Abstract
Background
Intracellular cytokine staining (ICS) by multiparameter flow cytometry is one of the primary methods for determining T cell immunogenicity in HIV-1 clinical vaccine trials. Data analysis requires considerable expertise and time. The amount of data is quickly increasing as more and larger trials are performed, and thus there is a critical need for high throughput methods of data analysis.
Methods
A web based flow cytometric analysis system, LabKey Flow, was developed for analyses of data from standardized ICS assays. A gating template was created manually in commercially-available flow cytometric analysis software. Using this template, the system automatically compensated and analyzed all data sets. Quality control queries were designed to identify potentially incorrect sample collections.
Results
Comparison of the semi-automated analysis performed by LabKey Flow and the manual analysis performed using FlowJo software demonstrated excellent concordance (concordance correlation coefficient >0.990). Manual inspection of the analyses performed by LabKey Flow for 8-color ICS data files from several clinical vaccine trials indicates that template gates can appropriately be used for most data sets.
Conclusions
The semi-automated LabKey Flow analysis system can analyze accurately large ICS data files. Routine use of the system does not require specialized expertise. This high-throughput analysis will provide great utility for rapid evaluation of complex multiparameter flow cytometric measurements collected from large clinical trials.
Keywords: Flow Cytometry, Intracellular Cytokine Staining, HIV-1, vaccine, T cell, immunogenicity, data analysis, automation
Introduction
Two primary methods currently are widely used for measuring vaccine-induced T-cell responses at the single cell level. One is the IFN-γ ELISpot assay (1), and the other the flow cytometric assay referred to as intracellular cytokine staining (ICS) (2), both of which enumerate antigen-specific cytokine-producing T cells. The ICS assay provides more information than the ELISpot assay because it identifies CD4+ or CD8+ responding cells and can examine multiple cytokines. When used in clinical HIV vaccine trials, the assays provide information about the immunogenicity of the regimen, which then guides decisions about proceeding to large-scale efficacy trials. Thus, standardized and validated assays performed in the good laboratory practices (GLP) setting are necessary to ensure accurate immunogenicity assessment.
As more vaccines are developed and advanced for clinical evaluation, the increased number of complex data from ICS assays demands implementation of high throughput analyses. The flow-based ICS assay requires a specialized analysis involving calculation of compensation for each data set, sequential gating and quality assessment. Due to the large number of assays performed daily, manual analysis and quality assessment for each data set are not feasible. Therefore, to address this problem, a web-based system, termed the LabKey flow analysis system, was developed to automate a large portion of the analysis. We established that once the performance of the assay and the collection of the samples on the flow cytometer are standardized, large datasets can be analyzed using this semi-automated analysis system. Based on our findings here, the LabKey analysis system will have broad utility in a wide variety of flow-based immunological investigations of both vaccine and therapeutic interventions.
Materials and Methods
8-color ICS assay
The 8-color ICS has been described previously (2). Briefly, previously frozen PBMC are thawed, placed in culture overnight (rested), washed and counted the next morning, and cultured for six hours with the superantigen Staphylococcal Enterotoxin B (SEB, positive control), with HIV-1 peptide pools, or without any stimulant (negative control). The HIV-1 peptide pools are 15 amino acids and are based on the global potential T-cell epitopes (PTE) for Env, Gag and Pol (3). Following this six-hour “stimulation”, cells are stained with a viability marker (Live/Dead Fixable Violet Dead Cell Stain, Invitrogen), fixed with FACSLyse (Beckton Dickinson, BD) and frozen. The following day or up to 4 weeks later, the cells are thawed, permeabilized with FACSPerm (BD) and stained intracellularly with fluorochrome-labeled monoclonal antibodies for CD3, CD4, CD8, IFN-γ, IL-2, TNF-α and IL-4. Following this, the samples are collected on a four-laser BD LSR II equipped with a High Throughput Sampler.
Before each sample collection, the LSR is standardized using single peak rainbow beads (Spherotech) (4). The PMT voltages for each of the fluorescence parameters and for forward and side scatter are adjusted so that the median fluorescence of the singlet-gated beads matches previously-determined target values.
