Abstract
Cyclin-dependent kinases (CDKs) control cell cycle transitions and progression. In addition to their activation via binding to cyclins, CDKs can be activated via binding to an unrelated class of cell cycle regulators termed Speedy/Ringo (S/R) proteins. Although mammals contain at least five distinct Speedy/Ringo homologues, the specific functions of members of this growing family of CDK activators remain largely unknown. We investigated the cell cycle roles of human Speedy/Ringo C in HEK293 cells. Down-regulation of Speedy/Ringo C by RNA interference delayed S and G2 progression whereas ectopic expression had the opposite effect, reducing S and G2/M populations. Double thymidine arrest and release experiments showed that overexpression of Speedy/Ringo C promoted late S phase progression. Using a novel three-color FACS protocol to determine the length of G2phase, we found that the suppression of Speedy/Ringo C by RNAi prolonged G2 phase by ~30 min whereas ectopic expression of Speedy/Ringo C shortened G2 phase by ~25 min. In addition, overexpression of Speedy/Ringo C disrupted the G2 DNA damage checkpoint, increased cell death and caused a cell cycle delay at the G1-to-S transition. These observations indicate that CDK-Speedy/Ringo C complexes positively regulate cell cycle progression during the late S and G2 phases of the cell cycle.
Keywords: cyclin-dependent kinase, speedy/ringo C, cell cycle, cyclin, S-G2 phase
Introduction
The temporal and spatial rise and fall of cyclin-dependent protein kinase (CDK) activities control eukaryotic cell cycles. CDK/cyclin activities are regulated by a variety of mechanisms including association with regulatory proteins (inhibitors and assembly factors), subcellular localization, transcriptional regulation, proteolysis and reversible protein phosphorylation on two inhibitory sites and one activating site.1–7 Binding of a cyclin to a CDK is a crucial step for its protein kinase activity and leads to its phosphorylation on multiple sites.8 Activating phosphorylation occurs on a conserved threonine residue within the T-loop corresponding to Thr-160 in human Cdk2 and to Thr-161 in human Cdc2. Inhibitory phosphorylations occur on Thr-14 and Tyr-15 in human Cdc2 and Cdk2 and are carried out by the Wee1 and Myt1 protein kinases.
Extensive studies have uncovered how CDK-cyclin complexes control various cell cycle transitions. In metazoans, Cdk4/6-cyclin D and Cdk2- (or Cdc2-) cyclin E control G1 progression and entry into S phase, respectively. Cdk2-cyclin A and Cdc2-cyclin A regulate S phase progression and the accumulation and activation of Cdc2-cyclin B at the G2/M transition. At the end of G2 phase, the activation of Cdc2-cyclin B drives progression into mitosis. Cdc2-cyclin B complexes accumulate during S phase and are kept inactive by inhibitory phosphorylation until they are abruptly activated by removal of inhibitory phosphorylations by Cdc25 phosphatases.6,7 Cdc2-cyclin B stimulates an elaborate network of positive feedback loops to facilitate its rapid activation.9 What triggers the initial activation of Cdc2-cyclin B is not fully understood. One of difficulties is distinguishing between factors involved in the initial activation of a small amount of Cdc2-cyclin B and those implicated in the amplification of Cdc2-cyclin B activity.
In vertebrates, CDKs are also activated by a distinct class of cell cycle regulators called Speedy or Ringo (Rapid inducer of G2/M progression in oocytes) proteins that were initially described in Xenopus based on their ability to induce the meiotic G2-to-M phase transition during oocyte maturation.10,11 Xenopus Speedy/Ringo can bind to and activate Cdc2 and Cdk2 directly, despite the lack of any primary sequence similarity between Speedy/Ringo and cyclins. Surprisingly, Speedy/Ringo can activate Cdc2 and Cdk2 in the absence of activating phosphorylation of the CDK and CDK-Speedy/Ringo complexes are less sensitive to inhibitory phosphorylations and inhibition by CDK inhibitory proteins.12,13 Thus, CDK-Speedy/Ringo complexes are in a strong position to function as the triggers of Cdc2-cyclin B activation during the G2-to-M transition.
In mammals, there are at least five different Speedy/Ringo homologues and all of them can bind to and activate CDKs, suggesting that Speedy/Ringo proteins might play versatile roles in cell cycle control.14,15 However, the specific functions of mammalian Speedy/Ringo proteins remain largely unknown except for Spy1 (also called Speedy/Ringo A), which is closely related to Xenopus Speedy/Ringo. Human Spy1 is involved in promoting the G1-to-S transition and in the DNA damage checkpoint.16–18 Overexpression of Spy1 accelerates S-phase entry and cell proliferation and its inhibition by RNAi causes a cell cycle arrest at G1/S in cultured cells.
Another close relative of Xenopus Speedy/Ringo is human Speedy/Ringo C. Unlike Spy1, which is widely expressed in tissues and cell lines, Speedy/Ringo C is only expressed in a limited set of tissues including testis and those that undergo polyploidization such as liver, kidney, placenta and bone marrow.15 In this study, we first examine the cell cycle role of Speedy/Ringo C in somatic cells using RNA interference (RNAi). Downregulation of Speedy/Ringo C inhibits cell growth, reduces the fraction of cells in G1, and increases the S and G2 populations. Conversely, ectopic expression of Speedy/Ringo C caused cell cycle arrest at the G1-to-S transition with elevated levels of p21 and p27, increases in cell death, and decreases in the S and G2/M populations. Consistent with its role as a CDK activator, FACS analysis indicated that overexpression of Speedy/Ringo C accelerated late S phase progression following double thymidine arrest and release, and can disrupt the G2 DNA damage checkpoint. By measuring the length of G2 phase in asynchronous HEK293 cells, we found that suppression of Speedy/Ringo C prolongs G2 phase by ~30 min; conversely, ectopic expression of Speedy/Ringo C shortens G2 phase by ~25 min. These observations indicate that Speedy/Ringo C regulates late S and G2 phase progression and suggest a potential role of Speedy/Ringo C as a trigger of mitotic entry.
Results
Reduction of speedy/ringo C by RNAi induces a cell cycle delay during S and G2 phases
We previously found that Speedy/Ringo C mRNA was expressed in a variety of tissues including testis, bone marrow, kidney, small intestine, liver and placenta.15 To see if Speedy/Ringo C was endogenously expressed in human cell lines derived from the above tissues, we used reverse transcriptase PCR (RT-PCR) on total RNAs isolated from several cell lines. Primers were designed from different exons to avoid amplifying genomic DNA. As shown in Figure 1A, Speedy/Ringo C is expressed in erythroleukemia cell line K562 and in human embryonic kidney cell line HEK293, but not in HeLa and A549 (lung carcinoma) cells. We chose HEK293 cells for further analysis since they can be transfected more efficiently than K562 cells and because K562 cells are grown in suspension.
Figure 1.
Reduction of Speedy/Ringo C by siRNA delays cell cycle progression through S and G2 phase. (A) Analysis of human Speedy/Ringo C and GAPDH expression in human cell lines by RT-PCR. RT-PCR was performed with total RNAs isolated from K562 (lane 1), HEK293 (lane 2), HeLa (lane 3), and A549 (lane 4) cells using PCR primers to Speedy/Ringo C (top) and GAPDH (bottom). (B) Inhibition of human Speedy/Ringo C expression in HEK293 cells by siRNAs. The expression of Speedy/Ringo C (top) and GAPDH (bottom) was analyzed by RT-PCR with total RNAs isolated from cells transfected with control siRNA (GL3) against firefly luciferase (lane 1), siRNA #1 against Speedy/Ringo C (lane 2), and siRNA #2 against Speedy/Ringo C (lane 3). (C) Reduction of Speedy/Ringo C inhibited cell growth. HEK293 cells were transfected with either control siRNA (GL3) or siRNAs against Speedy/Ringo C. The number of viable cells was measured at different times by trypan blue exclusion. Values represent the means ± S.E. from three separate experiments. (D) Downregulation of Speedy/Ringo C delayed cell cycle progression during S and G2 phases. HEK293 cells were transfected with control siRNAs (GL3) or siRNA #1 against Speedy/Ringo C. Cells were harvested after 48-h, fixed, and processed as described in EXPERIMENTAL PROCEDURES. DNA content was measured by flow cytometry after propidium iodide staining (panels). The percentages of cells in the different phases of the cell cycle were determined by using the Watson Pragmatic model in the FlowJo program (“Modeled”). To measure cell cycle distributions in the four cell cycle phases directly (“Measured”), cells were labeled with 10 μM BrdU for 1 h and processed for triple color FACS (DNA content, BrdU incorporation, staining with anti-phospho-histone H3) as described in EXPERIMENTAL PROCEDURES (see also Fig. 2). The actual durations of the cell cycle phases were calculated based on the measured cell cycle distributions and the length of G2 phase determined in Figure 6C.
