Abstract
Hospital-acquired bacterial infections are an increasingly important cause of morbidity and mortality worldwide. Staphylococcal species are responsible for the majority of hospital-acquired infections, which are often complicated by the ability of staphylococci to grow as biofilms. Biofilm formation by Staphylococcus epidermidis and Staphylococcus aureus requires cell-surface proteins (Aap and SasG) containing sequence repeats known as G5 domains; however, the precise role of these proteins in biofilm formation is unclear. We show here, using analytical ultracentrifugation (AUC) and circular dichroism (CD), that G5 domains from Aap are zinc (Zn2+)-dependent adhesion modules analogous to mammalian cadherin domains. The G5 domain dimerizes in the presence of Zn2+, incorporating 2–3 Zn2+ ions in the dimer interface. Tandem G5 domains associate in a modular fashion, suggesting a “zinc zipper” mechanism for G5 domain-based intercellular adhesion in staphylococcal biofilms. We demonstrate, using a biofilm plate assay, that Zn2+ chelation specifically prevents biofilm formation by S. epidermidis and methicillin-resistant S. aureus (MRSA). Furthermore, individual soluble G5 domains inhibit biofilm formation in a dose-dependent manner. Thus, the complex three-dimensional architecture of staphylococcal biofilms results from the self-association of a single type of protein domain. Surface proteins with tandem G5 domains are also found in other bacterial species, suggesting that this mechanism for intercellular adhesion in biofilms may be conserved among staphylococci and other Gram-positive bacteria. Zn2+ chelation represents a potential therapeutic approach for combating biofilm growth in a wide range of bacterial biofilm-related infections.
Keywords: bacterial pathogenesis, G5 domain, Aap, chelation, Staphylococcus
It has recently been estimated that hospital-acquired (nosocomial) infections are the fourth-leading cause of death in the United States, affecting 2 million patients per year and causing >100,000 annual deaths, with a total annual cost of over $30 billion (1, 2). Staphylococcal species such as Staphylococcus epidermidis and Staphylococcus aureus are responsible for the majority of nosocomial infections (3); treatment of these infections is often made much more challenging by the tendency of staphylococci to form biofilms. Biofilms are bacterial communities that adhere to biological or abiotic substrata, differentiate into micro- and macrocolonies, and produce an extracellular matrix typically comprised of polysaccharides and proteins (4, 5). Bacteria in biofilms are resistant to antibiotics and host immune responses (6, 7) and are extremely difficult to eradicate. For example, device-related infections resulting from staphylococcal biofilms often require surgical removal of the implanted device, debridement of the surrounding tissue, and prolonged antibiotic treatment (8).
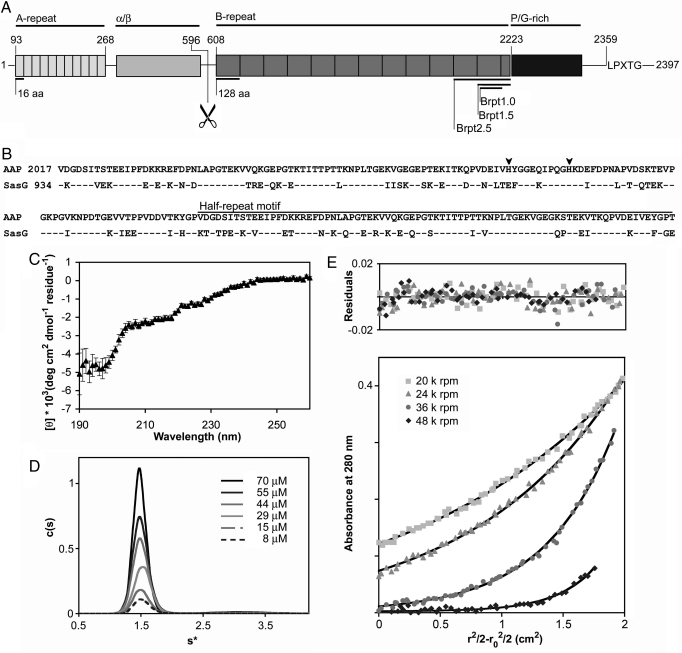
Staphylococcal biofilms are typically enmeshed within an extracellular polysaccharide matrix synthesized by proteins encoded by the ica operon (9); however, ica-negative staphylococcal biofilms have recently been described that rely on protein–protein interactions (10, 11). The S. epidermidis surface protein Aap (Accumulation-associated protein) has been implicated in both polysaccharide-based (12) and protein-based (10, 13) S. epidermidis biofilms. Aap contains an N-terminal A-repeat region with 11 degenerate 16-aa repeats, a putative globular domain (“α/β”), and a B-repeat region with a variable number (5 to 17) (13) of nearly identical 128-aa repeats terminating in a conserved “half repeat” motif (Fig. 1A). The repeated sequence element within the B-repeat region has recently been defined as a G5 domain, found in gram-positive surface proteins, Zn2+ metalloproteases, and other bacterial virulence factors [supporting information (SI) Fig. S1] (14). The B-repeat region in Aap is followed by a collagen-like repeat and an LPXTG cell wall anchor sequence. A similar domain arrangement exists in the S. aureus homolog SasG (Fig. 1B) (11).
