Abstract
Whole genome oligo-microarrays were used to characterize age-dependent and tissue-specific changes in gene expression in pancreatic lymph nodes, spleen, and peripheral blood cells, obtained from up to 8 individual NOD mice at 6 different time points (1.5 to 20 weeks of age), compared to NOD.B10 tissue controls. “Milestone Genes” are genes whose expression was significantly changed (~3 fold) as the result of splicing or changes in transcript level. Milestone Genes were identified among genes within type one diabetes (T1D) susceptibility regions (Idd). Milestone Genes showing uniform patterns of changes in expression at various time points were identified, but the patterns of distribution and kinetics of expression were unique to each tissue. Potential T1D candidate genes were identified among Milestone Genes within Idd regions and/or hierarchical clusters. These studies identified tissue- and age-specific changes in gene expression that may play an important role in the inductive or destructive events of T1D.
Keywords: Non-Obese Diabetic Mouse, Microarrays, Immune Pathogenesis of Type 1 Diabetes
INTRODUCTION
A strong association with the major histocompatibility complex (MHC) has been reported for all autoimmune diseases studied to date [1]. For many years a leading hypothesis for this association between the MHC and autoimmune disease has been that disease-associated MHC alleles bind autoantigens and present them to autoreactive T lymphocytes [2; 3; 4]. However, a major problem for any proposed relationship between the MHC and autoimmune disease is the concordance rate for autoimmune diseases seen among monozygotic siblings, frequently below 50% [5; 6; 7; 8].
Studies reported below examine the hypothesis that genes encoded as antigen-presenting molecules in the MHC allow “autoantigen” recognition, and subsequent progression of autoimmunity mainly through stochastic events driven by inflammation. Results presented in this manuscript examine the following scenario: exposure to a common environmental antigen(s) can be recognized by genetically at risk individuals expressing disease associated MHC molecules in either a disease-provoking manner (recognition and presentation of a disease-provoking antigenic epitope), or conversely, by recognition and presentation of a non-disease-provoking epitope. This hypothesis would allow an autoimmune disease concordance rate of 50% for monozygotic twins encountering a single environmental antigen that expresses two equally dominant epitopes, one of which is disease provoking. The larger the number of non-disease-provoking antigenic epitopes recognized on the inductive antigen(s), the less disease concordance, the more dominant the disease-provoking epitope, the greater the rate of disease concordance among monozygotic twins.
This hypothesis can be examined in an animal model of spontaneous autoimmune diabetes, through the use of MHC congenic mice, where imposition of the “non-permissive” H-2b haplotype onto NOD mice (NOD.B10) efficiently silences the NOD autoimmune disease [9]. In this hypothesis, NOD.B10 mice serve as surrogate to the non-disease expressing twin because they cannot present the disease provoking epitope of the causative antigen in a disease inductive fashion. By only allowing the disease related I-Ag7 MHC bearing NOD mice to recognize and respond to the putative disease provoking antigenic epitope, changes in gene expression seen in multiple tissues at multiple time points in these otherwise isogenic mice should allow identification of the genes whose expression (mRNA isoform or transcript level) is changed as a part of disease processes.