The assay is used to examine PBMC from participants enrolled in HIV candidate vaccine trials within the National Institutes of Health-sponsoned HIV Vaccine Trials Network (HVTN). Additionally, the assay is used to validate reagent peptide pools with PBMC from control donors. Typically, evaluations for a given donor PBMC are tested in a batch, such that eight or 16 PBMC samples are observed at one time (see plate layout, Figure 1). The processing of these samples and their collection on the LSR are referred to as an experiment, and resulting data are referred to as a dataset. Because each study includes large numbers of PBMC samples, multiple experiments are required. Within each experiment, each PBMC sample is divided into several different stimulation conditions, each of which is processed for ICS in a single well of a 96-well plate. These are referred to as sample wells, and the collection of each well results in one FCS file. These are in the “Flow Cytometry Standard” format and include fluorescence data for each cell as well as any sample-identifying keywords entered at the time of collection.
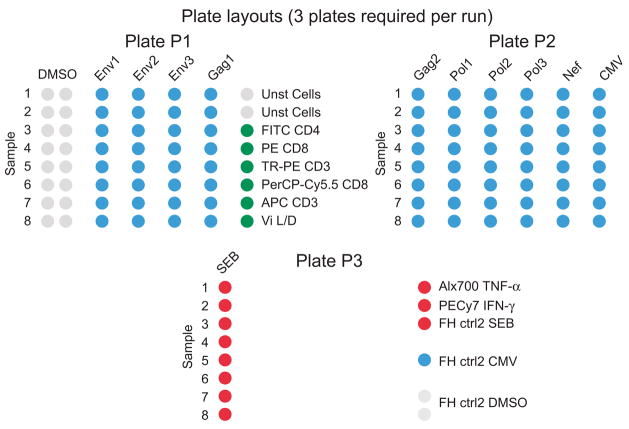
Figure 1.
Plate layout used for 8-color ICS assay. For the ICS assay used within the HVTN, each row on a plate is a different sample, labeled by number and identified by the keyword “sample order” in the collection template. Each column or set of columns is a different stimulation condition, such as DMSO indicating the negative control (cells cultured with the diluent for the peptide pools, DMSO, but no antigen specific stimulant). The wells stimulated with the positive control, SEB, are included on a separate plate to avoid potential contamination of the test wells. Before collection on the flow cytometer, PBS is added to the empty columns between stimulation conditions to reduce carry-over as the samples are sequentially collected. A PBMC sample with a known response to CMV (the FH ctrl2) is included in each experiment and is included on plate 3. Most compensation samples are included in one column on plate 1. Compensation samples that require stimulation with SEB are included on the plate with the other SEB-stimulated wells (plate 3).
LabKey Verification
For the verification of the LabKey Flow results in comparison with the FlowJo results, data from a peptide validation study were used. In this study, PBMC from 30 HIV seronegative and eight HIV seropositive individuals were examined by 8-color ICS for responses to nine HIV-1 global PTE peptide pools (three pools for Env, three pools for Pol, two pools for Gag and one pool for Nef).
Statistical testing
The Lin’s concordance correlation coefficient (CCC) was used to assess the agreement between the results from LabKey Flow and FlowJo (5). The CCC is the product of the coefficients measuring two sources deviation from the identity line: precision and accuracy. The coefficient of precision measures how far each pair of observations deviates from the best-fit line; the coefficient of accuracy measures how far the best-fit line deviates from the 45-degree line through the origin. A CCC value of 1 indicates a perfect agreement, −1 a perfect reverse agreement, and 0 no agreement. Acceptance criterion was chosen as the 95% lower (one-tailed) confidence limit ≥ 0.98 based on the observed median CCC value of 1.0 from an historical data set.
For comparison of the compensation matrices as calculated by LabKey Flow and by FlowJo, a CCC was computed for comparisons of all off-diagonal spillover coefficient values from the compensation matrices that are above 0.01 (1%) as determined in the FlowJo data set. For comparison of lineage event counts, the CCC was determined for each stimulation condition for the event counts in each lineage gate (singlets, live cells, lymphocytes, CD3+ T cells, CD4+ and CD8+ T cells). For the cytokine counts, CCC was performed for each stimulation condition for each cytokine subset only for data determined to be positive. To ensure proper statistical inferences, if fewer than 10 PBMC samples were positive for a particular antigen, then the analysis was not performed.