Fig. 1.
Characterization of the G5 domain from Aap. (A) The five regions of Aap are illustrated: the A-repeat region, the putative globular domain (α/β), the B-repeat region containing 5–17 tandem G5 domains, the collagen-like proline/glycine-rich region and a cell wall anchoring motif (LPXTG). The proteolysis site is illustrated with scissors. The domain boundaries of the Brpt1.0, Brpt1.5, and Brpt2.5 constructs are illustrated. (B) Sequence alignment of the terminal G5 domain and C-terminal half-repeat motif from Aap and SasG. Identical amino acids are indicated by dashes (–); histidines are highlighted with arrowheads. Blast alignment (41) shows 80% conservation and 65% identity. (C) Far-UV circular dichroism spectrum of Brpt1.5 in 20 mM Tris pH 7.4, 50 mM NaF. Deconvolution of the data reveals predominantly β-sheet and coil secondary structure elements (Table S1). (D) Sedimentation coefficient distribution plot for Brpt1.5 at varying concentrations. Molecular weight estimation indicates Brpt1.5 is monomeric. (E) Representative sedimentation equilibrium data for Brpt1.5, confirming a monomeric state (Table S3). Black lines show the global fits; residuals are shown above.
Proteolytic processing of Aap or SasG between the α/β and B-repeat regions induces the formation of protein-based biofilms in S. epidermidis and S. aureus (10, 11); both proteinaceous (10, 13, 15) and polysaccharide-based (12) S. epidermidis biofilms are inhibited by anti-Aap antisera. Furthermore, addition of a large soluble Aap fragment containing the G5 and collagen-like regions inhibited formation of protein-based S. epidermidis biofilms (10). In S. aureus, protein-based biofilm formation was dependent on the number of G5 domains in SasG; five or more G5 domains were required to support biofilm formation (11). These studies suggested that the C-terminal halves of Aap and SasG containing the G5 domains are involved in bacterial interactions within staphylococcal biofilms, but the precise molecular mechanism was unclear. Here, we report a detailed biophysical characterization of a G5 domain from Aap, revealing that it is a Zn2+-dependent adhesion module responsible for intercellular interaction in staphylococcal biofilms. Zn2+ chelation inhibits formation of both S. epidermidis and methicillin-resistant S. aureus (MRSA) biofilms, and supplementation with additional Zn2+ in the physiological range reverses the effect. Furthermore, addition of a soluble Aap fragment containing a single intact G5 domain inhibits biofilm formation in a dose-dependent manner, indicating that the G5 domain is indeed a required element for intercellular adhesion in staphylococcal biofilms.
Results
Solution Characterization of the Terminal G5 Domain from Aap.
On the basis of the role of Aap and SasG in mediating protein-based staphylococcal biofilms (10, 11), we sought to test the hypothesis that homophilic association of these proteins is mediated solely via self-association of their G5 domains. We expressed the C-terminal G5 domain from the B-repeat region of Aap both as an isolated domain (called Brpt1.0) and as the single G5 domain followed by the C-terminal 79-aa half repeat (called Brpt1.5; Fig. 1 A and B). This C-terminal half repeat is conserved in Aap homologs, suggesting that it may function as a terminal cap required for the stability of the B-repeat region, as seen with other proteins containing repeating structural motifs (16, 17). Solution characterization of Brpt1.0 by far-UV circular dichroism (CD) showed a preponderance of random coil with little evidence of regular structural elements (data not shown). In contrast, Brpt1.5 samples yielded consistent spectra corresponding to 39% β-sheet, 32% coil, 24% turn, and 4% α-helix (Fig. 1C, Table S1). These data support the role of the half repeat as a structural cap required for stability of the G5 domain. Sedimentation velocity analytical ultracentrifugation (AUC) experiments revealed that Brpt1.5 sedimented as a single elongated species with an estimated molecular weight consistent with a monomer (Fig. 1D, Table S2). No assembly to a higher order species was observed under these conditions, even at high (70 μM) concentrations. Sedimentation equilibium experiments verified that Brpt1.5 was monomeric under these conditions (Fig. 1E, Table S3).
Zn2+-Mediated Dimerization of G5 Domains.