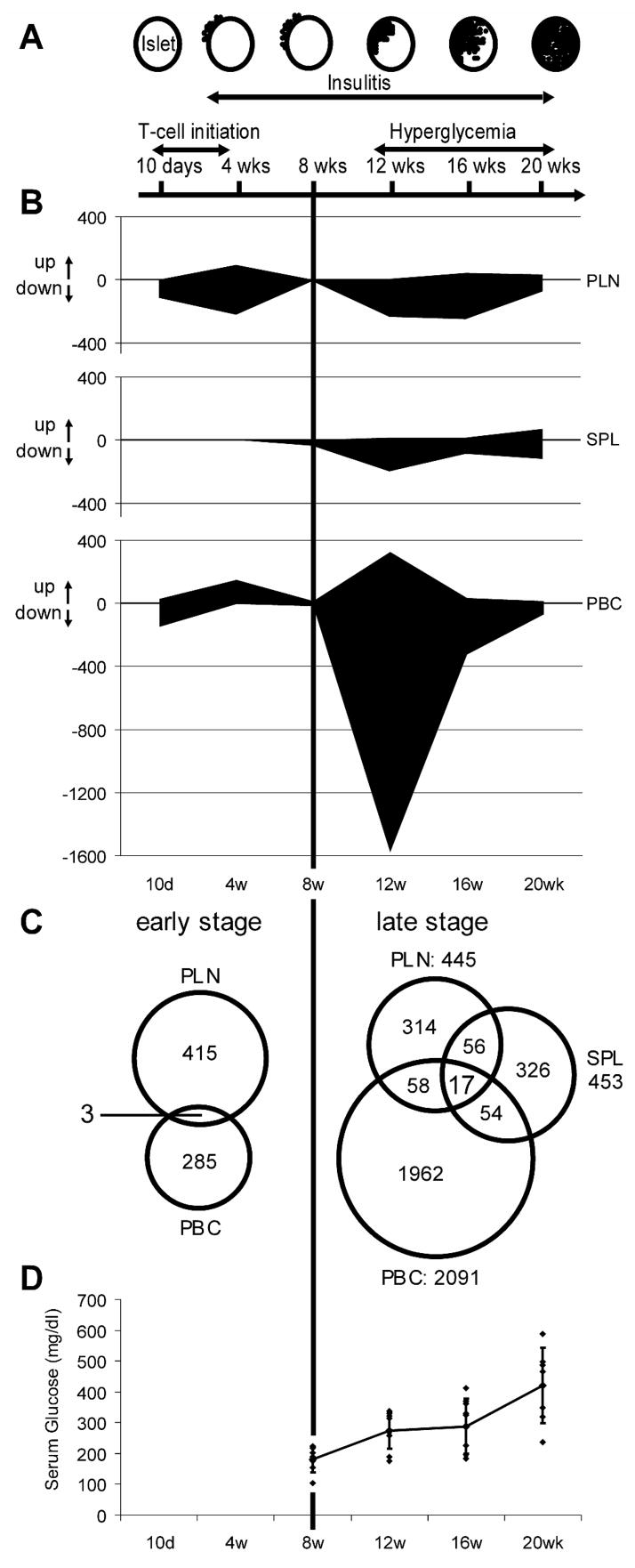
Here we report the initial results of a gene expression analysis examining three tissues [pancreatic lymph nodes (PLN), spleen (SPL) and peripheral blood cells (PBC)] at 6 different ages: 10 days and 4 weeks (disease initiation; peri-insulitis), and 8, 12, 16 and 20 weeks (disease onset; destructive insulitis) comparing tissues from NOD to NOD.B10 mice, using 41K Agilent mouse whole genome oligo arrays (Figure 1A & Table 1). These data were used to develop a “road map” of the “expressed genotype” of NOD mice in an attempt to identify tissue-specific and age-dependent changes in gene expression that might identify type 1 diabetes (T1D) candidate genes (or gene subsets) for disease induction or progression within previously identified high-resolution congenic Idd regions [10; 11; 12; 13] and/or in the hierarchical clusters of Milestone Genes.
Figure 1.
TABLE 1.
Numbers of NOD mice (= microarray chips) used for statistical data analysis.
| Age
|
||||||
|---|---|---|---|---|---|---|
| 10 days | 4 wks | 8 wks | 12 wks | 16 wks | 20 wks | |
| PLN | 7 | 6 | 4 | 7 | 6 | 5 |
| SPL | 5 | 5 | 3 | 5 | 4 | 5 |
| PBC | 8 | 3 | 7 | 8 | 7 | 4 |
RESEARCH DESIGN AND METHODS
Mice
NOD/LtJ (NOD), NOD.B10Sn-H2b/J (NOD.B10), NOD.B10 Idd9.2 (NOD Idd9.2) female mice of multiple ages were used for this study.
