecular Dynamics). The ratios of poly(A) site I versus site V usage for each lane are indicated at the bottom.
To evaluate the global phenotypes of our mutants, we analyzed the length distribution of cellular poly(A). The ts ysh1-32, ysh1-13, and ysh1-15 strains showed a marked reduction of poly(A) tail length after growth at 37°C (Fig. 3B, left panel; data not shown). In contrast, the poly(A) tail length distribution remained unchanged following shift to the restrictive temperature for ysh1-12 (Fig. 3B, right panel). Thus, global poly(A) length reflected the trends of mRNA levels observed in the Northern blot analyses for both the ysh1 ts strains (reduced mRNA) and the ysh1-12 cs strain (unchanged or increased mRNA).
To test whether the observed phenotypes were due to underaccumulation of Ysh1p or other subunits of the 3′ end processing complex, we carried out Western blot analyses (Fig. 3C). We observed reduced levels of Ysh1p and Ssu72p in the ysh1-32 strain after 1-h growth at 37°C (Fig. 3C, lanes 4–6), whereas other CPF components (Pfs2p, Pap1p, Fip1p) and Rna15p remained largely unchanged. Interestingly, the amounts of all analyzed proteins remained relatively stable in the ysh1-12 strain during a 6-h shift to 15°C (Fig. 3C, lanes 9–12). These results support the view that the ts and cs ysh1 mutations caused distinct functional deficiencies (see Discussion).
Ysh1p is required for correct poly(A) site recognition of ACT1 pre-mRNA
Several subunits of the CPF complex including Yhh1p and Ydh1p were previously shown to be required for poly(A) site recognition (Dichtl et al. 2002b; Kyburz et al. 2003). The well-characterized 3′-UTR of the ACT1 gene includes five alternative polyadenylation sites and is therefore useful to examine poly(A) site selection in 3′ end processing mutants (see scheme in Fig. 3D; Mandart and Parker 1995). Low-resolution Northern blot analysis revealed a change in the ACT1 hybridization pattern in ysh1 mutants compared to wild-type (Fig. 3D). To corroborate this phenotype, we further analyzed RNA obtained from two mutant strains (ysh1-13 and ysh1-15) on high-resolution Northern blots following RNase H treatment (Mandart and Parker 1995). In these experiments, total RNA was targeted by RNase H and oligonucleotides complementary to the 3′ end of ACT1 mRNA and to the poly(A) tail allowing the resolution of the five poly(A) sites (Fig. 3E). Quantification showed that the most proximal poly(A) site I was preferentially used in wild-type cells, as expected; downstream sites II–V were utilized four to seven times less frequently. In contrast, ysh1 mutant strains displayed a change in poly(A) site usage toward the minor site V, both at permissive and restrictive temperatures. Taken together, these results demonstrate the involvement of Ysh1p in correct poly(A) site recognition.
Ysh1p is required for RNAP II transcription termination on mRNA genes
To address the role of Ysh1p in RNAP II transcription termination, we carried out transcriptional run-on analysis (TRO) on a plasmid-borne CYC1 gene, which is placed under the control of the GAL1/10 promoter (Birse et al. 1998). Correct termination produces run-on signals over probes P1–P3, whereas defective termination results in increased signals also over probes P4–P6 (see scheme in Fig. 4A). TRO was performed with wild-type and ysh1-12 cells following growth in galactose-containing medium at permissive and restrictive conditions. Quantification of slot hybridizations showed that the ysh1-12 mutant displayed an approximately twofold to threefold increased run-on signal over probes P3–P6 (Fig. 4A,B). Similar results were obtained with ysh1-32 and ysh1-15 mutants (Fig. 4C; data not shown).
FIGURE 4.
Mutations in YSH1 impair correct RNAP II transcription termination on mRNA genes in vivo. (A) Slot hybridizations of run-on transcripts obtained after transcriptional run-on analysis (TRO) (Birse et al. 1998). The scheme represents the pGAL-CYC1 gene construct that was used for TRO analysis. The order of M13 probes spanning the CYC1 gene (P1–P6) and poly(A) site (position 506) are depicted. Wild-type and ysh1-12 cells transformed with pUGCYC1 plasmid were grown in synthetic medium lacking uracil and containing 2% galactose under permissive growth conditions (30°C) and after shift for 1 h to restrictive temperature (15°C). P1–P6 represent probes complementary to CYC1 transcripts as indicated on the scheme. Empty M13 was used as a background hybridization control. Hybridization of RNAP III transcripts to the tRNA probe is shown. (B) Quantitative analysis of transcriptional run-on profiles for the ysh1-12 mutant. Values obtained by PhosphorImager scanning (Molecular Dynamics) were corrected by subtraction of the M13 background signal and normalized to the value of P1, which was fixed at 100%. Results shown in the diagram represent the average value of three independent experiments. (C) Quantitative analysis of transcriptional run-on profiles for the ysh1-32 strain. Values were obtained as described in B. (D,E) Read-through mRNA transcripts accumulate in the ysh1-12 strain in vivo. Northern blot analysis of total RNA extracted from wild-type and ysh1-12 cells (lanes 1–3,7–9) grown in YPD medium at permissive temperature 30°C and (lanes 4–6,10–12) followed by a shift for 1, 3 or 6 h to 15°C. RNAs were resolved as described in Figure 3A. The filter was developed with random-primed labeled probe directed against (D) YSH1 or (E) ADH1, then subsequently washed and probed for the RNA of the downstream gene (DBP9 or YOL087C, respectively), as indicated. (*) Unidentified intermediate-sized transcripts. Probing to the RNAP III transcript SCR1 served as a loading control. (Bottom) Schematic representation of the (D) YSH1-DBP9 and the (E) ADH1-YOL087C gene loci.