We investigated the cell cycle functions of Speedy/Ringo C by reducing Speedy/Ringo C expression in HEK293 cells using small interfering RNAs (siRNAs). Two siRNA duplexes were designed to target two different sites in the coding region of human Speedy/Ringo C. Cells were transiently transfected with two siRNA duplexes against Speedy/Ringo C or with a control siRNA duplex, GL3, against firefly luciferase. Total RNAs were isolated after 2 days and the expression of Speedy/Ringo C mRNA was detected by RT-PCR. As shown in Figure 1B, both siRNA duplexes reduced the mRNA expression of Speedy/Ringo C, although siRNA #1 was more potent than siRNA #2. Downregulation of Speedy/Ringo C inhibited cell growth, as indicated by the number of viable cells in the days following transfection (Fig. 1C). Compared to the control siRNA, both siRNAs against Speedy/Ringo C slowed cell proliferation, although siRNA #1 was again more potent than siRNA #2.
To determine the cell cycle effects of reducing Speedy/Ringo C expression, we analyzed the cell cycle distributions of propidium iodide-stained cells by fluorescent-activated cell sorting two days following treatment of HEK293 cells with siRNA #1 against Speedy/Ringo C or a control siRNA (GL3). Single cell events were distinguished from debris and aggregates/doublets using FL2-A (total DNA fluorescence) vs. FL2-W (transit time of DNA content, a measure of the size of the cell or cell cluster) plots (see Fig. 2A). The percentage of cells in different phases of the cell cycle was determined using the Watson Pragmatic model in the FlowJo program. Compared to those treated with the control siRNA, cells treated with siRNA #1 against Speedy/Ringo C had a reduced G1 population and an increase in the fraction of S and G2/M cells (Fig. 1D, under Modeled).
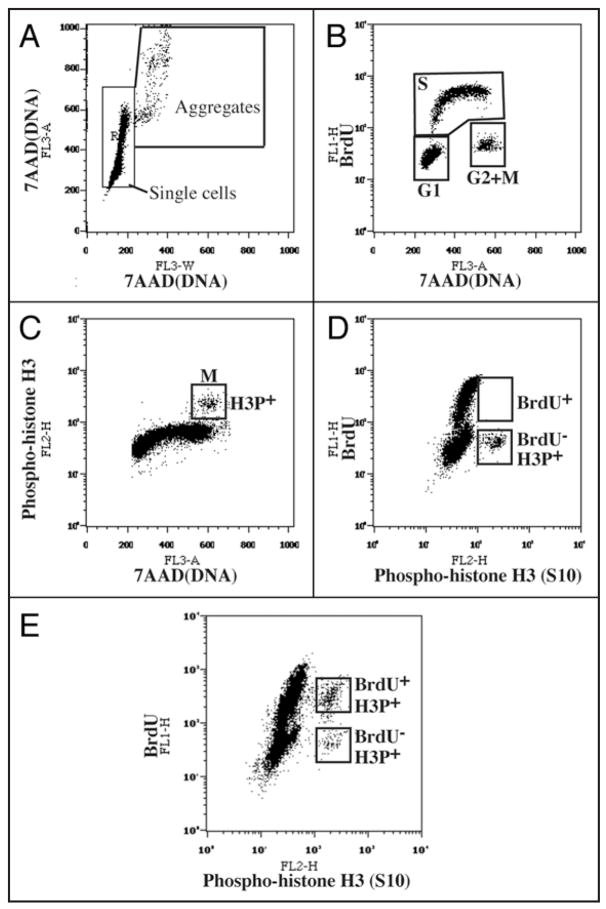
Figure 2.
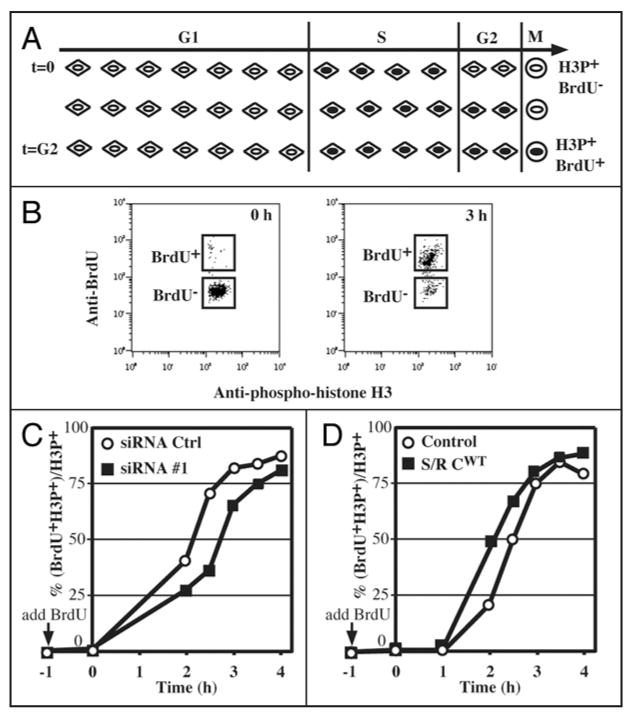
Flow cytometry analysis of cell cycle parameters after triple staining for BrdU incorporation (S phase cells), phosphohistone H3Ser10 (mitotic cells) and 7-AAD (DNA content). (A) Doublet discrimination was performed with 7-AAD fluorescence (FL3). Single cell events were distinguished from cell aggregates on a FL3-A vs. FL3-W plot of 7-AAD fluorescence. The FL3-A (“area”) value represents the total fluorescent intensity of an event whereas the FL3-W (“width” or “transit time”) represents the width of the fluorescent entity as it passes through the detector. Thus, events with larger values of FL3-W represent cell aggregates containing higher amounts of DNA (FL3-A). This plot is particularly useful for distinguishing, for instance, individual G2 cells from pairs of G1 cells. Only cells in the “Single cells” box were used for further analysis. (B) A BrdU incorporation vs. 7-AAD plot distinguishes cells in G1, S and G2/M. (C) An anti-phosphohistone H3Ser10 (mitotic cells) vs. 7-AAD plot distinguishes G2 cells from mitotic cells. (D and E) Measuring the duration of G2. Cells were incubated with BrdU and fixed and processed immediately (D) or after three hours (E). The anti-BrdU vs. anti-phosphohistone H3Ser10 plot distinguishes mitotic (H3P+) cells that have incorporated BrdU (i.e., that have had enough time to traverse G2) from those that do not contain BrdU and that have not yet had time to traverse G2. For details, see EXPERIMENTAL PROCEDURES.
To further discriminate G2 cells from mitotic cells, we developed a triple staining FACS protocol to measure the fraction of cells in each cell cycle stage directly, rather than relying on a computational analysis of a typical FACS profile of DNA content. Exponentially proliferating cells were labeled with BrdU before staining with anti-BrdU antibodies for S phase, anti-phospho-histone H3Ser10 antibodies for mitotic cells, and 7-Amino-Actinomycin D (7-AAD) for DNA content. As shown in Figure 2B, by plotting BrdU incorporation vs. DNA content, cells can be divided into three populations: G1, S and G2/M. G2/M cells can be further separated into phospho-histone H3-positive (mitotic cells) and phospho-histone H3-negative cells (G2 cells). The inhibition of Speedy/Ringo C reduced the G1 and M populations and increased the S and G2 populations (Fig. 1D, under Measured). In particular, the G2 population rose from 9.3% in control cells to 11.1% in Speedy/Ringo C-knocked down cells, a 19% increase.
Knowing the percentage of cells in G2 and the length of G2 in asynchronous cells under exponential growth conditions (see below) allowed us to calculate the lengths of the cell cycle phases. In comparison to cells treated with the siRNA control, cells treated with an siRNA against Speedy/Ringo C significantly prolonged the lengths of S phase and G2 and had a minor effect on the length of G1 (Fig. 1D, under Length). Because the cell cycle distributions derived by modeling and by direct measurement gave comparable results for the G1 and G2/M populations but the former procedure is more convenient and economical, most of our FACS experiments used propidium iodide staining and modeling.