Study Design
Six Groups of NOD mice were sacrificed at 10 days, and 4, 8, 12, 16, and 20 weeks of age (Figure 1A & Table 1) and PLN, SPL and PBC were removed and prepared for mRNA analysis. Two Groups of 10 NOD.B10 mice [10 days (n=10) + 20 weeks of age (n=10)] were used as a tissue specific control. Gene (mRNA) expression was analyzed using the 41K Whole Mouse Genome Oligo Microarray Kit (Agilent Technologies), and gene expression data are processed (cy3: NOD, cy5: NOD.B10) and reported as normalized expression ratios: Log 10 (NOD processed signal/NOD.B10 processed signal) (Log ratio). Selected mice at various ages were used for qPCR or FACS validation of candidate genes whose expression seemed changed in one or another tissue, at one or more time point, and the relevant tissue(s) was/were extracted for qPCR analysis and/or confirmatory microarray analysis or FACS analysis.
Statistical data analysis to detect Milestone Genes
The Welch t-test was used to compare changes in gene expression between 10 days and 4 weeks of age in the individual tissues. ANOVA was performed to detect genes whose expression was significantly changed between 8, 12, 16 and 20 weeks of age. The threshold used to consider whether gene expression was significantly changed was given by a false discovery rate; Q-value (<0.01) in order to control multiple comparisons. From these statistically-significant-gene expression lists, the genes whose expression had changed significantly and were well-expressed were extracted using criteria of mean Log Ratio > 0.45 or < −0.45 (≈3 fold change in expression) and mean signal above back ground (BGSub Signal) > 150 (Signal/average background signal of all spots > 3) at any time points. These genes whose expression was significantly changed (called Milestone Genes) were sorted and stratified into each time point where the gene’s expression was changed.
RESULTS
Milestone Genes (genes whose expression changes significantly) are Tissue-Specific and Age-specific
The overall results of 41K whole genome expression profile for PLN, SPL and PBC are summarized in Supplementary Material Figure S1. Genes whose apparent expression was significantly changed were selected as Milestone Genes (≥3 fold & false discovery rate: Q-value<0.01; see research design and methods), expressed in Figure 1B and Supplementary Material Table S1 as the number of genes whose expression was significantly changed. When the serum glucose level was used as an independent variable, and changes in gene expression in all three NOD tissues from mice over 8 weeks of age that were normoglycemic were compared to the changes in gene expression seen in the NOD mouse tissues from mice over 12 weeks of age that were hyperglycemic (serum glucose level > 300 mg/dL), no statistically relevant changes distinguished these two groups (data not shown). Thus, in this animal model of T1D, age is a more important determinant of changes in gene expression than serum glucose levels. In the PLN, >100 Milestone Genes were seen at 10 days of age, >300 at 4 weeks of age, >200 at 12 and ~300 at 16 weeks of age, while few changes in gene expression were seen at 8 weeks of age. In the SPL, very few Milestone Genes were identified before 12 weeks of age. In the PBC, ~1900 genes had significantly changed expression at 12 weeks of age and, although fewer in number, Milestone Genes were also identified in the PBC samples at 10 days, 4, 16 and 20 but not at 8 weeks of age. In all tissues and at most time points (except PBC at 4 weeks of age where the gene expression of Milestone Genes was up-regulated), expression of the great majority of Milestone Genes in NOD compared to NOD.B10 appeared down regulated. The magnitude and direction of the changes in expression in the Milestone Genes in the PBC 4 week sample is also noteworthy, as these are the only genes whose change in expression is predominantly increased compared to NOD.B10 (Supplementary Material Figure S1).
As one potential way to identify candidate disease related genes, we looked for genes whose expression changed coordinately in different tissues at the same time. Represented as Venn diagrams in Figure 1C are the data from this analysis, however this simple analysis did not reveal good candidates for disease-related genes, thus we analyzed the data to look for disease associated genes in another manner, by looking for Milestone Genes contained within the previously identified disease susceptibility regions, the Idd chromosomal segments
T1D Candidate Genes in Idd Regions
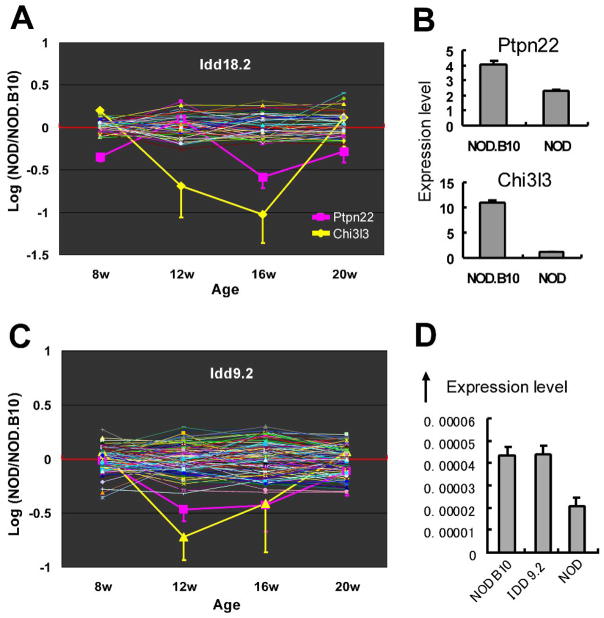
Most Idd candidate genes identified previously are also Milestone Genes. 58 Milestone Genes that were identified within known NOD congenic Idd intervals; the tissue in which the Milestone Gene was identified, and the age at which the change in expression was seen, are listed in Supplementary Material Figure S2.
