Abstract
Summary
We created a mouse model where conditional expression of an Mll-AF4 fusion oncogene induces B-precursor acute lymphoblastic (ALL) or acute myeloid leukemias (AML). Gene expression profile analysis of the ALL cells demonstrated significant overlap with human MLL-rearranged ALL. ChIP-chip analysis demonstrated histone H3 Lysine 79 (H3K79) methylation profiles that correlated with Mll-AF4 associated gene expression profiles in murine ALLs, and in human MLL-rearranged leukemias. Human MLL-rearranged ALLs could be distinguished from other ALLs by their H3K79 profiles and suppression of the H3K79 methyltransferase DOT1L inhibited expression of critical MLL-AF4 target genes. We have thus demonstrated that ectopic H3K79 methylation is a distinguishing feature of murine and human MLL-AF4 ALLs and is important for maintenance of MLL-AF4 driven gene expression.
Significance
The t(4;11) encodes an MLL-AF4 fusion protein, and predicts a particularly poor prognosis when found in acute lymphoblastic leukemias (ALL). Recent studies suggest certain MLL-fusion proteins enhance gene expression by recruitment of the histone H3 lysine79 (H3K79) methyltransferase DOT1L. We demonstrate that H3K79 methylation is enhanced at many loci in leukemia cells from a murine model of Mll-AF4 and in human MLL-AF4 leukemia cells and this elevation is correlated with enhanced gene expression. Furthermore, suppression of H3K79 methylation leads to inhibition of gene expression in MLL-AF4 cells. These data demonstrate that inhibition of DOT1L may be a therapeutic approach in this disease, and that this mouse model should be useful for assessment of therapeutic approaches for MLL-rearranged ALL.
Introduction
Leukemias mediated by MLL-rearrangements possess unique clinical and biological features. MLL-rearrangements can be found in Acute Lymphoblastic Leukemias (ALL), Acute Myeloid Leukemias (AML) and acute biphenotypic or Mixed Lineage Leukemias (MLL). MLL-translocations are present in over 70% of cases of infant leukemias (Biondi et al., 2000) and in general account for approximately 5% of ALL, 5–10% of AML, and a significant portion of acute biphenotypic or mixed-lineage leukemias (Huret et al., 2001). Patients with MLL-rearranged ALL have a particularly unfavorable prognosis as compared to other forms of ALL (Chen et al., 1993). To date more than 50 MLL fusion partner genes have been reported (Ayton and Cleary, 2001; Krivtsov and Armstrong, 2007). The t(4;11)(q21;q23) encodes MLL-AF4 and is the most frequent MLL-translocation found in ALL.
Multiple mouse models have been developed that recapitulate MLL-rearranged AML (reviewed in (Ayton and Cleary, 2001). However, development of models that faithfully recapitulate MLL-fusion mediated ALL has proven more difficult. Constitutive Mll-AF4 knock-in resulted in mixed lymphoid/myeloid hyperplasia and mature B-cell neoplasms in mice (Chen et al., 2006). Conditional expression of Mll-AF4 based on interchromosomal recombination in lymphoid lineages produced mature B-cell lymphomas (Metzler et al., 2006). Thus further development of murine models of MLL-rearranged ALL is needed.
MLL is a mammalian homologue of Drosophila trithorax and possesses multiple functional domains, including amino-terminal AT hooks that bind DNA and a carboxyl-terminal Su(var)3–9, Enhancer-of-zeste, Trithorax (SET) domain that methylates lysine 4 of histone H3 (H3K4). H3K4 methylation is associated with transcriptional activation (reviewed in (Shilatifard, 2006), and MLL positively regulates expression of clustered Homeobox (HOX) and other genes during development, at least in part via H3K4 methylation (Milne et al., 2002; Nakamura et al., 2002). Mll has important roles in development including hematopoietic development (Hess et al., 1997; Jude et al., 2007)
MLL-translocations invariably encode fusion proteins that have lost the H3K4 methyltransferase (SET) domain. However MLL-fusion proteins retain the ability to bind HOX genes and other promoter regions and are associated with enhanced gene expression (Armstrong et al., 2002; Guenther et al., 2005; Rozovskaia et al., 2001; Yeoh et al., 2002; Zeisig et al., 2004). Several mechanisms have been proposed as to how MLL-fusions may deregulate gene expression including recruitment of abnormal histone modification activities (Cheung et al., 2007; Krivtsov and Armstrong, 2007; Okada et al., 2005). For example, MLL-AF10 and MLL-ENL have been shown to recruit the non-SET domain methyltransferase DOT1L which promotes methylation of histone H3 lysine 79 (H3K79) on the HoxA9 promoter (Mueller et al., 2007; Okada et al., 2005; Zeisig et al., 2005). Since H3K79 methylation is linked to positive transcriptional regulation (Schubeler et al., 2004; Shilatifard, 2006), DOT1L mediated methylation of H3K79 may contribute to increased expression of HOXA9 in MLL-AF10 and MLL-ENL induced leukemias. Furthermore, AF10, ENL and other MLL-fusion partners such as AF4 and AF9 are normally found in nuclear complexes associated with DOT1L (Bitoun et al., 2007; Mueller et al., 2007; Okada et al., 2005; Zeisig et al., 2005; Zhang et al., 2006). Thus aberrant recruitment of DOT1L to the promoters of MLL target genes may be a common feature of many oncogenic MLL-fusion proteins. However, the extent of H3K79 methylation changes and the specificity of these epigenetic changes for MLL-rearranged leukemias have not been defined.
Here, we report the development of a murine model in which conditional expression of MLL-AF4 induces both ALL and AML. Genome wide assessment of gene expression and H3K79 methylation demonstrates that this model faithfully recapitulates human ALL resulting from MLL-AF4 translocation, and identifies ectopic H3K79 methylation as an important part of MLL-AF4 driven gene expression and transformation.
Results
Generation of a Conditional Mll-AF4 knock-in mouse
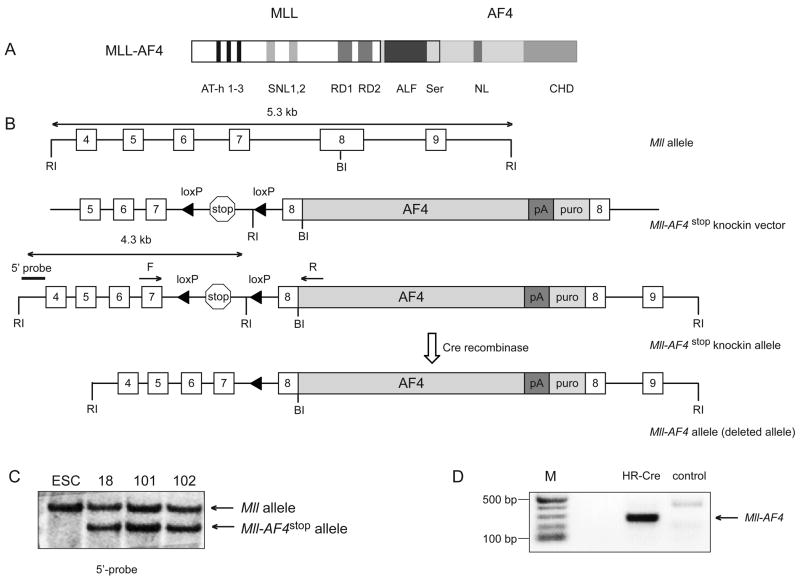
We used a conditional expression approach that proved successful for development of an Mll-Cbp myelodysplasia/AML model to create a model in which the Mll-AF4 fusion product was conditionally expressed from the endogenous Mll locus. We engineered a conditional Mll-AF4stop targeting construct by replacing the Cbp cDNA in the previously reported Mll-Cbpstop targeting vector (Wang et al., 2005) with a cDNA encoding the C-terminal portion of human AF4 (Figure 1A). This generated a targeting construct that placed the human AF4 sequence in the murine Mll exon 8, downstream of a transcriptional stop site flanked by LoxP sites which allows conditional expression of an Mll-AF4 fusion RNA upon expression of Cre recombinase (Figure 1B). The construct was electroporated into CJ7 mouse embryonic stem cells (Swiatek and Gridley, 1993), and clones possessing the targeted allele were selected by Southern blot (Figure 1C) and used to achieve germline transmission of the knock-in allele. Mice heterozygous for the Mll-AF4stop conditional allele were born at slightly less than Mendelian frequency presumably due to heterozygousity for Mll (Yu et al., 1995). Founder mice were backcrossed to C57BL6/129 F1 mice. We used Mll-AF4 specific primers to confirm the absence of the fusion RNA in bone marrow from mice heterozygous for the Mll-AF4stop allele.
Figure 1. Generation of mice with a conditional Mll-AF4stop allele.