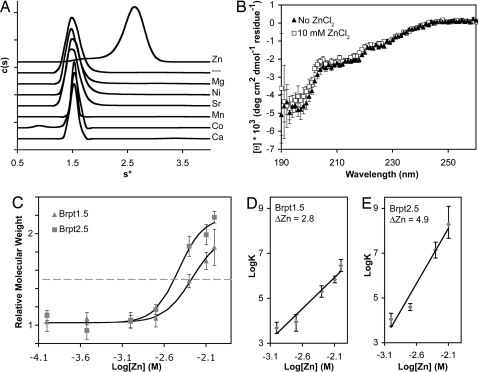
Because many mammalian adhesion proteins including cadherins, integrins, and neurexins require divalent cations for proper function (18, 19), we analyzed Brpt1.5 by AUC in the presence of Ca2+, Sr2+, Mg2+, Mn2+, Ni2+, Co2+, and Zn2+. Interestingly, Brpt1.5 formed dimers only in the presence of Zn2+ (Fig. 2A, Table S2 and Table S3). Sedimentation equilibrium analyses of Brpt1.5 in the presence of 2, 5, or 10 mM ZnCl2 revealed monomer–dimer equilibria with dissociation constants of 113 μM, 5.06 μM, and 0.353 μM, respectively (Table S3). CD spectra of Brpt1.5 in the presence or absence of 10 mM ZnCl2 were similar, suggesting that Zn2+-mediated Brpt1.5 dimerization occurs as a result of rigid-body association of two G5 domains mediated through coordination of one or more Zn2+ ions in trans rather than significant Zn2+-induced conformational changes (Fig. 2B, Table S1).
Fig. 2.
Characterization of Zn2+-dependent G5 dimerization. (A) Sedimentation velocity analyses of Brpt1.5 in the presence of divalent cations revealed a Zn2+-specific dimerization event. The dashed line label (––) represents cation-free Brpt1.5. (B) Far-UV CD spectra of Brpt1.5 in the presence (squares) and absence (triangles) of Zn. (C) Sedimentation equilibrium analysis of Zn2+-mediated dimerization of Brpt1.5 (triangles) and Brpt2.5 (squares). Brpt2.5 dimerization requires lower [Zn2+] (EC50 = 3.7 mM) compared to Brpt1.5 (EC50 = 5.4 mM) and shows a steeper slope for the monomer–dimer transition, suggesting enhanced cooperativity of Zn2+ binding with increasing numbers of tandem G5 domains. Error bars show 95% confidence intervals. (D) Linked equilibrium plot of Zn binding by Brpt1.5. Linear regression analysis of the slope indicates that the number of Zn2+ ions taken up upon Brpt1.5 dimerization (i.e., ΔZn2+) is approximately three. (E) Linked equilibrium plot of Zn binding by Brpt2.5. Linear regression analysis of the slope indicates that approximately five Zn2+ ions are bound upon Brpt2.5 dimerization.
In Aap and the related staphylococcal proteins SasG and Pls, G5 domains are found as tandem repeats ranging from 5 to 17 copies (10, 11, 13). SasG-dependent protein-based biofilm formation in S. aureus has been shown to require at least five tandem G5 repeats (11). To analyze the behavior of tandem G5 domains, we expressed a Brpt2.5 construct with two G5 domains and the C-terminal cap. Sedimentation equilibrium analysis of Brpt2.5 revealed a monomer–dimer equilibrium in the presence of Zn2+ as observed for Brpt1.5, but with a modest enhancement in Zn2+ affinity for Brpt2.5 compared to Brpt1.5, combined with a steeper transition between monomer and dimer as the repeat length increased (Fig. 2C). To analyze the linked equilibria between dimerization and Zn2+ binding, the logarithm of the dimerization association constant was plotted against the logarithm of the free Zn2+ concentration to reveal the net number of Zn2+ ions participating in the dimerization interface (20). There are two to three Zn2+ ions participating in the Brpt1.5 dimer interface, compared to approximately five Zn2+ ions for Brpt2.5. This is consistent with a modular arrangement in which each G5 domain contributes to an independent dimer interface (Fig. 2 D and E).
Zn2+ Coordination by the G5 Domain.
Structural Zn2+ ions are most commonly coordinated by cysteine, followed by histidine, aspartate, and glutamate (21). The entire sequence of Aap lacks cysteines; however, there are two histidine residues in the Brpt1.5 construct. We tested whether lowering the pH would inhibit Zn2+-mediated dimerization of Brpt1.5 via protonation of histidine side chains. AUC sedimentation velocity data revealed that Brpt1.5 with 10 mM Zn2+ transitions from a dimer at pH 7.4 to a monomer at pH 6.0 (Fig. S2A). Sedimentation equilibrium experiments verified these results; a plot of the relative molecular weight as a function of pH could be fitted to yield an apparent pKa of 6.7 (Fig. S2B), consistent with coordination by histidine. Linear regression analysis of the linked equilibria between protonation and dimerization by use of a double-log plot showed a ΔH+ value of 2.6, indicating that approximately three ionizable residues are linked to dimerization of Brpt1.5 (Fig. S2C). Site-directed mutagenesis confirmed the involvement of histidines, although other residues such as glutamates or aspartates are likely to contribute as well (21) (Fig. S2 D and E). Taken together, the CD and AUC results for Brpt1.5 and Brpt2.5 indicate that the G5 domains of Aap self-assemble in a modular fashion upon Zn2+ binding. The presence of tandem domain repeats and dependence upon a divalent cation for adhesion are reminiscent of calcium-dependent adhesion by mammalian cadherins (19).
