Abstract
Background
Human carcinogenesis is known to be initiated and/or promoted by exposure to chemicals that occur in the environment. Molecular cancer epidemiology is used to identify human environmental cancer risks by applying a range of effect biomarkers, which tend to be nonspecific and do not generate insights into underlying modes of action. Toxicogenomic technologies may improve on this by providing the opportunity to identify molecular biomarkers consisting of altered gene expression profiles.
Objectives
The aim of the present study was to monitor the expression of selected genes in a random sample of adults in Flanders selected from specific regions with (presumably) different environmental burdens. Furthermore, associations of gene expression with blood and urinary measures of biomarkers of exposure, early phenotypic effects, and tumor markers were investigated.
Results
Individual gene expression of cytochrome p450 1B1, activating transcription factor 4, mitogen-activated protein kinase 14, superoxide dismutase 2 (Mn), chemokine (C-X-C motif) lig-and 1 (melanoma growth stimulating activity, alpha), diacylglycerol O-acyltransferase homolog 2 (mouse), tigger transposable element derived 3, and PTEN-induced putative kinase1 were measured by means of quantitative polymerase chain reaction in peripheral blood cells of 398 individuals. After correction for the confounding effect of tobacco smoking, inhabitants of the Olen region showed the highest differences in gene expression levels compared with inhabitants from the Gent and fruit cultivation regions. Importantly, we observed multiple significant correlations of particular gene expressions with blood and urinary measures of various environmental carcinogens.
Conclusions
Considering the observed significant differences between gene expression levels in inhabitants of various regions in Flanders and the associations of gene expression with blood or urinary measures of environmental carcinogens, we conclude that gene expression profiling appears promising as a tool for biological monitoring in relation to environmental exposures in humans.
Keywords: biomarker, biomonitoring, environmental carcinogens, human blood, transcriptomics
Cancer is among the major causes of death in modern society (Boyle and Ferlay 2005). Carcinogenesis is initiated and/or promoted by exposure to chemicals with carcinogenic properties that occur in the human environment, for example, from ambient air, food, or lifestyle-related factors such as cigarette smoke (Czene et al. 2002; Higginson 1993; Le Marchand 2005; Luch 2005). Thus, in theory, a portion of cancer cases is preventable. Molecular epidemiology tools are used to identify human cancer risks posed by exposure to environmental carcinogens, thereby contributing to the scientific basis for environmental health policy measures. To this end, biomarkers are investigated that enable human risk assessment long before traceable, diagnostic health effects appear.
In monitoring environmental cancer risk among human populations, several markers of early biological effects [e.g., DNA damage markers such as micronuclei (MN) and DNA strand breaks] have been developed and applied over the last decades (Bonassi et al. 2005). However, these biomarkers tend to be nonspecific, raise questions about their sensitivity, and do not generate insights in underlying modes of action. Genomic technologies such as microarrays and quantitative polymerase chain reaction (PCR) provide the opportunity to explore altered expression of large numbers of genes simultaneously (Afshari et al. 1999; Gerhold et al. 2001; Lobenhofer et al. 2001; Lockhart and Winzeler 2000) and therefore may provide the opportunity to identify molecular biomarkers consisting of altered gene expression profiles representing environmental health risks. Studies on carcinogen-induced differential gene expression have been conducted in our laboratory previously. Besides experimental in vitro studies that were directed toward obtaining gene expression profiles from model carcinogens in human blood mono-nuclear cells (van Leeuwen et al. 2005), a concise human population of smoking-discordant monozygotic twin pairs has been investigated; genes were identified of which the expression significantly differed in smokers compared with their nonsmoking, genetically identical siblings (van Leeuwen et al. 2007). Furthermore, in a study of children from the Czech Republic, numerous gene expressions appeared relatively increased among children inhabiting a severely polluted area (van Leeuwen et al. 2006).
From these studies, eight genes have been identified as promising biomarkers for environmental carcinogenesis. They encompass genes of which the expression differed significantly between carcinogen-exposed and non-exposed individuals, in addition to genes that correlated significantly with an established biomarker of early biological effect (i.e., MN frequencies) (van Leeuwen et al. 2006, 2007). The aim of the present study was to monitor the expression of this set of genes in humans inhabiting specific regions in Flanders and to associate these with blood and urinary measures of established biomarkers of exposure and early biological effect. We measured the expression levels of these eight key genes—cytochrome P450 1B1 (CYP1B1), activating transcription factor 4 (ATF4), mitogen-activated protein kinase 14 (MAPK14), superoxide dismutase 2 (Mn) (SOD2), chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) (CXCL1), diacylglycerol O-acyltransferase homolog 2 (mouse) (DGAT2), tigger transposable element derived 3 (TIGD3), and PTEN-induced putative kinase-1 (PINK1) (van Leeuwen et al. 2006, 2007) (Table 1)—in peripheral blood cells by means of quantitative PCR. Furthermore, we explored associations with blood and urinary measures of biomarkers of exposure to certain environmental carcinogens, biomarkers of early biological effect (DNA strand breaks and MN frequencies), and serum levels of several tumor markers. The present study was conducted as part of the Flanders Environment and Health Study (FLEHS; Flemish Centre of Expertise on Environment and Health 2007). FLEHS is a multicenter program with the goal of researching the impact of the environment on human health across the general Flanders population.
Table 1.
Overview of genes monitored for expression in blood cells.
| Gene name (abbreviation)a | GenBank accession no.a | Biological summarya | Primers |
|---|---|---|---|
| Cytochrome P450 1B1 (CYP1B1) | NM_000104 | Catalysis of many reactions involved in drug and xenobiotic metabolism (e.g., metabolism of procarcinogens) | 5′-AGTGCAGGCAGAATTGGATCA-3′ (forward) 5′-GCGCATGGCTTCATAAAGGA-3′ (reverse) |
| Activating transcription factor 4 (ATF4) | NM_001675 | Encodes a transcription factor that belongs to a family of DNA-binding proteins, including the AP-1 and CREB families | 5′-CTCCAGCGACAAGGCTAAGG-3′ (forward) 5′GTTGTTGGAGGGACTGACCAA-3′ (reverse) |
| Superoxide dismutase 2 (SOD2) | NM_000636 | Associated with oxidative stress; converts superoxide to hydrogen peroxide and diatomic oxygen | 5′-ATCAGGATCCACTGCAAGGAA-3′ (forward) 5′-CGTGCTCCCACACATCAATC-3′ (reverse) |
| Mitogen-activated protein kinase 14 (MAPK14) | NM_001315 | Activated by various environmental stressors and proinflammatory cytokines; integration point for multiple biochemical signals and involved in a wide variety of cellular processes such as proliferation, differentiation, transcription regulation, and development | 5′-TGAAGACTGTGAGCTGAAGATTCTG-3′ (forward) 5′ CCACGTAGCCTGTCATTTCATC-3′ (reverse) |
| Chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) (CXCL1) | NM_001511 | Regulates cell trafficking of various types of leukocytes and has a role in development, homeostasis, and function of the immune system | 5′-CCACTGCGCCCAAACC-3′ (forward) 5′-GCAGGATTGAGGCAAGCTTT-3′ (reverse) |
| PTEN-induced putative kinase-1 (PINK1) | NM_032409 | Encodes a serine/threonine protein kinase that localizes to mitochondria; it is thought to protect cells from stress-induced mitochondrial dysfunction | 5′-AGCAGTCACTTACAGAAAATCCAAGA-3′ (forward) 5′-GGTGAAGGCGCGGAGAA-3′ (reverse) |
| Diacylglycerol O-acyltransferase homolog 2 (mouse) (DGAT2) | NM_032564 | Responsible for triglyceride synthesis | 5′-GCACAGAGGCCACAGAAGTG-3′ (forward) 5′-CCCTCAACACAGGCATTCG-3′ (reverse) |
| Tigger transposable element derived 3 (TIGD3) | NM_145719 | Belongs to the tigger subfamily of the pogo superfamily of DNA-mediated transposons in humans; exact function of gene is not known | 5′-GTGCTGGAACTCCTGGATGAG-3′ (forward) 5′-TTGCAGATGCGCGAGATCT-3′ (reverse) |
Materials and Methods
Study population
The study population consisted of 398 subjects from eight different regions of residence in Flanders, Belgium (Table 2). Participants were recruited within several communities or sectors of communities in each of the eight regions of interest, based on random sampling. Inclusion criteria were age 50–65 years, living in the region > 5 years, and being able to complete questionnaires in Dutch. Prior to blood collection, informed consent was obtained from all individuals. Study protocols were approved by the Institutional Review Board/Ethical Committee of Antwerp University. Participants completed a questionnaire covering age, sex, and smoking habits, among other items, and they donated a blood and urine sample for measurement of the biomarkers. The questionnaire was based partly on a questionnaire used in the pilot phase of the FLEHS project (Staessen et al. 2001) and supplemented with specific questions concerning the current study.