A Schematic representation of the MLL-AF4 fusion protein including a number of protein motifs. AT-hooks (AT-h 1–3); speckled nuclear localization sites (SNL1 and SNL2); transcriptional repression domain consisting of two functional subunits, (RD1) and (RD2); homologous regions among within the AF4/FMR2 family members (ALF); Serine-rich regions that contain a transactivation domain (Ser); nuclear localization signal (NL); C-terminal homology domain conserved between FMR2, AF4 and LAF4 family of transcription factors (CHD). B. A diagrammatic description of the Mll-AF4stop knockin allele. Mll exons are numbered 4 to 9. The poly-adenylation site (pA) and puromycin (puro) resistance cassette is shown. EcoRI (RI) and BamHI (BI) sites and Mll forward (F) and AF4 reverse (R) primers used for Mll-AF4 fusion RNA detection are shown. C. Southern blot analysis of EcoRI-digested genomic DNA from wild-type and targeted ESC, probed with the 5′-probe shown in B. D. RT-PCR analysis of Mll-AF4 transcript expression in total bone marrow from Mll-AF4stop mice retrovirally transduced either with “Hit and run Cre” (HR-Cre) or MSCV-MIG (control). (M) molecular weight marker in basepairs (bp).
Mll-AF4 expression enhances serial replating of lymphoid progenitors
First, we determined if expression of Mll-AF4 could transform lymphoid cells in vitro. We collected bone marrow from Mll-AF4stop (MA4) heterozygous mice (3 experiments, 5 mice each) 5-days after 5-fluorouracil (FU) treatment and transduced cells with retroviruses encoding either GFP control (MIG) or Cre-GFP (Cre) to initiate expression of Mll-AF4. Cells were then cultured in semisolid media supplemented with IL-7, SCF, and FLT3. After 14 days of culture, over 90% of cells in both Cre and MIG groups expressed CD19 and B220. Weekly replating of 1×104 MIG transduced cells exhausted their colony forming potential by the third week, while similar replating of Cre transduced cells did not exhaust their replating potential for at least 6 weeks (supplemental figure 1A). To assess for gene expression changes associated with Mll-AF4 expression, we extracted RNA from 1×105 cells at the end of second week of plating, amplified, labeled, and hybridized labeled RNA to Affymetrix mouse 430 A2.0 microarrays. Supervised analysis identified HoxA5, HoxA9, Runx2, Meis1, and Mylk among the 20 most up-regulated genes in the cells expressing Mll-AF4 (Supplemental figure 1B). These genes are also found as central members of early gene expression changes associated with MLL-AF9 expression in myeloid cells and in human MLL-rearranged lymphoblastic leukemias (Armstrong et al., 2002; Krivtsov et al., 2006). Thus, conditional expression of Mll-AF4 in lymphoid cells leads to in vitro transformation and gene expression changes associated with human MLL-fusion leukemias.
Mll-AF4 expression induces acute leukemias
The initial strategy we used to activate Mll-AF4 expression utilized a self-excising retrovirus that transiently expresses Cre-recombinase in transduced cells (Silver and Livingston, 2001). We collected bone marrow (BM) from heterozygous MA4 mice either 5 days after 5-FU treatment or used 5-FU untreated bone marrow depleted for cells expressing CD3, CD4, CD8α, F4/80, B220, Gr1, TER119 (Lin-) and transduced cells with retroviruses encoding either “Hit and Run Cre” (HR-Cre) or GFP (MIG) and transplanted the infected cells into lethally or sublethally irradiated syngeneic recipients. The expression of Mll-AF4 in HR-Cre but not MIG transduced cells was confirmed by RT-PCR (Figure 1D). Mice (n=43) expressing the Mll-AF4 allele as a result of retroviral transduction of Cre developed a fatal disease (Table 1, supplemental figure 2A, B) consistent with acute leukemia including bone marrow replacement, splenomegaly and variable lymphadenopathy (Table 1, figure 2A). Mice transplanted with BM from MA4 mice (n=20) transduced with MIG did not develop leukemia (Supplemental figure 2A).
Table 1.
| Latency(days) mean(range) | SP weight (mg) mean(range) NL (70–110) | WBC *1000/ul mean(range) NL (5–10) | HCT % mean(range) NL (45–47) | Platelet*1000/ul mean(range) NL (800–1400) | % of leukemia cells* in BM | |
|---|---|---|---|---|---|---|
| tBM tr (n=37) | ||||||
| ALL (n=11) | 122.8 (71–220) | 425.5 (200–670) | 138.0 (17–476) | 32.1(20–40.9) | 447.9(144–920) | 85.3(52–99) |
| AML (n=23) | 75.6 (25–221) | 404.8 (150–800) | 124.8 (8–389) | 24.7(8.9–41) | 310.3(70–824) | 86.0(54–98) |
| MLL (n=3) | 92 (81–105) | 666.7 (390–810) | 224.3(155–281) | 23.7(17.7–26) | 201(99–312) | 59(39–75) |
|
| ||||||
| Lin- tr (n=7) | ||||||
| ALL (n=4) | 67.5 (62–75) | 287.5 (200–450) | 459.8(236–748) | 25.5(16.1–39) | 373.5(251–496) | 88.3(67–98) |
| AML (n=2) | 103 (98–108) | 515 (470–560) | 72.5(23–122) | 33.6(29.9–37) | 394.5(367–422) | 83.5(80–87) |
| Alive (n=1) | ||||||
|
| ||||||
| Mx1-Cre (n=22) | ||||||
| ALL (n=8) | 152.3 (120–242) | 691 (260–2100) | 43.0 (5–92.6) | 30.3(5.2–44) | 640 (102–1683) | 46.6(24–86) |
| AML (n=6) | 143.5 (67–240) | 485.0 (300–910) | 84.3 (6–279) | 23.5(10.4–40) | 732 (400–1463) | 92.2(84–98) |
| No leukemia (n=3) | ||||||
| Alive (n=5) | ||||||
Characteristics of mice with Mll-AF4 induced leukemias. Normal ranges (NL) are given for reference.
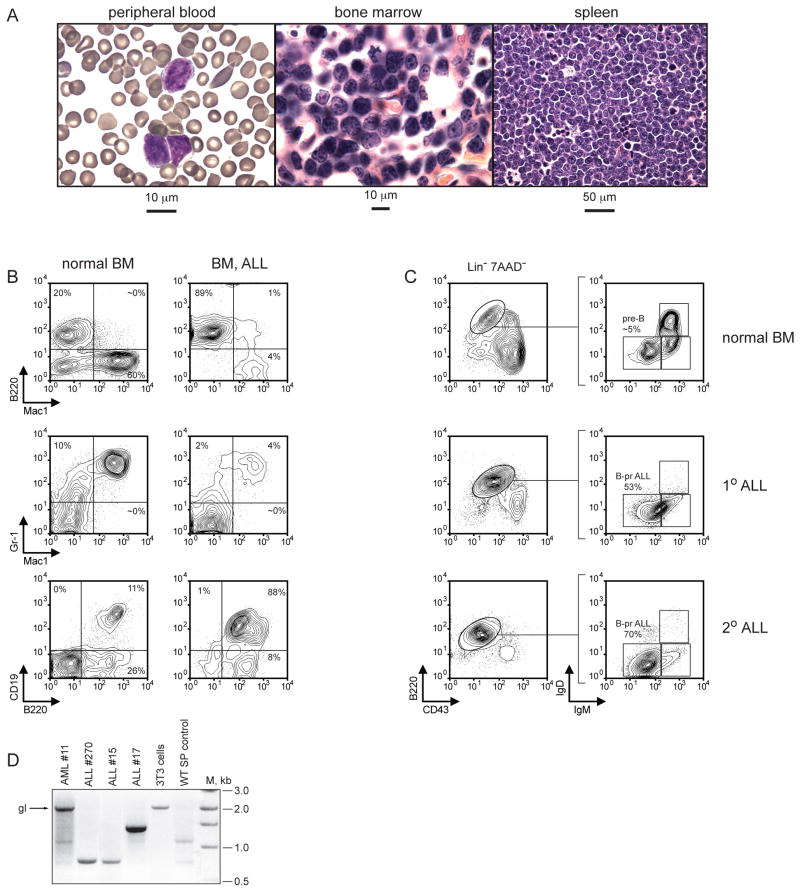
Figure 2. Characterization of Mll-AF4 mediated B-precursor ALLs.
A. Histopathologic analysis of B-pr ALL in Mll-AF4 mice. Cells consistent with lymphoblasts are found in the peripheral blood (left). The bone marrow (center) and spleen (right) are infiltrated with leukemia cells (right). B. FACS analysis of normal bone marrow (left) and leukemic bone marrow (right) using antibodies to B220, Gr1, Mac1, and CD19. C. Detailed immunophenotypic analysis of normal bone marrow (top) and leukemic bone marrow (middle) from a mouse with primary (1°) ALL and an ALL in a secondary (2°) recipient mouse (bottom). D. PCR based analysis showing germline or rearranged Immunoglobulin DH-JH heavy-chain loci in AML cells (11), ALL cells (15, 17, and 270), NIH3T3 cells or normal splenocytes. The arrow indicates germline (gl) DH-JH configuration. (M) molecular weight marker in basepairs (bp).