Zn2+ Chelation Inhibits Staphylococcal Biofilm Formation.
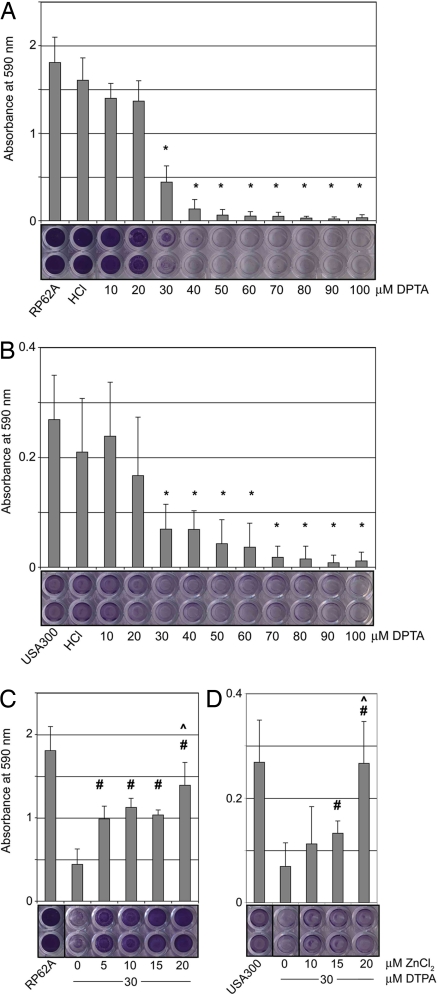
To test the physiological role of Zn2+-mediated G5 adhesion in the formation of staphylococcal biofilms, both S. epidermidis and S. aureus biofilms were grown in culture in the presence or absence of the Zn2+ chelator diethylenetriaminepentaacetic acid (DTPA). DTPA was nontoxic at all concentrations tested (up to 100 μM); N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN) and 1,10-phenanthroline, other Zn2+ chelators, were toxic to the bacteria and thus were not tested further. S. epidermidis strain RP62A formed robust biofilms when cultured in media alone, but biofilm growth was inhibited by nontoxic doses of DTPA (≥30 μM) (Fig. 3A). Under the conditions tested, DTPA did not disrupt preformed RP62A biofilms.
Fig. 3.
Zn2+ chelation inhibits biofilm formation by S. epidermidis and S. aureus. (A) S. epidermidis RP62A biofilms formed on polystyrene and were visualized with crystal violet; addition of the Zn2+ chelator DTPA (≥30 μM) inhibits biofilm formation. Representative wells have been scanned (Bottom). Untreated (“RP62A”) and vehicle (“HCl”) controls are shown. Error bars show standard deviation (*, P < 0.05 relative to untreated control; n = 3). (B) Methicillin-resistant S. aureus USA300 biofilms form on fibronectin-coated plates but are inhibited by addition of DTPA (≥30 μM). (*, P < 0.05; n = 4). (C) Addition of 5–20 μM ZnCl2 at a minimal inhibitory dose of DTPA (30 μM) rescues biofilm formation by RP62A. (#, P < 0.05 relative to the 0 μM ZnCl2/30 μM DTPA control; ^, data are statistically indistinguishable from untreated control RP62A; n = 3). (D) Addition of 15–20 μM ZnCl2 at a minimal inhibitory dose of DTPA (30 μM) rescues biofilm formation by USA300. (#, P < 0.05 compared to 0 μM ZnCl2/30 μM DTPA; ^, statistically indistinguishable from untreated control; n = 4).
Given the high sequence similarity between Aap and S. aureus proteins SasG and Pls, we tested whether Zn2+ chelation could also inhibit S. aureus biofilms. S. aureus strain USA300, which is responsible for recent epidemic outbreaks of community-acquired MRSA infections (22, 23), formed biofilms on fibronectin-coated plates. As observed for S. epidermidis, the MRSA biofilms were inhibited by Zn2+ chelation (≥30 μM DTPA) (Fig. 3B). Given that DTPA chelates several metals with high affinity, we tested whether the observed biofilm inhibition was specifically the result of zinc chelation. S. epidermidis biofilm formation could be rescued by the addition of 5–20 μM ZnCl2, indicating that the effect is Zn2+-specific (Fig. 3C). Addition of up to 60 μM CaCl2, MgCl2, and SrCl2 had no effect; MnCl2 effected partial rescue, but at concentrations nearly 1,000-fold above physiological levels (Fig. S3). Like S. epidermidis, the MRSA strain USA300 could be rescued by the addition of 15–20 μM ZnCl2 (Fig. 3D). Importantly, at the minimal inhibitory concentration of DTPA (30 μM), biofilm growth by both staphylococcal species could be rescued by the addition of 5–20 μM ZnCl2; these Zn2+ concentrations are similar to the physiological range in human plasma (12–16 μM resting concentration) (24).