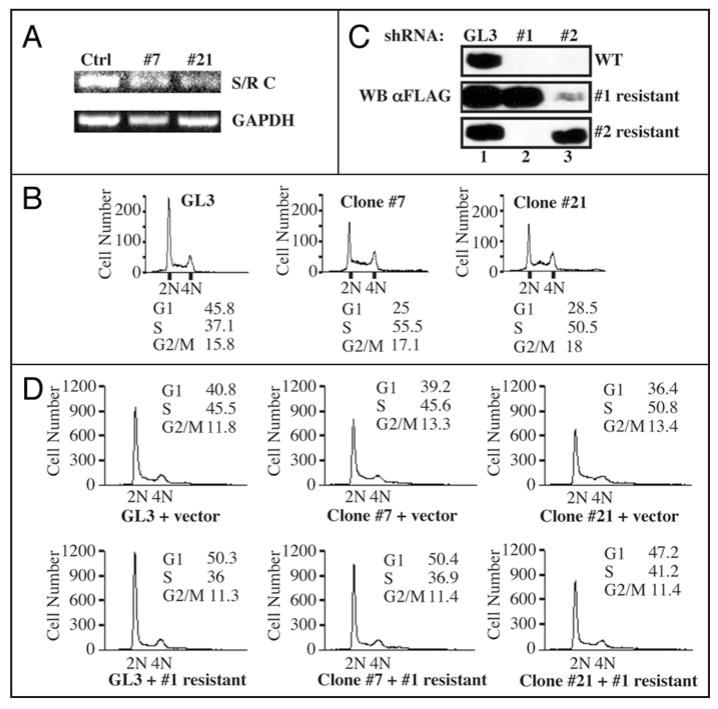
We generated stable cell lines in which Speedy/Ringo C expression was reduced by shRNA in order to study the long-term effects of reductions in Speedy/Ringo C. The shRNA expression vector targeted the same region in Speedy/Ringo C as siRNA #1. HEK293 cells were transfected with an shRNA expression plasmid against Speedy/Ringo C. Stable transfectants were selected and tested for their ability to express transiently transfected FLAG-tagged Speedy/Ringo CWT. We chose two stable clones expressing shRNA #1 for further analysis. As a control, we generated stable clones expressing shRNA against firefly luciferase (GL3). The mRNA expression of Speedy/Ringo C in one control clone and in two Speedy/Ringo C-knockdown clones (clone #7 and #21) were characterized by RT-PCR (Fig. 3A). Consistent with the growth inhibition caused by transient siRNA-knockdown (Fig. 1C), the inhibition of Speedy/Ringo C in cells stably expressing shRNA to Speedy/Ringo C slowed cell proliferation compared to cells expressing shRNA against firefly luciferase (not shown). One control clone expressing GL3 and two Speedy/Ringo-knockdown clones (#7 and #21) were plated at low density and grown for three days prior to analysis of cell cycle distributions by FACS. As shown in Figure 3B, stable reduction in Speedy/Ringo C resulted in a significant drop in the proportion of cells in G1 and an increase in the proportion of cells in S and G2, consistent with and more dramatic than the results from transient transfection experiments with siRNAs (Fig. 1D).
Figure 3.
Ectopic expression of RNAi-resistant forms of Speedy/Ringo C rescued the S and G2 delays caused by RNAi-knockdown of Speedy/Ringo C. (A) Analysis of Speedy/Ringo C and GAPDH expression in HEK293 clones expressing shRNA against firefly luciferase (Ctrl) and shRNA #1 against Speedy/Ringo C (clones #7 and #21) by RT-PCR. HEK293 cells were transfected with plasmids expressing shRNA and stable clones were selected against zeocin as described in EXPERIMENTAL PROCEDURES. The expression of Speedy/Ringo C (top) and GAPDH (bottom) were determined by RT-PCR. (B) The cell cycle distributions of HEK293 clones expressing mock shRNA (GL3) or shRNA #1 against Speedy/Ringo C (clone #7 and clone #21) were analyzed by FACS. DNA content was measured by flow cytometry of propidium iodide-stained cells. The percentage of cells in different phases of the cell cycle was determined using the Watson Pragmatic model in the FlowJo program. (C) Characterization of RNAi-resistant forms of Speedy/Ringo C. HEK293 cells were cotransfected with vectors expressing the GL3 control shRNA (lane 1) or with shRNA #1 (lane 2) or shRNA #2 (lane 3) against Speedy/Ringo C plus the indicated 3xFLAG-Speedy/Ringo C expression plasmid (WT, resistant to shRNA #1, or resistant to shRNA #2). The resistant Speedy/Ringo C constructs contained silent mutations rendering them resistant to the corresponding shRNAs. The expression of Speedy/Ringo C was analyzed by immunoblotting after 48 h. (D) Expression of RNAi-resistant Speedy/Ringo C rescued the cell cycle delay caused by Speedy/Ringo C knockdown. HEK293 clones expressing shRNA against firefly luciferase (GL3, left) and Speedy/Ringo C #1 (clones #7 and #21) were transfected with either empty vector (“vector”) or with shRNA #1-resistant 3xFLAG-Speedy/Ringo C. The cell cycle distributions of transfected cells were analyzed by flow cytometry.
Expression of an RNAi-resistant speedy/ringo C rescues the RNAi effects on the cell cycle
To rule out off-target effects of RNAi to Speedy/Ringo C, we created silent mutations in the RNAi target sites of Speedy/Ringo C to generate RNAi-resistant Speedy/Ringo Cs. To confirm these silent mutants were effective, wild-type and RNAi-resistant forms of Speedy/Ringo C were transiently cotransfected with shRNA expression vectors into HEK293 cells. Cells were harvested after 48 h and the expression of Speedy/Ringo C was determined by immunoblotting with anti-FLAG antibodies (Fig. 3C). Wild-type and two RNAi-resistant forms of Speedy/Ringo C were expressed at comparable levels in cells expressing the control shRNA (GL3) (lane 1). In contrast, the expression of wild-type Speedy/Ringo C was severely suppressed in cells expressing Speedy/Ringo C-targeting shRNAs (top, lanes 2 and 3). However, expression of both RNAi-resistant forms of Speedy/Ringo C was unaffected by their corresponding shRNA vectors but was suppressed by the other shRNA-expressing vector to Speedy/Ringo C.
We examined the cell cycle distributions following rescue of Speedy/Ringo C expression in clones #7 and #21, which express shRNA #1. Cells stably expressing shRNAs against firefly luciferase (GL3) and Speedy/Ringo C (clones #7 and #21) were transfected with either empty vector (“vector”) or vector expressing the RNAi-resistant form of Speedy/Ringo C. Cells were harvested after 48 h and subjected to FACS analysis of their cell cycle distributions. As shown in Figure 3D (top), Speedy/Ringo C-knockdown cells (Clone #7 + vector and Clone #21 + vector) had reduced G1 populations and increased S and G2/M percentages compared to control cells (GL3 + vector). Ectopic expression of Speedy/Ringo C in the control (GL3-expressing) cells decreased the S and G2/M populations (Fig. 3D, bottom, left). Similarly, ectopic expression of the RNAi-resistant form of Speedy/Ringo C overcame the effects of Speedy/Ringo C shRNA expression and decreased the S and G2/M phase populations in both shRNA-expressing clones (Fig. 3D bottom), indicating that the effects of Speedy/Ringo C knock-down on S and G2 phase progression were specific and not off-target effects.
Overexpression of speedy/ringo C inhibits cell growth and promotes cell death in HEK293 cells
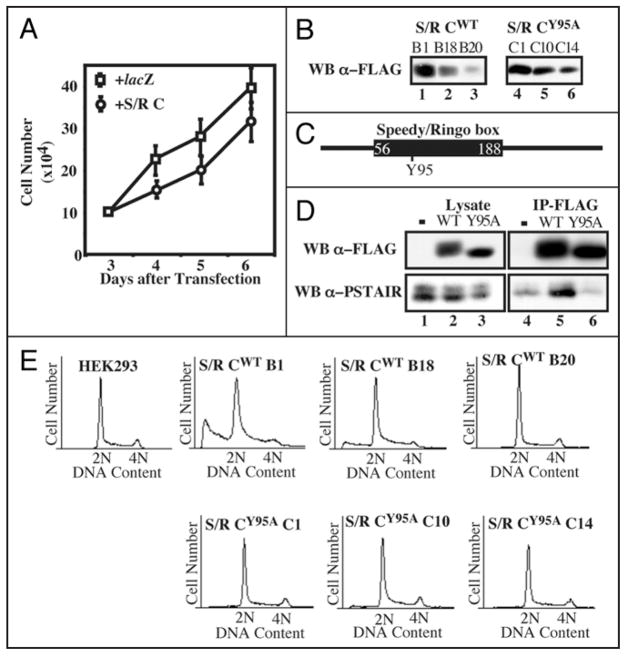
To determine if ectopic expression of Speedy/Ringo C could alter cell growth, plasmids expressing Speedy/Ringo C or lacZ (as a control) under the CMV promoter were transiently transfected into HEK293 cells. Cells were replated at low density and viable cells were counted daily. Since knockdown of Speedy/Ringo C by RNAi slowed cell growth, it was surprising that overexpression of Speedy/Ringo C also inhibited cell proliferation (Fig. 4A). However, in contrast to the effects of reducing Speedy/Ringo C expression (Fig. 3D), transient transfection of Speedy/Ringo C increased the fraction of cells in G1 and decreased the fraction in S and G2/M.