In order to confirm leukemia development in a purely genetic model, we crossed MA4 mice with mice transgenic for Mx1-Cre (Kuhn et al., 1995). Mx1-Cre × MA4 mice and control littermates received three intraperitoneal injections of pIpC. Mll-AF4 transcript expression was confirmed by RT-PCR. 14 of 22 Mx1-Cre × MA4 mice developed a disease consistent with acute leukemia, and three died from unknown causes within 250 days from the initial pIpC treatment. Mice that possess only Mx1-Cre or Mll-AF4stop did not develop leukemia (table 1, supplemental figure 2C). The median latency of leukemias originated using this approach was longer (131 days) than either of the transplant models (82 and 71 days), and these mice tended to have less bone marrow involvement than mice that received the retrovirally delivered cre (Table 1, supplemental figure 2A–C).
Histopathologic evaluation of tissues from moribund mice demonstrated bone marrow, splenic and liver infiltration with immature hematopoietic cells (Figure 2A). The 31 cases with a morphologic appearance most consistent with myeloid leukemia possessed immunophenotypes consistent with AML: Mac1+ Gr1+ B220− CD19− (n=26) (supplemental Figure 2D), Mac1− Gr1+ B220− CD19− (n=3), or Mac1+ Gr1− B220− CD19− (n=2). The 26 cases whose morphologic appearances were most consistent with a lymphoid leukemia possessed immunophenotypes that included expression of the lymphoid marker(s) B220 and/or CD19. 16 of 26 lymphoid appearing leukemias were B220+ CD19+ Mac1− (Figure 2B). Southern and PCR analyses of genomic DNA from spleen cells isolated from leukemic mice demonstrated that all CD19+ B220+ leukemias analyzed possessed a clonal DJH rearrangement (Figure 2D, supplemental figure 2E,F). This type of leukemia was therefore consistent with typical B-cell ALL. None of the AML analyzed possessed DJH rearrangements (Figure 2D, supplemental figure 2E,F). Three lymphoid-appearing leukemias were B220+ CD19− Mac1+ (Supplemental Figure 2D), and based on the co-expression of B220 and Mac1, these leukemias were termed Mixed Lineage Leukemias (MLL). One of 3 possessed a single DJH configuration as assessed by a PCR based approach (Supplemental Figure 2E). Three leukemias were B220+ CD19− Mac1−, one of which possessed a clonal DJH rearrangement in support of a very early lymphoid arrest, and are thus designated pro-B ALL (Supplemental figure 2D,E). Four lymphoid-appearing leukemias were CD19+ B220− Mac1−. All lymphoid leukemias expressed the Mll-AF4 fusion RNA (Supplemental figure 2H).
We assessed the ability of the leukemias to initiate disease in secondary recipient mice to functionally assess the presence of leukemia initiating cells (LIC). Bone marrow cells from mice with AML (n=1), MLL (n=2), or B-precursor (B-pr) ALL (n=4) could initiate leukemia in secondary recipient mice (n=21) when 3×104 cells were injected (data not shown). We performed limiting dilution transplant experiments to determine the frequency of LIC in the B220+ CD19+ B-pr ALLs (n=2) and found all (n=22) mice that received as few as 1×103 sorted B220+ CD19+ leukemia cells developed an identical ALL (Figure 2C) whereas 3 of 9 mice that received 100 cells developed leukemia. We also further characterized the MLLs (n=2) for LIC and found all (n=24) secondary recipient mice that received 5×104 sorted B220+ Mac1+ cells developed an identical mixed lineage leukemia (data not shown) whereas none of the mice (n=20) that received 1×103 cells developed leukemia. These data confirm the development of an acute leukemia in this model system and demonstrate the presence of leukemia initiating cells in leukemias with different immunophenotypes.
We further characterized the CD19+ B220+ leukemias and defined the precise stage of B-lymphoid development that most closely matched the leukemia. We performed immunophenotypic analysis that assessed B220, CD43, IgM, IgD expression to compare the ALL cells to various stages of B-cell development. Bone marrow cells taken from wild-type mice demonstrated appropriate percentages of B-cells at various stages of differentiation (Figure 2C). However, the majority of lymphoid cells present in BM from mice with primary or secondary B-pr ALL were B220+ CD43− IgM− IgD−, most consistent with mouse pre-B cells (Hardy, 1990). Thus, the leukemias represent an expansion of cells that by immunophenotypic analysis are arrested at the pre-B cell stage of development thus confirming their identity as B-pr ALL.
Murine Mll-AF4 ALL recapitulates human MLL-rearranged ALLs
To more definitively determine the developmental stage of leukemia cells, we compared the global gene expression pattern of the expanded leukemia cells to normal B-cells. We isolated RNA from 16 primary B-pr ALLs and normal lymphocyte populations (Hardy, 1990) including pro-B cells (Lin− B220+ CD43+), pre-B (Lin− B220+ CD43− IgM− IgD−), immature-B (Lin− B220+ CD43− IgM+ IgD−), and mature-B cells (Lin- B220+ CD43− IgM+ IgD+), amplified RNA and hybridized to Affymetrix 430 A 2.0 microarrays. Unsupervised hierarchical clustering analysis demonstrated that 14 ALLs were more similar to pre-B cells and that two ALLs were more similar to pro-B cells than to immature B or mature B-cells (Figure 3A) providing further support for leukemia cell expansion at an early stage of B-cell development. The relationship between the ALL samples and normal B-cells was further tested by consensus clustering, which demonstrated that ALLs 117 and 122 cluster separately and the other ALLs were most similar to pre-B cells (Supplemental figure 3A).
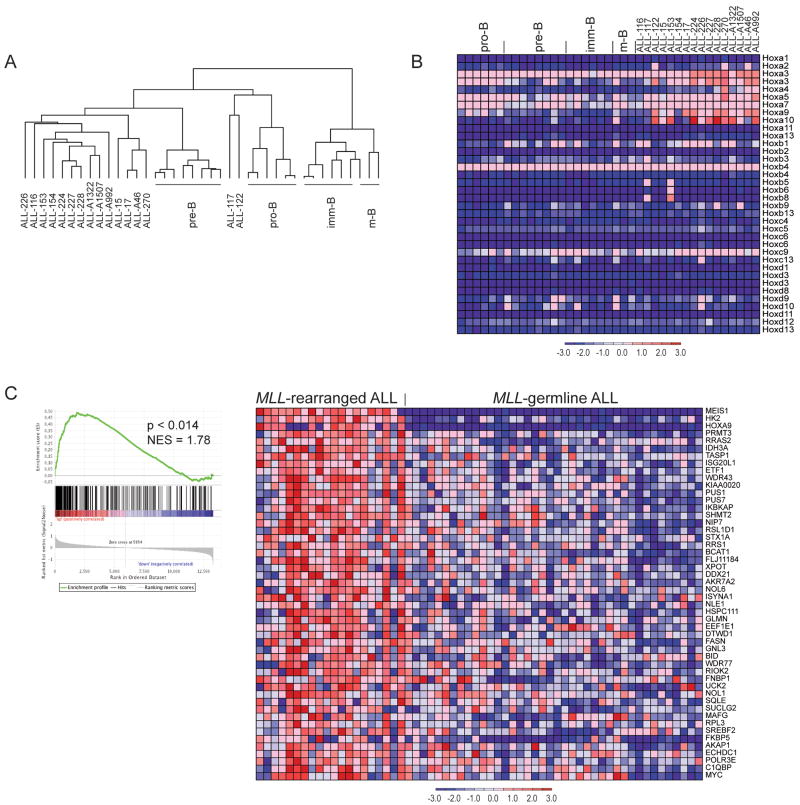
Figure 3. Gene expression in normal and malignant mouse B-cells and comparison to human leukemias.
A Hierarchical clustering using 10218 filtered (min fold 3; min delta 100, background 20; ceiling 20000) probe sets. B. Expression of homeobox A, B, C, and D cluster genes in normal developing B-lymphocytes and B-pr ALL cells. C. GSEA analysis of gene expression in human MLL-rearranged ALL (n=20) as compared to MLL-germline ALL (n=40) (Ross et al., 2003) using the top 386 genes identified as highly expressed in murine Mll-AF4 ALL as a gene set. GSEA enrichment plot (left) and the top 50 genes that show increased expression in human MLL-rearranged leukemias are shown (right).
Supervised gene expression analysis demonstrated that murine B-pr ALL cells express high levels of certain HoxA cluster genes but do not differentially express HoxB, HoxC or HoxD cluster genes (Figure 3B). The elevated expression of HoxA9 was confirmed by quantitative-PCR (qPCR) (Supplemental figure 3B). The murine ALL cells also express high levels of Meis1 as measured by microarrays and qPCR (Supplemental figure 3C) similar to human MLL-rearranged ALL (Armstrong et al., 2002).