Soluble G5 Domain Inhibits Biofilm Formation.
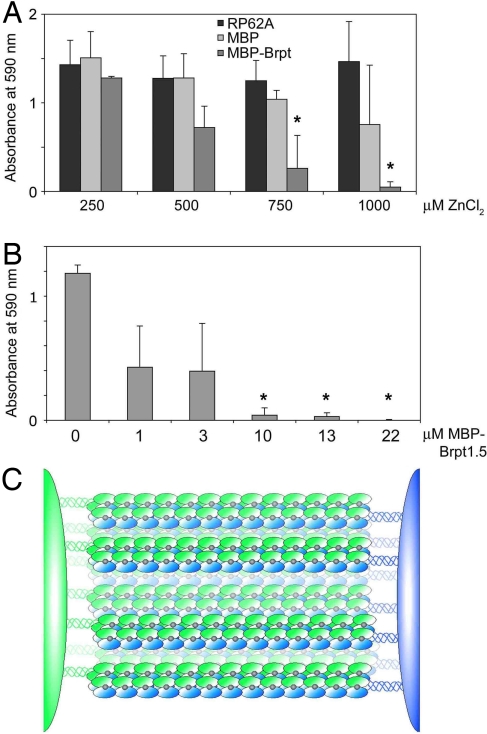
To determine whether Zn2+-dependent biofilm formation is specifically dependent upon G5 domain-mediated self-assembly, soluble Brpt1.5 was added to S. epidermidis RP62A biofilms as a maltose-binding protein (MBP) fusion. Because of the higher Zn2+ concentrations required to induce dimerization in the short construct compared to full-length Aap, inhibition by MBP–Brpt1.5 was tested at Zn2+ concentrations ranging up to 1 mM. RP62A biofilms were inhibited by soluble MBP–Brpt1.5 in the presence of 0.75 and 1 mM ZnCl2, whereas addition of MBP alone had no effect (Fig. 4A). Titration of MPB–Brpt1.5 at fixed Zn2+ concentrations showed that at 1 mM ZnCl2, addition of 10–22 μM MBP–Brpt1.5 completely abolished biofilm formation (Fig. 4B). Dose-dependent biofilm inhibition by MBP–Brpt1.5 also was observed at 750 μM ZnCl2 (Fig. S4). Thus, addition of even a single intact G5 domain in a soluble form is capable of inhibiting biofilm formation, highlighting the role of this domain as a Zn2+-dependent adhesion module in staphylococcal biofilms.
Fig. 4.
Soluble G5 domain inhibits biofilm formation in a dose-dependent manner. (A) Addition of soluble MBP–Brpt1.5 inhibits RP62A biofilms in a dose-dependent manner in the presence of 0.75–1 mM ZnCl2. MBP alone was statistically indistinguishable from untreated control at all concentrations tested. (*, P < 0.05 relative to RP62A control at the relevant Zn2+ concentration; n = 3). (B) Dose−response of biofilm inhibition by MBP–Brpt1.5 at a fixed 1 mM ZnCl2 concentration. (*, P < 0.0005; n = 3). (C) The zinc zipper model for intercellular adhesion in staphylococcal biofilms mediated by zinc-dependent self-association of G5 domains.
Discussion
We have demonstrated that the G5 domain, found in Aap, SasG, and related staphylococcal proteins such as Pls, mediates Zn2+-dependent staphylococcal biofilm formation. Multiple self-association events between stretches of tandem G5 domains in opposing Aap molecules would lead to an extensive adhesive contact between two cells, a “zinc zipper” (Fig. 4C). Aap was originally identified as a protein required for the formation of S. epidermidis microcolonies (12, 25). Subsequently, Aap and SasG were shown to undergo a proteolysis event near the beginning of the B-repeat region containing the tandem G5 domains, which leads to formation of protein-based staphylococcal biofilms (10, 11, 13). Our results suggest that the same mechanism for intercellular adhesion based upon Zn2+-dependent G5 self-association is responsible for early and late stages of biofilm formation. Zn2+-dependent intercellular adhesion in biofilms mediated by G5 domains has likely evolved as a defense mechanism against immune cell action. Zn2+ levels that are known to increase cytokine release and boost host immune responses (26) are sufficient to promote staphylococcal biofilm formation (5–20 μM Zn2+). Mast cells, basophils, and eosinophils contain high levels of Zn2+ in secretory granules that is released upon degranulation (27–29).