LabKey Flow System design
LabKey server is a web application implemented in Java running on a Tomcat web server, storing its data in either a PostgreSQL database, or Microsoft SQL Server. Experimental FCS data files are stored on a file server to which the web server has access. Keywords from FCS files, but not the FCS files themselves, are copied into the database. This database also stores the statistics and images of graphs that are calculated by LabKey Flow. Running on the web server in multiple background threads, the Flow analysis engine can apply polygon and interval gates to samples, and can calculate event counts, frequencies, mean, median, standard deviation and percentiles for parameters. LabKey Flow uses a hybrid log/linear axis to allow the display of negative values on a mostly logarithmic axis. Additionally, LabKey tracks the inputs (FCS file, compensation matrix, and gating template) used to perform the analysis, and allows properties of these to be displayed alongside the calculated statistics and graphs.
Results
Standardized assay procedure and FACS collection
To enable automated analysis, all assay procedures must be standardized. An 8-color ICS assay that examines production of four individual cytokines (IFN-γ, IL-2, TNF-α, IL-4) among peripheral blood cell populations expressing CD3, CD4, CD8 and a viability marker has been developed and validated (2). Standard Operating Procedures (SOP) were written for the assay, for the instrument set-up and for the collection of samples on the flow cytometer.
The assay uses 96-well plates with specified plate layouts, allowing for standardized templates to be used in the flow cytometer collection software at the time of collection. Templates include all the necessary pre-defined keywords, which circumvents the need for manual entry at the time of collection. The choice and assignment of keywords is essential for enabling automated analysis. For example, a keyword defined as “stim” contains values indicating the stimulus such as NegCtrl, SEB, CMV peptide pool. Another keyword includes values for “replicate”. Samples are placed in a fixed order in rows on the plates and the “sample order” keyword identifies these as sample order 1, 2, etc. Manual entry of a sample identifier is not required at the time of collection because there is a list that matches each sample with a sample order for a particular experiment. Eliminating manual entry has been beneficial because entry errors were common in the past.
The plate layout can accommodate eight samples per 96-well plate, as shown in Figure 1. Columns are the different experimental conditions for each sample. For example, positive and negative control conditions are included, and then stimulations for each peptide pool (such as Env-1, Gag-1). Since vaccine-induced responses are frequently low-level and the threshold for positivity is low, any amount of carry-over can result in false positive responses. Because wells containing SEB-stimulated PBMC have relatively high responses and may contaminate the test sample wells that often have very low-level responses, our positive control, SEB, is monitored on a separate plate. Also, to avoid another potential contamination problem, carryover from one sample well to the next at the time of collection, some wells contain PBS but not PBMC samples.
Overview
Analysis of flow cytometric data requires the sequential gating of populations of interest, i.e., using the expression level of the markers examined to identify the cells that express or do not express those markers. A typical gating sequence for the ICS assay includes gates to identify live cells, lymphocytes, CD3+ T cells, CD4+ or CD8+ T cells and then cytokine-producing cells. These gates need to be defined for each sample. The LabKey Flow analysis system was designed primarily for data for which standard gates can be used for the majority of samples in a study. Therefore, the system uses the same template gates for most samples. Gates unique to individual samples or sets of samples could possibly be assigned in this system, but this is not convenient. Since the primary goal is the objective analysis of large data sets and since manually adjusting gates introduces bias into the analysis, adjustment of gates as a major feature has not been included.
The design of the system is summarized in Figure 2 and is based on a multi-step analysis algorithm that is modeled on the standard analysis methods employed when analyzing data using flow cytometric analysis software. Briefly, for each experiment, a set of FCS files are uploaded into the system and compensation is calculated. Then, the template gates are applied to all the test samples and the appropriate statistics are calculated. Once the data set is analyzed, many different ways of viewing all the data or subsets of the data are available. In addition, there are methods for performing quality assessment and for requesting pre-defined statistical analyses or specialized data output formats. The system is available as open source for use in any laboratory. An installer is available for Microsoft Windows installations, and detailed instructions are available for non-windows installations. All help documentation is available online at www.labkey.org and the website also provides discussion boards for free support.
Figure 2.
Overview of LabKey Flow. The diagram illustrates the key features of LabKey Flow. Items indicated in gray boxes are external to the system and can be imported. Ovals represent function performed by the system. Clear boxes represent items created by or output from the system.
Beginning a new analysis
The starting or “home” page for a study summarizes the analysis steps and the analysis results and is referred to as the “flow dashboard”, as shown in Figure 3. The analysis steps are subdivided as: Load FCS files, Create Analysis Script, Provide Compensation Matrices, Calculate Statistics and Generate Graphs, Assign Additional Meanings to Keywords. On this home page, a section below lists the analysis results; a section below this lists the analysis scripts.