Next to assess globally whether mouse Mll-AF4 ALLs recapitulate gene expression profiles observed in human MLL-rearranged ALLs we compared gene expression signatures of mouse Mll-AF4 ALL to human MLL-rearranged ALL. We identified the top 500 probe sets (386 genes) with increased expression in murine Mll-AF4 B-pr ALL as compared to pre-B cells using a signal to noise statistic, and converted them to human homologues found on the Affymetrix U133 arrays (259 genes). Gene set enrichment analysis (GSEA) demonstrated strong enrichment (p<0.015) of the murine Mll-AF4 signature in human MLL-rearranged ALLs as compared to MLL-germline ALLs (Ross et al., 2003; Subramanian et al., 2005) (Figure 3C). These data demonstrate that the ALLs in our mouse model recapitulates gene expression programs found in human MLL-AF4 induced disease.
H3K79 methylation in murine Mll-AF4 ALL
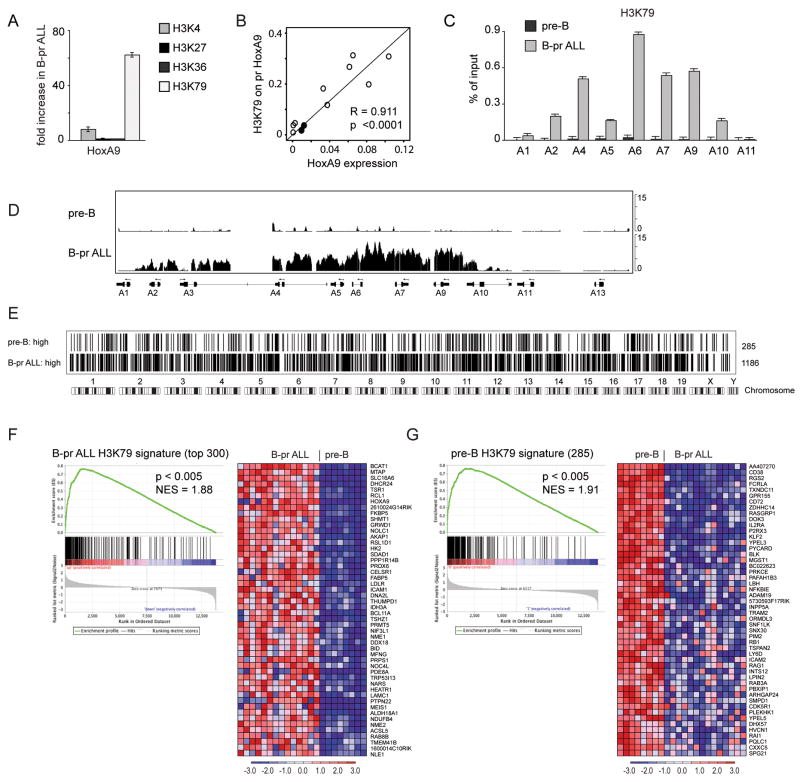
Since AF4 associates with the DOT1L methyltransferase (Bitoun et al., 2007), we hypothesized that Mll-AF4 may be associated with ectopic H3K79 methylation. First we determined that Dot1L is ubiquitously expressed in B cell progenitors and B-pr ALL (supplemental figure 4A). We used chromatin immunoprecipitation (ChIP) to assess histone methylation near the promoters of HoxA genes. We sorted pre-B cells and leukemia cells with a similar immunophenotype, and performed ChIP with antibodies directed against dimethylated H3K79 (H3K79me2), trimethylated lysine 27 (H3K27me3), trimethylated lysine 36 (H3K36me3) or trimethylated lysine 4 (H3K4me3). We assessed the ChIP DNA by q-PCR using primers that spanned the promoter region of HoxA9 and compared the amount of precipitated DNA as a percentage of input in normal vs. leukemia cells (supplemental figure 4B). We found slightly elevated H3K4me3, no obvious change in H3K27me3 or H3K36me3, and dramatically enhanced H3K79me2 associated with HoxA gene promoters in the leukemia cells (Figure 4A,C). Next, we assessed expression level of HoxA9 in pre-B ALLs and normal B-cell progenitors and found a strong correlation between the amount of H3K79me2 and HoxA9 expression (Figure 4B). We expanded our assessment and found enhanced H3K79me2 across much of the HoxA cluster (Figure 4C). Given the data demonstrating a potential role for DOT1L in some MLL-rearranged leukemias, and this data demonstrating enhanced H3K79me2 in Mll-AF4 ALL cells we focused further assessment on H3K79 methylation.
Figure 4. ChIP analysis of histone methylation in mouse Mll-AF4 B-precursor ALL and normal pre-B cells.
A Fold increase in trimethyl-H3K4, trimethyl-H3K27, trimethyl-H3K36, and dimethyl H3K79 marks associated with Hoxa9 promoter in B-pr ALL over pre-B cells as assayed by ChIP-qPCR. Two tailed T-test: H3K4 p=0.05; H3K79 p=0.005. Error bars represent +/− standard deviation (SD) between 4 independent experiments. B. Correlation of HoxA9 expression with H3K79me2 content on the HoxA9 promoter. HoxA9/Gapdh ratio (HoxA9 expression) was assessed by RT-qPCR and H3K79me2 content (percent of input) on HoxA9 promoter was assessed by ChIP-qPCR in pre-B (filled circles, n=2) and B-pr (open circles, n=9) ALL samples. Pearson correlation R=0.911, two tailed t-test p<0.0001. C. Enrichment of H3K79me2 associated with HoxA cluster promoter regions in B-pr ALL and normal pre-B cells expressed as a percentage of input. Error bars represent +/− SD of triplicates in one of 3 independent experiments. D. Identically scaled average tracks from ChIP-chip analysis representing HoxA cluster loci in normal pre-B (n=3) and B-pr ALL (n=3) cells. E. Graphical (bar) representation of regions associated with H3K79 me2 in normal pre-B and B-pr ALL cells. Averaged MAT scores from genome-wide promoter analysis of 3 pre-B cell and 3 B-pr ALL samples were used to define genomic regions possessing significant H3K79 signal (p<10−5) over background. The bars represent 285 genes that were identified to be associated with H3K79me2 in pre-B cells but not in B-pr ALL as compared with 1186 genes associated with H3K79me2 in B-pr ALL but not pre-B cells. F. Correlation of H3K79me2 marks with gene expression in B-pr ALL. GSEA was performed using the top (by MAT score) 300 genes (of 1186 genes) in the B-pr ALL H3K79 signature as a gene set and assessed for enrichment in those genes more highly expressed in B-pr ALL as compared to normal mouse pre-B cells. GSEA enrichment plot (left) and expression of genes (right) corresponding to the top 50 differentially expressed genes from the B-pr ALL H3K79 signature NES 1.88; p<0.005. G. Correlation of H3K79me2 marks with gene expression in pre-B cells. GSEA was performed using 285 genes of pre-B H3K79 signature as a gene set. GSEA enrichment plot (left) and expression of genes (right) corresponding to the top 50 differentially expressed genes from the B-pr ALL H3K79 signature NES=1.91; p<0.005.
Genome-wide assessment of DNA binding of MLL through the use of ChIP followed by microarray analysis (ChIP-chip) demonstrated association of MLL with the promoter regions in thousands of genes (Guenther et al., 2005). This prompted the question whether the MLL-AF4 fusion influences a select set of genes such as HoxA genes, or if there are more widespread histone methylation abnormalities in Mll-AF4 leukemias. We performed ChIP-chip analysis on 3 mouse Mll- AF4 B-pr ALLs and 3 normal pre-B samples using an antibody against H3K79me2 and Affymetrix 1R mouse promoter arrays. These studies verified an increase in H3K79me2 across the HoxA cluster in leukemia cells (Figure 4D). A second group of 6 murine Mll-AF4 ALLs confirmed H3K79 me2 across the HoxA loci, but not the HoxB, HoxC, or HoxD clusters (supplemental figure 5A–C). An expanded view across the genome revealed thousands of loci with significant H3K79me2 in both normal pre-B cells and ALL cells respectively, as would be expected given a normal role for H3K79me2 in epigenetic regulation. However, on a comparative basis, there were 1186 promoter regions in which H3K79me2 was increased in the leukemia cells by comparison to normal pre-B cells (B-pr ALL H3K79 signature), whereas only 285 promoter regions had H3K79 me2 that was higher in normal pre-B cells (pre-B H3K79 signature) (Figure 4E).
As H3K79 me2 has been associated with positive regulation of transcription (Schubeler et al., 2004; Shilatifard, 2006), we used GSEA to determine whether H3K79me2 was correlated with gene expression. Genes associated with increased H3K79me2 in Mll-AF4 induced leukemia cells (B-pr ALL H3K79 signature) were highly enriched for genes with elevated mRNA expression in leukemia cells (p<0.005, Figure 4F). Likewise, genes with elevated H3K79me2 within normal pre-B cells (pre-B H3K79 signature) were associated elevated expression in normal pre-B cells (p<0.005, Figure 4G). These analyses demonstrate ectopic H3K79me2 in Mll-AF4 leukemias is associated with enhanced gene expression.