It has previously been shown that defective termination can result in production of polycistronic transcripts (Connelly and Manley 1988). In line with this notion, we observed read-through RNAs in ysh1-12 cells at restrictive temperature. For example, the YSH1 transcript extended into the downstream gene DBP9 and ADH1 mRNA extended into the downstream gene YOL087C (Fig. 4E). The read-through transcripts were not detectable in the ts ysh1 mutant strains (ysh1-13, ysh1-15, ysh1-32) (data not shown), which is presumably resulting from a reduced stability of read-through RNAs in these strains. Taken together, the above results support a role for Ysh1p in termination of RNAP II transcription on mRNA genes.
snoRNA 3′ end formation is impaired in the ysh1-12 strain
To address the question whether Ysh1p is required for the 3′ end processing of snoRNA, we performed primer extension analysis on total RNA; in these experiments, the appearance of an extension signal is an indication of a defect in 3′ end formation of these noncoding RNAs. In the ysh1-12 strain, we detected accumulation of 3′ extended transcripts of snR13, snR33, snR39B, snR46, snR47, and snR128 snoRNAs (Fig. 5A, lanes 3–6). In contrast, no extended RNAs were observed for the ysh1-32 strain, when snR13, snR33, and snR47 were analyzed (data not shown), suggesting that the snoRNA processing defects in the ysh1 strains are allele-specific. However, we were unable to detect any defects in the 3′ end formation of several other snoRNAs such as snR3, snR50, snR71, and snR45 neither for ysh1-12 nor for the positive control strains nrd1-102 and sen1-1 (data not shown); this could be due to limitations of the primer extension procedure.
FIGURE 5.
Mutations in ysh1-12 impair snoRNA 3′ end formation and transcription termination. (A) 3′ end extended snoRNA transcripts are produced in the ysh1-12 mutant strain. Primer extension (PE) analysis of extended transcripts of several snoRNAs in the wild-type and ysh1-12 strains was carried out with radioactively labeled oligonucleotides complementary to sequences located downstream from the mature 3′ ends of the indicated snoRNAs. An oligonucleotide complementary to the mature U3 snoRNA was used as control. (B) Transcription termination on the SNR3 snoRNA gene is impaired in ysh1-12. Slot hybridizations of run-on transcripts were obtained after TRO. (Top) The scheme represents the SNR3-YJR129c genomic locus and location of the TRO probes A and B. Note that SNR3 and YJR129c are transcribed from opposite strands of the DNA, as indicated by the arrows. (Lanes 1,2) Wild-type and ysh1-12 cells were grown in YPD medium containing 2% glucose under permissive growth conditions (30°C) and (lanes 3,4) after the shift to restrictive temperature (15°C) for 1 h. M13 slots are single-stranded phagemids with no insert and were used as a background hybridization control. The ACT1 probe served as a positive control for hybridization. (C) Quantitative analysis of transcriptional run-on profiles for the ysh1-12 mutant. Values obtained by PhosphorImager scanning (Molecular Dynamics) were corrected by subtraction of the M13 background signal and normalized to the value of probe A, which was set at 100%. Results shown in the diagram represent the average value of three independent experiments. (D) Western blot analysis of isogenic wild-type (FY23) and brr5-td degron strain extracts prepared from cells grown at 25°C and shifted for 1, 2, or 3 h to 37°C. Equal amounts of total protein were loaded in each lane. The blot was probed with antibody directed against Ysh1p and subsequently re-probed with antibodies directed against other CPF subunits, as indicated. (E) Primer extension analysis of extended transcripts of several snoRNAs in the wild-type FY23, brr5-td (both shifted for 2 h to 37°C), and ysh1-12 (shifted for 6 h to 15°C) strains was carried out with radioactively labeled oligonucleotides complementary to sequences located downstream from the mature 3′ ends of the indicated snoRNAs. (Lanes 1,3) Levels of the RNAP III transcript U6 served as a loading control and showed that lower amounts of total RNA extracted from the FY23 and brr5-td strains were used in this experiment.
Next, we used transcription run-on analysis to measure RNAP II termination efficiency on the endogenous SNR3 locus (Steinmetz et al. 2001). Two probes were used, one spanning the SNR3 gene and its terminator sequences (probe A), and one downstream from this position (probe B) (Fig. 5B). These single-stranded DNAs do not hybridize with mRNA derived from the downstream ORF YJR129c; however, we cannot exclude the possibility that they could hybridize to antisense transcripts that may originate from this gene locus. Interestingly, ysh1-12 mutant cells revealed up to sixfold higher density of transcribing RNAP II over the downstream probe compared to wild type (Fig. 5B,C), indicating defective termination on this transcription unit. In contrast, termination on the SNR3 gene was unaffected in the ysh1-32 strain (data not shown), which is consistent with the primer extension analysis above.
Conditional depletion of Ysh1p in the brr5-td degron strain was reported not to affect termination at snoRNA genes, as determined by RNAP II chromatin immunoprecipitation (ChIP) experiments (Kim et al. 2006). As these data conflicted with our observations, we included the brr5-td strain in our analysis. The degron-Ysh1p levels were significantly reduced after 1-h growth at 37°C, and almost completely diminished after 3 h (Fig. 5D). In contrast, the levels of Fip1p, Ssu72p, and Rna15p remained unchanged following depletion of Ysh1p. We compared snoRNA 3′ end formation in brr5-td, its isogenic wild type (FY23), and ysh1-12 strains by primer extension analysis as described above. 3′-Extended forms of the tested snoRNAs (snR13, snR33, snR39B, and snR47) were readily detected in the ysh1-12 strain (Fig. 5E) but not following depletion of Ysh1p. These observations indicated that defects in snoRNA 3′ end formation were apparent only with distinct ysh1 mutant strains on a subset of snoRNA transcription units (see Discussion).
Putative role for Ysh1p in mediating premature termination within the NRD1 gene
Nrd1p controls its own expression through regulated premature termination, and levels of NRD1 mRNA are increased in nrd1, nab3, and sen1 mutant strains (Steinmetz et al. 2001; Arigo et al. 2006). Interestingly, we observed increased levels of NRD1 mRNA in ysh1-12 cells at the non-permissive temperature (Fig. 6A), which resulted in increased amounts of Nrd1p protein (Fig. 6B). Such a phenotype was not observed with the three ysh1 ts mutant strains (data not shown). Since a similar increase in Nrd1p protein was observed in the nrd1-102 strain, this indicated that Ysh1p is involved in regulating Nrd1p levels. Moreover, some NRD1 transcripts extended into the downstream gene MRPL17, resulting in a dicistronic NRD1-MRPL17 transcript (Fig. 6A, lanes 4–6). We also observed an ∼1-kb RNA that corresponded to 3′-truncated NRD1 RNA in the wild-type strain as previously reported (Arigo et al. 2006). We detected this RNA with a probe directed against the 5′ end of NRD1 mRNA but not with a probe directed against the 3′ end (data not shown). Interestingly, we found that this RNA was absent in ysh1-12 (Fig. 6A).