Table 2.
Environmental characteristics of the study regions.
| Region | Population (no.) | Description |
|---|---|---|
| Antwerp | 404,241 | City and suburbs with exclusion of harbor area |
| Harbor | 130,064 | Harbor areas of Antwerp and Gent combined (petrochemical and steel industries) |
| Fruit | 95,829 | Eight municipalities with > 10 ha/km2 apple and pear cultivation |
| Olen | 68,068 | Influenced by the presence of a nonferro industrial company |
| Gent | 213,025 | City and suburbs with exclusion of harbor area |
| Incinerators | 56,405 | Surroundings of 11 incinerators; municipalities with their center < 6 km from an incinerator (or < 12 km northeast of incinerator) |
| Rural | 153,770 | Twenty-four municipalities with population density < 250 persons/km2; no registered emission source; > 5% industry and no highway in the territory |
| Canal | 64,763 | Presence of six chemical companies (BP-Chembel, Exxon Mobil, Dow Chemical, Borealis, T.C. Ham, Tessenderlo Chemie-LVM) |
Fruit, fruit cultivation region.
Gene expression analysis
For gene expression analyses, blood samples of 2.5 mL were collected from each subject into PAXgene Blood RNA vacutainer tubes (PreAnalytix; Qiagen, Hilden, Germany), a system that accounts for immediate ex vivo preservation of blood RNA. Total RNA was isolated and purified using the PAXgene Blood RNA kit (PreAnalytix) according to the manufacturer’s instructions. cDNA was synthesized from 2 μg total RNA using the BioRad iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer’s instructions. Aliquots were used for quantitative PCR on the BioRad MyiQ iCycler Single Color quantitative detection system using iQ SYBR Green Supermix (both from Bio-Rad) according to the manufacturer’s instructions. Reactions were initiated for 3 min at 95°C, followed by 40 cycles of 15 sec at 95°C and 45 sec at 60°C. After each run, we performed a melting curve analysis starting at 60°C with stepwise temperature elevations of 0.5°C every 10 sec to check for nonspecific products. We included β-actin (ACTB) and cyclophillin A (PPIA) as reference genes (internal controls). Primers were as follows: ACTB, 5′-CCTGGCACCCAGCA-CAAT-3′ (forward) and 5′-GCCGATCCA-CACGGAGTACT-3′ (reverse); and PPIA, 5′-T T C C T G C T T T C A C A G A A T T ATTCC-3′ (forward) and 5′-GCCACCAG-TGCCATTATGG-3′ (reverse). These genes perform best in terms of most stable expression and best resemblance to microarray-derived results in our previous analyses (data not shown). All reactions were performed in duplicate. In each run, negative controls (not containing template) and positive controls (a dilution series of a pooled sample, consisting of cDNA reverse-transcribed from total RNA of 20 randomly selected subjects) were included to estimate PCR efficiency. Primer sequences are shown in Table 1.
Exposure analysis
We measured whole blood, serum, or urine levels of multiple environmental carcinogens or their metabolites by various methods: heavy metals (cadmium and lead) in whole blood as described by Schroijen et al. (2008); dioxins and furans in serum as described by Van Wouwe et al. (2004); p,p′-dichlorodiphenyldichloro-ethylene (p,p′-DDE) and non–dioxin-like polychlorinated biphenyls (PCBs) in serum as described by Covaci et al. (2002) and Gomara et al. (2002); 1-OH-pyrene (a metabolite of polycyclic aromatic hydrocarbons) and t,t-muconic acid (t,t-MA; a metabolite of benzene) in urine as described by Angerer and Schaller (1997, 1998). Smoking status was derived from questionnaires instead of cotinine measurements.
Measurement of early biological effect and tumor markers
We evaluated the induction of DNA strand breaks as a measure for DNA damage using the alkaline COMET assay as described by Singh et al. (1988). We analyzed the slides (200 cells per individual) using an image analysis system from Metasystems (Altslussheim, Germany). Median percentages of DNA migration in the tail areas were determined and used as a measure of DNA damage. As a positive control, one slide with nuclei from deep-frozen whole blood was added to each electrophoresis chamber. DNA migration in positive controls had to be > 30%. To investigate the MN frequencies, we performed the cytokinesis-block micronucleus assay on whole blood cultures using standard procedures according to Fenech (2000). For each individual, we evaluated 1,000–2,200 cells for the presence of micronuclei using the Metafer automatic program (Metasystems). We measured 8-hydroxydeoxyguanosine (8-OH-dG) in urine by means of ELISA using the competitive immunosorbent assay (Gentaur, Brussels, Belgium) according to the manufacturer’s instructions. Serum protein levels of the tumor marker p53 were analyzed using the enzyme immunometric assay and Titerzyme EIA p53 (Assay Designs, Ann Arbor, MI, USA). Carcino-embryonic antigen (CEA) and prostate-specific antigen (PSA) levels were measured using a solid-phase chemiluminescent immunometric assay and Immulite 2000 (DPC, Los Angeles, CA, USA). Samples were analyzed for PSA within 24 hr after collection.
Data analysis
Ct values [concentration of DNA molecules (in moles) multiplied by time] per subject and per gene were normalized by subtraction of the mean Ct value of ACTB and PPIA. Subsequently, we calculated Δ ΔCt values per subject and per gene relative to the normalized pooled reference sample within each quantitative PCR run using the following formula:
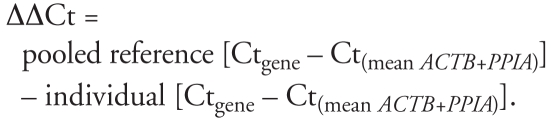 |
This generated one expression value per gene per individual study participant. All gene expression data are reported as ΔΔCt values on log (base = 2) scale (Livak and Schmittgen 2001) and presented as average group values. Next to gene expression for the eight individual genes, an integrative expression value was calculated as the mean of the eight individual genes.