Staphylococcal infections are a substantial reemerging threat to human health. Starting in the 1980s a dramatic increase in hospital-acquired infections has been observed, primarily associated with S. epidermidis and other coagulase-negative staphylococci (30). S. epidermidis alone is responsible for up to two-thirds of all central nervous system shunt or catheter-related infections and up to half of all infections of prosthetic heart valves or artificial joints (31). The nosocomial infections caused by staphylococci are often persistent and recurring, particularly in the case of infections of indwelling medical devices (5, 31). Furthermore, device-related S. epidermidis biofilms can lead to endocarditis or septicemia (32). Likewise, S. aureus biofilm-related infections can become disseminated to other regions, which is a concern because of the large number of toxins and tissue-degrading enzymes released by S. aureus (5).
Given the global emergence of staphylococcal biofilm-related infections, improved methods for prevention are needed. Targeting G5 self-association either by Zn2+ chelation or direct inhibition with small molecule compounds could provide a new therapeutic avenue for combating the formation of biofilms by a broad range of staphylococcal species, including epidemic MRSA strains. The application to MRSA infections is of particular interest, given that MRSA was recently reported to cause more deaths per year than AIDS (23). Finally, genes encoding surface proteins with multiple tandem G5 domains are found in other gram-positive bacterial species (Fig. S1), suggesting that the zinc zipper mechanism for intercellular adhesion may be conserved across a wide range of bacteria.
Materials and Methods
Strains and Media.
S. epidermidis RP62A was purchased from the American Type Culture Collection (ATCC). S. aureus USA300 was generously provided by Gerald Pier (Harvard University). Bacteria were cultured in trypticase soy broth (TSB).
Brpt Cloning.
Brpt1.5 (amino acids 2017–2223) and Brpt2.5 (amino acids 1889–2223) were PCR amplified from purified S. epidermidis strain RP62A genomic DNA using pfu polymerase and primers designed for use in the Gateway cloning system (Invitrogen). Clones were inserted into destination vector pHisMBP-DEST, kindly provided by Artem Evdokimov. The fusions resulted in Brpt constructs containing an N-terminal hexahistidine-tagged maltose binding protein (His-MBP) tag.
Protein Expression and Purification.
Tuner DE3 Escherichia coli (Calbiochem) was transformed with Brpt plasmids. Cultures were grown to an OD600 of 1.0–1.4 and then cooled to 8 °C in an ice water bath. Isopropyl β-d-thiogalactopyranoside (IPTG) and ethanol were added to final concentrations of 200 μM and 2% (vol/vol), respectively, and the cultures were shaken at 20 °C overnight (33). After centrifugation, the pellets were resuspended, frozen and thawed three times, and sonicated. The Brpt proteins were purified by Ni affinity chromatography using a Hi-Trap Chelating HP column (Amersham Biosciences). The fusion protein was cleaved using TEV protease, then purified using gravity-flow Ni-NTA and Superdex 75 gel filtration columns.
Analytical Ultracentrifugation.
All AUC experiments were performed at 20 °C in a Beckman XL-I analytical ultracentrifuge using absorbance optics. Sedimentation velocity was performed at 47,000 rpm overnight and the data were analyzed using Sedfit (34). Sedimentation equilibrium data were collected at speeds ranging from 15,000 to 37,000 rpm and analyzed globally using WinNonlin (www.rasmb.bbri.org/). The weight-averaged reduced buoyant molecular weight, σw, was determined followed by analysis of the monomer–dimer equilibrium. Calculation of the experimental molecular weights from σ, determination of the monomer–dimer equilibrium constants, and conversion of equilibrium constants into molar units were carried out as described (35, 36). Complete experimental procedures are described in SI Text.
Circular Dichroism.
Far-UV CD spectra were measured using an Aviv 215 circular dichroism spectrophotometer. Brpt1.5 at 0.5 mg·mL−1 was measured in 20 mM Tris 7.4, 50 mM NaF with and without 10 mM ZnCl2. The Brpt1.5 concentration was determined using a molar extinction coefficient of 4,470 M−1·cm−1, as calculated by Sednterp (37). Data were analyzed using Dichroweb (38) using the CDSSTR module (39) with reference set four. Mean residue ellipticity [θ] was determined according to the equation [θ] = where θ is the measured ellipticity in millidegrees, l is the path length in centimeters, c is concentration in mg·mL−1, and MRW is the mean residue weight (molecular weight divided by the number of residues). The MRW values used were 107.14 g·mol−1·residue−1 for wild-type Brpt1.5 and 106.45 g·mol−1·residue−1 for the H75A/H85A mutant.