FIGURE 6.
Involvement of Ysh1p in the autoregulation of NRD1 mRNA levels. (A) Increased levels of NRD1 mRNA accumulate in the ysh1-12 mutant strain. Northern blot analysis of NRD1 mRNA in wild-type and ysh1-12 strains was performed as in Figure 3A, with probes directed against the 5′ end of NRD1 mRNA, or against MRPL17, as indicated on the left. (Top) The scheme represents the NRD1-MRPL17 gene loci; transcripts originating from the NRD1 transcription start are indicated. Hybridization to the RNAP III transcript SCR1 served as a loading control. (B) The amount of Nrd1 protein increases in ysh1-12 cells. Western blot analysis of wild-type and mutant extracts prepared from cells grown in YPD at 25°C and shifted for 3 h to restrictive temperatures (15°C or 37°C, respectively). Equal amounts of total protein were loaded in each lane. Blots were probed with antibodies directed against the proteins indicated on the left. Act1p levels served as control for equal loading. (C) NRD1 premature termination occurs via an endonucleolytic cleavage of the NRD1 pre-mRNA. (Upper part) Schematic representation of the NRD1 gene locus. Positions of the oligonucleotides U (+300 nt) and D (+1300 nt; relative to the ORF start) used in the primer extension analyses, the full-length and prematurely terminated NRD1 mRNA transcripts, and of the putative 3′ end cutoff product are shown. (Below, right panel) Primer extension analysis of total RNA extracted from wild-type (W303) and rat1-1, xrn1Δ strains (grown for 2 h at 37°C), carried out with radioactively labeled oligonucleotide D complementary to sequences located downstream from the predicted premature termination region. (Left panel) Oligonucleotide U complementary to the 5′ end of NRD1 transcript was used as an internal control.
The above observations on the ysh1-12 phenotype suggested that Ysh1p might be involved in the regulation of NRD1 expression. We speculated that Ysh1p could facilitate premature termination through endonucleolytic cleavage of the NRD1 transcript. Direct evidence for occurrence of cleavage within NRD1 would be the identification of downstream 3′ cutoff products. Since such intermediates are predicted to be rapidly degraded by 5′-3′ exonucleases, we analyzed total RNA extracted from the rat1-1, xrn1Δ double-mutant strain (Henry et al. 1994). Primer extension analysis was carried out with oligonucleotide D, which is complementary to sequences located downstream from the predicted premature termination region (see scheme in Fig. 6C). Most interestingly, a primer extension stop was obtained that corresponded in size to a predicted downstream 3′ cutoff product (Fig. 6C, lane 4). An equivalent primer extension stop was not detected with wild-type RNA, where a higher molecular weight band was detectable instead (Fig. 6C, lane 3); this stop most likely represented the 5′ end of the NRD1 mRNA. As a control, we used oligonucleotide U, which is complementary to the 5′ end of the NRD1 transcript and which gave primer extension stops that corresponded to the 5′ end of NRD1 mRNA, as expected (Fig. 6C, lanes 1,2). These data are consistent with the proposal that premature termination within the NRD1 gene involves endonucleolytic cleavage of the primary transcript and that the putative endonuclease Ysh1p may provide this activity.
Splicing is impaired in the ysh1-12 strain
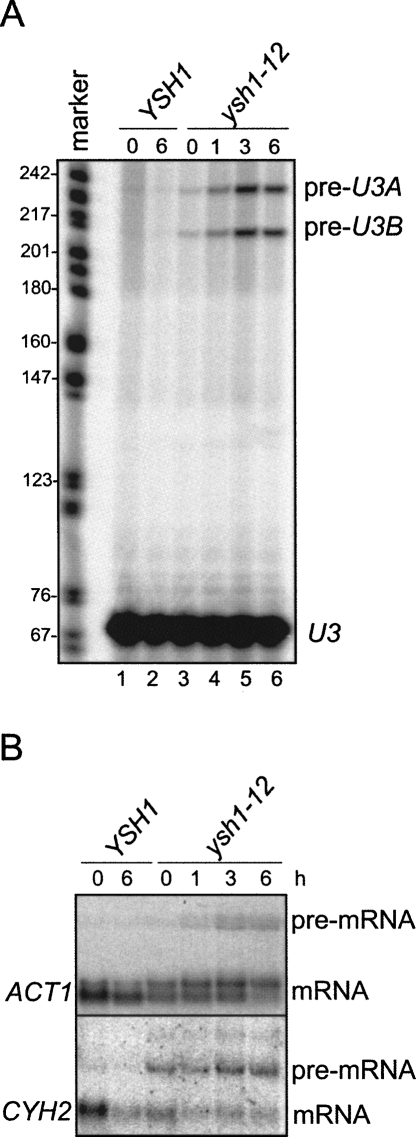
The brr5-1 allele of YSH1 originated from a screen for factors defective in RNA splicing (Noble and Guthrie 1996). To determine whether our ysh1 mutant strains were defective in splicing, we analyzed SNR17A and SNR17B pre-snoRNAs using primer extension analysis (Noble and Guthrie 1996). We found that the ysh1-12 strain exhibited a gradual accumulation of unspliced precursors derived from both U3 genes following shift to nonpermissive temperature (Fig. 7A, lanes 3–6). The rse1-1 mutant strain was analyzed as control and found to exhibit a similar splicing defect, as expected (Chen et al. 1998; data now shown). In contrast, the ysh1-32 ts strain was fully competent in splicing (data not shown). Moreover, Northern blot analysis of intron-containing ACT1 and CYH2 transcripts in the ysh1-12 strain detected extended species that were likely to represent unspliced pre-mRNAs (Fig. 7B). This was confirmed by primer extension analysis with an oligonucleotide complementary to the ACT1 exon 2 (data not shown). The above results suggest a function for Ysh1p in pre-mRNA and pre-snoRNA splicing and may reflect a functional coupling of 3′ end processing and splicing events.