Representative examples of three of the 58 Milestone Genes that lie within Idd intervals are presented in Figure 2. This Roadmap analysis allowed identification of candidate genes by looking at array probe expression for all genes within an Idd region and selecting those genes in that region that had a change in expression identified by one or more oligo probe. Among over 200 genes represented by array oligos in the chromosomal segment encompassed by the Idd18.2 interval, the PLN expression of oligos representing only two Milestone Genes, Chi3l3 and Ptpn22, was significantly changed (Figure 2A). The changes in expression detected by the oligos representing these two candidate genes represent diminished transcript level, demonstrated by qPCR (Figure 2B). Data presented in Figure 2C demonstrate another potential Idd candidate gene, Fbxo2, in the Idd 9.2 interval. Although there are over 100 genes represented by array oligos in the Idd 9.2 interval, only two probes for Fbxo2 show significantly changed expression (Figure 2C). If the C57Bl10 interval containing this gene is involved in the checkpoint between early induction and late destruction, as suggested elsewhere [13], the Idd 9.2 interval NOD congenic (NOD.Idd 9.2) mouse should express Fbxo2 in the NOD.Idd 9.2 PLN in a manner similar to that of the NOD.B10 PLN. The expression of Fbxo2 in the PLN of congenic NOD.Idd 9.2 at 12 weeks of age, both by microarray analysis (Supplementary Material Figure S3) and by qPCR (Figure 2D), is more like the expression seen in the NOD.B10 PLN tissue than that seen in the NOD PLN.
Figure 2.
T1D Candidate Genes
In order to begin to identify genes that might be associated with T1D disease induction (10 days to 4 weeks) or later beta cell destruction (12–20 weeks), we looked for clusters of Milestone Genes whose expression was changed in a uniform pattern over time by using hierarchical clustering. Although there were Milestone Genes whose changes in expression were clustered by tissue and age, none of these clusters were shared among tissues (Supplementary Material Figure S4), and less than half of the Milestone Genes were included in such age-dependent, and tissue-specific patterns of expression (Supplementary Material Table S4). In the PLN, there were five distinct patterns of Milestone Genes identified by hierarchical clustering (Supplementary Material Figure S4, and Supplementary Material Table S5A–E). The expression of one subset of 31 genes (N pattern) was down regulated at 10 days, up regulated at 4 weeks, down regulated at 12 and 16 weeks of age, and back toward the NOD.B10 expression level at 20 weeks (Supplementary Material Table S5A). Some of the genes within this cluster represent peripheral tissue antigens (PTAs), and the change in expression of these genes was confined to the PLN. The expression profile for Ins2 was chosen as a representative example of all of the PTA genes and the change in expression for Ins2 was clearly both tissue-specific and age-dependent (Figure 3A).
Figure 3.
