Abstract
Pseudomonas aeruginosa has two complete quorum-sensing systems. Both of these systems have been shown to be important for Pseudomonas virulence in multiple models of infection. Thus, these systems provide unique targets for novel antimicrobial drugs.
Quorum sensing in Pseudomonas aeruginosa
Pseudomonas aeruginosa is a Gram-negative organism that is commonly found in soil and water. Although P. aeruginosa can survive under multiple harsh conditions, it is an opportunistic pathogen and is only able to infect hosts with defective immune system function, such as that observed in individuals with cystic fibrosis, burns, and HIV (1). To facilitate the establishment of infection, P. aeruginosa produces an impressive array of both cell-associated and extracellular virulence factors. Several of these virulence factors have been demonstrated to be regulated by quorum sensing (QS). QS is the mechanism whereby an individual bacterium produces small diffusible molecules that can be detected by surrounding organisms. In P. aeruginosa, and most Gram-negative bacteria, these signal molecules are acyl homoserine lactones (AHLs). Only when the concentration of AHLs in the environment increases, potentially because of increasing numbers of bacteria, are intracellular levels of AHLs sufficient to maximally induce the activation of transcriptional regulators. This mechanism of communication enables bacteria to act as a community in the coordinated regulation of gene expression. This regulated expression of virulence genes is thought to give the bacteria a selective advantage over host defenses and thus is important for the pathogenesis of the organism.
There are two QS systems in P. aeruginosa, which have been extensively studied. The las system consists of the LasR transcriptional regulator and the LasI synthase protein. LasI is essential for the production of the AHL signal molecule N-(3-oxododecanoyl)-L-homoserine lactone (3O-C12-HSL) (2, 3). LasR requires 3O-C12-HSL in order to become an active transcription factor. It was recently demonstrated that, in the presence of 3O-C12-HSL, LasR forms multimers, and that only the multimeric form of this protein is able to bind DNA and regulate the transcription of multiple genes (Figure 1a) (4). A second QS system in P. aeruginosa consists of the RhlI and RhlR proteins. The RhlI synthase produces the AHL N-butyryl-L-homoserine lactone (C4-HSL), and RhlR is the transcriptional regulator (5, 6). Only when RhlR is complexed with C4-HSL does it regulate the expression of several genes. Both 3O-C12-HSL and C4-HSL have been shown to freely diffuse out of bacterial cells; however, 3O-C12-HSL diffusion is significantly slower than that of C4-HSL. Removal of 3O-C12-HSL from bacteria is most efficiently accomplished via the MexAB-OprM efflux system (7). Recently, a third LuxR homologue termed QscR was identified, which has been shown to regulate the transcription of both lasI and rhlI (8). Although QscR exhibits significant homology to LasR and RhlR, it is currently unknown whether an AHL or similar molecule is needed to stimulate QscR function. Data indicate that qscR is important in regulating the production of several virulence factors but that this regulation may occur through control of the expression of both the las and the rhl systems.
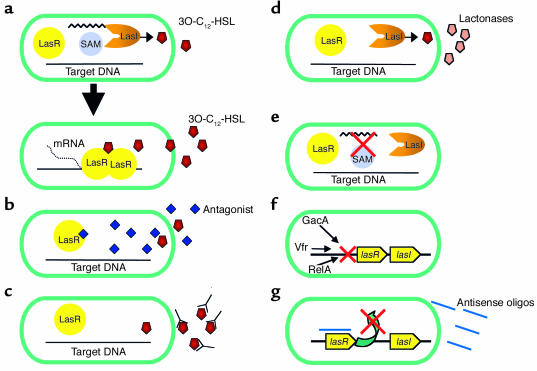
Figure 1.