FIGURE 7.
The ysh1-12 mutant is defective in splicing. (A) Primer extension analysis of spliced and unspliced U3 transcripts in wild-type and ysh1-12 mutant strains grown in YPD for up to 6 h at 15°C. Analysis was carried out with a radioactively labeled oligonucleotide complementary to sequences located in U3 exon 2. HpaII-digested pBR322 fragments were 5′ end labeled and served as markers. (Right) Bands corresponding to pre-U3A, pre-U3B, and mature U3 snoRNA are indicated. (B) Northern blot analysis of total RNA extracted from wild-type and mutant ysh1-12 cells, performed as in Figure 3A. Filters were developed with random-primed labeled probes directed against RNA transcripts as indicated on the left.
DISCUSSION
In this study, we investigated the role of Ysh1p in mRNA and snoRNA metabolism. Our results are consistent with the view that Ysh1p plays an essential role in 3′ end formation, probably through catalysis of endonucleolytic cleavage of the pre-mRNA. In addition to this, we provide evidence that Ysh1p is involved in termination of RNAP II both at mRNA and snoRNA genes, that it may contribute to the accumulation of distinct mRNAs independent of its role in 3′ end formation, and that it may act in pre-mRNA splicing.
A striking outcome of our study is that distinct sets of phenotypes were associated with specific groups of ysh1 mutant strains. Table 1 summarizes a broad range of phenotypic observations, which were either resulting from our work or were previously described by others. Based on biochemical phenotypes two groups, class I and class II, can be formed by the ts strains (ysh1-13, ysh1-15, ysh1-32, and brr5-td) and the cs strains (ysh1-12 and brr5-1), respectively. Extracts prepared from strains of both classes displayed defects in 3′ end processing in vitro. The only exception is the brr5-1 extract, which was reported not to have a clear cleavage defect (Chanfreau et al. 1996). Moreover, all analyzed ts and cs alleles of YSH1 showed an aberrant pattern of ACT1 poly(A) site usage. These observations are consistent with the proposal that Ysh1p indeed represents the 3′ endonuclease. However, defects in 3′ end cleavage and polyadenylation are generally associated with reduced mRNA stability. Consistently, class I mutants show under-accumulation of several mRNAs and a reduction of the global poly(A) tail length distribution. As this, in turn, leads to reduced protein levels of certain factors (such as, for example, Ssu72p), it cannot be excluded that defects observed in this class of strains are indirect. In stark contrast, class II mutations did not cause a general loss of mRNA levels. Moreover, phenotypes in snoRNA 3′ end formation and termination, in the control of NRD1 mRNA accumulation, and in RNA splicing were observed in the ysh1-12 cs mutant strain. Notably, these phenotypes were not seen with class I mutants. Such striking differences of mutant phenotypes cannot easily be explained by a simple loss-of-function model and thus they complicate the mechanistic interpretation of the role of Ysh1p in different pathways of cellular RNA synthesis. Consistent with this idea, it was suggested previously that cold-sensitive phenotypes can unravel physiologic features that may be resistant to mutation to temperature sensitivity (Moir et al. 1982; Noble and Guthrie 1996).
TABLE 1.
ysh1/brr5 phenotypes
In the torpedo model of RNAP II termination, cleavage at the poly(A) site is an obligatory step to provide an entry point for the Rat1/Xrn2 exonuclease, which contributes to termination through exonucleolytic degradation of the RNA fragment that remains associated with the polymerase following 3′ end cleavage (Kim et al. 2004; West et al. 2004, 2008). Recently, this model has been challenged by the observation that although Rat1p associates with 3′ end factors and may contribute to the assembly of the processing machinery, degradation of the downstream RNA appears not to be essential for termination (Luo et al. 2006). Interestingly, it was also shown that degradation of the 3′ cleavage fragment is stimulated when coupled to 3′ end cleavage in mammalian cells (Kaneko et al. 2007). Because it was demonstrated previously that exonuclease activity is required for Rat1p termination activity (Kim et al. 2004), the role of Rat1p in this process remains paradoxical (Rosonina et al. 2006). We found that class I and class II ysh1 mutants were defective in transcription termination when assayed on the CYC1 gene via transcriptional run-on. These results suggest that Ysh1p, the endonuclease acting in the 3′ end processing of pre-mRNA transcripts, is also essential for RNAP II termination. Unlike other 3′ end processing factors previously shown to be involved in termination, Ysh1p interacts neither with the CTD of RNAP II nor with RNA (Kyburz et al. 2003; Sadowski et al. 2003; M. Garas, B. Dichtl, and W. Keller, unpubl.). We, therefore, predict that it is the endonucleolytic cleavage that contributes mechanistically to transcription termination, possibly by providing the entry site for exonucleases. Moreover, transcriptional read-through products were detected in the class II ysh1-12 and brr5-1 strains but not in the class I strains. As both classes of mutants were defective in 3′ end cleavage and in RNAP II termination, it seems that aberrant transcripts were somehow stabilized in class II mutants, pointing to a possible role of Ysh1p in RNA quality control.