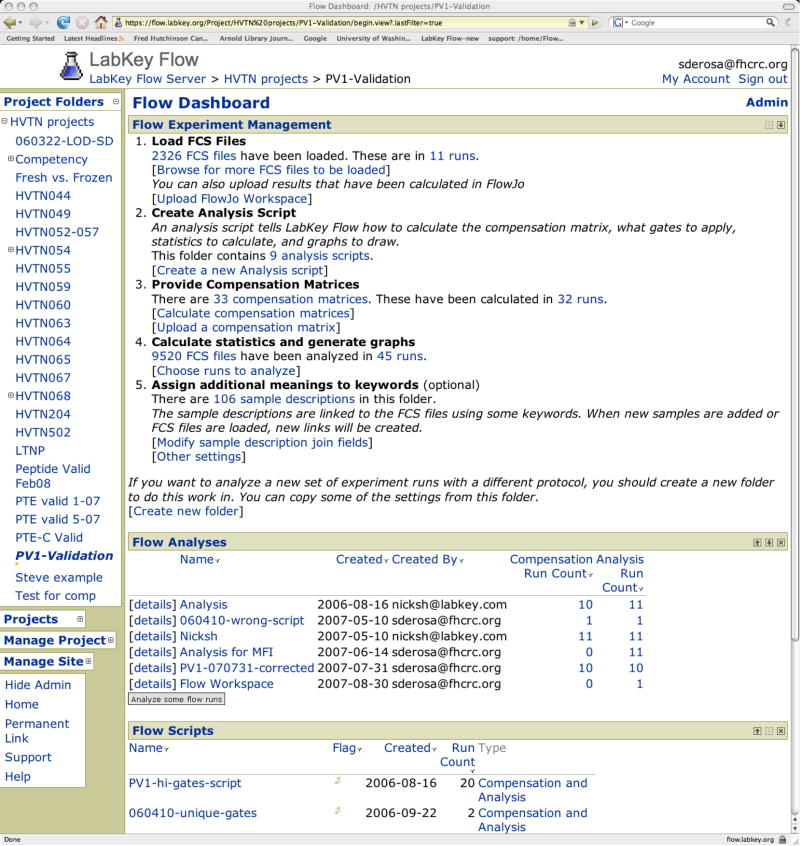
Figure 3.
Home web page for a study. The starting web page for a study is referred to as the Flow Dashboard. The first section, Experiment Management, includes 5 parts referring to the main functional features of the system. The second section, Flow Analyses, list analysis folders. The same FCS file cannot be analyzed more than once within an analysis folder; however, the same FCS file can be analyzed in different folders, e.g., using different analysis scripts. Within an analysis folder, all experiments may be analyzed using a single analysis script. Alternatively, different analysis scripts may be used for different experiments, e.g., if one experiment requires specialized gating. Results of analyses are viewed by selecting an analysis folder. The third section, Flow Analysis Scripts, lists analyses scripts.
Upload FCS files
FCS files must be stored in a standardized format on a server in order for LabKey to access them. Standard nomenclature is used for naming experiments and storing the data in separate folders for each study. In setting up a new analysis for a study, or for analyzing a new set of data for an existing study, the first step is to identify where FCS files are located. Folders containing FCS files are selected by browsing though a web interface. The analysis system extracts the keyword value and accesses the FCS files as needed, but does not make a copy of the FCS files.
Gating template
A user defines initially a set of template gates to be applied for all samples in that study. Once the template is defined, each dataset is automatically analyzed using that template. These gates are defined using flow cytometric analysis software. Currently, the system has been designed to extract the gating specifications from workspaces created in the FlowJo analysis software. Two gating specifications are required: one for the compensation samples, and one for the test samples; and these can be specified in the same or different workspaces. For each compensation sample, the named gates identify the positive and negative populations. For the test samples, the sequential gates identifying the populations of interest are defined.
Within FlowJo, samples should optimally be separated into two distinct groups: the compensation samples and the test samples. Standard naming conventions for gates (e.g., CD4+, CD8+) will ensure standardized analyses across experiments and across studies. The FlowJo workspaces are saved in XML format prior to upload to LabKey. Note that if the data are compensated at the time of collection and exported as compensated data, compensation samples are not required. Since the FlowJo workspace is used solely for defining template gates, only one set of data is required in the workspace. However, when defining template gates, several different experiments need to be examined to define a set of gates appropriate for application across all experiments. Defining template gates can be an iterative process, and as larger datasets are examined in LabKey Flow using the template gates, the definition of the gates can be re-evaluated in terms of the applicability across samples and across experiments. Template gates can be adjusted as needed, and all the experiments can be re-analyzed with the new set of gates.