H3K79 methylation in Human MLL-rearranged ALLs
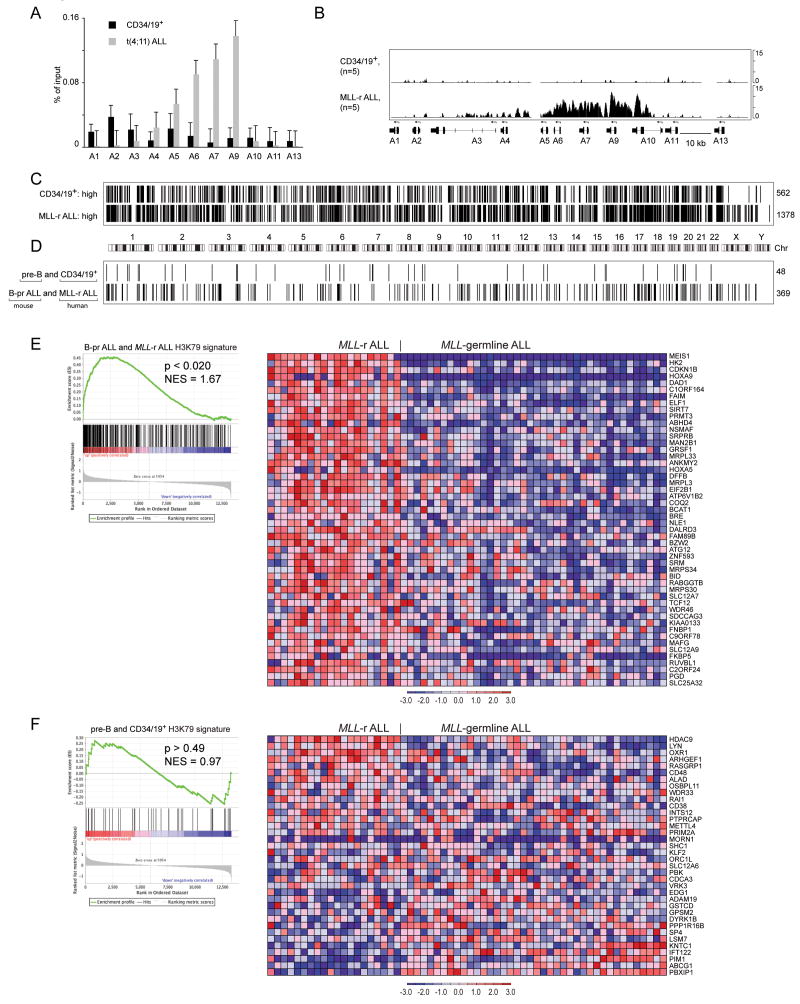
To assess whether similar abnormalities in H3K79me2 exist in human MLL-rearranged ALLs we assessed histone methylation in MLL-rearranged ALLs as compared to Lin− CD34+ CD19+ bone marrow cells. First we performed ChIP-qPCR to assess H3K79me2 associated with HOXA cluster loci. We found elevated H3K79me2 associated with multiple HOXA genes (Figure 5A). Next, we performed ChIP-chip analyses on 5 MLL-rearranged ALLs and 5 bone marrow Lin− CD34+ CD19+ cell samples using Affymetrix 1R human promoter arrays. The data demonstrates elevation of H3K79me2 associated with multiple HOXA cluster genes similar to that seen in the mouse leukemias (Figure 5B, Supplemental figure 6A). We assessed genome wide differences in H3K79me2 in MLL-rearranged ALL as compared to normal CD34/CD19+ cells just as we did for murine cells. There were many genomic regions associated with H3K79me2 in both cell types. We found elevated H3K79me2 associated with 1378 promoter regions in the leukemias and elevated H3K79me2 associated with 562 promoter regions in the normal samples thus demonstrating more genes associated with increased H3K79me2 in the MLL-rearranged leukemias as compared to normal cells (Figure 5C). Since the general trend in H3K79me2 appeared similar in the mouse and human cells, we were interested if there was overlap between the genes associated with H3K79me2 in the human and mouse cells. We compared the B-pr ALL H3K79 signature derived from mouse and human cells and identified 369 genes associated with increased H3K79me2 in both mouse and human MLL-AF4 leukemias (Figure 5D). We also identified 48 genes associated with increased H3K79me2 in normal B-pr cells in mouse and human (Figure 5D). These data show that the epigenetic differences between MLL-rearranged ALL and normal B-pr cells are similar between our mouse model and the human disease with more genes associated with enhanced H3K79me2 in the leukemia cells than in normal cells.
Figure 5. ChIP-chip analysis of histone methylation in human MLL-rearranged ALL and normal pre-B cells.
A Enrichment of H3K79-dimethyl marks associated with HOXA cluster promoters in t(4;11) ALL and human bone marrow Lin− CD34+ CD19+ (CD34/19+) cells expressed as percentage of input as assessed by ChIP-qPCR. Error bars represent +/− SD of triplicates in one of 3 independent experiments. B. Identically scaled average tracks from ChIP-chip analysis of H3K79me2 modifications associated with HOXA cluster loci in human CD34/19+ cells (n=5) or MLL-rearranged ALL (MLL-r ALL) (n=5). C. Graphical (bar) representation of regions associated with H3K79me2 in normal CD34/CD19+ and MLL-rearranged ALL cells. 562 genes were identified in CD34/19+ cells but not in MLL-rearranged ALL (CD34/19+: high) whereas 1378 genes with significant H3K79 signal were present in MLL-rearranged ALL but not CD34/19+ cells (MLL-r ALL: high). D. 48 genes had higher H3K79 ChIP-chip signals in both mouse pre-B and human CD34/19+ cells as compared to mouse and human leukemias. 369 genes had higher H3K79 ChIP-chip signal in both mouse Mll-AF4 and human MLL-rearranged ALL as compared to normal B-cells. Fisher test p<0.0001. E. GSEA analysis of the 369-gene signature with elevated H3K79me2 found in both human MLL-rearranged ALL and mouse B-pr ALL found enrichment of the gene expression signature in MLL-rearranged ALL as compared to MLL-germline ALL (NES=1.63, p<0.019). GSEA enrichment plot (left) and heatmap (right) of expressio n for the top 50 probe sets in MLL-rearranged ALL as compared to MLL-germline ALL (Ross et al., 2003). F. GSEA analysis of the 48 genes with elevated H3K79me2 found in both human bone marrow CD34/CD19+ and mouse pre-B cells found no enrichment of the signature in MLL-rearranged ALL as compared to MLL-germline ALL (NES=0.97, p>0.49). GSEA enrichment plot (left) and heatmap (right) of expression for 36 probe sets in MLL-rearranged ALL as compared to MLL-germline ALL (Ross et al., 2003).
Next, we reasoned that genes found associated with H3K79me2 in both mouse and human MLL-AF4 leukemias may represent a set of MLL-AF4 genes and should be functionally associated with increased gene expression in human MLL-rearranged ALLs as compared to ALLs that are not driven by MLL-fusion oncoproteins. We performed GSEA using the 369 genes found in the mouse/human H3K79me2 overlap as a gene set and gene expression data from MLL-rearranged and germline ALL samples (Ross et al., 2003). The analysis demonstrated significant enrichment of the H3K79me2 associated gene set in genes with elevated mRNA expression in MLL-rearranged ALL in comparison to other ALLs (p<0.02, Figure 5E). The 48 genes that were common to mouse and human normal pre-B cells were not associated with gene expression changes in either MLL-rearranged or other ALLs (p<0.49, Figure 5F). These data demonstrate that ectopic H3K79me2 is associated with gene expression in human leukemias just as is the case in mouse leukemias.