Figure 4.
Effects of overexpression of Speedy/Ringo C on cell growth and the cell cycle. (A) Ectopic expression of Speedy/Ringo C inhibited cell growth. HEK293 cells were transfected with plasmids encoding lacZ or 3xFLAG-Speedy/Ringo C. The cell number was measured at different times. Values represent the means ± S.E. from three separate experiments. (B) Isolation of HEK293 clones expressing different levels of Speedy/Ringo CWT or Speedy/Ringo CY95A. HEK293 cells were transfected with pcDNA3-3xFLAG-Speedy/Ringo CWT and pcDNA3-3xFLAG-Speedy/Ringo CY95A. Stable clones were selected using G418 and cell lysates were immunoblotted for 3xFLAG-Speedy/Ringo C. (C) Schematic illustration of the conserved Speedy/Ringo box and the position of Tyr-95 in Speedy/Ringo C. (D) The Speedy/Ringo C Y95A mutant was unable to interact with CDKs. HEK293 cells were lysed 48 hours after transfection with pcDNA3, pcDNA3-3xFLAG-Speedy/Ringo CWT, and pcDNA3-3xFLAG-Speedy/Ringo CY95A. Cell lysates were immunoprecipitated with anti-FLAG M2 agarose. Cell lysates (lanes 1–3) and immunoprecipitates (lanes 4–6) were analyzed by immunoblotting with antibodies against FLAG to detect Speedy/Ringo C (top) and PSTAIRE to detect Cdc2 and Cdk2 (bottom). (E) DNA content in HEK293 cells and selected clones from (D) was determined by flow cytometry of propidium iodide-stained cells.
HEK293 cells were stably transfected with 3xFLAG-Speedy/Ringo C to study the long-term effect of overexpression of Speedy/Ringo C. Clones with different Speedy/Ringo C expression levels were isolated (Fig. 4B, left). As with transiently-transfected cells, stable clones of Speedy/Ringo C displayed a dose-dependent slow growth phenotype with a corresponding increase of G1 phase cells and decrease of S and G2/M cells (data not shown). Overexpression of Speedy/Ringo C also promoted cell death, as indicated by an increase of cells possessing sub-G1 DNA content (Fig. 4E). We noticed that high-level expression of Speedy/Ringo C resulted in the detachment of many cells from the tissue culture dish. Removal of these “floaters” from the culture medium eliminated the sub-G1 population from the FACS analysis. Therefore, the sub-G1 population appeared to consist of dead cells resulting from the toxicity of overexpressed Speedy/Ringo C in HEK293 cells.
As a control for these cell cycle effects, we constructed and used a mutant form of Speedy/Ringo C with substantially reduced ability to bind to CDKs. We previously found that the interaction between Speedy/Ringo proteins and CDKs depended on a highly conserved region termed the Speedy/Ringo box (S/R box).13 Mutagenesis of a single tyrosine (Tyr-106) to alanine in the Speedy/Ringo A S/R box diminished binding to CDKs without altering Speedy/Ringo A protein expression.15 We mutated the corresponding residue (Tyr-95) in Speedy/Ringo C to alanine (Fig. 4C) and examined if Speedy/Ringo CY95A could associate with CDKs. HEK293 cells were transiently transfected with an empty vector, 3xFLAG-Speedy/Ringo CWT and 3xFLAG-Speedy/Ringo CY95A. After 48 h, the expression of Speedy/Ringo C and its ability to coimmunoprecipitate CDKs was analyzed by immunoblotting (Fig. 4D). Speedy/Ringo CY95A migrated as a single band and migrated faster than wild-type Speedy/Ringo C on SDS-PAGE, likely due to autophosphorylation of Speedy/Ringo CWT within CDK-Speedy/Ringo C complexes. Although the expression level and immunoprecipitation efficiency of Speedy/Ringo CWT and CY95A were comparable, Cdc2 and Cdk2, detected with anti-PSTAIRE antibodies, associated efficiently with wild-type Speedy/Ringo C but not with the Y95A mutant (Fig. 4D, bottom).
As with wild-type Speedy/Ringo C, we created stably-transfected cell lines expressing three different levels of Speedy/Ringo CY95A (Fig. 4B, right). Though expressed at similar levels as wild-type Speedy/Ringo C, expression of the mutant form of Speedy/Ringo C did not slow cell proliferation (data not shown) and caused no changes in cell cycle distribution or cell toxicity (Fig. 4E, lower row). Thus, the inhibitory effects of Speedy/Ringo C expression on cell proliferation are dependent on CDK-binding.
Overexpression of speedy/ringo C promoted late S phase progression and caused cell cycle arrest at the G1/S boundary
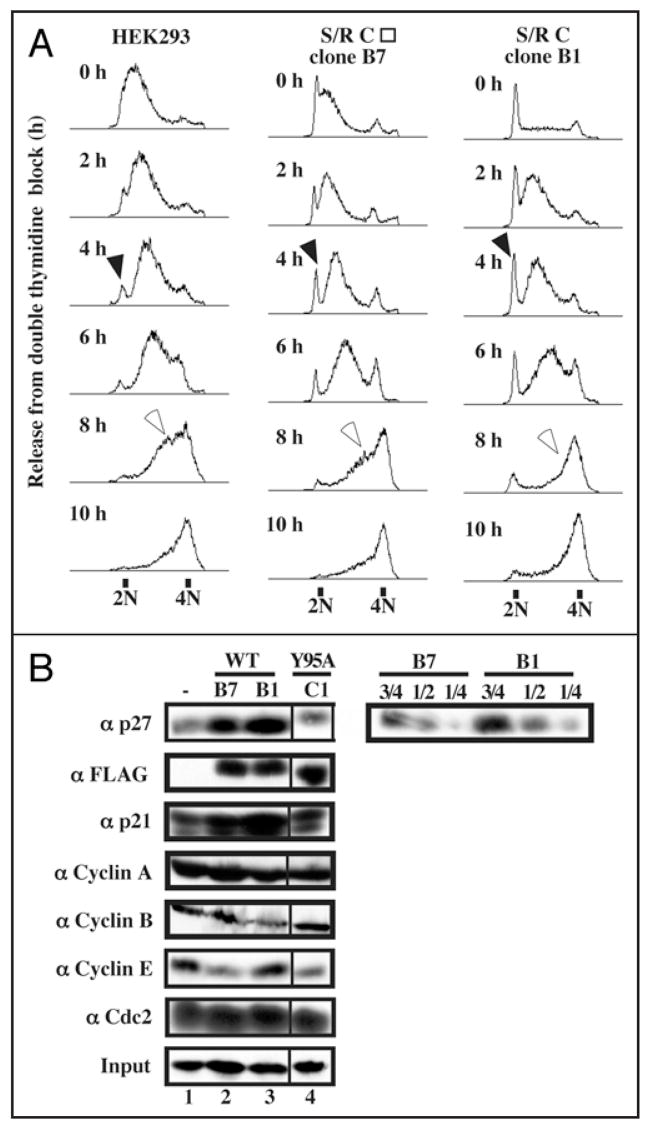
Since the reduction of Speedy/Ringo C increased the fraction of cells in S phase (Figs. 1D and 3B) and the overexpression of Speedy/Ringo C reduced the S phase population (Figs. 3D and 4E), we investigated whether Speedy/Ringo C overexpression directly affected S phase progression. HEK293 cells and two Speedy/Ringo C-expressing clones (clone B1, which expresses Speedy/Ringo C at a high level (Fig. 4B) and clone B7, which expressed Speedy/Ringo C at a slightly lower level than clone B1) were synchronized at G1/S using a double thymidine arrest protocol and released into fresh medium containing nocodazole to accumulate released cells in mitosis. The kinetics of cell passage through S phase was examined by FACS (Fig. 5A). The initial rates of passage through S phase were nearly identical in control cells and in Speedy/Ringo C-overexpressing cells. However, the fraction of cells in late S phase was significantly less in the Speedy/Ringo C-overexpressing cells (Fig. 5A, 8 h as indicated by open arrows), indicating that these cells traverse the end of S phase more rapidly than normal cells. In addition, while the vast majority of control cells enter into S phase quickly after the release, a significant proportion of Speedy/Ringo C-overexpressing cells showed a delayed entry into S phase (Fig. 5A, filled arrows), indicating that these cells either arrested at the G1-to-S transition or entered into G0 stages. Since these 2N cells eventually entered S phase, it appears that they were delayed at the G1-to-S transition.