The mechanism of 3′ end processing and transcription termination on independently transcribed snoRNA genes remains unclear. Several lines of evidence support the notion that the canonical 3′ end processing complex is involved in pre-snoRNA 3′ end formation and that an endonucleolytic cleavage step may be associated with this process. Specific cleavage sites were identified on SNR13 and SNR47 snoRNAs as primer extension stops in rat1-1 Δxrn1 double-mutant cells (Fatica et al. 2000). Several subunits of CF IA and CPF complexes are important for snoRNA termination (see Introduction), and ChIP experiments clearly show that poly(A) factors and Rat1p exonuclease are recruited to snoRNA genes (Kim et al. 2006). ChIP analysis of elongating RNAP II suggested that degron-mediated depletion of Ysh1p did not affect snoRNA transcription termination (Kim et al. 2006); however, these experiments did not reveal whether or not cleavage occurs at the snoRNA 3′ ends. In our analyses, the brr5-td strain did not accumulate 3′ extended snoRNA transcripts following depletion of Ysh1p, suggesting that the protein may not play an essential role in this process. Nevertheless, our analysis of the class II ysh1-12 allele demonstrated that Ysh1p has a function in snoRNA 3′ end formation and transcription termination. One potential explanation for this apparent paradox apart from other possible indirect effects could be that for reasons not known, the 3′ extended snoRNA transcripts are stabilized in the ysh1-12 cs mutant, but not in the ysh1 ts/td strains. Canonical 3′ end processing factors and Rat1p are recruited to genes transcribed by RNAP II regardless of whether they encode mRNA or snoRNA (Kim et al. 2006). It is possible that an early stage of termination is similar for both types of transcripts and that in a later phase of termination a combination of factors used by RNAP II for 3′ end formation may vary depending on the specific snoRNA gene (Kim et al. 2006). A speculative scenario therefore is that the phenotype associated with the cs ysh1-12 strain might unravel an early stage of the snoRNA transcription termination that requires a certain function of Ysh1p/CPF.
Whereas a mechanistic understanding of RNAP II termination at the end of transcription units is emerging, there is little information available on regulated premature termination within a transcription unit. Indeed, the case of NRD1 autoregulation is one of the best documented examples (Arigo et al. 2006). It has been found that the RNA-binding proteins Nrd1p and Nab3p recognize sequence elements within the NRD1 mRNA to trigger RNAP II termination prematurely, i.e., before the “regular” poly(A)-dependent terminator in the 3′-UTR is transcribed (Arigo et al. 2006). In agreement with these observations, the early NRD1 terminator resembles Nrd1p/Nab3p-dependent snoRNA terminators (Steinmetz et al. 2001; Arigo et al. 2006). It was shown that this autoregulation involves the exosome and the TRAMP complex (Arigo et al. 2006), but it remains unclear whether endonucleolytic cleavage and possibly a torpedo-like termination mechanism may play a role as well. We observed elevated levels of the full-length NRD1 mRNA and of the Nrd1p protein in the ysh1-12 strain, suggesting a negative role for Ysh1p in the regulation of NRD1 expression. Therefore it is possible that the increased amounts of the Nrd1p protein cause the snoRNA formation defect in the ysh1-12 mutant due to an imbalance in the stoichiometry of the components that form the Nrd1 complex. As Nrd1p bound to Nab3p and Sen1p associates with the RNAP II CTD, the excess free Nrd1 could displace this complex from the CTD, resulting in termination defects. Importantly, we were also able to demonstrate the existence of a NRD1 3′ cutoff product in rat1-1, Δxrn1 cells, which is consistent with a mechanism involving endonucleolytic cleavage. These results imply a role for Ysh1p in NRD1 premature termination and thus for regulation of NRD1 mRNA levels, possibly via its cleavage activity. Interestingly, it was previously suggested that the Nrd1p/Nab3p/Sen1p pathway acts primarily on transcripts that are 500 nucleotides (nt) or shorter (Steinmetz et al. 2001, 2006b). The observation that the prematurely terminated NRD1 RNA is at least 1000 nt long is consistent with the involvement of Ysh1p, which is a component of CPF that preferentially mediates termination of longer transcripts. In addition to Ssu72p (Ganem et al. 2003; Steinmetz and Brow 2003) and Hrp1p (Kuehner and Brow 2008), Ysh1p may therefore represent another example of a 3′ end processing factor involved in regulated premature RNAP II termination.
Interestingly, Northern blot analyses showed that the levels of several other mRNAs were elevated in the ysh1-12 strain as well. Among the messages accumulating in the ysh1-12 mutant, we identified NAB2, which is believed to autoregulate the levels of its own mRNA in a process that requires the nuclear exosome component Rrp6p (Roth et al. 2005). This post-transcriptional mechanism involves a sequence of 26 adenosines (A26) in the NAB2 3′-UTR, which represses NAB2 3′ end formation and subsequently directs the transcript for degradation by Rrp6p (Roth et al. 2005). The increased concentration of NAB2 mRNA in the ysh1-12 strain suggests a contribution of Ysh1p to the autogenic control of NAB2 levels. Interestingly, we found that YSH1 genetically interacts with the nuclear exosome subunit Rrp6p, as deletions of RRP6 suppressed the cs growth phenotype of ysh1-12 cells (data not shown). However, further investigations of the involvement of Ysh1p in NAB2 autoregulation are required. Our results indicate that Ysh1p directly or indirectly influences the nuclear turnover of mRNAs. Class II mutations in ysh1 alleles could inhibit correct mRNP assembly, which in turn might lead to impaired mRNA export and retention of the mRNA in the nucleus. An alternative and more intriguing possibility would be that the endonucleolytic cleavage activity of Ysh1p directly contributes to RNA surveillance. In this case, we would predict that also class I mutations result in some up-regulation of NAB2 and/or NRD1 mRNAs; however, such phenotypes were clearly not observed in these mutant strains. Taken together, our results support the idea that Ysh1p is involved in the negative regulation of certain mRNAs and that it may contribute to RNA surveillance.
Our analyses of the ysh1-12 allele revealed a splicing defect that correlates well with the phenotype observed previously for the cs brr5-1 allele (Chanfreau et al. 1996; Noble and Guthrie 1996). However, the molecular basis of these defects remains unclear. snRNA synthesis appeared normal in ysh1 mutant strains (data not shown), and this excludes the possibility that snRNA depletion underlies the defect. We cannot rule out the possibility that secondary effects resulting from under-accumulation of critical splicing components contribute to the phenotype. Nevertheless, the possibility remains that Ysh1p acts in the coupling of splicing and 3′ end processing. Evidence for such coupling is ample in mammalian cells but so far was not reported in yeast. Mechanisms of coupling include physical interactions between components of the 3′ end formation and splicing machineries, e.g., an interaction between CPSF and the U2 snRNP (Kyburz et al. 2006), and between U2AF 65 and CF Im (Millevoi et al. 2006). Interestingly, Rse1p, the yeast homolog of SF3b130 and a component of U2 snRNP, genetically interacts with different subunits of CPF (A. Kyburz and W. Keller, unpubl.). Future experiments will have to address the question whether there is, indeed, a functional interdependence of splicing and poly(A) addition in yeast.