H3K79me2 profiles distinguish human MLL-rearranged ALL from other ALL
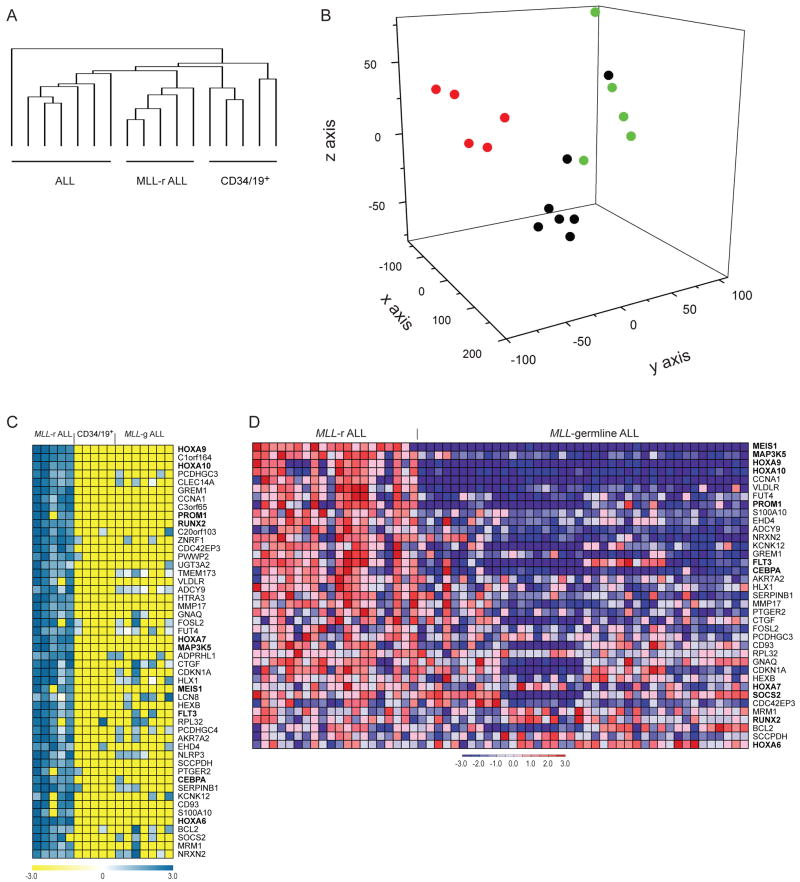
Having demonstrated that we can identify histone methylation abnormalities associated with the presence of MLL-AF4 in mouse as well as human MLL-rearranged leukemias, we wondered if the histone methylation abnormalities were sufficiently different between MLL-rearranged and MLL-germline leukemias such that global patterns of H3K79me2 could be used to distinguish MLL-rearranged ALLs from MLL-germline ALLs and normal B-cell progenitors. We assessed genome wide patterns of H3K79me2 in 5 MLL-rearranged ALL samples, 7 MLL-germline ALL samples and 5 normal controls. Similar to normal pre-B cells and MLL-rearranged leukemia, ALL samples with germline MLL had thousands of loci with H3K79me2. We developed a method to use histone methylation ChIP-chip data to cluster samples. After individually processing each promoter array with an enrichment P-value cut-off of 10−5, we associated the resulting ChIP-chip regions to RefSeq genes if the site fell within a window extending from −500 to 2500bp of the transcriptional start site. We retained only those genes that had H3K79 me2 in at least 3 of the 17 samples. This filtering identified 5438 distinct genes with H3K79me2. Remarkably, the H3K79 epigenetic profile of MLL-rearranged leukemia samples were clearly distinct from normal CD34/CD19+ cells and from MLL-germline ALLs when clustered with hierarchical clustering (Figure 6A) or when projected using principal component analysis (PCA) (Figure 6B). Thus, MLL-rearranged ALLs have a global H3K79me2 profile that is sufficiently distinct to distinguish MLL-rearranged ALLs from other lymphoblastic leukemias, just as profiles of mRNA expression can distinguish these classes (Armstrong et al., 2002). Next, we identified the genes that had the highest mean histone methylation score in MLL-rearranged samples compared to normal CD34/CD19+ cells and MLL-germline ALL cells. This analysis identified a number of genes previously associated with MLL-rearranged leukemias including HOXA genes, MEIS1, RUNX2, PROM1, and FLT3 (Figure 6C,D, supplemental figure 6B) (Armstrong et al., 2003; Armstrong et al., 2002; Krivtsov and Armstrong, 2007; Krivtsov et al., 2006; Rozovskaia et al., 2001). A number of other genes of interest including BCL2, and CEBPα have previously been implicated in leukemias but not specifically those with MLL rearrangements. CEBPα is of particular interest given its role in specification of myeloid cell fate and the propensity for myeloid gene expression in MLL-AF4 ALLs. These data demonstrate a unique H3K79me2 profile for MLL-rearranged leukemias as compared to other acute lymphoblastic leukemias, which includes a number of loci previously identified as playing important roles in MLL-rearranged leukemias.
Figure 6. Unsupervised and supervised analysis of ChIP-chip data for H3K79 methylation in MLL-rearranged ALL, MLL-germline ALL, and CD34/19+ cells.
A. Hierarchical clustering of 5438 genes associated with H3K79me2 demonstrates that MLL-rearranged ALL samples cluster together and are separate from other MLL-germline ALLs and normal B-pr cells. B. Principal component analysis (PCA) of the same 5438 genes associated with H3K79me2 separates MLL-rearranged ALL (red) from MLL-germline ALL (black) and CD34/19+ (green). C. The top 50 genes associated with H3K79me2 in MLL-rearranged ALL compared to the rest of the samples are shown. Genes were ranked based on difference of means of MAT scores between MLL-rearranged (MLL-r) ALL samples and MLL-germline (MLL-g) ALL plus CD34/19+ cells. D. Expression of genes identified in figure 6C. 37 of the 50 genes in 6D had corresponding probesets on the U133A arrays.
DOT1L suppression inhibits H3K79me2 and HOXA expression in MLL-AF4 cells
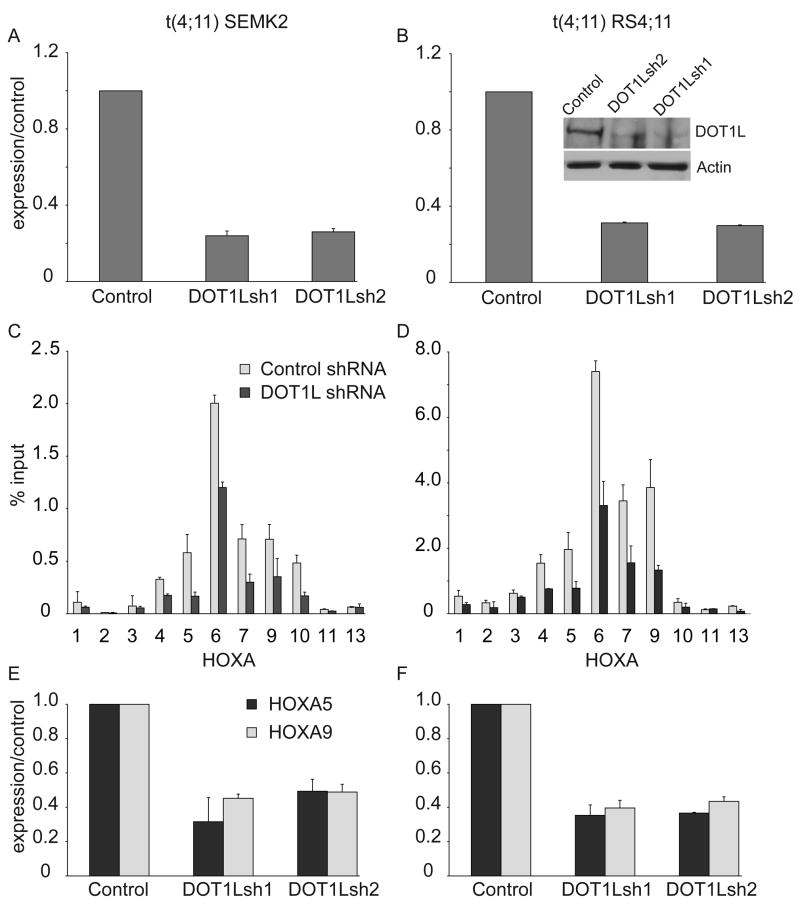
We have demonstrated a strong correlation between H3K79me2 and aberrant HOXA gene expression in MLL-AF4 cells. Thus we were interested to know if continued expression of the H3K79 methyltransferase DOT1L was required to maintain aberrant HOX gene expression, as a first step toward assessing DOT1L as a potential therapeutic target in this disease. We developed 2 lentiviral-based shRNA constructs directed toward DOT1L, and transduced 2 different t(4;11) (MLL-AF4) cell lines. Forty-eight hours after transduction with a viral titer sufficient to transduce >90% of cells, DOT1L expression was assessed by quantitative RT-PCR, demonstrating suppression of DOT1L RNA to approximately 20–30% of control levels (Figure 7A,B). Suppression of DOT1L protein expression was confirmed in one of the two cell lines (Figure 7B). Seventy-two hours after transduction, we assessed H3K79me2 across the HOXA cluster and found diminished H3K79me2 in cells transduced with the DOT1L directed shRNA (Figure 7C,D). This reduction of H3K79 across the HOXA locus resulted in decreased expression of both HOXA5 and HOXA9 (Figure 7E,F). These data demonstrate that continued expression of DOT1L is required for H3K79me2 and HOXA gene expression in MLL-AF4 cells.
Figure 7. shRNA mediated DOT1L suppression in human MLL-AF4 cell lines.
SEMK2 (A) or RS4;11 (B) ALL cell lines possessing a t(4;11) were transduced with either a control shRNA or one of 2 different shRNAs that target DOT1L, and DOT1L RNA expression was assessed by quantitative RT−PCR and western blot for RS4;11 cells 72 hours after transduction (Inset). Error bars represent +/− SD of duplicates in one of two independent experiments. C, D. Chromatin Immunoprecipitation assessment of H3K79 associated with HOXA cluster genes 96 hours after treatment of SEMK2 (C) cells or RS4;11 Cells (D) with either a control or DOT1L-directed shRNA (DOT1Lsh1) shRNA. Error bars represent +/− SD of duplicates in one of two independent experiments. E, F. Assessment of HOXA5 and HOXA9 expression in SEMK2 (E) or RS4;11 (F) cells 96 hours after lentiviral transduction with either control or one of two different shRNA constructs. Error bars represent +/− SD of duplicates in one of two independent experiments.