A second subset of 111 Milestone Genes in the NOD PLN (V pattern) represents genes whose expression was down regulated only at 4 weeks of age (Supplementary Material Table S5B). Some of these genes encode a subset of 7 (about 50%) of the non-classical MHC Class 1b products. Although the change in expression of the subset of MHC Class 1b genes was unique to the PLN; not seen in the PBC or the SPL at 4 weeks of age, down regulated expression of the identical subset of 7 MHC Class 1b genes was observed in the PBC, at 12 weeks of age (Supplementary Material Table S6 & Figure 3B). These array data were validated by FACS analysis where the mean fluorescence intensity of Qa-2 expression (the product of MHC Class 1b genes H2-Q5 and H2-Q7, 2 of the 7 MHC Class 1b genes whose expression was changed) was lower in the 4 week old NOD PLN CD4 T cells than in the same aged NOD.B10 PLN CD4 T cells, and also lower in the PBC from 12 week old NOD than the 12 week old NOD.B10 (Figure 3C & D).
A third pattern of Milestone Gene clustering is represented by 32 Milestone Genes whose apparent expression was up regulated, only at 4 weeks of age (reverse V pattern), and similar to the NOD.B10 PLN expression at other time points (Supplementary Material Table S5C). There was also a subset of 66 Milestone Genes identified in the NOD PLN whose expression was down regulated only at 10 days of age while similar to that seen in the NOD.B10 PLN at other time points (10-flat pattern) (Supplementary Material Table S5D). Finally, there was a subset of 127 Milestone Genes in the PLN (U pattern) whose apparent expression was down regulated at 12 and 16 weeks of age and normal at other time points (Supplementary Material Table S5E).
In the NOD SPL, two distinct Milestone Gene clusters were identified (Supplementary Material Table S5F & S5G). There was little overlap among the genes associated with these clusters in the NOD SPL and that in the NOD PLN (Supplementary Material Figure S4). In the PBC, two distinct clusters were also seen (Supplementary Material Table S5H & S5I). Importantly, 58 Milestone Genes within Idd congenic intervals whose expression changed in the PBC during the two disease stages (induction, 10 days to 4 weeks and beta cell destruction, 12 to 20 weeks) before the onset of hyperglycemia, represent potential biomarkers, and are listed in Supplementary Material Figure S2.
Mechanisms of Changes in Gene Expression
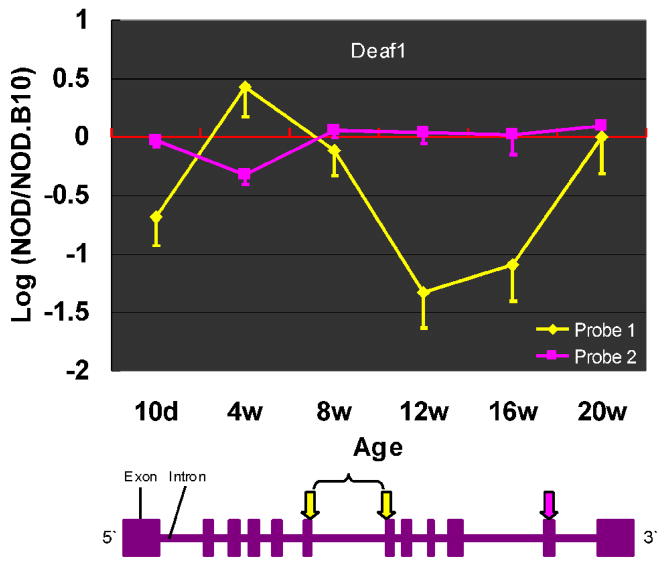
Since the strategy employed during these studies held the two strains (NOD and NOD.B10) isogenic outside of the MHC complex, mechanisms to explain changes in expression of the non-MHC genes seen over time in various tissues must include promoter and splicing elements that respond differently to inflammatory signals that in turn correspond to the response of the I-Ag7 mouse (NOD) to exogenous or endogenous stimuli. These differences in expression could result in changes in the total transcript level, as described above, or to alternate splice forms. In support of the expression of alternate splice forms, Milestone Genes that potentially express more than one isoform were initially identified by examining the microarray data for genes that had more than one oligo on the array and for whom the oligo array data were discordant. We documented that these inconsistencies represented splice variants of some genes whose expression was described as changed by one oligo and as normal or changed in another manner by one or more additional oligo. One example is Fbxo2, the single gene in the Idd 9.2 interval whose expression was changed, where among the three oligos on the array representing this gene, two indicated that the gene was down regulated at 12 weeks in the NOD PLN whereas its expression as determined by the third oligo was similar to the NOD.B10 PLN (Figure 2C). A second example of multiple isoform expression is seen in the PLN N pattern (Supplementary Material Table S5A), for Deaf1. The expression of Deaf1 is characterized as down regulated using one of the two Deaf1 oligos, but normally expressed using the other oligo (Figure 4). We have cloned several variants of Fbxo2 and Deaf1 mRNA from NOD PLN of 12 week-old NOD mice (data not presented). These data support the hypothesis of alternate splicing to explain the apparent down regulation of these two genes and presumably other genes where apparent expression seen by different gene-specific oligos was discordant.