Potential QS targets for the inhibition of P. aeruginosa virulence. For simplicity, only the las QS system is shown; however, similar mechanisms could be used to inhibit the rhl system as well. (a) P. aeruginosa LasI synthase utilizes S-adenosyl methionine (SAM) and acyl-ACP to form 3O-C12-HSL. As the density of the bacteria, and thus of 3O-C12-HSL, increases, the 3O-C12-HSL molecule binds to the LasR regulator, resulting in dimerization, DNA binding, and transcription of multiple genes. (b) Antagonistic analogues of cognate AHLs compete for binding to LasR but do not result in activation of the protein. (c) Specific antibodies bind to AHLs as they exit the bacteria, inhibiting their re-entry and thus inhibiting activation of LasR as well as their interaction with host cells. (d) Lactonases degrade AHLs as they leave the bacteria, thus inhibiting their activation of LasR and host cells. (e) Targeting the expression of LasI substrates would prevent the production of 3O-C12-HSL, and thus QS activation. (f) Multiple factors have been shown to regulate lasR and lasI. Drugs that inhibit these factors would result in altered QS activation. (g) Specific antisense oligonucleotides (oligos) pair with lasR or lasI RNA and inhibit gene translation and thus protein production.
Role of QS in the global regulation of P. aeruginosa genes
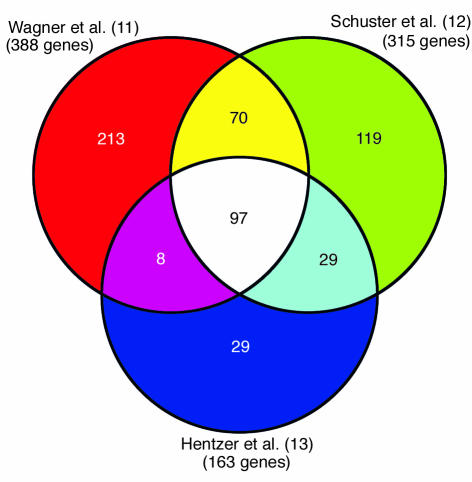
The importance of tight regulation of QS gene expression and AHL production has become evident with our increase in knowledge regarding QS-regulated genes. Several studies have identified numerous genes regulated by QS in P. aeruginosa, many of which are virulence factors (9, 10). With the sequencing of the P. aeruginosa genome and the availability of microarray technologies, a more comprehensive evaluation of QS regulation was recently undertaken. Three individual research groups have used microarray experiments to analyze the QS-regulated transcriptome of P. aeruginosa (11–13). All three studies used independently derived mutant lasI/rhlI strains of P. aeruginosa PAO1. Gene-expression levels were determined for this mutant bacterium when grown with or without exogenous 3O-C12-HSL and C4-HSL. Schuster et al. (12) also used a P. aeruginosa PAO1 strain in which lasR and rhlR had been deleted. In all three studies, an overwhelmingly large number of genes were shown to be regulated by QS, with 3–7% of all P. aeruginosa open reading frames affected. Data from Hentzer et al. (13) represent only those genes that were induced fivefold or greater in comparison with control cultures, while the studies of Schuster et al. (12) and Wagner et al. (11) report all genes induced by QS. Although in many instances several genes were identified by only one of the three groups, a large number of genes were identified in two or more of the studies, and 97 induced genes were identified in all three studies (Figure 2).
Figure 2.
Analyses of the QS-regulated transcriptome of P. aeruginosa. A comparison of QS-induced genes from microarray experiments performed by three different research groups is shown. The data reported by Hentzer et al. (13) represent only those genes that were induced fivefold or more. The studies by Wagner et al. (11) and Schuster et al. (12) include all induced genes. A total of 97 QS-regulated genes were common to all three studies.
Wagner et al. also identified 222 genes that showed statistically significant repression by the addition of AHLs to cultures (11), while Schuster et al. found only 38 such genes (12). Interestingly, there was little overlap between these two data sets. The disparity between these studies may in part be due to experimental design, including the choice of growth media, aeration of cultures, and the concentration of added AHLs. Not surprisingly, we found that the induction of various genes was greatly affected by the growth conditions used. Changes in media composition and oxygen concentration appeared to have significant effects on the genes up- or downregulated by QS (11). Therefore, when evaluating variation in gene expression, it is important to consider the influence of growth medium, oxygen concentration, and other environmental conditions that may impact QS-regulated gene expression. Additionally, a similar more stringent statistical analysis of all three data sets may reveal additional overlap between experiments. Despite some differences in the modes of analysis of the QS-regulated transcriptome, a large group of genes was common to all three reports that appeared to be regulated by QS.