Analysis scripts
Within LabKey Flow, analysis scripts specify how data are analyzed. Two types of scripts are available: one for compensation and one for analysis of test samples. These are separated because typically compensation is only performed once for data from one experiment, whereas many different types of analyses can be performed on the compensated data. Multiple analysis scripts can be quickly created without the need to repeat the process of defining the compensation specifications.
Compensation
Uploading the FlowJo workspace containing the template gates is the first step in creating a compensation script. Next, a web interface (the compensation editor, Figure 4) identifies the compensation samples and chooses the gates that define the positive population for each sample. The compensation editor requires a pre-defined keyword that identifies the compensation samples. In Table 1, the keyword “Comp” is used for this purpose. Once each compensation sample is identified, the gates previously defined in the FlowJo template for each sample are listed, and the appropriate gate defining the positive cells for that comp sample is chosen. Gates defining the negative cells for each sample can also be chosen, or alternatively, the negative cells from one sample can be used as the negative population for all compensation samples (universal negative). Note that this process is performed only once for a study (Figure 4). All other datasets (i.e., experiments) within a study are compensated based on these specifications, using the compensation samples for that specific experiment.
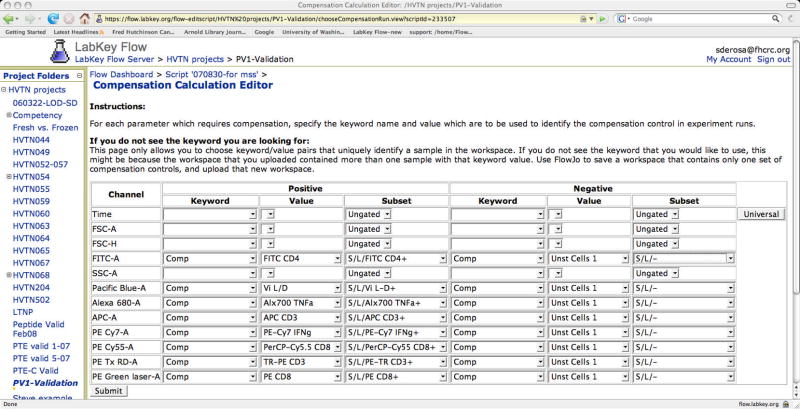
Figure 4.
The compensation editor. This figure shows the web page used to define the compensation analysis script. Each parameter included in the FCS files is listed. One column allows the user to choose which keyword contains the compensation sample label. Another column allows the user to choose the value of that keyword for that particular compensation sample. A third column allows the user to choose which of the predefined gates for that sample to use as the positive population for the purpose of compensation calculation. On the right side of the window, individually identifying negative populations for each sample is possible. Alternatively, one sample may be chosen and the negative population from that sample used for all the compensations samples (i.e., a universal negative).
Test samples
As for the compensation script, an analysis script is created by uploading a FlowJo workspace that includes the gates to be used for the test samples. Typically, this is a set of sequential gates. An option is included for defining which statistics to calculate for each gated population, e.g., cell count, percentage of the parent gate, median fluorescence value. Otherwise, all the analyses requirements are specified from the FlowJo workspace.
Performing analyses
Once the scripts are defined, analyses are requested by choosing datasets to analyze, which are selected through a web interface that lists all uploaded experiments. The results of the analysis are saved in an analysis folder that is created by the user. If an analysis folder for a study has already been created, analysis results from new data sets for that study can be added to this folder. Alternatively, new analysis folders can be created and used for additional analyses of the same datasets. At the time the analysis is requested, several options are related to compensation. The default is for the compensation to be calculated based on the compensation script. If the dataset to be analyzed had been previously analyzed using a different analysis script, using the previously-calculated compensation matrix is an available alternative. Another option uses a compensation matrix from a different experimental dataset that was previously analyzed, which is helpful when one or more compensation samples for a particular experiment are incorrect. Rather than repeating the entire experiment, the user may decide to use an alternate compensation matrix and then later verify that the resulting compensation is appropriate. After requesting analyses, the user can close the web browser, and the analysis will continue to completion.