Figure 4.
DISCUSSION
Here we report for the first time, a temporal analysis of gene expression in three separate tissues from NOD mice, PLN, SPL and PBCs, compared to gene expression in tissue matched samples from congenic NOD.B10 mice, beginning from a newborn setting (10 days old) to 20 weeks of age. The purpose of this analysis was to look for tissue-specific and time-dependent changes in gene expression during disease progression in NOD mice in order to attempt to identify tissues in which one could begin to study disease pathophysiology and identify disease associated candidates and potential biomarkers. Many previous studies had demonstrated that there are multiple genes involved in disease progression in T1D, and that disease relevant genes can be identified through polymorphisms or mutations (e.g. SNP association studies) (10; 11). We chose to study another possibility, that changes in gene expression (splice variants or transcript levels) in different tissues and at different times might be a way to identify potential disease associated genes.
Most of the previously characterized NOD diabetes candidate genes, often detected by SNP analysis, were detected as Milestone Genes (Supplementary Material Figure S2) and the 58 Milestone Genes identified in NOD Idd congenic intervals could be considered possible Idd candidate genes. One example of a potential new Idd candidate gene was the F box (a subunit of the Skip-Cullin-Fbox (SKF) complex ubiquitin E3 ligase) gene (Fbxo2), located within the Idd9.2 chromosomal segment (Figure 2C). One splice variant we identified (data not shown) contains an alternative exon that encodes the c-terminal half of the substrate-binding domain of Fbxo2. Since Fbxo2 is a subunit of a SKF E3 ligase of the endoplasmic reticulum-associated degradation (ERAD) complex and the ERAD is responsible in part for the generation of MHC class I binding peptides, it can be hypothesized that the expression, or lack of expression of an Fbxo2 variant could modify the universe of epitopes available for MHC Class I loading, of potential relevance to autoimmune processes.
A second example of a potential new Idd candidate gene (in Idd18.2) is Chitinase 3-like 3 (Chi3l3) (Figure 2A). It has been reported that Chi3l3 produced by DCs exerts effects on T cells to promote Th2 differentiation [14] and Chi3l3 expression was strongly induced in macrophages by Th2 cytokines [15]. This gene’s down-regulation in the NOD PLN at the late stage of disease progression may have effects in inducing a high Th1/Th2 ratio resulting in the onset of destructive insulitis in NOD mice.
When hierarchical clustering was used to look for uniform patterns of changes in gene expression that was tissue-specific and age-dependent, several clearly distinguishable “patterns” were observed in all three tissues (Supplementary Material Figure S4). One of the patterns identified by hierarchical clustering is the “N” pattern in the NOD PLN. Several of the genes in the N pattern represent pancreas-specific peripheral tissue antigens (PTAs) [Insulin 1 and 2, Glucagon, Pap (or Reg3b or Ingap)] (Ins 2, Figure 3A). Expression of these pancreas-specific genes in the PLN indicates that either pancreatic precursor cells do regularly visit or inhabit the PLN, or that the PLN gets certain cues, perhaps from pancreatic soluble factors that induce PTA gene expression by PLN cells. The expression of Aire is not significantly changed during disease progression in the NOD PLN. However, there is one very interesting gene, Deaf1 that shares the N expression pattern with the PTAs and coincidently, a functional domain with the Aire protein. In one possible scenario, under inflammatory stimuli, the regional lymph nodes alter gene expression and, in the early stage of disease, produce a markedly diminished PLN antigenic representation of self, primarily there in normal tissue to induce or reinforce immune tolerance through negative selection of effector T cells or positive selection of Tregs. Perhaps, because of changes in transcript or the induction of splice variants in PTA’s early during disease progression, tolerance turns into priming, followed by islet infiltration and eventual destruction. Alternatively, the loss of PTA expression seen in the later stages of disease, from 12 to 16 weeks of age, prevents “regulatory” phenomena such as clonal deletion, or Treg education or re-education in the peripheral tissue environment.