The majority of the genes regulated by QS were found to be hypothetical or of unknown function. However, the remaining genes encoded, for example, membrane proteins, putative enzymes, transcription factors, and secreted enzymes involved in a broad range of cellular functions, including two-component regulatory systems, energy metabolism, small-molecule transport, and export apparatus. In all three studies, a large number of the known genes were found to be probable virulence factors; however, several other identified genes may also fall into this category. To evaluate the interactions between QS factors and QS-regulated genes, an analysis was performed to identify QS-regulated genes that contained consensus DNA sequences for the binding of LasR and RhlR. Surprisingly, only approximately 7% of the QS-regulated genes possessed upstream DNA sequences homologous to known LasR- and RhlR-binding domains indicating that QS regulates many genes via indirect mechanisms (11, 12). This observation correlated with the demonstration that a large number of the QS-regulated genes were classified as transcriptional regulators or as members of a two-component regulatory system (11). By inducing these regulatory factors, QS may indirectly affect multiple mechanisms that regulate gene expression, and this may explain the low occurrence of specific DNA-binding domains for the QS regulators.
Overall, the data from these three studies demonstrate the global effects of QS in P. aeruginosa; however, the complete QS-regulated transcriptome may yet be incomplete. Although many genes were identified to be QS regulated, it is important to keep in mind that under different experimental conditions additional genes may be differentially regulated by QS.
Role of P. aeruginosa QS in pathogenesis
Considering that QS regulates such a wide range of factors that play such diverse roles in the function of P. aeruginosa, it is important to examine how QS affects the pathogenesis of this bacterium. Many studies examining QS during infection have used strains of P. aeruginosa that have deletions of one or more of the QS-related genes, in addition to wild-type strains. Studies of the role of P. aeruginosa QS in infection using a burnt-mouse model, a murine model of acute pneumonia, and a rat model of chronic lung infection have all demonstrated that deletions of one or more QS genes result in reduced P. aeruginosa virulence compared with wild-type P. aeruginosa (14–17). These data confirm that both the las and the rhl QS systems are important for P. aeruginosa to disseminate, which leads to septicemia, induces both acute and chronic lung infections, and causes pathology and mortality. QS has also been shown to be functional during P. aeruginosa infections in humans. In sputum samples from cystic fibrosis patients colonized with P. aeruginosa, levels of transcripts for QS genes were found to correlate with those of QS-regulated genes, indicating that QS was regulating their expression during infection (18, 19). Furthermore, AHLs were directly measured in the sputum of cystic fibrosis patients chronically colonized with P. aeruginosa (19). Collectively, these studies indicate that functional QS systems significantly affect the severity of both acute and chronic P. aeruginosa infections.
Additional studies have demonstrated that AHLs produced by P. aeruginosa are able to interact with eukaryotic cells and to stimulate the production of various factors that may affect the pathogenesis of this bacterium. In vitro experiments have shown that purified 3O-C12-HSL stimulates the production of the inflammatory cytokine IL-8 from human lung bronchial epithelial cells (20, 21). It was subsequently demonstrated that 3O-C12-HSL could stimulate a broad-spectrum response in vivo by inducing several inflammatory cytokines and chemokines (15). It has also been shown that 3O-C12-HSL can inhibit the production of IL-12 and TNF-α from LPS-activated mouse peritoneal exudate cells or human PBMCs (22, 23). These data demonstrate that, under certain conditions, 3O-C12-HSL also acts as an immunosuppressor. In additional studies characterizing this response in leukocytes, it was observed that the structure of the AHL molecule was important for regulation of cytokine production. AHLs with a 3-oxo or 3-hydroxy substitution and an acyl chain of 12–14 carbons were the most active molecules in these experiments; however, AHLs with acyl side chains shorter than eight carbons were inactive (23).