Once the analyses are complete, the results can be viewed in several ways. The default displays a list of experiments that have been analyzed. The results for each sample well within an individual experiment can then be viewed, or the results for each sample well for all experiments can be selected. For each view, options are available for choosing keywords, statistics and graphs to display. The displayed results can be sorted and filtered based on any variable in the display, and can be exported as text or as an Excel file. Any individual sample can be examined in more detail simply by selecting the sample. This detailed view displays pre-defined pseudocolor contour plots and allows visual inspection of the staining patterns. Figure 5 shows an example of a data view customized to show a few of the gates of the gating tree and filtered only for samples stimulated with SEB.
Figure 5.
View of analysis results. Shown is an example of a view of analysis results. These views are customized by the user. In this view, four plots are shown. The first shows lymphocytes and indicates the CD3+ gate, the second shows CD3+ cells and indicates the CD4+ and CD8+ gates. The third and fourth show the CD4+ and CD8+ cells indicating the gate for IFN-γ+ cells. Only SEB-stimulated samples are listed.
Linking ancillary data
Often additional data (meta-data), that are not included as keywords within the FCS files, are also available concerning samples. For our experiments in which sample identifiers are not entered at the time of collection, these ancillary data include a sample identifier or identifiers (such as a subject code along with a visit code indicating when the blood sample was drawn). Other examples of ancillary data include clinical laboratory results for that particular blood sample and PBMC processing information such as cell yields and viability, treatment codes, etc. These data can be linked to the analysis results within LabKey Flow. Having appropriate keywords both within the FCS file and the ancillary data table that uniquely identify each sample is the only requirement. This can be one keyword or multiple keywords. For example, we use the combination of experiment name (that identifies a set of plates for an experiment) and sample order (that identifies the position of the sample on the plates). This ancillary table (as a text file) is uploaded through a web interface, which includes a method for choosing which keywords should be used for linking the data. Creating and defining the appropriate keywords is critical when planning an experiment, to ensure they will be included as part of the FCS file.
Quality assessment
For manual analysis of flow cytometric data, quality assessment is performed at each step of the analysis process, based on the experience of the person performing the analysis. However, when analysis is automated, quality cannot be assessed manually; and the creation of automated methods to assess quality is crucial. Within LabKey Flow, these methods are referred to as queries and can be designed generically for most types of flow cytometric analyses or for the specific needs of the study. If one or more samples fail the quality assessment, “flags” are automatically assigned at the individual file level, the sample level or the experiment level. The flow cytometric graphs for those samples can quickly be examined by selecting the link to the sample. If examination reveals that gates need to be adjusted, this can be performed within LabKey or by creating a new gating template in FlowJo to be used only for those samples.
One such query examines the compensation samples to identify potentially incorrect samples. Identifying problems with compensation is especially critical because the results of the compensation are applied to the entire dataset and incorrect compensation would corrupt all the analyzed data from the dataset. To identify individual compensation samples that may be incorrect, we have developed a very simple query which establishes a minimum acceptable percentage of positive cells for each compensation sample. Since each compensation sample uses biological markers (such as CD4), PBMC samples show considerable variability. Nevertheless, lower limits that are well below the expected frequency for that marker can be established, and samples that fall below this value are identified with a flag. A selected flagged sample can then be viewed in more detail to determine if the sample is appropriate to use. A sample that fails is likely due to an assay error; if this is the case, the compensation cannot be performed accurately.
For our 8-color ICS assay performed to evaluate T cell responses in clinical vaccine trials, the SOP defines acceptance criteria for the analysis results for each sample. A query that identifies samples that fail these acceptance criteria has been developed. The first of three criteria is that the minimum number of CD4 or CD8 cells collected is 10,000, which ensures enough cells to perform the statistical positivity testing and also identifies samples that may not have been correctly stained with the CD4 or CD8 reagents. The second is that the maximum background cytokine response is 0.1% of CD4 or CD8 cells; and the third requires that the minimum response for the positive control, SEB, is 1.2% of CD4 or CD8 cells. Samples that fail must be repeated. This query, which can be performed after each data set is analyzed, quickly identifies those samples that need to be repeated.
We are developing more sophisticated quality control algorithms based on our particular assay procedures. For example, multiple stimulation conditions exist for each PBMC sample; and therefore, multiple FCS collection files also exist for each. Each of these is stained with the same set of markers. The frequency of cells stained with the lineage markers should be similar across these conditions, and a quality control algorithm can identify outliers based on these frequencies. Likely causes for failure include incorrect staining or an incorrect PBMC sample used for that condition. In addition, for each experiment, one control sample is included, and the responses to CMV and SEB, which should be similar over time, are evaluated. A query can be created that monitors this control sample over time and reports any outliers. Complex statistical computations can also be performed within queries because R scripts can be run on the server using its powerful statistics functions. LabKey hosts and runs R directly from the web server.