We performed statistical testing using SPSS 14.0 (SPPS Inc., Chicago, IL, USA). We used one-way analysis of variance (ANOVA) with post hoc Bonferroni correction to test for significance of differences in gene expression between groups of individuals inhabiting different regions and between groups of individuals with different smoking status (current vs. former, current vs. never-smoker, and former vs. never-smoker) and other variables, as well as blood and urinary measures of exposure and effect biomarkers. For examination of intervariable correlations at the level of all individuals (no division based on region of inhabitance), we applied Pearson correlation analysis. p-Values <0.05 are considered statistically significant.
Results
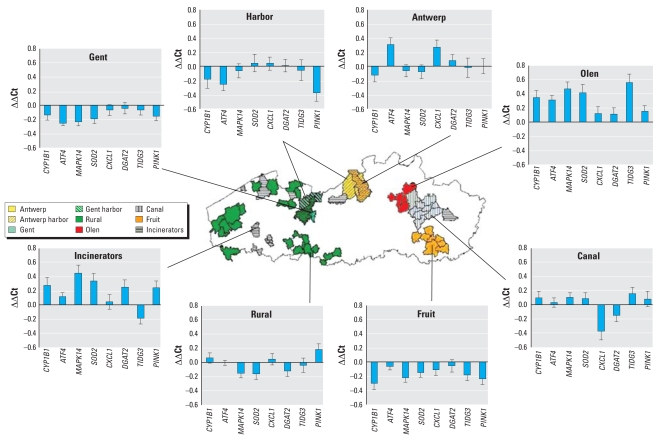
Tables 3–5 describe the characteristics of the study population according to the region of inhabitance and of the population as a whole. For seven individuals (1.8%), the smoking status was unknown; therefore, these were excluded from the analyses. Across regions, participant groups did not differ significantly with respect to age, sex, and smoking status. Statistical analysis of differences in gene expression between current, former, and never-smokers revealed CYP1B1 expression levels to be significantly different between current and former smokers (p = 0.029) and between current and never-smokers (p < 0.001). Because of the apparent confounding effect of smoking, we further investigated gene expression in non-smokers (i.e., never and former smokers only), changing the size of the total population to 319 individuals. Per region, at least 29 individuals remained in the analyses; therefore, we consider the populations still of adequate size in terms of power. A map of Flanders with bar charts of the average gene expressions among habitants per region is shown in Figure 1. Compared with the total population average, subjects with the most distinct gene expression profiles live in Olen (expressions well above the population average) as well as in the fruit cultivation region and in Gent (both with expressions well below the population average). In a one-way ANOVA analysis with a post hoc Bonferroni test, all genes appeared to significantly differ in expression between inhabitants from two or more regions (p < 0.003), except for DGAT2 (p = 0.06). Based on the individual gene expression as well as the mean/sum of all gene expressions, inhabitants from Olen show the most significant differences compared with subjects living in the fruit cultivation region (Fruit) and Gent (p < 0.001). We performed Pearson correlation analyses to investigate associations between individual gene expression (i.e., gene expression values per study participant regardless of region of inhabitance) and blood and urinary measures of biomarkers of exposure, early biological effect, and tumor markers. These analyses were carried out for the total nonsmoking population (composed of never and former smokers) or separately for female and male participants. Significant correlations between gene expression and blood and urinary measures of biomarkers of exposure are presented in Table 6 (all currently nonsmoking individuals, females only, and males only).
Table 3.
Characteristics of the study population by region of residence.
| Characteristic | Antwerp | Harbor | Fruit | Olen | Gent | Incinerator | Rural | Canal | Total |
|---|---|---|---|---|---|---|---|---|---|
| Subjects (no.) | 50 | 41 | 35 | 39 | 72 | 50 | 76 | 35 | 398 |
| Age (years) | 58.6 ± 0.6 | 57.1 ± 0.7 | 57.0 ± 0.7 | 58.9 ± 0.7 | 58.9 ± 0.6 | 59.0 ± 0.6 | 59.3 ± 0.5 | 60.1 ± 0.6 | 59.2 ± 0.2 |
| Sex [no. (%)] | |||||||||
| Males | 30 (60) | 20 (48.8) | 19 (54) | 17 (44) | 38 (53) | 25 (50) | 42 (55.3) | 16 (46) | 207 (52) |
| Females | 20 (40) | 21 (51.2) | 16 (46) | 22 (56) | 34 (47) | 25 (50) | 34 (44.7) | 19 (54) | 191 (48) |
| Smokers [no. (%)] | |||||||||
| Nonsmokers | 19 (38) | 19 (46.3) | 19 (54.3) | 17 (43.6) | 38 (52.8) | 24 (48) | 43 (56.5) | 17 (48.6) | 196 (49.2) |
| Former | 16 (32) | 11 (26.8) | 12 (34.3) | 14 (35.9) | 23 (31.9) | 15 (30) | 20 (26.3) | 12 (34.3) | 123 (30.9) |
| Current | 15 (30) | 6 (14.6) | 4 (11.4) | 7 (17.9) | 11 (15.3) | 11 (22) | 12 (15.8) | 6 (17.1) | 72 (18.1) |
| Unknown | 0 (0) | 5 (12.2) | 0 (0) | 1 (2.6) | 0 (0) | 0 (0) | 1 (1.3) | 0 (0) | 7 (1.8) |
Fruit, fruit cultivation region. Values shown for age are mean ± SE.
Table 5.
Measurements [mean (range)] of early biological effect markers and tumor markers by region of residence.
| Antwerp | Harbor | Fruit | Olen | Gent | Incinerator | Rural | Canal | Total | |
|---|---|---|---|---|---|---|---|---|---|
| Cells with MN (no.) | 8.48 (1.8–20.0) | 5.54 (0.8–15.2) | 7.72 (0.3–14.5) | 6.70 (1.5–16.4) | 7.02 (0.9–18.3) | 9.41 (0.0–35.9) | 8.71 (1.0–22.7) | 7.62 (1.8–28.8) | 7.76 (0.0–35.9) |
| MN ‰a | 9.41 (1.9–21.9) | 5.99 (0.8–15.9) | 8.45 (3.0–15.4) | 7.23 (1.5–18.6) | 7.75 (0.9–20.4) | 10.57 (0.0–38.9) | 9.14 (1.0–28.2) | 8.84 (2.3–45.9) | 8.63 (0.0–45.9) |
| 8-OH-dGb | 15.37 (5.1–32.8) | 14.93 (5.9–23.0) | 17.21 (7.3–32.4) | 15.15 (5.7–40.3) | 15.76 (7.2–29.6) | 18.87 (3.7–57.2) | 15.02 (3.7–42.2) | 15.79 (7.9–24.5) | 15.95 (3.7–57.2) |
| COMET (P90)c | 8.90 (5.4–15.5) | 12.97 (6.7–21.7) | 10.06 (6.6–13.6) | 8.82 (5.0–13.5) | 8.43 (5.4–11.5) | 8.06 (3.7–13.7) | 8.63 (5.4–12.0) | 8.58 (5.9–12.1) | 8.82 (3.7–21.7) |
| COMET (median)c | 1.75 (0.1–5.5) | 3.85 (0.4–7.5) | 2.83 (1.2–4.3) | 1.65 (0.0–4.3) | 2.33 (0.5–7.6) | 1.79 (0.0–3.5) | 2.29 (0.4–4.5) | 1.95 (0.1–4.4) | 2.14 (0.0–7.6) |
| PSAd | 1.48 (0.3–4.9) | 1.47 (0.3–3.9) | 1.12 (0.3–4.8) | 1.27 (0.2–4.1) | 2.06 (0.3–27.1) | 0.95 (0.2–4.4) | 1.20 (0.0–3.8) | 1.32 (0.4–4.0) | 1.40 (0.0–27.1) |
| CEAd | 2.65 (0.4–22.2) | 1.96 (0.4–4.8) | 2.03 (0.3–5.6) | 2.45 (0.4–8.7) | 2.37 (0.5–15.5) | 2.66 (0.6–13.8) | 2.61 (0.6–14.0) | 1.63 (0.7–4.0) | 2.39 (0.3–22.2) |
| p53e | 41.1 (0.01–748.0) | 176.6 (0.01–1378.0) | 84.5 (0.01–804.0) | 71.6 (0.1–1396.0) | 86.9 (0.01–1531.0) | 71.9 (0.01–1327.0) | 95.9 (0.01–1620.0) | 35.9 (0.01–507.0) | 76.6 (0.01–1620.0) |
Fruit, fruit cultivation region. Individuals for whom smoking status was not known or reported are not included.