These data demonstrate the importance of AHL production to P. aeruginosa. Not only are these molecules important for cell-to-cell communication between bacteria and the regulation of multiple bacterial factors, but they may also directly act as virulence factors. These data also suggest that AHL interactions with various host cells induce different responses. This diverse stimulation may result in different immune responses during various stages of P. aeruginosa infection. Therefore it is important to acknowledge that AHLs may have several different effects on the pathogenesis of P. aeruginosa.
Inhibition of QS as a therapeutic approach to P. aeruginosa infections
Based on the facts that QS regulates such an array of P. aeruginosa factors and that deletion of QS regulators attenuates P. aeruginosa virulence, it is conceivable that QS would be an ideal target for the inhibition of Pseudomonas infections. Multidrug-resistant P. aeruginosa are becoming more prevalent, and current antimicrobial treatments for cystic fibrosis are unable to eradicate P. aeruginosa infections. Therefore, alternative mechanisms for targeting P. aeruginosa have been the focus of much research. Therapeutics that target and inhibit QS in P. aeruginosa would attenuate the virulence of the bacterium and thus potentially assist the host immune response in clearing the infection.
Several components of the QS networks represent ideal targets for potential therapeutics: (a) LasR and RhlR activation (b) AHL formation and activity, and (c) lasR/lasI and rhlR/rhlI expression.
LasR and RhlR activation.
The most promising mechanism for inhibiting LasR or RhlR activation is the use of AHL analogues that act as antagonists for 3O-C12-HSL and C4-HSL. These molecules would most likely be similar in structure to natural AHLs produced by P. aeruginosa and would compete for binding to LasR or RhlR proteins (Figure 1b). Structural variations of the 3O-C12-HSL and C4-HSL molecules have revealed epitopes that are important for the activation and inhibition of LasR activity (24, 25). A recent study used a reporter assay to identify a group of compounds containing a common aniline-ring structure with a hydrogen-bond acceptor that were able to compete with 3O-C12-HSL and subsequently inhibit the activation of LasR and elastase production (25). These data demonstrate that use of a high-throughput assay for the screening of large libraries of AHL analogues may be fruitful in the identification of additional QS inhibitors. Although neither LasR nor RhlR has been purified and structurally analyzed, the structure of TraR, a homologous transcriptional regulator in Agrobacterium tumefaciens, has been extensively studied (26, 27). Data from the crystal structure of this protein have been immensely important in understanding how TraR and similar proteins may interact with AHLs and DNA. Similar studies in P. aeruginosa would be extremely useful in elucidating potential antagonists and also for enhancing our understanding of how cognate AHLs interact with LasR and RhlR.
Additional studies have identified a synthetic halogenated-furanone compound that is able to inhibit the production of many QS-induced factors (13). Microarray analysis of P. aeruginosa following exposure to this compound revealed that 80% of the genes repressed were QS regulated, thus revealing the specificity of the furanone for the QS system. In a mouse model of chronic P. aeruginosa lung infection, animals treated with 0.7 μg/g body weight of this furanone compound demonstrated a 3-log decrease in the number of bacteria that could be isolated from their lungs. These data are very exciting, as QS inhibitors have not previously been shown to be effective during in vivo infections. This compound and other similar analogues hold significant promise as potential treatments for patients fighting P. aeruginosa infection. The most significant advantage to this method of QS inhibition is in the small size and ease of delivery of these molecules.
AHL formation and activity.
As previously discussed, these molecules diffuse or are actively pumped out of the bacterial cell. Therefore, once they appear in the extracellular environment, they are potential targets for destruction or inactivation. We have evaluated the potential of using AHL-specific antibodies to inhibit AHL activity. After AHLs diffuse out of the bacterial cell into the extracellular environment, they can potentially be bound by these antibodies and prevented from interacting with eukaryotic cells or from re-entering the bacterial cell and activating their cognate transcriptional regulators (Figure 1c). Preliminary experiments in our laboratory have demonstrated that mAb’s made from a 3O-C12-HSL–protein conjugate were able to substantially inhibit 3O-C12-HSL activation of a lasB transcriptional reporter in P. aeruginosa as well as the production of IL-8 from epithelial cells (unpublished observations). Although additional studies are needed to further evaluate this approach, these data suggest that antibodies specific for AHLs inhibit QS and may be useful as therapeutics.