The examples of quality control queries described above are performed on data after analysis by LabKey Flow. Currently they are not automatically performed, but are easily requested by the user as often as desired, e.g., after each new data set is analyzed. LabKey has a general purpose configurable “analysis pipeline”, and this allows for incorporation of quality control queries directly into the flow pipeline. Thus, there are many options for expanding quality control capabilities.
Statistical analyses
The final step in the analysis of the ICS data collected from clinical trials is statistical analysis. One analysis is the determination of positive responses, i.e., those cytokine responses for a particular PBMC sample for a particular stimulation condition that are large enough to be considered positive. This is currently not performed in LabKey Flow due to the complexity of the statistical procedure. A query is available that outputs all the statistics and keywords required to perform this analysis and formats this in the standard format requested by the statisticians. In addition, any data view as customized by the user can be exported as a text file or directly into Excel. Other statistical processing can be performed by LabKey Flow if programmed into specialized queries. Results of these queries can be obtained quickly and in “real-time” throughout a study, and depending on the design of the query, can serve as another beneficial method of assessing quality during the study.
Verification of the system
All processes performed for GLP studies require validation, and software requires specialized validation methods. A major part of the validation of LabKey Flow has been completed and is verifying the results produced by the software. This verification has been performed by comparing the results from LabKey Flow with a manual analysis of the same data set using FlowJo, with the same gating template being used for both analyses. Both the results for the compensation and the results for the analysis of the test samples were compared. The results of the compensation calculations were evaluated by comparing the matrix spillover percentages. For this analysis, only values of 1% or greater were used, because values below this likely do not reflect true compensation requirements. The concordance correlation coefficient (CCC) was calculated as 1.000 and the analysis passed the acceptance criterion. The results of the computed lineage counts for each of the gated populations and the computed counts for cytokine-producing cells in the test samples were also compared and passed the acceptance criteria. All CCC were >0.988. Examples of the correlations of the cell counts are shown in Figure 6. These analyses demonstrate close agreement between the two analysis methods.
Figure 6.
Comparison of cell counts as determined by FlowJo and by LabKey Flow. The number of CD3+ T cells as determined by FlowJo (x-axis) correlate with the number of cells as determined by LabKey Flow (y-axis) for analysis of ICS data from PBMC samples from 41 adult individuals (left graph). The right graph shows this correlation for CD4+ T cells producing both IFN-γ and IL-2 in response to stimulation with SEB.
Finally, the data exported from LabKey Flow and the data as determined by the FlowJo manual analysis where independently analyzed by SCHARP to identify sample data that do not meet the SOP-defined acceptance criteria and to perform the positivity testing. No discordant results were observed for the SOP acceptance criteria testing. One discordant result was observed for the positivity testing. For this sample, the number of cytokine-producing cells was low and the significance value resulting from the positivity testing was at the threshold of positivity. Therefore, the small difference in cell counts between LabKey Flow and FlowJo (43 vs. 42 cells respectively) resulted in a difference in the positivity testing. Overall, this comparison with FlowJo verifies comparability between the two analysis methods, but does not substitute for validation. As the system is further developed, testing for other validation parameters will be performed.
Discussion
As polychromatic flow cytometry is becoming more commonplace and as it has now been introduced into the clinical setting, new approaches to the analysis of these complex large data sets are required. A semi-automated web-based system for the analyses of standardized flow cytometric ICS data has been developed. Although developed particularly for analysis of 8-color ICS data in the context of HIV-1-candidate vaccine trials (2), the system can be used for analysis of any type of standardized flow cytometric data. The assay procedures and sample collection must necessarily be standardized, as the aim of the system is to objectively analyze large data sets with minimal manual intervention. The system is not designed as a means of exploring flow cytometric data. Numerous software products perform this function well. Ours is an alternative method especially suitable for analysis of the large clinical data sets.
LabKey Flow has several unique features that optimize standard analyses. Although some expertise is required when establishing the analysis template at the beginning of a study, routine use of LabKey Flow does not require specialized expertise in flow cytometric data analysis. Although the manual definition of the analysis template is subjective, the subsequent analysis of the entire data set using the standard template is objective and un-biased. The process of requesting an analysis is quick, with calculations performed off-line. Quality control queries and other specialized queries are not generally available in other flow cytometric analysis software. Thus, LabKey Flow streamlines the high-throughput analysis of standardized data.