Figure 5.
Overexpression of Speedy/Ringo C slowed the G1-to-S transition and promoted rapid progression through late S phase. (A) HEK293 cells and two Speedy/Ringo C-expressing clones (B1 and B7, whose Speedy/Ringo C level was slightly lower than that of clone B1) were synchronized by a double thymidine block protocol. The cells were released from the thymidine block into medium containing nocodazole and harvested at the indicated times. Cell cycle distributions were analyzed by flow cytometry of propidium iodide-stained cells. Cells delayed at the G1/S boundary are indicated with solid arrowheads; cells in late S phase are indicated with open arrowheads. (B) Immunoblotting analysis of cell cycle regulators in HEK293 cells (lane 1), Speedy/Ringo CWT clone B7 (lane 2), clone B1 (lane 3), and Speedy/Ringo CY95A clone C1 (lane 4). Serial dilutions of proteins from Speedy/Ringo CWT clones B1 and B7 were made to facilitate quantitation of p27 levels between HEK293 cells and Speedy/Ringo CWT clones. Note that the lane 4 samples in the left-hand panels are from a different part of the same autoradiographs as lanes 1–3.
Overexpression of speedy/ringo C increased the levels of p21 and p27
To explore the molecular mechanism of G1/S delay caused by ectopic expression of Speedy/Ringo C, we examined the levels of several G1-to-S cell cycle regulators—including Cdc2, Cdk2, cyclin A, cyclin E, p21, p27—as well as cyclin B in asynchronous cells by immunoblotting (Fig. 5B). Consistent with the cell cycle delay at the G1/S boundary, both p21 and p27 levels were significantly upregulated in Speedy/Ringo C-expressing cells (rows 1 and 3). We estimated that the level of p27 in these cells was about twice as much as in control cells, even though there were only 10% more G1 phase cells.
Speedy/ringo C regulates the duration of G2 phase
We explored whether Speedy/Ringo C might regulate progression through G2 phase. Support for this possibility comes from the initial identification of Speedy/Ringo as an inducer of the G2-to-M transition during the maturation of Xenopus oocytes, and our observation that RNAi-mediated knockdown of Speedy/Ringo C increases the proportion of cells in G2 (Fig. 1D). To obtain quantitative information on the effect of Speedy/Ringo C expression on the duration of G2 phase, we determined the length of G2 phase by tracking S phase cells into mitosis using triple staining FACS as described above (see Fig. 2). An outline of this assay is shown in Figure 6A. First, proliferating cells are incubated briefly with BrdU to label cells in S phase. At the end of this labeling phase, all mitotic cells, which are identified based on their reaction with an antibody to phospho-histone H3, are negative for BrdU staining. As BrdU-labeled S phase cells pass through G2 phase and enter mitosis, mitotic cells became BrdU-positive. The time it takes for BrdU-positive mitotic cells to appear is a measure of the length of G2.
Figure 6.
Speedy/Ringo C controls G2 length and the G2/M transition. (A) Diagram of the strategy to measure the length of G2 phase. Asynchronous cells were incubated with 10 μM BrdU to label S phase cells. Samples were taken at various times and processed to detect BrdU, phosphohistone H3Ser10 (HH3P; mitotic marker), and DNA content by flow cytometry. Initially, all HH3P-positive cells (HH3P+) are BrdU-negative (BrdU-). As BrdU-labeled cells pass through G2 and enter into mitosis, an increasing fraction of HH3P-positive cells become BrdU-positive (BrdU+). The length of G2 phase was defined as the time until half of the HH3P-positive cells are also BrdU-positive. (B) Plots of BrdU-positive vs. HH3P-positive cells. See EXPERIMENTAL PROCEDURES and Figure 2 for details of this analysis. Representative examples of such plots at times 0 h and 3 h are shown. (C) Analysis of G2 length in HEK293 cells treated with siRNA control (GL3, open circle ○) siRNA #1 against Speedy/Ringo C (solid square ▪). Cells were labeled with BrdU two days following transfection, harvested at the indicated times, and processed for flow cytometry as described above. The percent-age of HH3P+ cells that were also BrdU+ was plotted against time. (D) Analysis of G2 length in HEK293 cells overexpressing Speedy/Ringo C. HEK293 control cells (open circle ○) and cells overexpressing Speedy/Ringo C (clone B1, solid square ▪) were labeled with BrdU, and analyzed as in (C).
To measure the length of G2, asynchronous cells were grown at low density, labeled with BrdU, and collected at different times for analysis. Cells were stained for BrdU, phospho-histone H3Ser10 (HH3P), and DNA content (see Fig. 2). The movement of BrdU-positive cells into mitosis (HH3P-positive) was monitored on anti-BrdU (FL2-H) vs. anti-HH3P (FL1-H) plots (Fig. 6B). The G2 length was defined as the time until 50% of the HH3P-positive cells became BrdU-positive. It is noteworthy that this measurement was unaffected by the presence of tetraploid (4N) G1 cells since, as G1 cells, they were BrdU- and HH3P-negative.
We first investigated the effects of downregulation of Speedy/Ringo C on the length of G2. Cells were transiently transfected with siRNAs against either Speedy/Ringo C or control (GL3) for 2 days before labeling with BrdU and subsequent analysis as described above. The movement of S phase cells into mitosis shows that G2 phase in cells treated with siRNA against Speedy/Ringo C was about 30 minutes longer than that of cells treated with the control siRNA (Fig. 6C). We then used the same approach to determine the length of G2 in cells overexpressing Speedy/Ringo C (clone B1). In contrast to the lengthening of G2 upon downregulation of Speedy/Ringo C, ectopic expression of Speedy/Ringo C reduced the length of G2 phase by about 25 minutes (Fig. 6D). Thus, our results demonstrate that Speedy/Ringo C is a positive regulator of progression through G2.
Suppression of the G2 DNA damage checkpoint by ectopic expression of speedy/ringo C
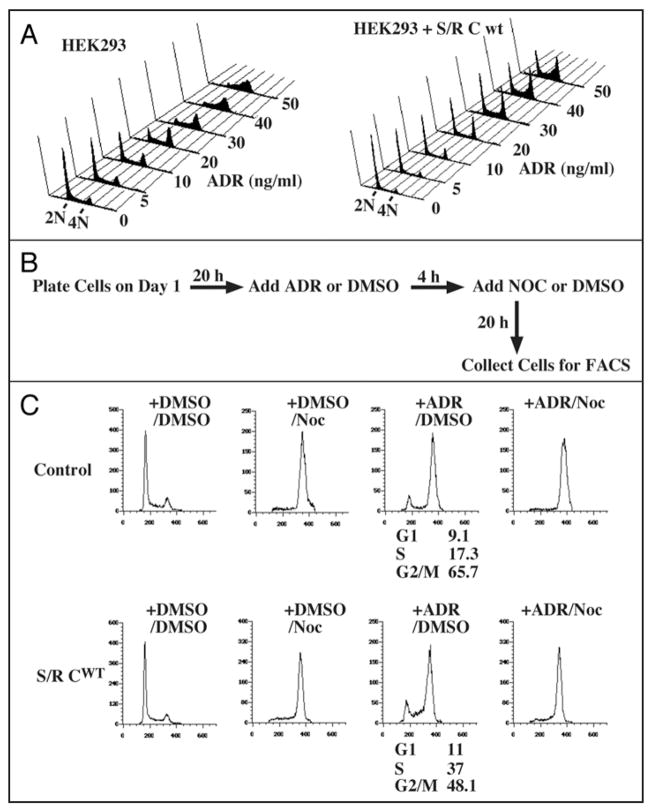
DNA damage can arrest cell cycle progression in G2 phase by increasing inhibitory phosphorylation of Cdc2 and Cdk2. Because Speedy/Ringo C regulates progression through G2, we examined whether its overexpression could over-ride the DNA damage-induced G2 arrest. To activate the G2 DNA damage checkpoint, HEK293 cells were treated with adriamycin (ADR) for 18 h. The cell cycle distribution was analyzed by FACS after staining with propidium iodide (Fig. 7A). Parental cells treated with ADR at 30 ng/ml were almost completely arrested in S and G2 phases (Fig. 7A, left). In contrast, the same concentration of ADR had only modest effects on cells expressing Speedy/Ringo C (Fig. 7A, right), indicating that overexpression of Speedy/Ringo C can suppress the G2 DNA damage checkpoint.
Figure 7.