Number of micronuclei per 1,000 binucleated cells.
μg/g creatinine.
COMET was not measured in all study participants, but it was measured in at least 10 individuals per region; COMET P90 and COMET median in % DNA in the comet tail.
ng/mL.
pg/mL.
Figure 1.
Gene expression (mean ± SE) per region, relative to the total population averages, for all nonsmokers (former and never-smokers). Fruit, fruit cultivation region.
Table 6.
Correlation coefficients (CCs) of the correlations of any of the gene expressions with blood or urinary measures of biomarkers of exposure, markers of early biological effect, or tumor markers, regardless of regions of residence, among current nonsmokers (never-smokers and former smokers combined) for all individuals, female participants only, and male participants only.
| CYP1B1 | ATF4 | MAPK14 | SOD2 | CXCL1 | DGAT2 | TIGD3 | PINK1 | CYP1B1 | ATF4 | MAPK14 | SOD2 | CXCL1 | DGAT2 | TIGD3 | PINK1 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All individuals | Females only | ||||||||||||||||
| Dioxins | 1-OH-pyrene | ||||||||||||||||
| CC | −0.021 | −0.058 | −0.008 | −0.006 | 0.136 | 0.119 | −0.069 | 0.068 | CC | −0.033 | 0.147 | −0.069 | −0.066 | 0.001 | −0.144 | 0.146 | −0.041 |
| p-Value | 0.723 | 0.323 | 0.898 | 0.918 | 0.021* | 0.043* | 0.242 | 0.248 | p-Value | 0.682 | 0.065 | 0.387 | 0.411 | 0.986 | 0.071 | 0.066 | 0.605 |
| PCBs | COMET (median) | ||||||||||||||||
| CC | 0.021 | 0.053 | 0.127 | 0.145 | 0.076 | 0.089 | 0.143 | 0.125 | CC | 0.150 | 0.041 | 0.085 | 0.138 | 0.124 | 0.255 | 0.106 | 0.134 |
| p-Value | 0.704 | 0.345 | 0.023* | 0.010* | 0.177 | 0.111 | 0.010* | 0.026* | p-Value | 0.106 | 0.662 | 0.358 | 0.136 | 0.182 | 0.005* | 0.252 | 0.147 |
| HCB | Cells with MN | ||||||||||||||||
| CC | −0.017 | 0.083 | 0.116 | 0.079 | 0.046 | 0.046 | 0.045 | 0.086 | CC | 0.085 | 0.065 | −0.143 | −0.230 | −0.153 | −0.084 | −0.118 | 0.009 |
| p-Value | 0.762 | 0.141 | 0.039* | 0.161 | 0.41 | 0.416 | 0.424 | 0.127 | p-Value | 0.341 | 0.464 | 0.109 | 0.009* | 0.085 | 0.347 | 0.185 | 0.921 |
| p,p′-DDE | MN ‰ | ||||||||||||||||
| CC | 0.022 | 0.049 | 0.137 | 0.124 | −0.063 | 0.03 | 0.104 | 0.175 | CC | 0.081 | 0.038 | −0.138 | −0.216 | −0.147 | −0.098 | −0.124 | 0.017 |
| p-Value | 0.701 | 0.38 | 0.014* | 0.027* | 0.265 | 0.599 | 0.063 | 0.002* | p-Value | 0.364 | 0.670 | 0.122 | 0.015* | 0.098 | 0.271 | 0.165 | 0.851 |
| Cd (blood) | 8-OH-dG | ||||||||||||||||
| CC | 0.058 | −0.07 | 0.113 | 0.101 | 0 | 0.05 | 0.072 | −0.035 | CC | −0.004 | 0.061 | 0.04 | 0.044 | −0.017 | 0.022 | 0.109 | 0.054 |
| p-Value | 0.302 | 0.215 | 0.045* | 0.072 | 0.996 | 0.371 | 0.201 | 0.534 | p-Value | 0.960 | 0.447 | 0.621 | 0.583 | 0.831 | 0.782 | 0.175 | 0.504 |
| Cd (urine) | p53 | ||||||||||||||||
| CC | 0.023 | 0.029 | 0.118 | 0.131 | 0.051 | 0.103 | 0.071 | −0.085 | CC | −0.042 | −0.106 | −0.111 | −0.129 | −0.059 | −0.153 | −0.002 | −0.141 |
| p-Value | 0.676 | 0.604 | 0.034* | 0.019* | 0.363 | 0.066 | 0.207 | 0.13 | p-Value | 0.625 | 0.214 | 0.191 | 0.128 | 0.490 | 0.071 | 0.977 | 0.096 |
| Pb | CEA | ||||||||||||||||
| CC | 0.042 | 0.005 | 0.013 | −0.057 | −0.045 | −0.105 | −0.037 | 0.003 | CC | 0.180 | −0.119 | −0.059 | −0.186 | −0.008 | 0.031 | −0.094 | −0.097 |
| p-Value | 0.455 | 0.923 | 0.819 | 0.307 | 0.424 | 0.061 | 0.509 | 0.961 | p-Value | 0.041* | 0.178 | 0.507 | 0.034* | 0.927 | 0.730 | 0.286 | 0.271 |
| t,t-MA | Males only | ||||||||||||||||
| CC | −0.158 | −0.019 | −0.056 | −0.021 | −0.016 | −0.056 | 0.051 | −0.086 | Dioxins | ||||||||
| p-Value | 0.007* | 0.744 | 0.341 | 0.718 | 0.782 | 0.334 | 0.379 | 0.14 | CC | 0.012 | 0.029 | 0.067 | 0.054 | 0.227 | 0.181 | −0.011 | 0.138 |
| 1-OH-pyrene | p-Value | 0.889 | 0.729 | 0.424 | 0.523 | 0.006* | 0.030* | 0.895 | 0.098 | ||||||||
| CC | −0.012 | 0.134 | −0.012 | 0.041 | −0.004 | 0.003 | 0.147 | −0.021 | PCBs | ||||||||
| p-Value | 0.825 | 0.016* | 0.837 | 0.461 | 0.946 | 0.962 | 0.008* | 0.706 | CC | 0.089 | 0.092 | 0.228 | 0.258 | 0.150 | 0.213 | 0.146 | 0.233 |
| COMET (median) | p-Value | 0.262 | 0.247 | 0.004* | 0.001* | 0.058 | 0.007* | 0.065 | 0.003* | ||||||||
| CC | 0.096 | 0.102 | 0.019 | 0.095 | 0.052 | 0.104 | 0.049 | 0.115 | HCB | ||||||||
| p-Value | 0.148 | 0.123 | 0.774 | 0.151 | 0.431 | 0.117 | 0.464 | 0.082 | CC | 0.156 | 0.081 | 0.225 | 0.124 | 0.135 | 0.062 | 0.058 | 0.245 |
| Cells with MN | p-Value | 0.049* | 0.310 | 0.004* | 0.118 | 0.088 | 0.438 | 0.469 | 0.002* | ||||||||
| CC | −0.001 | 0.093 | −0.017 | −0.036 | −0.017 | 0.071 | −0.002 | −0.008 | p,p′-DDE | ||||||||