Biofilm Inhibition Assay.
Biofilm formation was assayed using a semiquantitative plate assay (40). Overnight cultures of S. epidermidis RP62A or S. aureus USA300 grown in TSB were diluted 1:200 into fresh TSB (S. epidermidis) or BHI + 0.5% (wt/vol) glucose (S. aureus) and grown in polystyrene (S. epidermidis) or fibronectin-coated polystyrene (S. aureus) 96-well plates. Suspensions (200 μl) of that solution were added to wells containing no additive, 0.5 mM HCl (buffer control for DTPA) or DTPA. ZnCl2, MgCl2, CaCl2, SrCl2, or MnCl2 was added to the media used for the dilution as noted. Biofilms were grown overnight by static incubation at 37 °C. Bacteria were removed by flicking (S. epidermidis) or aspiration (S. aureus), and the wells were rinsed with 150 mM NaCl before drying by inversion for 1 h. Wells were then stained with 1% (wt/vol) crystal violet, rinsed, and the absorbance at 590 nm was read in a SpectraMax Plus 384 microplate spectrophotometer (Molecular Devices). Statistical significance was determined using the two-tailed, two-sample unequal variance Student's t test in Microsoft Excel. Additive toxicity was assessed by growing 3 ml planktonic cultures in the presence of DTPA or other additives overnight with shaking. The cultures were grown to stationary phase and the OD600 measured. DTPA was nontoxic at all doses tested, up to 100 μM; likewise, 0.5 mM HCl included as a buffer control was nontoxic.
Supplementary Material
Acknowledgments.
We thank Mike Flagler, Sara Murdock, Rhett Kovall, Tom Thompson, and members of the Herr laboratory for helpful discussions. This work was supported in part by funds from the State of Ohio Eminent Scholar Program (to A.B.H.). D.G.C. was supported by a National Institutes of Health training grant and C.C.B. was supported by an American Heart Association postdoctoral fellowship.
Footnotes
Conflict of interest statement: D.G.C., C.C.B., and A.B.H. have filed a provisional patent application based on the work in this paper.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807717105/DCSupplemental.
References
- 1.McCaughey B. Unnecessary deaths: The human and financial costs of hospital infections. Committee to Reduce Infection Deaths. 2006:1–67. [Google Scholar]
- 2.Klevens RM, et al. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep. 2007;122:160–166. doi: 10.1177/003335490712200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.National Nosocomial Infections Surveillance (NNIS) System Report. National Center for Infectious Diseases, Center for Disease Control and Prevention. 1998:1–26. [Google Scholar]
- 4.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 5.Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel R. Biofilms and antimicrobial resistance. Clin Orthop Relat Res. 2005;437:41–47. doi: 10.1097/01.blo.0000175714.68624.74. [DOI] [PubMed] [Google Scholar]
- 7.Vuong C, et al. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004;6:269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 8.Darouiche RO. Treatment of infections associated with surgical implants. N Engl J Med. 2004;350:1422–1429. doi: 10.1056/NEJMra035415. [DOI] [PubMed] [Google Scholar]
- 9.Heilmann C, Gerke C, Perdreau-Remington F, Gotz F. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect Immun. 1996;64:277–282. doi: 10.1128/iai.64.1.277-282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rohde H, et al. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol. 2005;55:1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x. [DOI] [PubMed] [Google Scholar]
- 11.Corrigan RM, Rigby D, Handley P, Foster TJ. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology. 2007;153:2435–2446. doi: 10.1099/mic.0.2007/006676-0. [DOI] [PubMed] [Google Scholar]
- 12.Hussain M, Herrmann M, von Eiff C, Perdreau-Remington F, Peters G. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun. 1997;65:519–524. doi: 10.1128/iai.65.2.519-524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rohde H, et al. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28:1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 14.Bateman A, Holden MT, Yeats C. The G5 domain: A potential N-acetylglucosamine recognition domain involved in biofilm formation. Bioinformatics. 2005;21:1301–1303. doi: 10.1093/bioinformatics/bti206. [DOI] [PubMed] [Google Scholar]