Numerous studies have recently identified lactonase enzymes, which are produced by several bacteria including Bacillus sp., Variovorax paradoxus, Arthrobacter sp., and A. tumefaciens (28–31). These enzymes cleave the lactone ring and thus produce nonfunctional molecules that are unable to activate their cognate transcriptional regulators (Figure 1d). When one of these lactonases was expressed in P. aeruginosa, there was a significant decrease in AHL production and virulence factor expression (32). Although these enzymes may not be ideal therapeutics, because of the difficulty in delivering active enzymes to the site of infection, they should prove to be strong tools for the study of QS. Identification of other bacterial strains that produce these enzymes may also provide insight into bacterial interactions in various environments.
Since the discovery of QS in P. aeruginosa, extensive research has evaluated the mechanisms involved in AHL production. LasI and RhlI synthase proteins utilize components of the amino acid and fatty acid biosynthesis pathways to produce AHLs. The homoserine lactone rings are typically derived from S-adenosyl methionine (SAM) and the acyl chains from the acyl-acyl carrier protein (acyl-ACP) pools in the bacteria (33, 34). Although LasI has a preference for 12-carbon acyl chains, it is able to utilize chains of varying lengths. Recent in vitro studies have shown that alterations in fatty acid biosynthesis that result in increased pools of short-chain acyl-ACPs saturate LasI and result in decreased production of the active AHL, 3O-C12-HSL (35). These data demonstrate that LasI is very sensitive to environmental conditions and that control of LasI substrates may have a significant effect on functional AHL production (Figure 1e). Therefore, drugs that target this pathway or inhibit LasI or RhlI functions would be ideal candidates to achieve inhibition of virulence factor production.
lasR/lasI and rhlR/rhlI expression.
Given that QS regulates such a broad spectrum of factors in P. aeruginosa, it is not surprising that the regulation of QS genes is itself controlled. Several factors, including GacA, Vfr, and RelA, have been demonstrated to positively regulate the expression of LasR (Figure 1f) (1, 9). Deletion of vfr virtually eliminated all expression of lasR and reduced the production of virulence factors. Additionally, a deletion of GacA, part of a two-component regulatory system, resulted in reduced production of many virulence factors and a decrease in pathogenesis in a burnt-mouse model of P. aeruginosa infection. These data reveal the significance of these regulatory factors in controlling P. aeruginosa QS and virulence production. The importance of these other factors was also alluded to in transcriptional-profile studies in which the addition of exogenous AHLs to low-density cultures was unable to prematurely induce gene expression, suggesting that other factors need to be produced prior to QS induction (12, 13). Based on these data, various factors involved in the regulation of QS expression, many of which may not yet be known, would be ideal targets for potential disruption of QS virulence induction.
An alternative approach to the inhibition of QS is the use of antisense oligonucleotides that specifically bind to lasR/lasI or rhlR/rhlI transcripts and inhibit gene expression (Figure 1g). The use of antisense technologies to specifically inhibit translation of particular genes has been comprehensively studied in eukaryotic systems, and many of these technologies are currently being evaluated in clinical trials (36). However, the efficacy of these techniques for the inhibition of bacterial genes has not been extensively evaluated. Studies in Staphylococcus aureus, Mycobacterium tuberculosis, and Escherichia coli have successfully demonstrated that antisense oligonucleotides can specifically bind to target transcripts and inhibit gene expression (37–39). Although such an approach has multiple obstacles, such as cell wall permeability and efficacy of the mode of delivery, studies using modified oligonucleotides have shown significant promise (37, 38). The advantages of antisense technology are that antisense regulation is a common phenomenon in bacteria and that this mechanism of regulation is substantially different from most antimicrobials (39).