Ectopic expression of Speedy/Ringo C can suppress the DNA damage checkpoint. (A) Effects of Adriamycin (ADR) on HEK293 cells and Speedy/Ringo C-expressing cells. HEK293 cells and Speedy/Ringo C clone B1 were treated with the indicated concentrations of ADR for 18 h. Cells were harvested, stained with propidium iodide, and subjected to FACS. (B) Diagram of Adriamycin/nocodazole experiments to assess the ability of cells to overcome the DNA damage checkpoint. Cells were treated with either mock (DMSO) or ADR for 4 hours, then treated with mock or nocodazole (Noc) for an additional 20 hours to block cell cycle progression at mitosis. (C) Overexpression of Speedy/Ringo C overcame the G2 DNA damage checkpoint induced by ADR. HEK293 cells and Speedy/Ringo C clone B1 were treated with 30 ng/ml ADR and/or 50 ng/ml Noc as described above. Cells were harvested and analyzed by FACS. Data were plotted as cell number vs. DNA content. The percentage of cells in different phases of the cell cycle was determined using the Watson Pragmatic model in the FlowJo program. All G1 and S phase cells following ADR treatment resulted from passage through the G2 DNA damage checkpoint (compare third panels on each row with the fourth panels).
We next addressed whether the significant population of G1 cells following ADR treatment of cells expressing Speedy/Ringo C results from a delay in entry to S phase (Fig. 5A), or from cells that have bypassed the DNA damage checkpoint and begun a subsequent cell cycle. To distinguish these possibilities, cells were exposed to adriamycin followed by treatment for an additional 20 hours with nocodazole (Noc) (Fig. 7B) to prevent entry into a second cell cycle. As shown in Figure 7C, both control cells and Speedy/Ringo C clone B1 cells were able to pass through G1 and S phase to reach G2/M (+ADR/Noc) during the experiment. This result suggests that the G1 and S phase cells in the ADR-treated samples had already bypassed the G2 DNA damage check-point induced by ADR. About 48% (11% G1 and 37% S) of Speedy/Ringo C clone B1 cells bypassed the G2 DNA damage checkpoint; however, only 26% (9.1% G1 and 17.3% S) of the parental cells went through the G2 arrest induced by ADR.
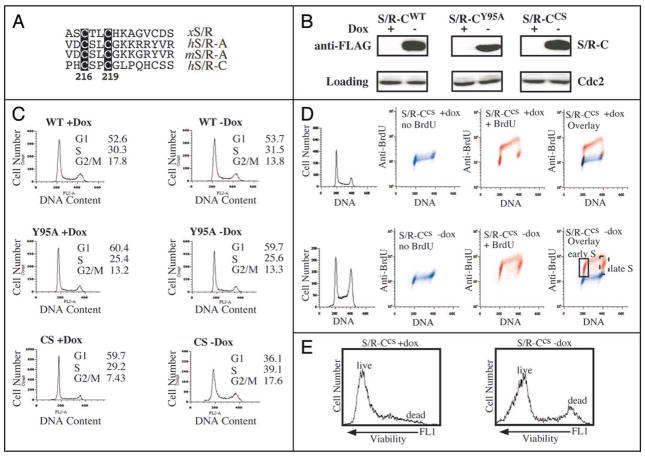
Overexpression of speedy/ringo CCS converts the HeLa somatic cell cycle to an “embryonic/stem cell-like” type of cell cycle with minimal gap phases
We created a Speedy/Ringo C mutant with dominant-negative-like properties. Unlike the highly conserved Speedy/Ringo box, which is necessary and sufficient for CDK-binding, the N- and C-termini of Speedy/Ringo proteins are less well conserved. However, we noticed that there is a conserved CxxC motif in the C-termini of Speedy/Ringo A and C (Fig. 8A). Since CxxC motifs are frequently involved in protein-protein interactions or metal-binding, we mutated the CxxC motif to SxxS (CS mutant, C217S/C219S) and examined the properties of the resulting protein. HeLaTet-Off cells were transiently transfected with Speedy/Ringo CWT, Speedy/Ringo CY95A, and Speedy/Ringo CCS vectors in the presence or absence of doxycycline, a tetracycline derivative. After 48 h, Speedy/Ringo C expression was analyzed by immunoblotting (Fig. 8B) and cell cycle profiles were analyzed by FACS (Fig. 8C). As shown in Figure 8B, doxycycline suppressed protein expression of Speedy/Ringo C, while removal of doxycycline induced Speedy/Ringo C. As observed previously, cells induced to express Speedy/Ringo CWT (wt-dox) showed a decreased population of G2/M cells (Fig. 8C, top) and cells expressing the Cdk-binding deficient Speedy/Ringo CY95A had no effects on the cell cycle distribution (Fig. 8C, middle). Interestingly, expression of Speedy/Ringo CCS significantly increased the G2/M population (Fig. 8C, bottom). This phenotype, which was similar to that following Speedy/Ringo C knockdown by RNAi, suggested that Speedy/Ringo CCS acted as a dominant-negative protein.
Figure 8.
Overexpression of Speedy/Ringo CCS induces an embryonic cell-like cell cycle in HeLa cells. (A) Illustration of the conserved CxxC motif in Speedy/Ringo proteins. Numbers denote the position of the CxxC in human Speedy/Ringo C. (B) Expression of FLAG-tagged Speedy/Ringo CWT, Speedy/Ringo CY95A and Speedy/Ringo CCS in HeLa Tet-Off cells. Transiently transfected cells were grown in the presence (Off) or the absence (On) of doxycycline for three days. The expression of Speedy/Ringo C was detected by immunoblotting. (C) Cell cycle profiles of HeLa cells expressing Speedy/Ringo proteins. Cells were grown as described in (B) above. Cell cycle profiles were determined by FACS after cells were stained with propidium iodide. (D) Cell cycle profiles of HeLa cells expressing Speedy/Ringo CCS were measured by BrdU incorporation. HeLa Tet-Off cells expressing Speedy/Ringo CCS were grown in the presence (Off) or the absence (On) of doxycycline for three days and then incubated for one hour with either BrdU (red) to label replicating DNA or with PBS (blue) as a negative control. Samples were stained with anti-BrdU antibodies and propidium iodide. The overlap between BrdU-negative (blue) and BrdU-positive (red) signals indicates cells that did not undergo DNA replication during the labeling period and hence were in G1, G2 and M. Red (BrdU-positive) signals that did not overlap with the blue (BrdU-negative) signals indicate that the majority of cells underwent DNA replication (S phase) and very few cells were in G1 and G2. (E) Effect of Speedy/Ringo CCS expression on cell viability. Viabilities of the cells in (D) were measured with a LIVE/DEAD fixable green dead cell stain kit from Invitrogen, Inc. Samples were incubated with fluorescent reactive dye for 30 min on ice, then cells were fixed and processed prior to FACS analyses. Dead cells have strong signals in the FL1 (green fluorescence) channel. 10,000 cells were analyzed for each sample.
To investigate the cell cycle effects of Speedy/Ringo CCS expression in greater detail, we generated stable HeLaTet-Off clones expressing Speedy/Ringo CCS. Stable clones were plated at low density in the presence or absence of doxycycline. These cells expressed high levels of Speedy/Ringo CCS two days after removal of doxycycline (not shown). Five days after plating, cells were incubated with or without BrdU for 1 h to label cells in S phase. Cell cycle profiles were determined by FACS after cells were stained with anti-BrdU antibodies and propidium iodide (Fig. 8D). In parallel, cell viability was assessed with LIVE/DEAD Viability/Cytotoxicity Kit (Invitrogen) staining and FACS (Fig. 8E). In the absence of Speedy/Ringo CCS expression, cells showed a typical HeLa somatic cell cycle (Fig. 8D, top row). The prominent red spots in the third panel reflect the high percentage of non-S phase (BrdU−) cells with G1 and G2 DNA contents. In contrast, expression of Speedy/Ringo CCS resulted in most cells being in S phase (BrdU+), with very substantial numbers of cells in very early and very late S phase. This pattern is reminiscent of embryonic cell cycles that lack G1 and G2 phases. Overexpression of Speedy/Ringo CCS had only minor effects on cell growth for the first five days; after that time cells started to exhibit decreased proliferation and reduced viability (Fig. 8E).
Discussion
We have explored the cell cycle function of Speedy/Ringo C and found that it promotes cell cycle progression during late S and G2 phases. This role is in contrast to its most closely related paralog, Speedy/Ringo A, which functions during the G1/S transition and S phase progression.16 In particular, RNAi-mediated knockdown of Speedy/Ringo C slowed cell proliferation, increased the proportion of cells in S phase and G2, and resulted in slower passage through G2. Conversely, overexpression of Speedy/Ringo C reduced the proportion of cells in S phase and resulted in faster progression through the late stages of S phase and through G2. Consistent with a role in promoting progression through G2, Speedy/Ringo C overexpression also caused bypass of the G2 DNA damage checkpoint.