MATERIALS AND METHODS
Yeast growth and strain construction
The S. cerevisiae strains used in this study and their genotypes are listed in Table 2. Construction and isolation of temperature-sensitive ysh1 alleles was done with an error-prone PCR and in vivo gap–repair approach as described previously (Dichtl et al. 2002b); details are available upon request. Manipulations and growth of S. cerevisiae were carried out by established procedures. Typically, for temperature shift experiments, yeast cultures were grown to midexponential phase at 23°C (ts strains) or 30°C (cs strains), and transferred to a water bath at 37°C or 15°C, respectively. Yeast cells were grown in rich YPD medium (2% glucose, 2% bacto-tryptone, 1% yeast extract) or in synthetic drop-out medium (2% glucose or galactose, respectively, 0.67% yeast nitrogen base, 1× amino acids) as indicated in the figure legends. For the droplet test, strains were grown overnight and diluted to an OD600 of 0.1, 0.01, 0.001, and 0.0001. Five microliters of each dilution were spotted on five different YPAD plates and incubated at 15°C, 23°C, 30°C, 33°C, and 37°C.
TABLE 2.
Yeast strains used in this study
RNA analyses
Yeast total RNA was extracted with the hot phenol method. Northern analyses and RNase H experiments were carried out as described (Dichtl et al. 2002b, 2004). Oligonucleotides used as probes in Northern analyses were routinely labeled with 30 μCi of [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs). Random-primed Northern probes were as published previously (Dichtl et al. 2002a,b, 2004) or were obtained using the following oligonucleotides:
ASN1 (ASN1-For, 5′-ATGTGTGGTATTTTCGCCGC; ASN1-Rev, 5′-GAATCGTATACGTGGCCCGG);
DBP9 (DBP9-For, 5′-AGCTTGGAAGGCGAGCCTG; DBP9-Rev, 5′-TCATTTGAAGTTCTTCAACG);
RPB1 (RPB1-For, 5′-CAGAGGCTAAAAAGAAAGTTTTG; RPB1-Rev, 5′-GATCTATGTGGAAAGTTTGTTG);
SCR1 (SCR1-For, 5′-AGGCTGTAATGGCTTTCTGGTGGG; SCR1-Rev, 5′-TATGGTTCAGGACACACTCCATCCC);
SEN1 (SEN1-For, 5′-CTCAATACGTCGCAGGCTGAGGC; SEN1-Rev, 5′-GAAAGAACAGTTGGTGGTAGTTG);
YOL087C (YOL087C-For, 5′-GTCCTGATAATGTACACGATGG; YOL087C-Rev, 5′-GTTCATGGTGATGATGATGATGG); and
YSH1 (YSH1-For, 5′-GTCAATGGTATCAAATTTACGGC; YSH1-Rev, 5′-GGTTATTTCCGGATTATTTATGG).
Total poly(A) tail labeling was performed as described previously (Minvielle-Sebastia et al. 1998). The poly(A) tails were separated on a 10% denaturing polyacrylamide/8.3 M urea/TBE gel; images were generated by a PhosphorImager (STORM). Transcriptional run-on (TRO) analyses on the CYC1 gene with the multi-copy plasmid pUGCYC were performed according to Birse et al. (1998). TRO analysis on the endogenous SNR3 gene was done as described previously (Steinmetz et al. 2001) with the probes A and B. Primer extension analysis (PE) was carried out as described previously (Beltrame and Tollervey 1992) with SuperScript III reverse transcriptase (Invitrogen) and 6 μg of total RNA. Extension primers were as published (Steinmetz et al. 2001) and additionally as the following:
SNR46 (5′-ATCGACCAGCTCTTTAGCATCC);
U3exon2 (5′-GAGCCACTGAATCCAACTTGGTTGAT);
U6 (5′-GGGGAACTGCTGATCATCTCTGTATTG);
NRD1 U (5′-CGTGTTTATGGCATGGGCACAAG); and
NRD1 D (5′-CAGCGTCCGTGAGCCTGTGCATAG).
Extract preparation and in vitro cleavage and polyadenylation assays
3′ End processing extracts were made following the procedure described previously (Ohnacker et al. 2000). Cleavage and polyadenylation assays were carried out according to Minvielle-Sebastia et al. (1994). When cleavage only was assayed, EDTA replaced magnesium acetate (MgAc), and ATP was omitted. Internally [32P]-labeled CYC1 and CYC1-pre RNA substrates were produced by in vitro run-off transcription. For reactions at restrictive temperatures, both the reaction mix and protein extracts were preincubated separately for 10 min at the respective restrictive temperatures, combined, and assayed for 1 h. Each of the processing extracts was prepared independently at least three times and subsequently tested for cleavage and polyadenylation activity in vitro in order to confirm the specific processing defect. CPF was obtained by affinity purification as described previously (Ohnacker et al. 2000).
Western blotting analysis
Proteins were separated on 10% SDS polyacrylamide gels, transferred to nitrocellulose membranes, and incubated according to standard procedures with the antibodies indicated in the legends to the figures. The sources of the antibodies used in this work were the following: anti-Ysh1p (Jenny et al. 1996); anti-Ssu72p (Dichtl et al. 2002a); anti-Rna15p (Minvielle-Sebastia et al. 1994); anti-Fip1p and anti-Pap1p (Preker et al. 1995); anti-Pfs2p (Ohnacker et al. 2000); anti-Nrd1p (Steinmetz and Brow 1998); anti-Act1p (Chemicon International). Peroxidase-conjugated swine anti-rabbit and rabbit anti-mouse immunoglobulins (DAKO) served as secondary antibodies for detection of the primary antibodies.
ACKNOWLEDGMENTS
We thank Drs. Claire Moore and Jeffry Corden for generous gifts of yeast strains, Drs. David Brow and Eric Steinmetz for anti-Nrd1p antibody, and Dr. Bertrand Paguet for purified CPF. We also thank the reviewers for constructive suggestions. We are grateful to Drs. Stepanka Vanacova and Bertrand Paguet for comments and discussions on the project and for improving the manuscript. W.K. was supported by the University of Basel, the Swiss National Science Fund, and, as part of the European Science Foundation EUROCORES Programme EuroDYNA, from funds of the EC Sixth Framework Programme, under contract ERAS-CT-2003-980409. B.D. was supported by the Swiss National Science Fund (Grant no. PP00A-102941).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1293008.