QS is an attractive therapeutic target because of the role that it plays in the global regulation of multiple P. aeruginosa factors and the importance of this role for the virulence of the organism in multiple different infections. As with any drug that is used to inhibit bacterial infections, there is a concern that resistant mutants may arise during the course of treatment. Inhibition of QS is not immune from such possibilities. It was previously demonstrated that las-deficient P. aeruginosa strains grown under selective pressure gave rise to spontaneous mutations that restored production of certain las-regulated virulence factors; however, such mutations could not be found when both las and rhl were deleted (1). These data demonstrate the importance of targeting both QS systems by therapeutics to effectively inhibit virulence factor production. Because QS can be inhibited in numerous ways, several different approaches may be used to achieve a more complete inhibition. Additionally, in concert with currently used antipseudomonal therapies, these inhibitors may be potent drugs for the eradication of P. aeruginosa infections.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: quorum sensing (QS); acyl homoserine lactone (AHL); N-(3-oxododecanoyl)-l-homoserine lactone (3O-C12-HSL); N-butyryl-l-homoserine lactone (C4-HSL); acyl–acyl carrier protein (acyl-ACP).
References
- 1.Van Delden C, Iglewski BH. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gambello MJ, Iglewski BH. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearson JP, et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. U. S. A. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiratisin P, Tucker KD, Passador L. LasR, a transcriptional activator of Pseudomonas aeruginosa virulence genes, functions as a multimer. J. Bacteriol. 2002;184:4912–4919. doi: 10.1128/JB.184.17.4912-4919.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochsner UA, Koch AK, Fiechter A, Reiser J. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J. Bacteriol. 1994;176:2044–2054. doi: 10.1128/jb.176.7.2044-2054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearson JP, Van Delden C, Iglewski BH. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 1999;181:1203–1210. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chugani SA, et al. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Kievit TR, Iglewski BH. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 2000;68:4839–4849. doi: 10.1128/iai.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whiteley M, Lee KM, Greenberg EP. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner VE, Bushnell D, Passador L, Brooks A, Iglewski B. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuster M, Lostroh P, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hentzer M, et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rumbaugh KP, Griswold JA, Iglewski BH, Hamood AN. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect. Immun. 1999;67:5854–5862. doi: 10.1128/iai.67.11.5854-5862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith RS, Harris SG, Phipps R, Iglewski BH. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J. Bacteriol. 2002;184:1132–1139. doi: 10.1128/jb.184.4.1132-1139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearson JP, Feldman M, Iglewski BH, Prince A. Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect. Immun. 2000;68:4331–4334. doi: 10.1128/iai.68.7.4331-4334.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu H, et al. Pseudomonas aeruginosa mutations in lasI and rhlI quorum sensing systems result in milder chronic lung infection. Microbiology. 2001;147:1105–1113. doi: 10.1099/00221287-147-5-1105. [DOI] [PubMed] [Google Scholar]
- 18.Storey DG, Ujack EE, Rabin HR, Mitchell I. Pseudomonas aeruginosa lasR transcription correlates with the transcription of lasA, lasB, and toxA in chronic lung infections associated with cystic fibrosis. Infect. Immun. 1998;66:2521–2528. doi: 10.1128/iai.66.6.2521-2528.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erickson DL, et al. Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infect. Immun. 2002;70:1783–1790. doi: 10.1128/IAI.70.4.1783-1790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DiMango E, Zar HJ, Bryan R, Prince A. Diverse Pseudomonas aeruginosa gene products stimulate respiratory epithelial cells to produce interleukin-8. J. Clin. Invest. 1995;96:2204–2210. doi: 10.1172/JCI118275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith RS, et al. IL-8 production in human lung fibroblasts and epithelial cells activated by the Pseudomonas autoinducer N-3-oxododecanoyl homoserine lactone is transcriptionally regulated by NF-kappaB and activator protein-2. J. Immunol. 2001;167:366–374. doi: 10.4049/jimmunol.167.1.366. [DOI] [PubMed] [Google Scholar]