| p-Value | 0.984 | 0.144 | 0.785 | 0.567 | 0.788 | 0.261 | 0.981 | 0.905 | CC | 0.109 | 0.061 | 0.236 | 0.193 | 0.028 | 0.117 | 0.105 | 0.287 |
| MN ‰ a | p-Value | 0.170 | 0.444 | 0.003* | 0.015* | 0.727 | 0.141 | 0.185 | 0.000* | ||||||||
| CC | −0.027 | 0.075 | −0.032 | −0.037 | −0.009 | 0.048 | −0.022 | −0.011 | Cd (blood) | ||||||||
| p-Value | 0.669 | 0.235 | 0.616 | 0.558 | 0.891 | 0.452 | 0.731 | 0.859 | CC | 0.114 | −0.091 | 0.133 | 0.067 | −0.015 | 0.075 | 0.060 | −0.064 |
| 8-OH-dG | p-Value | 0.152 | 0.255 | 0.094 | 0.403 | 0.853 | 0.346 | 0.452 | 0.426 | ||||||||
| CC | 0.033 | −0.008 | 0.071 | 0.055 | 0.013 | 0.086 | 0.011 | 0.031 | Cd (urine) | ||||||||
| p-Value | 0.554 | 0.884 | 0.209 | 0.327 | 0.811 | 0.127 | 0.839 | 0.579 | CC | 0.198 | 0.093 | 0.224 | 0.182 | 0.076 | 0.205 | 0.061 | 0.009 |
| PSA | p-Value | 0.012* | 0.240 | 0.004* | 0.022* | 0.337 | 0.009* | 0.440 | 0.910 | ||||||||
| CC | −0.115 | −0.039 | −0.063 | −0.111 | 0.019 | 0.002 | 0.073 | −0.008 | Pb | ||||||||
| p-Value | 0.146 | 0.625 | 0.429 | 0.164 | 0.808 | 0.983 | 0.358 | 0.924 | CC | 0.110 | 0.049 | 0.092 | 0.033 | 0.020 | −0.035 | −0.061 | 0.051 |
| p53 | p-Value | 0.168 | 0.537 | 0.247 | 0.680 | 0.807 | 0.665 | 0.443 | 0.522 | ||||||||
| CC | −0.022 | −0.044 | −0.067 | −0.105 | −0.084 | −0.125 | 0.02 | −0.065 | t,t-MA | ||||||||
| p-Value | 0.719 | 0.459 | 0.263 | 0.08 | 0.161 | 0.037* | 0.734 | 0.279 | CC | −0.115 | −0.041 | −0.060 | −0.057 | −0.139 | −0.082 | 0.058 | −0.055 |
| CEA | p-Value | 0.166 | 0.625 | 0.470 | 0.495 | 0.093 | 0.321 | 0.485 | 0.512 | ||||||||
| CC | 0.113 | −0.107 | −0.015 | −0.097 | −0.108 | 0.018 | −0.106 | −0.018 | 1-OH-pyrene | ||||||||
| p-Value | 0.07 | 0.085 | 0.815 | 0.121 | 0.082 | 0.771 | 0.09 | 0.779 | CC | 0.038 | 0.135 | 0.029 | 0.104 | −0.026 | 0.089 | 0.144 | 0.029 |
| Females only | p-Value | 0.633 | 0.089 | 0.712 | 0.189 | 0.745 | 0.265 | 0.069 | 0.712 | ||||||||
| Dioxins | COMET (median) | ||||||||||||||||
| CC | −0.055 | −0.154 | −0.086 | −0.067 | 0.056 | 0.051 | −0.129 | −0.007 | CC | 0.039 | 0.174 | −0.055 | 0.044 | −0.037 | −0.038 | −0.017 | 0.101 |
| p-Value | 0.510 | 0.066 | 0.307 | 0.426 | 0.508 | 0.542 | 0.124 | 0.931 | p-Value | 0.684 | 0.068 | 0.566 | 0.645 | 0.702 | 0.692 | 0.860 | 0.293 |
| PCBs | Cells with MN | ||||||||||||||||
| CC | −0.065 | 0.008 | 0.028 | 0.050 | 0.013 | −0.042 | 0.145 | −0.015 | CC | 0.003 | 0.165 | 0.095 | 0.089 | 0.117 | 0.156 | 0.140 | 0.081 |
| p-Value | 0.413 | 0.920 | 0.730 | 0.531 | 0.874 | 0.596 | 0.068 | 0.854 | p-Value | 0.973 | 0.068 | 0.298 | 0.326 | 0.198 | 0.085 | 0.123 | 0.374 |
| HCB | MN ‰ | ||||||||||||||||
| CC | −0.099 | 0.122 | 0.002 | −0.044 | −0.079 | −0.038 | 0.017 | 0.041 | CC | −0.048 | 0.157 | 0.053 | 0.065 | 0.128 | 0.116 | 0.100 | 0.069 |
| p-Value | 0.213 | 0.124 | 0.978 | 0.584 | 0.325 | 0.633 | 0.831 | 0.611 | p-Value | 0.602 | 0.083 | 0.560 | 0.473 | 0.158 | 0.202 | 0.271 | 0.449 |
| p,p′-DDE | 8-OH-dG | ||||||||||||||||
| CC | −0.050 | 0.042 | 0.028 | 0.037 | −0.163 | −0.092 | 0.099 | 0.074 | CC | 0.105 | −0.069 | 0.098 | 0.040 | 0.031 | 0.126 | −0.092 | 0.048 |
| p-Value | 0.532 | 0.598 | 0.723 | 0.648 | 0.040* | 0.249 | 0.214 | 0.354 | p-Value | 0.185 | 0.383 | 0.216 | 0.618 | 0.696 | 0.112 | 0.245 | 0.544 |
| Cd (blood) | PSA | ||||||||||||||||
| CC | 0.060 | −0.032 | 0.083 | 0.092 | −0.015 | −0.030 | 0.077 | 0.084 | CC | −0.115 | −0.039 | −0.063 | −0.111 | 0.019 | 0.002 | 0.073 | −0.008 |
| p-Value | 0.452 | 0.688 | 0.300 | 0.251 | 0.854 | 0.704 | 0.334 | 0.294 | p-Value | 0.146 | 0.625 | 0.429 | 0.164 | 0.808 | 0.983 | 0.358 | 0.924 |
| Cd (urine) | p53 | ||||||||||||||||
| CC | −0.062 | −0.015 | 0.005 | 0.014 | −0.011 | −0.073 | 0.070 | −0.098 | CC | −0.031 | 0.172 | 0.020 | −0.097 | −0.169 | −0.122 | −0.109 | 0.101 |
| p-Value | 0.439 | 0.849 | 0.952 | 0.863 | 0.886 | 0.361 | 0.378 | 0.220 | p-Value | 0.846 | 0.281 | 0.900 | 0.545 | 0.290 | 0.448 | 0.497 | 0.530 |
| Pb | CEA | ||||||||||||||||
| CC | −0.045 | −0.045 | −0.065 | −0.132 | −0.099 | −0.178 | −0.009 | −0.074 | CC | 0.016 | −0.106 | 0.039 | 0.028 | −0.230 | 0.018 | −0.123 | 0.041 |
| p-Value | 0.575 | 0.572 | 0.415 | 0.098 | 0.216 | 0.025* | 0.910 | 0.354 | p-Value | 0.859 | 0.235 | 0.665 | 0.754 | 0.009* | 0.838 | 0.166 | 0.643 |
| t,t-MA | |||||||||||||||||
| CC | −0.164 | 0.023 | −0.055 | −0.019 | 0.093 | −0.061 | 0.036 | −0.074 | |||||||||
| p-Value | 0.045* | 0.782 | 0.508 | 0.815 | 0.259 | 0.458 | 0.662 | 0.371 | |||||||||
Abbreviations: HCB, hexachlorobenzene; MN ‰, number of micronuclei per 1,000 binucleated cells.