As part of our analysis, we employed a powerful three-color FACS analysis of the length of G2. The overall logic of the method—to measure how long it takes cells that incorporate a label during S phase to progress into mitosis—is a classic approach. The three parameters used were BrdU fluorescence (to label cells in S phase), total DNA, and phosphorylation of histone H3 (a mitotic marker). We also performed a simple gating to eliminate cell aggregates and polyploid cells. The use of FACS permitted facile quantitation on an unsynchronized cell population.
The cell cycle phases accelerated by Speedy/Ringo C—late S and G2—are also phases progression through which involves the activities of Cdk2-cyclin A and Cdc2-cyclin A. This finding raises the possibility that CDK-Speedy/Ringo C complexes might contribute to the activation of CDK-cyclin A complexes much as Xenopus Speedy/Ringo is thought to help jumpstart activation of Cdc2. The resistance of CDK-Speedy/Ringo complexes to inhibitory phosphorylation of the CDK and to CDK-inhibitory proteins place these complexes in a good position to serve as the trigger for the activation of CDK-cyclin complexes. Such activation could involve titration of CDK inhibitors such as p27.21 Alternatively, CDK-Speedy/Ringo C complexes may work side-by-side with CDK-Cyclin A complexes, possibly by providing the initial activation of Cdc25 and inactivation of Wee1/Myt1 proteins necessary for full activation of the CDK-cyclin complexes.
We found that ectopic expression of Speedy/Ringo C could shorten the lengths of S and G2 phases, override the G2 DNA damage checkpoint, and induce a G1/S arrest in HEK293 cells. Interestingly, these phenotypes are similar to those caused by ectopic expression of a Cdc2 mutant immune to inhibitory phosphorylations (T14A/Y15F, Cdc2AF).19,20 For example, ectopic expression of Cdc2AF can bypass the G2 DNA damage checkpoint. Like Speedy/Ringo C overexpression, overexpression of Cdc2AF or Cdk2AF is cytotoxic and overexpression of Cdc2AF causes a G1 arrest.19,20
A possible explanation for this anti-proliferative effect is that Cdc2AF, by speeding up S and G2 progression, might promote mitosis even in the presence of damaged DNA. Thus, the cytotoxicity could be caused by unrepaired damage from the previous cell cycle, which might induce the expression of CDK inhibitors such as p21 and p27 to induce G1/S arrest. Similar reasoning might explain why overexpression of Speedy/Ringo C also results in a delay in G1 progression. It is particularly noteworthy that this overexpression can cause a bypass of the G2 DNA damage checkpoint, which could result in G1 cells with damaged DNA. In support of this model, we found that overexpression of Speedy/Ringo C had little effect on cell viability in HeLa cells, which have a longer G2 phase than HEK293 cells (unpublished results).
Interestingly, overexpression of Speedy/Ringo CCS can convert the somatic cell cycle into an embryonic-like cell cycle. In contrast, there is no evidence to suggest that overexpression of cyclins can do the same. It is possible that Speedy/Ringo proteins play important roles in specialized cell cycles. In this regard, it is worth noting that while Speedy/Ringo C is expressed in only a limited set of tissues, this set is highly enriched for tissues such as testis, liver, placenta and bone marrow that contain cells that constantly undergo self renewal.
Materials and Methods
Reagents
Tissue culture media, fetal bovine serum, pcDNA3, pTracer-EF, lipofectamine-2000, TRIzol, oligofectamine, SUPERSCRIPT II RNase H-reverse transcriptase, Taq DNA poly-merase, 5-Bromo-2′-Deoxyuridine (BrdU), anti-BrdU antibody (PRB-1), Alexa Fluor 488-conjugated goat anti-mouse IgG (H + L) antibody, R-Phycoerythrin (RPE)-conjugated goat anti-rabbit IgG (H + L) antibody, 7-Amino-Actinomycin D (7-AAD), LIVE/DEAD fixable green dead cell stain kit, and all antibiotics were purchased from Invitrogen (Carlsbad, CA). Rabbit anti-phospho-histone H3Ser10 (HH3P) antibody was obtained from Millipore (Charlottesville, VA). HRP-conjugated secondary antibodies and SuperSignal ECL reagents were from Pierce (Rockford, IL). Antibodies against p21 (sc-397), p27 (sc-528), cyclin A (sc-751), cyclin B (sc-7393), cyclin E (sc-247), and Cdc2 (sc-954) were from Santa Cruz Biotechnology (Santa Cruz, CA). All other chemicals were from Sigma (St. Louis, MO) unless indicated otherwise. 1x protease inhibitor mix (PI) contained 1 mM PMSF and 10 μg/ml each of leupeptin, chymostatin, and pepstatin. RIPA buffer contained 50 mM Tris-HCl, pH 7.4, 0.5% (v/v) Nonidet P-40, 0.1% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 1 mM NaF, 1 mM Na3VO4 and 1xPI.
Plasmids and siRNAs
Triple FLAG epitope (MDYKDDDDK) tags were added to the N-termini of human Speedy/Ringo C by PCR followed by cloning into the KpnI/XhoI sites of pcDNA3. A CDK-binding mutant of Speedy/Ringo C (Y95A) was generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). siRNA duplexes against Speedy/Ringo C and against firefly luciferase (GL3) were obtained from Dharmacon (Lafayette, CO). Speedy/Ringo C mRNA sequences targeted by siRNA #1 and siRNA #2 were 5′-TTAAGCCTGTGTCATCCA-3′ and 5′-AGATTGGTGTTTACGAGTG-3′, respectively. To generate RNAi-resistant expression vectors for Speedy/Ringo C containing silent mutations, the siRNA targeting region of the Speedy/Ringo C expressing vector was mutagenized from 5′-TTAAGCCTGTGTCATCCA-3 to 5′-TTAAGCCTGTCAGCTCCA-3′ for siRNA #1 and from 5′-AGATTGGTGTTTACGAGTG-3′ to 5′-AGATTGGTGCCTGAGAGTG-3′ for siRNA #2 by using QuikChange mutagenesis. To construct shRNA vectors against Speedy/Ringo C, the EF1a promoter, multiple cloning site, and poly-A signal in the pTracer-EF vector were replaced with a mouse U6 promoter, human microRNA-30 (mir-30; 360-bp), and stop sequence for RNA Pol III, which yielded an shRNA vector with a GFP-zeocin marker. shRNA plasmids used in this study were Speedy/Ringo C #1 (5′-TTAAGCCTGTGTCATCCA-3′), Speedy/Ringo C #2 (5′-AGATTGGTGTTTACGAGTG-3′), and GL3 (5′-CTGACGCGGAATACTTCGA-3′).
Cell lines, transfection and selection procedures
HEK293, HeLa and A549 cells were obtained from ATCC and maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum, 2 mM glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml in a 5% CO2 environment. Cells were routinely passaged when 90~95% confluent. For transient transfection, 5 × 105 cells were seeded in a 10 cm-diameter dish with 10 ml of medium lacking antibiotics. After 48 h, cells were transfected with 8 μg of plasmids using 40 μl Arrest-In reagent (Open biosystems, Huntsville, AL). After an additional 48 h, cells were trypsinized and washed with ice-cold phosphate-buffered saline (PBS) prior to RT-PCR, immunoblotting, and FACS analysis. To make stable cell lines, 4 μg FspI-cut plasmids were transfected into HEK293 cells at 80% confluency in a 6 cm-diameter plate using 16 μl lipofectamine-2000 reagent. Clones were selected in the presence of G418 at 250 μg/ml for Speedy/Ringo C or in the presence of zeocin at 400 μg/ml for shRNA against Speedy/Ringo C. For maintenance of stable clones, cells were grown in medium supplemented with appropriate antibiotics (100 μg/ml of G418 or zeocin).
HEK293 cells were synchronized using a double-thymidine block. Briefly, cells were grown to a subconfluent density in DMEM plus 10% FCS. Thymidine was added to 2 mM and cells grown for 16 hours. Cells were washed three times with serum-free DMEM and grown in fresh DMEM plus 10% FCS for 10 hours. Thymidine was again added and the cells incubated for 14 hours, at which time they were washed three times with serum-free DMEM and refed with DMEM plus 10% FCS to begin the time courses.