REFERENCES
- Allmang C., Kufel J., Chanfreau G., Mitchell P., Petfalski E., Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L. An evolutionary classification of the metallo-β-lactamase fold proteins. In Silico Biol. 1999;1:69–91. [PubMed] [Google Scholar]
- Arigo J.T., Carroll K.L., Ames J.M., Corden J.L. Regulation of yeast NRD1 expression by premature transcription termination. Mol. Cell. 2006;21:641–651. doi: 10.1016/j.molcel.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Beltrame M., Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D.L. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Birse C.E., Minvielle-Sebastia L., Lee B.A., Keller W., Proudfoot N.J. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Butler J.S. The yin and yang of the exosome. Trends Cell Biol. 2002;12:90–96. doi: 10.1016/s0962-8924(01)02225-5. [DOI] [PubMed] [Google Scholar]
- Callebaut I., Moshous D., Mornon J.P., de Villartay J.P. Metallo-β-lactamase fold within nucleic acids processing enzymes: The β-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G., Noble S.M., Guthrie C. Essential yeast protein with unexpected similarity to subunits of mammalian cleavage and polyadenylation specificity factor (CPSF) Science. 1996;274:1511–1514. doi: 10.1126/science.274.5292.1511. [DOI] [PubMed] [Google Scholar]
- Chanfreau G., Legrain P., Jacquier A. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 1998;284:975–988. doi: 10.1006/jmbi.1998.2237. [DOI] [PubMed] [Google Scholar]
- Chen E.J., Frand A.R., Chitouras E., Kaiser C.A. A link between secretion and pre-mRNA processing defects in Saccharomyces cerevisiae and the identification of a novel splicing gene, RSE1. Mol. Cell. Biol. 1998;18:7139–7146. doi: 10.1128/mcb.18.12.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., He X., Moore C. The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3. Mol. Cell. Biol. 2004;24:2932–2943. doi: 10.1128/MCB.24.7.2932-2943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S., Manley J.L. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes & Dev. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- Conrad N.K., Wilson S.M., Steinmetz E.J., Patturajan M., Brow D.A., Swanson M.S., Corden J.L. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics. 2000;154:557–571. doi: 10.1093/genetics/154.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daiyasu H., Osaka K., Ishino Y., Toh H. Expansion of the zinc metallo-hydrolase family of the β-lactamase fold. FEBS Lett. 2001;503:1–6. doi: 10.1016/s0014-5793(01)02686-2. [DOI] [PubMed] [Google Scholar]
- Dheur S., Vo L.T.A., Voisinet-Hakil F., Minet M., Schmitter J.-M., Lacroute F., Wyers F., Minvielle-Sebastia L. Pti1p and Ref2p found in association with the mRNA 3′ end formation complex direct snoRNA maturation. EMBO J. 2003;22:2831–2840. doi: 10.1093/emboj/cdg253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B., Blank D., Ohnacker M., Friedlein A., Roeder D., Langen H., Keller W. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell. 2002a;10:1139–1150. doi: 10.1016/s1097-2765(02)00707-4. [DOI] [PubMed] [Google Scholar]
- Dichtl B., Blank D., Sadowski M., Hübner W., Weiser S., Keller W. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 2002b;21:4125–4135. doi: 10.1093/emboj/cdf390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B., Aasland R., Keller W. Functions for S. cerevisiae Swd2p in 3′ end formation of specific mRNAs and snoRNAs and global histone 3 lysine 4 methylation. RNA. 2004;10:965–977. doi: 10.1261/rna.7090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A., Morlando M., Bozzoni I. Yeast snoRNA accumulation relies on a cleavage- dependent/polyadenylation-independent 3′-processing apparatus. EMBO J. 2000;19:6218–6229. doi: 10.1093/emboj/19.22.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem C., Devaux F., Torchet C., Jacq C., Quevillon-Cheruel S., Labesse G., Facca C., Faye G. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J. 2003;22:1588–1598. doi: 10.1093/emboj/cdg141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D.S., Fan R. Derailing the locomotive: Transcription termination. J. Biol. Chem. 2008;283:661–664. doi: 10.1074/jbc.R700032200. [DOI] [PubMed] [Google Scholar]
- Henry Y., Wood H., Morrissey J.P., Petfalski E., Kearsey S., Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A., Minvielle-Sebastia L., Preker P.J., Keller W. Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science. 1996;274:1514–1517. doi: 10.1126/science.274.5292.1514. [DOI] [PubMed] [Google Scholar]
- Kaneko S., Rozenblatt-Rosen O., Meyerson M., Manley J.L. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes & Dev. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Krogan N.J., Vasiljeva L., Rando O.J., Nedea E., Greenblatt J.F., Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Kim M., Vasiljeva L., Rando O.J., Zhelkovsky A., Moore C., Buratowski S. Distinct pathways for snoRNA and mRNA termination. Mol. Cell. 2006;24:723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Kuehner J.N., Brow D.A. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol. Cell. 2008;31:201–211. doi: 10.1016/j.molcel.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Kyburz A., Sadowski M., Dichtl B., Keller W. The role of the yeast cleavage and polyadenylation factor subunit Ydh1p/Cft2p in pre-mRNA 3′ end formation. Nucleic Acids Res. 2003;31:3936–3945. doi: 10.1093/nar/gkg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyburz A., Friedlein A., Langen H., Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol. Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Logan J., Falck-Pedersen E., Darnell J.E., Jr, Shenk T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse β maj-globin gene. Proc. Natl. Acad. Sci. 1987;84:8306–8310. doi: 10.1073/pnas.84.23.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Bentley D. A ribonucleolytic rat torpedoes RNA polymerase II. Cell. 2004;119:911–914. doi: 10.1016/j.cell.2004.11.041. [DOI] [PubMed] [Google Scholar]