p < 0.05.
Discussion
The FLEHS project was initiated by the Flemish government in 2001, with the plan to use the forthcoming study results in environmental risk assessment and environmental health policy making. Flanders typically comprises a range of environmental burdens such as urban areas, regions with dense traffic, intensive agriculture, and industry. In the present cross-sectional study, we investigated the expression of eight key genes in peripheral blood sampled from the adult Flanders population by means of quantitative PCR.
Except for DGAT2, all gene expression levels differed significantly between current nonsmoking (i.e., never-smoking and formerly smoking) inhabitants from two or more regions. Based on individual gene expression as well as the mean of all gene expressions, the most deviating region is Olen, because its inhabitants returned the most significant differences compared with inhabitants of the other regions, in particular Harbor, Fruit, Gent, and Rural (Figure 1).
Many significant correlations of gene expression with endogenous levels of relevant environmental carcinogens were observed among all currently nonsmoking individuals; Cd in blood or urine, PCBs, dioxins and furans, hexachlorobenzene (HCB), p,p′-DDE, t,t-MA, and 1-OH-pyrene (Table 6). The majority of these associations have not yet been described in literature, whereas reported biological functions of these gene expressions have been linked to carcinogenesis. All but one of these correlations were positive. The only negative correlation was found between CYP1B1 expression levels and urinary levels of t,t-MA. Up to now, CYP1B1 gene expression in vivo has not been reported to be influenced by benzene or its metabolites such as t,t-MA. CYP1B1 gene expression is known to be inducible by dioxin and PAH. This was not demonstrated in this population, possibly because of heterogeneity of the general population for blood levels of this environmental carcinogen. SOD2 expression correlated significantly with blood or urine measures of three exposure markers: PCBs, p,p′-DDE, and Cd in urine. Acknowledging SOD2 for its function in oxidant scavenging, these correlations indicate elevated oxidative stress as a result from these exposures. Although this association has been reported extensively for Cd exposure (Bertin and Averbeck 2006), it has not for exposure to PCBs and DDT or DDE, although these compounds are metabolized through oxidative processes. ATF4 expression levels showed a significant correlation with urinary levels of 1-OH-pyrene, a metabolite well known for representing PAH exposure. Although not previously reported, this might indicate that PAH exposure influences transcription. Furthermore, ATF4 expression correlated with DNA strand breaks, expressed as COMET P90. ATF4 is also known as CREB2, a member of CREB family, which is known as a key regulator in the control of cellular gene expression, regulating cell cycle and growth factor genes of which aberrant expression is observed in certain cancers (Cheng et al. 2007). MAPK14 gene expression returned correlations with most of the environmental carcinogens; PCBs, HCB, p,p′-DDE, and Cd in blood as well as in urine. Upon environmental carcinogenic exposure, reported for PCB-47, MAPK14 functions as a mediator of COX2 gene expression (Bezdecny et al. 2007). In turn, COX2 expression is known to be deregulated in certain tumors. Considering the substrates of this MAP kinase, ATF2, MEF2C, MAX, CDC25B, and p53, it is suggested to be associated with stress-related transcription and cell cycle regulation, as well as genotoxic stress response (Cuenda and Rousseau 2007). MAPK is also known for its extensive role in cell differentiation and proliferation, as well as its involvement in the SOS-Ras-Raf-MAPK cascade, which plays a central role in acquired growth signal autonomy, considered a key event on the route of cell normalcy to malignancy, and at other levels of the carcinogenic process (Cuenda and Rousseau 2007; Hanahan and Weinberg 2000). PINK1 expression correlated significantly with endogenous levels of PCBs and p,p′-DDE. PINK1 is a PTEN-induced putative kinase. PTEN is a tumor suppressor, functioning as an inhibitor of the AKT/PKB signaling pathway, and is mutated in a large number of cancers (Kim and Mak 2006). CXCL1 is a chemokine whose gene expression was found in the present study to correlate significantly with blood measures of dioxins and furans. It has been associated with tumor growth and metastasis (Wang et al. 1998). TIGD3 expression, a gene encoding a DNA-transposable element, correlated significantly with the blood or urinary measures of PCBs and with 1-OH-pyrene in urine. TIGD3 expression also appeared to correlate with serum levels of p53. Furthermore, a positive significant correlation was found between dioxins/furans and DGAT2 expression, a gene involved in triglyceride synthesis. The functions of these two genes as described in literature, however, do not indicate a relationship with environmental carcinogenesis.
When we assessed the influence of sex on the correlations of gene expressions and bio-markers of exposure, early effect, or tumor markers, we observed substantial differences between females and males. The correlation profile, as observed in males only, reflects the total population profile more than that of females only. This suggested unique correlation profile in females includes significant associations of gene expression with mostly biomarkers of early effect, such as SOD2 expression correlating with the number of cells with MN, the number of MN per 1,000 binucleated cells, and the serum level of CEA. CYP1B1 expression correlated with CEA levels. In addition, DGAT2 expression correlated significantly with blood measures of Pb and median COMET values. Differences in kinetics, dynamics, and biotransformation of xenobiotics between the sexes (Beierle et al. 1999; Schwartz 2003) may explain the differences observed in the present study. These observations warrant more in-depth analysis of this effect in the future.
In general, the contributions of MAPK, CREB, and PTEN to the cell circuitry involved in carcinogenesis are also comprehensively summarized by Hanahan and Weinberg (2000) in their review on the essential alterations in cell physiology that establish malignant growth. Furthermore, observed effects on CXCL1 may be linked with the process of environmental carcinogenesis. These gene expression markers appear promising in their qualitative use because of their biologically relevant modes of action and consequential broadening of the insights into cancer risks due to environmental carcinogen exposure. We therefore suggest that the observed dose–response relationships between these modified gene expressions and well-known environmental carcinogens as present in the Flemish population deepen our understanding in environmental cancer risks and clearly present the value of gene expression analysis as a tool for biological monitoring purposes.