Discussion
Translocations involving the MLL gene result in both AML and ALL in humans. MLL-AF9 and MLL-AF4 are the most common translocations resulting in AML and ALL respectively. Although a number of retroviral and genetically engineered mouse models of MLL-fusion mediated AML have been developed, models recapitulating MLL-AF4 mediated ALL have been more elusive. Ectopic expression of MLL-AF4 through retroviral transduction has been difficult and other genetically engineered mouse models have resulted in myelodysplasia or mature B-cell lymphomas. We report the generation of a conditional knock-in model in which the MLL-AF4 fusion product is expressed within the context of the endogenous MLL locus. Upon conditional activation, mice develop AML or ALL, the latter with an immunophenotype and gene expression profile consistent with acute B-precursor cell leukemia. The gene expression signature of murine Mll-AF4 ALL cells was found to be highly enriched in the gene expression profile of human ALL samples with MLL rearrangements as compared to ALL samples with germline MLL. Therefore, this murine model recapitulates human MLL-AF4 mediated ALL both in terms of disease phenotype, as well as overall patterns of gene expression. We note that our mice have a propensity toward AML development which differs from the strong association between t(4;11) and ALL in humans. Future studies will determine if this is due to expression of Mll-AF4 in different cells of origin or microenvironmental influences as recently shown for MLL-AF9 (Wei et al., 2008). Also, the genetic background may influence the leukemia incidence and phenotype. The development of a faithful model of MLL-AF4 ALL will allow detailed characterization of the mechanisms of MLL-AF4 mediated leukemogenesis, including the mechanisms of transformation, cell(s) of origin, the phenotypes of cancer stem cells in leukemias expressing various lineage markers, and mechanisms associated with the development of mixed myeloid/lymphoid (mixed lineage) disease. Also, a faithful model of MLL-AF4 ALL will allow for assessment of putative therapeutics in a well-defined model system.
Recent studies have identified association of multiple MLL-fusion partners including AF4, AF9, and AF10 with DOT1L, a histone H3K79 methyltransferase (Bitoun et al., 2007; Mueller et al., 2007; Okada et al., 2005; Zeisig et al., 2005; Zhang et al., 2006). We used our murine model to ask whether abnormalities in H3K79me2 were present in Mll-AF4 mediated ALL. Genome-wide assessment revealed enhanced H3K79me2 (in comparison to matched normal cells) at many loci across the genome, including across the HOXA cluster. This prompted a similar analysis in human ALL samples, where we confirmed widespread abnormalities in H3K79me2 as a characteristic that distinguished MLL-rearranged from MLL-germline ALLs. These results demonstrate that in both murine and human disease mediated by MLL rearrangement, there are widespread changes in H3K79me2, including on genes that are critical for leukemogenesis such as HOXA9 and MEIS1. Future studies will determine which subset of these genes are direct MLL-AF4 target genes. The results described here further underscore the utility of this murine model system for providing mechanistic insight into human disease.
In both human and murine ALL, we found that ectopic H3K79me2 was highly associated with increased mRNA expression. Wildtype MLL possesses H3K4 methyltransferase activity, a modification associated with transcriptional “priming” of the promoters of genes important for appropriate developmental cell-fate decisions (Guenther et al., 2007). Since the methyltransferase domain of MLL is invariably lost in MLL-fusion proteins, including MLL-AF4, the question arises as to how MLL-fusions promote enhanced expression of its target genes such as HOXA genes. We found that H3K4me3 is still associated with HOX genes in our murine MLL-AF4 model and preliminary experiments suggest this to also be the case in human ALL with MLL-fusions (A.K. unpublished data). In both human disease and our murine model, the second MLL allele remains germline, and thus wildtype MLL protein, or perhaps other H3K4 methyltransferases, may continue to regulate H3K4me3 at these loci. However, H3K4me3 is insufficient to fully active transcription, which appears to require the presence of other histone modifications such as H3K79me2 for transcriptional elongation to proceed (Guenther et al., 2007). This leads to a model where MLL-AF4 recruits DOT1L to MLL target genes, and promotes methylation of H3K79 at loci with existing H3K4 methylation (i.e., by wildtype MLL or other H3K4 methyltransferases) thus stimulating transcriptional elongation of genes that are normally primed but not fully transcribed.
Prior studies have established that various cancer sub-types can be distinguished on the basis of specific oncoproteins or global mRNA expression patterns. In this study, we demonstrate that widespread differences in an epigenetic histone modification can likewise distinguish different cancer subtypes, and malignant vs. normal cells. The ability to distinguish cancer sub-types on the basis of epigenomic profiles does not necessarily offer any advantages over mRNA profiling in terms of disease diagnosis. However, the tight linkage between oncoprotein (MLL-AF4), specific epigenetic changes (H3K79me2) and altered gene expression has important therapeutic implications. Specifically, many oncoproteins are DNA binding proteins, and are generally not readily amenable to targeting by small molecules or biologics. Pharmacologic inhibition of transcription factor function, including disrupting protein-protein or protein-DNA interactions, remains a largely experimental undertaking with few examples of success. A common strategy to bypass this impasse has been to look for downstream transcriptional targets that may be more druggable (e.g., enzymes, receptors). Since oncoproteins may alter the expression of hundreds or thousands of genes, in most cases inhibiting the activity of a single druggable downstream target may not fully reverse the malignant phenotype. Inhibition of epigenetic modifying enzymes offers an alternative approach that is facilitated by the fact that enzymes are generally more amenable to drug development. Targeting the epigenetic link between an oncoprotein and downstream gene expression changes may more broadly impact the multitude of gene expression changes that in aggregate contribute to the malignant phenotype. In the case of diseases driven by MLL-fusion proteins, development of a DOT1L inhibitor may impact expression of a diversity of critical genes including HOXA9, MEIS1, FLT3, and BCL2. Indeed our data demonstrate that suppression of DOT1L expression leads to decreased expression of HOXA cluster genes. Furthermore, a DOT1L inhibitor might be applicable to other diseases characterized by ectopic H3K79me2. In this regard, the ability to identify disease sub-types that have widespread ectopic H3K79me2 would have important implications for patient selection and stratification in testing a DOT1L inhibitor. In as much as epigenetic modifications are likely to be linked to the maintenance of gene expression in many human malignancies, these strategies may be applicable to a wide variety of cancer subtypes.
Experimental Procedures
Human samples
Normal bone marrow samples as well as diagnostic leukemia samples (peripheral blood or bone marrow) were obtained with informed consent from individuals treated on protocols approved by the Institutional Review Board at the Dana Farber Cancer Institute between 2000 and 2007. For details see supplemental data.
Generation of Conditional Mll-AF4 knockin mice
Animals were maintained in the Animal Resourses at Children’s Hospital. Animal experiments were approved by the Institutional Animal Care and Use Committee. The Mll-AF4stop targeting vector was assembled in the plasmid vector Litmus 28 (NEB). We removed Cbp cDNA from a previously reported (Wang et al., 2005) Mll-Cbpstop targeting construct using BamHI digestion and ligated in a portion of AF4 (aa 347–1210, gene bank #L13773) using 5′ BamHI and 3′ blunt ends. For further details see supplemental data Mx1-Cre × MA4 mice were generated by crossing MA4 males with females transgenic for Mx1-Cre. Mx1-Cre × MA4 mice (6–8 week old) and control littermates were injected intraperioneally with 400μg of pIpC (Sigma-Aldrich) 3 times over 6-day interval.
Bone marrow reconstitution assay and retroviral infections
Bone marrow was collected from tibias and femurs of MA4 mice either treated or untreated with 5-FU (Sigma-Aldrich). Red blood cells were lysed using RBC lysis buffer (Purgene) the remaining cells were washed twice in PBS (Invitrogen) and transduced with the “Hit-and-run-Cre” (Silver and Livingston, 2001) retrovirus (produced as previously described (Krivtsov et al., 2006)) overnight in IMDM media supplemented with 10% FBS (Invitrogen) 1/100 penicillin/streptomycin (Invitrogene) IL-3, IL-6, IL-7 each 10 ng/ml, SCF, and FLT3 50 ng/ml each (PeproTech) and polybrene 7 μg/ml (Sigma-Aldrich). Syngeneic recipient mice were lethally (2×700 Rads) irradiated or sub-lethally (600 Rads) irradiated. The retrovirally transduced cells were transplanted retroorbitally. Detection of Mll-AF4 described in supplemental methods.
Flow cytometry
Labeling of cells for FACS analysis and cell sorting was performed on ice in PBS supplemented with 0.5% horse serum (Invitrogen) using conjugated antibodies purchased either from BD Pharmingen, eBiosciences (San Diego, CA) or Invitrogen (Carlsbad, CA) unless otherwise indicated. Details of the antibodies used can be found in supplemental data.