- 15.Sun D, Accavitti MA, Bryers JD. Inhibition of biofilm formation by monoclonal antibodies against Staphylococcus epidermidis RP62A accumulation-associated protein. Clin Diagn Lab Immunol. 2005;12:93–100. doi: 10.1128/CDLI.12.1.93-100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Main ER, Lowe AR, Mochrie SG, Jackson SE, Regan L. A recurring theme in protein engineering: The design, stability, and folding of repeat proteins. Curr Opin Struct Biol. 2005;15:464–471. doi: 10.1016/j.sbi.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Forrer P, Stumpp MT, Binz HK, Pluckthun A. A novel strategy to design binding molecules harnessing the modular nature of repeat proteins. FEBS Lett. 2003;539:2–6. doi: 10.1016/s0014-5793(03)00177-7. [DOI] [PubMed] [Google Scholar]
- 18.Juliano RL. Signal transduction by cell adhesion receptors and the cytoskeleton: Functions of integrins, cadherins, selectins, and immunoglobulin-superfamily members. Annu Rev Pharmacol Toxicol. 2002;42:283–323. doi: 10.1146/annurev.pharmtox.42.090401.151133. [DOI] [PubMed] [Google Scholar]
- 19.Shapiro L, Love J, Colman DR. Adhesion molecules in the nervous system: Structural insights into function and diversity. Annu Rev Neurosci. 2007;30:451–474. doi: 10.1146/annurev.neuro.29.051605.113034. [DOI] [PubMed] [Google Scholar]
- 20.Wyman J, Gill SJ. Binding and Linkage: Functional Chemistry of Biological Macromolecules. Mill Valley, CA: University Science Books; 1990. [Google Scholar]
- 21.Auld DS. Zinc coordination sphere in biochemical zinc sites. Biometals. 2001;14:271–313. doi: 10.1023/a:1012976615056. [DOI] [PubMed] [Google Scholar]
- 22.King MD, et al. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann Intern Med. 2006;144:309–317. doi: 10.7326/0003-4819-144-5-200603070-00005. [DOI] [PubMed] [Google Scholar]
- 23.Klevens RM, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. J Am Med Assoc. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 24.Wellinghausen N, Rink L. The significance of zinc for leukocyte biology. J Leukoc Biol. 1998;64:571–577. doi: 10.1002/jlb.64.5.571. [DOI] [PubMed] [Google Scholar]
- 25.Gotz F. Staphylococcus and biofilms. Mol Microbiol. 2002;43:1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- 26.Driessen C, Hirv K, Kirchner H, Rink L. Zinc regulates cytokine induction by superantigens and lipopolysaccharide. Immunology. 1995;84:272–277. [PMC free article] [PubMed] [Google Scholar]
- 27.Sobotka AK, Malveaux FJ, Marone G, Thomas LL, Lichtenstein LM. IgE-mediated basophil phenomena: Quantitation, control, inflammatory interactions. Immunol Rev. 1978;41:171–185. doi: 10.1111/j.1600-065x.1978.tb01464.x. [DOI] [PubMed] [Google Scholar]
- 28.Pihl E, Gustafson GT, Josefsson B, Paul KG. Heavy metals in the granules of eosinophilic granylocytes. Scand J Haematol. 1967;4:371–379. doi: 10.1111/j.1600-0609.1967.tb01639.x. [DOI] [PubMed] [Google Scholar]
- 29.Gustafson GT. Heavy metals in rat mast cell granules. Lab Invest. 1967;17:588–598. [PubMed] [Google Scholar]
- 30.Rupp ME, Archer GL. Coagulase-negative staphylococci: Pathogens associated with medical progress. Clin Infect Dis. 1994;19:231–243. doi: 10.1093/clinids/19.2.231. [DOI] [PubMed] [Google Scholar]
- 31.O'Gara JP, Humphreys H. Staphylococcus epidermidis biofilms: Importance and implications. J Med Microbiol. 2001;50:582–587. doi: 10.1099/0022-1317-50-7-582. [DOI] [PubMed] [Google Scholar]
- 32.Arber N, et al. Pacemaker endocarditis. Report of 44 cases and review of the literature. Medicine (Baltimore) 1994;73:299–305. doi: 10.1097/00005792-199411000-00003. [DOI] [PubMed] [Google Scholar]
- 33.Lima CD, Wang LK, Shuman S. Structure and mechanism of yeast RNA triphosphatase: An essential component of the mRNA capping apparatus. Cell. 1999;99:533–543. doi: 10.1016/s0092-8674(00)81541-x. [DOI] [PubMed] [Google Scholar]
- 34.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herr AB, White CL, Milburn C, Wu C, Bjorkman PJ. Bivalent binding of IgA1 to FcαRI suggests a mechanism for cytokine activation of IgA phagocytosis. J Mol Biol. 2003;327:645–657. doi: 10.1016/s0022-2836(03)00149-9. [DOI] [PubMed] [Google Scholar]
- 36.Herr AB, Ornitz DM, Sasisekharan R, Venkataraman G, Waksman G. Heparin-induced self-association of fibroblast growth factor-2. Evidence for two oligomerization processes. J Biol Chem. 1997;272:16382–16389. doi: 10.1074/jbc.272.26.16382. [DOI] [PubMed] [Google Scholar]
- 37.Laue TM, Shah BD, Ridgeway TM, Pelletier SM. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding SE, Rowe A J, Horton JC, editors. London: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 38.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson WC. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins. 1999;35:307–312. [PubMed] [Google Scholar]
- 40.Christensen GD, et al. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.