- 22.Telford G, et al. The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-L-homoserine lactone has immunomodulatory activity. Infect. Immun. 1998;66:36–42. doi: 10.1128/iai.66.1.36-42.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chhabra SR, et al. Synthetic analogues of the bacterial signal (quorum sensing) molecule N-(3-oxododecanoyl)-l-homoserine lactone as immune modulators. J. Med. Chem. 2003;46:97–104. doi: 10.1021/jm020909n. [DOI] [PubMed] [Google Scholar]
- 24.Passador L, Tucker K, Guertin K, Kende A, Iglewski B. Functional analysis of the Pseudomonas aeruginosa autoinducer PAI. J. Bacteriol. 1996;178:5595–6000. doi: 10.1128/jb.178.20.5995-6000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith KM, Bu Y, Suga H. Library screening for synthetic agonists and antagonists of a Pseudomonas aeruginosa autoinducer. Chem. Biol. 2003;10:563–571. doi: 10.1016/s1074-5521(03)00107-8. [DOI] [PubMed] [Google Scholar]
- 26.Zhang RG, et al. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature. 2002;417:971–974. doi: 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- 27.Vannini A, et al. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 2002;21:4393–4401. doi: 10.1093/emboj/cdf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong Y-H, Gusti AR, Zhang Q-L, Xu J, Zhang L-H. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appl. Environ. Microbiol. 2002;68:1754–1759. doi: 10.1128/AEM.68.4.1754-1759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leadbetter JR, Greenberg EP. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 2000;182:6921–6926. doi: 10.1128/jb.182.24.6921-6926.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park SY, et al. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology. 2003;149:1541–1550. doi: 10.1099/mic.0.26269-0. [DOI] [PubMed] [Google Scholar]
- 31.Carlier A, et al. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-Acyl homoserine lactonase activity. Appl. Environ. Microbiol. 2003;69:4989–4993. doi: 10.1128/AEM.69.8.4989-4993.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reimmann C, et al. Genetically programmed autoinducer destruction reduces virulence gene expression and swarming motility in Pseudomonas aeruginosa PAO1. Microbiology. 2002;148:923–932. doi: 10.1099/00221287-148-4-923. [DOI] [PubMed] [Google Scholar]
- 33.Parsek MR, Val DL, Hanzelka BL, Cronan JE, Greenberg EP. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. U. S. A. 1999;98:4360–4365. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoang TT, Schweizer HP. Characterization of the Pseudomonas aeruginosa enoyl-acyl carrier protein reductase: a target for triclosan and its role in acylated homoserine lactone synthesis. J. Bacteriol. 1999;181:5489–5497. doi: 10.1128/jb.181.17.5489-5497.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoang TT, Sullivan SA, Cusick JK, Schweizer HP. β-Ketoacyl acyl carrier protein reductase (FabG) activity of the fatty acid biosynthetic pathway is a determining factor of 3-oxo-homoserine lactone acyl chain lengths. Microbiology. 2002;148:3849–3856. doi: 10.1099/00221287-148-12-3849. [DOI] [PubMed] [Google Scholar]
- 36.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 37.Harth G, Zamecnik PC, Tang J-Y, Tabatadze D, Horwitz M. Treatment of Mycobacterium tuberculosis with antisense oligonucleotides to glutamine synthetase mRNA inhibits glutamine synthetase activity, formation of the poly-L-glutamate/glutamine cell wall structure, and bacterial replication. Proc. Natl. Acad. Sci. U. S. A. 2000;97:418–423. doi: 10.1073/pnas.97.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Good L, Awasthi SK, Dryselius R, Larsson O, Nielsen P. Bactericidal antisense effects of peptide-PNA conjugates. Nat. Biotechnol. 2001;19:360–364. doi: 10.1038/86753. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Ji Y, et al. Identification of critical Staphylococcal genes using conditional phenotypes generated by antisense RNA. Science. 2001;293:2266–2269. doi: 10.1126/science.1063566. [DOI] [PubMed] [Google Scholar]