In a few previous studies, gene expression has also been monitored in peripheral blood from human populations. Wu et al. (2003) described gene expression profiling in a Taiwanese population exposed to arsenic pollution through drinking water. They found differential expression between groups with low-, intermediate-, and high-arsenic exposure, in particular, inflammation-related genes showing up-regulation with increasing exposure. In addition, gene expression profiling has been used to examine the risks of occupational exposures to benzene and metal fumes (Forrest et al. 2005; Wang et al. 2005). However, these studies did not include as many study subjects as the present study.
Conclusions
In this large cross-sectional study, we have demonstrated the potential of gene expression analysis for monitoring in relation to environmental carcinogenic exposures in humans. We found that a gene expression profile differed significantly between human populations according to their environmental exposure. Many correlations between gene expression and blood or urinary measures of biomarkers of exposure to environmental carcinogens were observed. Furthermore, we found evidence for the contribution of xenobiotic exposure to environmental carcinogenesis at the molecular level in terms of impact on the expression of genes related to metabolism, stress response, signaling pathways, and tumorigenesis. We therefore conclude that gene expression profiling appears promising for application to the analysis of environmental health risks. This should be considered with respect to the increased human biomonitoring activities as foreseen under the European Environment Action Programme (European Commission 2008).
Table 4.
Blood or urine concentrations [mean (range)] of environmental pollutants by region of residence.
| Antwerp | Harbor | Fruit | Olen | Gent | Incinerators | Rural | Canal | Total | |
|---|---|---|---|---|---|---|---|---|---|
| Cd (urine)a | 0.73 (0.2–2.2) | 0.83 (0.3–2.4) | 0.72 (0.3–1.9) | 0.86 (0.3–2.1) | 0.61 (0.2–2.2) | 0.82 (0.2–6.5) | 0.61 (0.1–1.7) | 0.78 (0.3–1.7) | 0.72 (0.1–6.5) |
| Cd (blood)b | 0.61 (0.1–1.7) | 0.68 (0.2–1.6) | 0.50 (0.1–1.8) | 0.72 (0.2–3.0) | 0.59 (0.1–3.4) | 0.80 (0.2–1.8) | 0.61 (0.1–2.4) | 0.77 (0.3–2.0) | 0.65 (0.1–3.4) |
| Pb (blood)b | 44.60 (10.7–181.5) | 38.41 (12.7–74.1) | 39.04 (2.5–129.6) | 43.58 (21.0–96.8) | 45.24 (15.7–106.9) | 46.07 (9.3–99.6) | 51.19 (18.1–133.2) | 37.78 (12.0–106.6) | 44.41 (2.5–181.5) |
| 1-OH-pyrenea | 0.34 (0.02–2.3) | 0.27 (0.01–2.2) | 0.37 (0.02–2.0) | 0.32 (0.02–1.8) | 0.20 (0.01–1.2) | 0.26 (0.01–1.8) | 0.24 (0.01–1.3) | 0.22 (0.01–1.0) | 0.27 (0.01–2.3) |
| HCBc | 71.19 (22.9–189.0) | 63.00 (13.7–157.3) | 62.18 (12.6–125.8) | 82.74 (30.1–177.2) | 63.52 (20.7–216.7) | 74.40 (14.6–242.7) | 73.95 (20.3–242.6) | 74.71 (27.5–188.7) | 70.62 (12.6–242.7) |
| t,t-MAa | 0.16 (0.01–0.59) | 0.12 (0.01–0.50) | 0.16 (0.01–0.65) | 0.14 (0.01–0.63) | 0.14 (0.01–0.58) | 0.12 (0.01–0.61) | 0.13 (0.01–1.34) | 0.09 (0.01–0.36) | 0.13 (0.01–1.34) |
| PCBsd | 382.4 (183.1–875.4) | 363.2 (62.1–600.0) | 368.9 (67.4–1061.6) | 445.8 (126.5–919.3) | 388.9 (152.4–949.5) | 378.7 (42.3–710.7) | 391.6 (75.5–765.4) | 407.1 (211.5–1317.6) | 390.4 (42.3–1317.6) |
| p,p′-DDEc | 692.7 (11.4–3014.2) | 527.2 (1.8–1926.2) | 1189.2 (1.8–16967.0) | 1444.3 (126.7–7575.8) | 600.3 (61.8–4458.6) | 813.1 (1.1–5208.4) | 905.1 (30.8–3846.2) | 1446.7 (89.0–8614.9) | 900.8 (1.1–16967.0) |
| Dioxinse | 28.85 (4.2–64.9) | 17.68 (3.5–51.9) | 41.67 (7.3–69.2) | 16.84 (4.2–70.9) | 20.08 (4.2–103.4) | 20.65 (4.4–44.3) | 25.80 (3.6–114.8) | 19.58 (3.5–52.2) | 23.26 (3.5–114.8) |
Abbreviations: Fruit, fruit cultivation region; HCB, hexachlorobenzene. Individuals for whom smoking status was not known or reported are not included.
mg/g creatinine.
μg/L.
ng/g fat.
Sum of PCBs 138, 153, and 180; expressed as ng/g fat in serum.
pg TEQ/gr fat in serum.
Footnotes
We thank the volunteers for participating and the nurses for the sample collections.
This study was conducted as part of the Flemish Centre of Expertise of Environment and Health and was commissioned, financed, and steered by the Ministry of the Flemish Community (Department of Economics, Science and Innovation; Flemish Agency for Care and Health; and Department of Environment, Nature and Energy). The sponsor was not involved in analysis or interpretation of the data.