Transduction with small hairpin RNA (shRNA) constructs
ShRNA cloned into hairpin pLKO.1-lentiviral vector were synthesized by the Harvard/MIT Broad Institute RNA consortium utilizing a complex selection algorithm to minimize off-target effects (http://www.broad.mit.edu/genome_bio/trc/). The target sequences for suppression of human DOT1L (NM_032482) were: DOT1LshRNA1: 5′-CGCCAACACGAGTGTTATATT-3′, DOT1LshRNA2: 5′-CCGCAAGAAGAAGCTAAACAA-3′, non-targeting GFP control shRNA: 5′-GCAAGCTGACCCTGAAGTTCA-3′. Details of lentiviral transduction can be found in supplemental data.
RNA amplification and microarray data analysis
1–5×105 cells of different populations were used to isolate RNA. 20 ng of total RNA was amplified and labeled using Ovation RNA amplification system V2 (NuGEN). Expression data were analyzed with GenePattern release 3.1 and GSEA 2.0 software package (http://www.broad.mit.edu/tools/software.html). Details of the gene expression analysis can be found in supplemental data.
Chromatin Immunoprecipitation, DNA amplification, data analysis, and PCR analysis
3–10×105 (FACS sorted) cells were fixed in 1% formaldehyde with gentle rotation for 10 minutes and the reaction was stopped by addition of Glycine (final conc. 125mM). The fixed cells were washed twice with PBS, re-suspended in SDS buffer and further processed as described (Bracken et al., 2006) using anti-dimethyl H3K79 (#ab3594, AbCam) it has been shown that this antibody recognize also trimethyl H3K79 (Steger et al., 2008), anti- trimethyl H3K4 (#05-745, Upstate Biotech), anti-trimethyl H3K36 (Abcam 9050), or anti-trimethyl H3K27 (Upstate Biotech 07-449). Eluted DNA fragments were either used directly for qPCR with HoxA promoters specific primers (Bracken et al., 2006) and CybergreenER qPCR mix using ABI 7700 Sequence Detection System (ABI). Alternatively, ChIP DNA fragments were amplified using LM-PCR as described (http://chiponchip.org/protocol_itm3.html) (Experiment 1 in figure 4) or using the WGA1 whole genome amplification kit (Sigma) (experiment 2 in Supplemental figure 5) and subjected to hybridization with Affymetrix mouse 1R or 1R human promoter arrays.
Identification of ChIP-chip Regions
The Model-based Analysis of Tiling-array (MAT) algorithm (Johnson et al., 2006) was used to identify genomic regions (“hits”) and highest mean histone methylation scores (MAT scores) associated with those regions on Affymetrix promoter tiling arrays enriched by ChIP. MAT was run with the following parameters to capture regions of increased signal intensity: bandwidth=300, maximum gap=300, minimum probes=10, and window P-value cutoff=1×10−5. The resulting hits were all FDR < 5 so no additional filtering was used. The MAT library and mapping files were based on the March 2006 Human Genome Assembly (HG18) or on the February 2006 Mouse Genome Assembly (MM8), as appropriate. Hits flagged by MAT as mapping to repeat regions were excluded from consideration in all cases.
Linking ChIP Hits to RefSeq Genes
The University of California Santa Cruz RefGene tables (ftp://hgdownload.cse.ucsc.edu/goldenPath) for human and mouse genomes (HG18 and MM8, respectively), which contain transcription start site (TSS) and end position information for RefSeq genes, were linked with the gene_info table (24 May 2007 download) from the National Center for Biotechnology Information (NCBI) (NCBI, ftp://ftp.ncbi.nih.gov/gene/DATA/) via reference RNA accession number. Chromosomal positions were then used to associate ChIP hits with RefSeq gene IDs. Specifically, hits falling within a window of −0.5kb to +2.5kb of a given RefSeq TSS were annotated as being associated with that gene. This window is more restrictive on the 5′ side than the regions tiled on the promoter arrays (7.5kb–10kb 5′ to 2.5kb 3′ of TSS).
Linking Human and Mouse Genes
Homologous human and mouse genes were linked via the NCBI HomoloGene database. Mouse and human gene symbols and IDs were extracted from each homology group that included both human and mouse homologs.
Clustering and Principal Components Analysis of ChIP hits
Individual samples were processed using MAT as above and the resulting ChIP hits linked to RefSeq genes as before. A list of represented genes, G, was generated from the non-redundant union of all genes associated with a ChIP hit in any sample. A matrix, M, was constructed such that for each gene i in G and for each sample j, Mi,j = 0 if the jth sample had no ChIP hit associated with gene i or Mi,j = MAT score of the ChIP hit in sample j associated with gene i. Hierarchical clustering by sample of the resulting matrix was done using the clustering tool of GenePattern 2.0. Principal Components Analysis (PCA) was performed using an R (http://ww.r-project.org) script based on the pca Methods library from the Bioconductor project (http://bioconductor.org). PCA images were generated using Origin 7.5. All data has been deposited to GEO (GSE12363).
Supplementary Material
Acknowledgments
We would like to thank Yuko Fujiwara for help with ES cell modification and blastocysts injections. This work was supported in part by the National Cancer Institute (K08CA92551, 5P01CA0684841), the Leukemia and Lymphoma Society and the Damon Runyon Cancer Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armstrong SA, Kung AL, Mabon ME, Silverman LB, Stam RW, Den Boer ML, Pieters R, Kersey JH, Sallan SE, Fletcher JA, et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell. 2003;3:173–183. doi: 10.1016/s1535-6108(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- Biondi A, Cimino G, Pieters R, Pui CH. Biological and therapeutic aspects of infant leukemia. Blood. 2000;96:24–33. [PubMed] [Google Scholar]
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CS, Sorensen PH, Domer PH, Reaman GH, Korsmeyer SJ, Heerema NA, Hammond GD, Kersey JH. Molecular rearrangements on chromosome 11q23 predominate in infant acute lymphoblastic leukemia and are associated with specific biologic variables and poor outcome. Blood. 1993;81:2386–2393. [PubMed] [Google Scholar]
- Chen W, Li Q, Hudson WA, Kumar A, Kirchhof N, Kersey JH. A murine Mll-AF4 knock-in model results in lymphoid and myeloid deregulation and hematologic malignancy. Blood. 2006;108:669–677. doi: 10.1182/blood-2005-08-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung N, Chan LC, Thompson A, Cleary ML, So CW. Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9:1208–1215. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci U S A. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RR. Development of murine B cell subpopulations. Semin Immunol. 1990;2:197–206. [PubMed] [Google Scholar]
- Hess JL, Yu BD, Li B, Hanson R, Korsmeyer SJ. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood. 1997;90:1799–1806. [PubMed] [Google Scholar]
- Huret JL, Dessen P, Bernheim A. An atlas of chromosomes in hematological malignancies. Example: 11q23 and MLL partners. Leukemia. 2001;15:987–989. doi: 10.1038/sj.leu.2402135. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci U S A. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1:324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Metzler M, Forster A, Pannell R, Arends MJ, Daser A, Lobato MN, Rabbitts TH. A conditional model of MLL-AF4 B-cell tumourigenesis using invertor technology. Oncogene. 2006;25:3093–3103. doi: 10.1038/sj.onc.1209636. [DOI] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007 doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK, Liu HC, Mahfouz R, Raimondi SC, Lenny N, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102:2951–2959. doi: 10.1182/blood-2003-01-0338. [DOI] [PubMed] [Google Scholar]
- Rozovskaia T, Feinstein E, Mor O, Foa R, Blechman J, Nakamura T, Croce CM, Cimino G, Canaani E. Upregulation of Meis1 and HoxA9 in acute lymphocytic leukemias with the t(4 : 11) abnormality. Oncogene. 2001;20:874–878. doi: 10.1038/sj.onc.1204174. [DOI] [PubMed] [Google Scholar]
- Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Silver DP, Livingston DM. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatek PJ, Gridley T. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox20. Genes Dev. 1993;7:2071–2084. doi: 10.1101/gad.7.11.2071. [DOI] [PubMed] [Google Scholar]
- Wang J, Iwasaki H, Krivtsov A, Febbo PG, Thorner AR, Ernst P, Anastasiadou E, Kutok JL, Kogan SC, Zinkel SS, et al. Conditional MLL-CBP targets GMP and models therapy-related myeloproliferative disease. Embo J. 2005;24:368–381. doi: 10.1038/sj.emboj.7600521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Wunderlich M, Fox C, Alvarez S, Cigudosa JC, Wilhelm JS, Zheng Y, Cancelas JA, Gu Y, Jansen M, et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell. 2008;13:483–495. doi: 10.1016/j.ccr.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- Zeisig BB, Milne T, Garcia-Cuellar MP, Schreiner S, Martin ME, Fuchs U, Borkhardt A, Chanda SK, Walker J, Soden R, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisig DT, Bittner CB, Zeisig BB, Garcia-Cuellar MP, Hess JL, Slany RK. The eleven-nineteen-leukemia protein ENL connects nuclear MLL fusion partners with chromatin. Oncogene. 2005;24:5525–5532. doi: 10.1038/sj.onc.1208699. [DOI] [PubMed] [Google Scholar]
- Zhang W, Xia X, Reisenauer MR, Hemenway CS, Kone BC. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. J Biol Chem. 2006;281:18059–18068. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.