LabKey Flow requires some integration with flow cytometric analysis software for the initial defining of template gates. Enhanced integration, as currently in development, will allow further exploration of selected samples as flagged in LabKey Flow. Also, selected samples that require specialized gates can best be examined in external software with the altered analysis imported into LabKey Flow. The altered analysis can be imported as a new gating template. A new feature also allows the import of results as computed in FlowJo as an alternative to analysis performed within LabKey Flow using template gates. Enhanced integration also enables access to other features, such as creation of layouts and printing, that work well in analysis software.
A comparison between the results of analyses performed by LabKey Flow and by FlowJo demonstrates excellent concordance. As for other software, LabKey Flow will continue to be modified, and this comparison will need to be performed whenever new versions of the LabKey Flow system are released. Since the initial release of the system for analysis of HVTN clinical trial samples, the system has been used successfully to analyze datasets from several clinical trials. The SOP acceptance criteria query quickly identified those samples requiring repeated analysis, and template gates were appropriate for nearly all of the samples. This experience has demonstrated that the same gates can be used for large datasets collected over many months. Note that only one instrument in one laboratory was used. When multiple instruments are used, further standardization may be required to allow for the use of template gates across instruments. For the few samples that to date required unique gates, causes that justified the use of unique gates were identified (e.g., an incorrect PMT voltage for one parameter, resulting in a different position for one of the gated populations).
Continued development of the LabKey Flow system is planned. One key feature is to enhance annotation and to track who performs the analyses and how the analysis is performed. More complex assignments of permissions will be added. The system is not currently 21 CFR part 11 compliant, but work toward compliance is under way. As noted above, enhanced integration with flow cytometry analysis software is another goal. In addition, the ability to examine more complex datasets that include more than one staining panel and more than one compensation matrix per experiment will be added. Finally, enhanced quality control queries will be developed and implemented.
There have been major advances in flow cytometry in recent years. The ability to examine more than 8 parameters simultaneously, which had previously been restricted to a limited group of research laboratories, has now become widely available due to the introduction of new instrumentation and reagents. Multi-parameter assays have thus become more common and have also been introduced as validated assays in clinical studies. The performance of the assays has been well characterized and standardized. However, analysis methods have not been standardized. LabKey Flow represents a major step forward in the standardization and automation of the increasing volume of complex data sets generated from these complex assays.
Acknowledgments
HIV Vaccine Trials Network Laboratory Program (U01 AI 068618-02), funded by NIH/DAIDS. University of Washington Center for AIDS Research (NIH P30 AI27757).
We thank Mario Roederer for his work in pioneering many of the analysis methods included in LabKey Flow and for his continuing insights that influence our approach to flow cytometry. We would like to acknowledge the technicians in the HVTN repository, endpoints and research and development laboratories who have processed blood samples and performed the ICS assays. We thank Phyllis Stegall for help with editing. We also thank the clinicians and staff at the Seattle Vaccine Trials Unit. Special appreciation goes to the study volunteers for donating their time and blood as part of the global effort to develop an HIV vaccine.
References
- 1.Russell ND, Hudgens MG, Ha R, Havenar-Daughton C, McElrath MJ. Moving to human immunodeficiency virus type 1 vaccine efficacy trials: defining T cell responses as potential correlates of immunity. J Infect Dis. 2003;187(2):226–42. doi: 10.1086/367702. [DOI] [PubMed] [Google Scholar]
- 2.Horton H, Thomas EP, Stucky JA, Frank I, Moodie Z, Huang Y, Chiu YL, McElrath MJ, De Rosa SC. Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J Immunol Methods. 2007;323(1):39–54. doi: 10.1016/j.jim.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li F, Malhotra U, Gilbert P, Hawkins N, Duerr A, McElrath M, Corey L, Self S. Peptide selection for human immunodeficiency virus type 1 CTL-based vaccine evaluation. Vaccine. 2006;24(47–48):6893–904. doi: 10.1016/j.vaccine.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Perfetto SP, Ambrozak D, Nguyen R, Chattopadhyay P, Roederer M. Quality assurance for polychromatic flow cytometry. Nat Protoc. 2006;1(3):1522–30. doi: 10.1038/nprot.2006.250. [DOI] [PubMed] [Google Scholar]
- 5.Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45(1):255–68. [PubMed] [Google Scholar]