The potential validation of Milestone Genes as biomarkers of disease is possible by first identifying Milestone Genes or clusters of these genes whose expression changes in the PBCs before the onset of hypeglycemia (see Supplemental Material S2). One such example is the cluster of 7 non-classical MHC Class 1b genes whose expression changes in a coordinate manner but at different ages in the PLN and the PBC. This subset of 7 MHC Class 1b genes is down regulated at 4 weeks in the PLN but normal in the PBC at 4 weeks while the same subset is down regulated at 12 weeks in the NOD PBC but not the PLN (Supplementary Material Table S6 & Figure 3B). Recent interest in the potential action of these MHC Class 1b gene products in CD8 Treg phenomena raises the real possibility that Qa-1 gene products may be involved in regulating autoimmunity [16]. Current studies are underway to ask whether MHC Class 1b genes serve as biomarkers in T1D in man.
In summary, the large number of genes that demonstrate changes in expression between these two isogenic mouse strains that differ only by the disease permissive H2g7 of NOD mice compared to the non-disease permissive H2b of NOD.B10 mice is remarkable. Equally remarkable is the tissue-specificity and age-dependence of these changes in gene expression (Figure 3). The underlying message from this analysis is that there may not be a simple genome wide gene or set of genes outside the MHC that control the pathophysiology of T1D in NOD mice, rather that many “disease-associated” genes are either tissue-specific or age-dependent (or both) and represent splice variants or changes in transcript levels that occur as stochastic events, driven by inflammation, and do not represent mutations or polymorphisms in coding regions of “disease associated” genes. Thus the identification and characterization of disease-associated genes may well require a similar age-dependent tissue-specific analysis of gene expression, both in tissues and in specific cells from those tissues, to accurately identify and characterize those genes that are involved in disease pathogenesis of complex autoimmune diseases like T1D.
Supplementary Material
Acknowledgments
We wish to thank Claire Holness and Demi Dang for invaluable technical assistance and Linda S Wicker for providing valuable information for Idd regions and Pearl Chang for FACS analysis. This work was funded by NIH grant U19- DK 61934 & U19- AI050864.
Abbreviations
- ANOVA
One factor analysis of variance
- NOD
Non-obese diabetic
- T1D
type 1 diabetes
Footnotes
Data deposition footnote: Microarray database (http://fathmanlab.stanford.edu/)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jones EY, Fugger L, Strominger JL, Siebold C. MHC class II proteins and disease: a structural perspective. Nat Rev Immunol. 2006;6:271–82. doi: 10.1038/nri1805. [DOI] [PubMed] [Google Scholar]
- 2.Nepom GT, Erlich H. MHC class-II molecules and autoimmunity. Annu Rev Immunol. 1991;9:493–525. doi: 10.1146/annurev.iy.09.040191.002425. [DOI] [PubMed] [Google Scholar]
- 3.Todd JA, Acha-Orbea H, Bell JI, Chao N, Fronek Z, Jacob CO, McDermott M, Sinha AA, Timmerman L, Steinman L, et al. A molecular basis for MHC class II--associated autoimmunity. Science. 1988;240:1003–9. doi: 10.1126/science.3368786. [DOI] [PubMed] [Google Scholar]