References
- Afshari CA, Nuwaysir EF, Barrett JC. Application of complementary DNA microarray technology to carcinogen identification, toxicology, and drug safety evaluation. Cancer Res. 1999;59(19):4759–4760. [PubMed] [Google Scholar]
- Angerer J, Schaller KH. Analyses of Hazardous Substances in Biological Materials. Vol. 5. New York: Wiley-VCH; 1997. pp. 125–141. [Google Scholar]
- Angerer J, Schaller KH. Analyses of Hazardous Substances in Biological Materials. New York: Wiley-VCH; 1998. pp. 170–187. [Google Scholar]
- Beierle I, Meibohm B, Derendorf H. Gender differences in pharmacokinetics and pharmacodynamics. Int J Clin Pharmacol Ther. 1999;37(11):529–547. [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin G, Averbeck D. Cadmium: cellular effects, modifications of biomolecules, modulation of DNA repair and genotoxic consequences (a review) Biochimie. 2006;88(11):1549–1559. doi: 10.1016/j.biochi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Bezdecny SA, Karmaus P, Roth RA, Ganey PE. 2,2′,4,4′-Tetrachlorobiphenyl upregulates cyclooxygenase-2 in HL-60 cells via p38 mitogen-activated protein kinase and NF-kappaB. Toxicol Appl Pharmacol. 2007;221(3):285–294. doi: 10.1016/j.taap.2007.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonassi S, Ugolini D, Kirsch-Volders M, Stromberg U, Vermeulen R, Tucker JD. Human population studies with cytogenetic biomarkers: review of the literature and future prospectives. Environ Mol Mutagen. 2005;45(2–3):258–270. doi: 10.1002/em.20115. [DOI] [PubMed] [Google Scholar]
- Boyle P, Ferlay J. Cancer incidence and mortality in Europe, 2004. Ann Oncol. 2005;16(3):481–488. doi: 10.1093/annonc/mdi098. [DOI] [PubMed] [Google Scholar]
- Cheng L, Li L, Qiao X, Liu J, Yao X. Functional characterization of the promoter of human kinetochore protein HEC1: novel link between regulation of the cell cycle protein and CREB family transcription factors. Biochim Biophys Acta. 2007;1769(9–10):593–602. doi: 10.1016/j.bbaexp.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Covaci A, Koppen G, Van Cleuvenbergen R, Schepens P, Winneke G, van Larebeke N, et al. Persistent organochlorine pollutants in human serum of 50–65 years old women in the Flanders Environmental and Health Study (FLEHS). Part 2: Correlations among PCBs, PCDD/PCDFs and the use of predictive markers. Chemosphere. 2002;48(8):827–832. doi: 10.1016/s0045-6535(02)00112-1. [DOI] [PubMed] [Google Scholar]
- Cuenda A, Rousseau S. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta. 2007;1773(8):1358–1375. doi: 10.1016/j.bbamcr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Czene K, Lichtenstein P, Hemminki K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int J Cancer. 2002;99(2):260–266. doi: 10.1002/ijc.10332. [DOI] [PubMed] [Google Scholar]
- European Commission. The Sixth Environment Action Programme of the European Community 2002–2012. 2008. [accessed 24 September 2008]. Available: http://ec.europa.eu/environment/newprg/intro.htm.
- Fenech M. The in vitro micronucleus technique. Mutat Res. 2000;455(1–2):81–95. doi: 10.1016/s0027-5107(00)00065-8. [DOI] [PubMed] [Google Scholar]
- Flemish Centre of Expertise on Environment and Health. Milieu & Gezondheid. 2007. [accessed 23 September 2008]. Available: http://www.milieu-en-gezondheid.be.
- Forrest MS, Lan Q, Hubbard AE, Zhang L, Vermeulen R, Zhao X, et al. Discovery of novel biomarkers by microarray analysis of peripheral blood mononuclear cell gene expression in benzene-exposed workers. Environ Health Perspect. 2005;113:801–807. doi: 10.1289/ehp.7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhold D, Lu M, Xu J, Austin C, Caskey CT, Rushmore T. Monitoring expression of genes involved in drug metabolism and toxicology using DNA microarrays. Physiol Genomics. 2001;5(4):161–170. doi: 10.1152/physiolgenomics.2001.5.4.161. [DOI] [PubMed] [Google Scholar]
- Gomara B, Ramos L, Gonzalez MJ. Determination of poly-chlorinated biphenyls in small-size serum samples by solid-phase extraction followed by gas chromatography with micro-electron-capture detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;766(2):279–287. doi: 10.1016/s0378-4347(01)00508-4. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Higginson J. Environmental carcinogenesis. Cancer. 1993;72(suppl 3):971–977. doi: 10.1002/1097-0142(19930801)72:3+<971::aid-cncr2820721304>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Kim RH, Mak TW. Tumours and tremors: how PTEN regulation underlies both. Br J Cancer. 2006;94(5):620–624. doi: 10.1038/sj.bjc.6602994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Marchand L. The predominance of the environment over genes in cancer causation: implications for genetic epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14(5):1037–1039. doi: 10.1158/1055-9965.EPI-04-0816. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lobenhofer EK, Bushel PR, Afshari CA, Hamadeh HK. Progress in the application of DNA microarrays. Environ Health Perspect. 2001;109:881–891. doi: 10.1289/ehp.01109881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Winzeler EA. Genomics, gene expression and DNA arrays. Nature. 2000;405(6788):827–836. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- Luch A. Nature and nurture—lessons from chemical carcinogenesis. Nat Rev Cancer. 2005;5(2):113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information. GenBank. 2008. [accessed 23 September 2008]. Available: http://www.ncbi.nlm.nih.gov/Gene.
- Schroijen C, Baeyens W, Schoeters G, Den Hond E, Koppen G, Bruckers L, et al. Internal exposure to pollutants measured in blood and urine of Flemish adolescents in function of area of residence. Chemosphere. 2008;71(7):1317–1325. doi: 10.1016/j.chemosphere.2007.11.053. [DOI] [PubMed] [Google Scholar]
- Schwartz JB. The influence of sex on pharmacokinetics. Clin Pharmacokinet. 2003;42(2):107–121. doi: 10.2165/00003088-200342020-00001. [DOI] [PubMed] [Google Scholar]
- Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175(1):184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- Staessen JA, Nawrot T, Hond ED, Thijs L, Fagard R, Hoppenbrouwers K, et al. Renal function, cytogenetic measurements, and sexual development in adolescents in relation to environmental pollutants: a feasibility study of biomarkers. Lancet. 2001;357(9269):1660–1669. doi: 10.1016/s0140-6736(00)04822-4. [DOI] [PubMed] [Google Scholar]
- van Leeuwen DM, Gottschalk RW, van Herwijnen MH, Moonen EJ, Kleinjans JC, van Delft JH. Differential gene expression in human peripheral blood mononuclear cells induced by cigarette smoke and its constituents. Toxicol Sci. 2005;86(1):200–210. doi: 10.1093/toxsci/kfi168. [DOI] [PubMed] [Google Scholar]
- van Leeuwen DM, van Agen E, Gottschalk RW, Vlietinck R, Gielen M, van Herwijnen MH, et al. Cigarette smoke-induced differential gene expression in blood cells from monozygotic twin pairs. Carcinogenesis. 2007;28(3):691–697. doi: 10.1093/carcin/bgl199. [DOI] [PubMed] [Google Scholar]
- van Leeuwen DM, van Herwijnen MH, Pedersen M, Knudsen LE, Kirsch-Volders M, Sram RJ, et al. Genome-wide differential gene expression in children exposed to air pollution in the Czech Republic. Mutat Res. 2006;600(1–2):12–22. doi: 10.1016/j.mrfmmm.2006.05.032. [DOI] [PubMed] [Google Scholar]
- van Wouwe N, Eppe G, Xhrouet C, Windal I, Vanderperren H, Debacker N, et al. Validation of the CALUX bioassay for PCDD/F analyses in human blood plasma and comparison with GC-HRMS. Talanta. 2004;63:1157–1167. doi: 10.1016/j.talanta.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Wang JM, Deng X, Gong W, Su S. Chemokines and their role in tumor growth and metastasis. J Immunol Methods. 1998;220(1–2):1–17. doi: 10.1016/s0022-1759(98)00128-8. [DOI] [PubMed] [Google Scholar]
- Wang Z, Neuburg D, Li C, Su L, Kim JY, Chen JC, et al. Global gene expression profiling in whole-blood samples from individuals exposed to metal fumes. Environ Health Perspect. 2005;113:233–241. doi: 10.1289/txg.7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MM, Chiou HY, Ho IC, Chen CJ, Lee TC. Gene expression of inflammatory molecules in circulating lymphocytes from arsenic-exposed human subjects. Environ Health Perspect. 2003;111:1429–1438. doi: 10.1289/ehp.6396. [DOI] [PMC free article] [PubMed] [Google Scholar]