RNAi-knock down and rescue experiment
For transient transfection, 5 × 105 cells were seeded in a 10 cm-diameter dish with 10 ml of medium. After 48 h, cells were transfected with 100 nM siRNA duplexes using 12.5 μl oligofectamine reagent. After an additional 48 h, the cells were trypsinized, washed with ice-cold PBS, and subjected for RT-PCR and FACS analysis. For rescue experiments, 5 × 105 cells were seeded in a 10 cm-diameter dish with 10 ml of medium lacking antibiotics. After 48 h, cells were transfected with 8 μg RNAi-resistant plasmids using 40 μl Arrest-In reagent. After an additional 48 h, cells were trypsinized and washed with ice-cold PBS prior to immunoblotting and FACS analysis. The expression of human Speedy/Ringo C and GAPDH by RT-PCR was performed as described previously.15
Flow cytometry
For cell cycle analysis, 5 × 105 cells were seeded in 10 cm-diameter dishes with 10 ml of medium. After 48 h, cells were trypsinized, washed with ice-cold PBS, fixed in 75% ethanol, and stored at −20°C. Cells were treated with 1 ml RNase A (0.2 mg/ml) for 0.5 h at 37°C and stained with propidium iodide at 20 μg/ml. The stained cells were analyzed in a fluorescence-activated cell sorter (FACSCalibur, Becton Dickinson) using the CellQuest program. Single cell events were distinguished from cell aggregates and polyploid cells using on FL2-A (“area”) vs. FL2-W (“width” or “transit time”) plots (see Fig. 2A). The FL3-A value represents the total fluorescent intensity of an event whereas the FL3-W represents the time it takes the fluorescent entity to pass through the detector. Thus, events with larger values of FL3-W represent cell aggregates containing higher amounts of DNA (FL3-A). The percentage of cells in different phases of the cell cycle was determined using the Watson Pragmatic model in the FlowJo program.
For BrdU-labeling, 1–2 × 106 cells were labeled with 10 μM BrdU for 1 h, harvested, fixed and stored as above. Cells were stained with anti-BrdU FITC-coupled antibody (BD Bioscience) according to the manufacturer’s protocol.
We used a three-color FACS protocol to measure cells in all four stages of the cell cycle. 1–2 × 106 cells were labeled with 10 μM BrdU for 1 h, harvested, fixed and stored as above. Cells were re-fixed in 2% paraformaldehyde/0.1% Triton X-100 and treated with 5 μg DNase I per sample for 20 min (Worthington Biochemical, DPFF grade). Samples were incubated with anti-BrdU antibodies (PRB-1, 1 μg per sample) and anti-phospho-histone H3Ser10 (HH3P) antibodies (1 μg per sample) at 4°C overnight. Samples were then stained with Alexa Fluor 488-conjugated goat anti-mouse secondary antibody and RPE-conjugated goat anti-rabbit secondary antibody. DNA was stained with a 7-AAD/RNase A solution. Samples were analyzed by flow cytometry using the CellQuest program. BrdU and HH3P signals were recorded on a logarithmic scale in the FL1 (green) and FL2 (red) channels, respectively. DNA was recorded on a linear scale in the FL3 channel. Representative FACS data is shown in Figure 2. Single cell events were distinguished from cell aggregates on a FL3-A vs. FL3-W plot of 7-AAD fluorescence. By plotting DNA content vs. BrdU-incorporation, cells could be divided into three populations: G1, S and G2/M. G2/M cells could be further separated into phospho-histone H3-positive (mitotic cells) and -negative cells (G2 phase cells). To measure the length of G2 phase, asynchronous cells were labeled with 10 μM BrdU. At different times following labeling, cells were harvested, fixed, stored and processed according to the three-color FACS protocol described above. Single cell events were distinguished from cell aggregates on FL3-A vs. FL3-W plots of 7-AAD fluorescence. Single cell evens were then plotted onto FL1 (BrdU signal) vs. FL2 (HH3P signal) plots. As S phase (BrdU-positive, HH3P-negative) cells passed through G2 phase and entered mitosis, they became both BrdU-positive and HH3P-positive. The length of G2 phase was determined as the time when half of the HH3P-positive cells were also BrdU-positive.
Acknowledgments
We thank Janet Burton and Denis Ostapenko for helpful discussions. This work was supported by grant GM47830 to M.J.S. from the National Institutes of Health, grant 0455851T to M.J.S. from the American Heart Association, and support from the Robert Leet and Clara Guthrie Patterson Trust. Aiyang Cheng was a special fellow of the Leukemia & Lymphoma Society.
References
- 1.Pines J. Cyclins and cyclin-dependent kinases: a biochemical view. Biochem J. 1995;308:697–711. doi: 10.1042/bj3080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sherr CJ, Roberts JM. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–63. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 3.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 4.King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–9. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 5.Morgan DO. The dynamics of cyclin dependent kinase structure. Curr Opin Cell Biol. 1996;8:767–72. doi: 10.1016/s0955-0674(96)80076-7. [DOI] [PubMed] [Google Scholar]
- 6.Morgan DO. Cyclin-dependent kinases: engines, clocks and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–91. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 7.Solomon MJ, Kaldis P. Regulation of CDKs by phosphorylation. In: Pagano M, editor. Results and Problems in Cell Differentiation Heidelberg: Springer. Vol. 22. 1998. pp. 79–109. [DOI] [PubMed] [Google Scholar]
- 8.Solomon MJ, Glotzer M, Lee TH, Philippe M, Kirschner MW. Cyclin activation of p34cdc2. Cell. 1990;63:1013–24. doi: 10.1016/0092-8674(90)90504-8. [DOI] [PubMed] [Google Scholar]
- 9.Ferrell JE., Jr Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol. 2002;14:140–8. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- 10.Lenormand JL, Dellinger RW, Knudsen KE, Subramani S, Donoghue DJ. Speedy: a novel cell cycle regulator of the G2/M transition. EMBO J. 1999;18:1869–77. doi: 10.1093/emboj/18.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferby I, Blazquez M, Palmer A, Eritja R, Nebreda AR. A novel p34cdc2-binding and activating protein that is necessary and sufficient to trigger G2/M progression in Xenopus oocytes. Genes Dev. 1999;13:2177–89. doi: 10.1101/gad.13.16.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karaiskou A, Perez LH, Ferby I, Ozon R, Jessus C, Nebreda AR. Differential regulation of Cdc2 and Cdk2 by RINGO and cyclins. J Biol Chem. 2001;276:36028–34. doi: 10.1074/jbc.M104722200. [DOI] [PubMed] [Google Scholar]
- 13.Cheng A, Gerry S, Kaldis P, Solomon MJ. Biochemical characterization of Cdk2-Speedy/Ringo A2. BMC Biochem. 2005;6:19. doi: 10.1186/1471-2091-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinarina A, Perez LH, Davila A, Schwab M, Hunt T, Nebreda AR. Characterization of a new family of cyclin-dependent kinase activators. Biochem J. 2005;386:349–55. doi: 10.1042/BJ20041779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng A, Xiong W, Ferrell JE, Jr, Solomon MJ. Identification and comparative analysis of multiple mammalian Speedy/Ringo proteins. Cell Cycle. 2005;4:155–65. doi: 10.4161/cc.4.1.1347. [DOI] [PubMed] [Google Scholar]
- 16.Porter LA, Dellinger RW, Tynan JA, Barnes EA, Kong M, Lenormand JL, Donoghue DJ. Human Speedy: a novel cell cycle regulator that enhances proliferation through activation of Cdk2. J Cell Biol. 2002;157:357–66. doi: 10.1083/jcb.200109045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnes EA, Porter LA, Lenormand JL, Dellinger RW, Donoghue DJ. Human Spy1 promotes survival of mammalian cells following DNA damage. Cancer Res. 2003;63:3701–7. [PubMed] [Google Scholar]
- 18.Gastwirt RF, Slavin DA, McAndrew CW, Donoghue DJ. Spy1 expression prevents normal cellular responses to DNA damage: inhibition of apoptosis and checkpoint activation. J Biol Chem. 2006;281:35425–35. doi: 10.1074/jbc.M604720200. [DOI] [PubMed] [Google Scholar]
- 19.Jin P, Gu Y, Morgan DO. Role of inhibitory CDC2 phosphorylation in radiation-induced G2 arrest in human cells. J Cell Biol. 1996;134:963–70. doi: 10.1083/jcb.134.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chow JPH, Siu WY, Ho HTB, Ma KHT, Ho CC, Poon RYC. Differential contribution of inhibitory phosphorylation of CDC2 and CDK2 for unperturbed cell cycle control and DNA integrity checkpoints. J Biol Chem. 2003;278:40815–28. doi: 10.1074/jbc.M306683200. [DOI] [PubMed] [Google Scholar]
- 21.Porter LA, Kong-Beltran M, Donoghue DJ. Spy1 interacts with p27Kip1 to allow G1/S progression. Mol Biol Cell. 2003;14:3664–74. doi: 10.1091/mbc.E02-12-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]