- 4.Vaysburd M, Lock C, McDevitt H. Prevention of insulin-dependent diabetes mellitus in nonobese diabetic mice by immunogenic but not by tolerated peptides. J Exp Med. 1995;182:897–902. doi: 10.1084/jem.182.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaprio J, Tuomilehto J, Koskenvuo M, Romanov K, Reunanen A, Eriksson J, Stengard J, Kesaniemi YA. Concordance for type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes mellitus in a population-based cohort of twins in Finland. Diabetologia. 1992;35:1060–7. doi: 10.1007/BF02221682. [DOI] [PubMed] [Google Scholar]
- 6.Kumar D, Gemayel NS, Deapen D, Kapadia D, Yamashita PH, Lee M, Dwyer JH, Roy-Burman P, Bray GA, Mack TM. North-American twins with IDDM. Genetic, etiological, and clinical significance of disease concordance according to age, zygosity, and the interval after diagnosis in first twin. Diabetes. 1993;42:1351–63. doi: 10.2337/diab.42.9.1351. [DOI] [PubMed] [Google Scholar]
- 7.Matsuda A, Kuzuya T. Diabetic twins in Japan. Diabetes Res Clin Pract. 1994;24(Suppl):S63–7. doi: 10.1016/0168-8227(94)90229-1. [DOI] [PubMed] [Google Scholar]
- 8.Ikegami H, Ogihara T. Genetics of insulin-dependent diabetes mellitus. Endocr J. 1996;43:605–13. doi: 10.1507/endocrj.43.605. [DOI] [PubMed] [Google Scholar]
- 9.Todd JA, Aitman TJ, Cornall RJ, Ghosh S, Hall JR, Hearne CM, Knight AM, Love JM, McAleer MA, Prins JB, et al. Genetic analysis of autoimmune type 1 diabetes mellitus in mice. Nature. 1991;351:542–7. doi: 10.1038/351542a0. [DOI] [PubMed] [Google Scholar]
- 10.Lyons PA, Armitage N, Argentina F, Denny P, Hill NJ, Lord CJ, Wilusz MB, Peterson LB, Wicker LS, Todd JA. Congenic mapping of the type 1 diabetes locus, Idd3, to a 780-kb region of mouse chromosome 3: identification of a candidate segment of ancestral DNA by haplotype mapping. Genome Res. 2000;10:446–53. doi: 10.1101/gr.10.4.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyons PA, Armitage N, Lord CJ, Phillips MS, Todd JA, Peterson LB, Wicker LS. Mapping by genetic interaction: high-resolution congenic mapping of the type 1 diabetes loci Idd10 and Idd18 in the NOD mouse. Diabetes. 2001;50:2633–7. doi: 10.2337/diabetes.50.11.2633. [DOI] [PubMed] [Google Scholar]
- 12.Wicker LS, Chamberlain G, Hunter K, Rainbow D, Howlett S, Tiffen P, Clark J, Gonzalez-Munoz A, Cumiskey AM, Rosa RL, Howson JM, Smink LJ, Kingsnorth A, Lyons PA, Gregory S, Rogers J, Todd JA, Peterson LB. Fine mapping, gene content, comparative sequencing, and expression analyses support Ctla4 and Nramp1 as candidates for Idd5.1 and Idd5.2 in the nonobese diabetic mouse. J Immunol. 2004;173:164–73. doi: 10.4049/jimmunol.173.1.164. [DOI] [PubMed] [Google Scholar]
- 13.Lyons PA, Hancock WW, Denny P, Lord CJ, Hill NJ, Armitage N, Siegmund T, Todd JA, Phillips MS, Hess JF, Chen SL, Fischer PA, Peterson LB, Wicker LS. The NOD Idd9 genetic interval influences the pathogenicity of insulitis and contains molecular variants of Cd30, Tnfr2, and Cd137. Immunity. 2000;13:107–15. doi: 10.1016/s1074-7613(00)00012-1. [DOI] [PubMed] [Google Scholar]
- 14.Arora M, Chen L, Paglia M, Gallagher I, Allen JE, Vyas YM, Ray A, Ray P. Simvastatin promotes Th2-type responses through the induction of the chitinase family member Ym1 in dendritic cells. Proc Natl Acad Sci U S A. 2006;103:7777–82. doi: 10.1073/pnas.0508492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welch JS, Escoubet-Lozach L, Sykes DB, Liddiard K, Greaves DR, Glass CK. TH2 cytokines and allergic challenge induce Ym1 expression in macrophages by a STAT6-dependent mechanism. J Biol Chem. 2002;277:42821–9. doi: 10.1074/jbc.M205873200. [DOI] [PubMed] [Google Scholar]
- 16.Sarantopoulos S, Lu L, Cantor H. Qa-1 restriction of CD8+ suppressor T cells. J Clin Invest. 2004;114:1218–21. doi: 10.1172/JCI23152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.