- Luo W., Johnson A.W., Bentley D.L. The role of Rat1 in coupling mRNA 3′ end processing to transcription termination: Implications for a unified allosteric-torpedo model. Genes & Dev. 2006;20:954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandart E., Parker R. Effects of mutations in the Saccharomyces cerevisiae RNA14, RNA15, and PAP1 genes on polyadenylation in vivo. Mol. Cell. Biol. 1995;15:6979–6986. doi: 10.1128/mcb.15.12.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel C.R., Kaneko S., Zhang H., Gebauer D., Vethantham V., Manley J.L., Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′ end processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S., Loulergue C., Dettwiler S., Karaa S.Z., Keller W., Antoniou M., Vagner S. An interaction between U2AF 65 and CF Im links the splicing and 3′ end processing machineries. EMBO J. 2006;25:4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Preker P.J., Keller W. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′ end processing factor. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Beyer K., Krecic A.M., Hector R.E., Swanson M.S., Keller W. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J. 1998;17:7454–7468. doi: 10.1093/emboj/17.24.7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir D., Stewart S.E., Osmond B.C., Botstein D. Cold-sensitive cell-division-cycle mutants of yeast: Isolation, properties, and pseudoreversion studies. Genetics. 1982;100:547–563. doi: 10.1093/genetics/100.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M., Greco P., Dichtl B., Fatica A., Keller W., Bozzoni I. Functional analysis of yeast snoRNA and snRNA 3′ end formation mediated by uncoupling of cleavage and polyadenylation. Mol. Cell. Biol. 2002;22:1379–1389. doi: 10.1128/mcb.22.5.1379-1389.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedea E., He X., Kim M., Pootoolal J., Zhong G., Canadien V., Hughes T., Buratowski S., Moore C.L., Greenblatt J. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′ ends. J. Biol. Chem. 2003;278:33000–33010. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- Nedea E., Nalbant D., Xia D., Theoharis N.T., Suter B., Richardson C.J., Tatchell K., Kislinger T., Greenblatt J.F., Nagy P.L. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol. Cell. 2008;29:577–587. doi: 10.1016/j.molcel.2007.12.031. [DOI] [PubMed] [Google Scholar]
- Noble S.M., Guthrie C. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996;143:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnacker M., Barabino S.M., Preker P.J., Keller W. The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3′ end processing complex. EMBO J. 2000;19:37–47. doi: 10.1093/emboj/19.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco I.J., Kim S.J., Martinson H.G. The poly(A) signal, without the assistance of any downstream element, directs RNA polymerase II to pause in vivo and then to release stochastically from the template. J. Biol. Chem. 2002;277:42899–42911. doi: 10.1074/jbc.M207415200. [DOI] [PubMed] [Google Scholar]
- Preker P.J., Lingner J., Minvielle-Sebastia L., Keller W. The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell. 1995;81:379–389. doi: 10.1016/0092-8674(95)90391-7. [DOI] [PubMed] [Google Scholar]
- Preker P.J., Ohnacker M., Minvielle-Sebastia L., Keller W. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 1997;16:4727–4737. doi: 10.1093/emboj/16.15.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J., Furger A., Dye M.J. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Rosonina E., Kaneko S., Manley J.L. Terminating the transcript: Breaking up is hard to do. Genes & Dev. 2006;20:1050–1056. doi: 10.1101/gad.1431606. [DOI] [PubMed] [Google Scholar]
- Roth K.M., Wolf M.K., Rossi M., Butler J.S. The nuclear exosome contributes to autogenous control of NAB2 mRNA levels. Mol. Cell. Biol. 2005;25:1577–1585. doi: 10.1128/MCB.25.5.1577-1585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K., Calvo O., Manley J.L. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA. 2004;10:565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski M., Dichtl B., Hübner W., Keller W. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 2003;22:2167–2177. doi: 10.1093/emboj/cdg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon K.E., Mauger D.M., Arndt K.M. A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol. Cell. 2005;20:225–236. doi: 10.1016/j.molcel.2005.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E.J., Brow D.A. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc. Natl. Acad. Sci. 1998;95:6699–6704. doi: 10.1073/pnas.95.12.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E.J., Brow D.A. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol. Cell. Biol. 2003;23:6339–6349. doi: 10.1128/MCB.23.18.6339-6349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E.J., Conrad N.K., Brow D.A., Corden J.L. RNA-binding protein Nrd1 directs poly(A)-independent 3′ end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- Steinmetz E.J., Ng S.B., Cloute J.P., Brow D.A. Cis- and trans-acting determinants of transcription termination by yeast RNA polymerase II. Mol. Cell. Biol. 2006a;26:2688–2696. doi: 10.1128/MCB.26.7.2688-2696.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E.J., Warren C.L., Kuehner J.N., Panbehi B., Ansari A.Z., Brow D.A. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell. 2006b;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Thomas B.J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Ursic D., Himmel K.L., Gurley K.A., Webb F., Culbertson M.R. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 1997;25:4778–4785. doi: 10.1093/nar/25.23.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A., Lennertz P., Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S., Gromak N., Proudfoot N.J. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- West S., Proudfoot N.J., Dye M.J. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol. Cell. 2008;29:600–610. doi: 10.1016/j.molcel.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M., Gonzalez T.N. Knives, accomplices, and RNA. Science. 2004;306:1299–1300. doi: 10.1126/science.1100137. [DOI] [PubMed] [Google Scholar]
- Winston F., Dollard C., Ricupero-Hovasse S.L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Gilmour D.S. Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol. Cell. 2006;21:65–74. doi: 10.1016/j.molcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Fu J., Gilmour D.S. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′ end processing factor, Pcf11. Genes & Dev. 2005;19:1572–1580. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Hyman L., Moore C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelkovsky A., Tacahashi Y., Nasser T., He X., Sterzer U., Jensen T.H., Domdey H., Moore C. The role of the Brr5/Ysh1 C-terminal domain and its homolog Syc1 in mRNA 3′ end processing in Saccharomyces cerevisiae . RNA. 2006;12:435–445. doi: 10.1261/rna.2267606. [DOI] [PMC free article] [PubMed] [Google Scholar]