Abstract
Background
Extensive epidemiological data indicate that vulnerabilities to nicotine dependence (ND) are influenced by genes, environmental factors, and their interactions. Although it has been documented from molecular experiments using in vitro and animal models that brain-derived neurotrophic factor (BDNF) exerts its functions via neurotrophic tyrosine kinase receptor 2 (NTRK2) and both alpha 4 (CHRNA4) and beta 2 (CHRNB2) subunits are required to form functional α4β2-containing nicotinic receptors (nAChRs), no study is reported demonstrating that there exist gene-gene interactions among the four genes in affecting ND.
Methods
To determine if gene-gene interactions exist among the four genes, we genotyped 6 SNPs for CHRNA4 and BDNF, 9 SNPs for NTRK2, and 4 SNPs for CHRNB2 in a case-control sample containing 275 unrelated smokers with a FTND score of 4.0 or more and 348 unrelated nonsmokers.
Results
By using a generalized multifactor dimensionality reduction (GMDR) algorithm recently developed by us, we found highly significant gene-gene interactions for the gene pairs of CHRNA4 and CHRNB2, CHRNA4 and NTRK2, CHRNB2 and NTRK2, and BDNF and NTRK2 (P<0.01 for all four gene pairs), and significant gene-gene interaction between CHRNA4 and BDNF (P=0.031) on ND. No significant interaction was detected for the gene pair CHRNB2 and BDNF (P=0.068).
Conclusion
Our study provides first evidence on the presence of gene-gene interaction among the four genes in affecting ND. Although CHRNB2 alone was not significantly associated with ND in several previously reported association studies on ND, we found it affects ND through interactions with CHRNA4 and NTRK2.
Keywords: Interaction, tobacco smoking, nicotine dependence, epistasis, GMDR
Introduction
Despite increasing public awareness of the health risks of using tobacco products and legislation that reduces the availability of cigarettes and prohibits smoking in many public facilities, little reduction in smoking prevalence has occurred in this country since 1990. In 2006, an estimated 45.3 million adults in the United States were current smokers; of them, an estimated 36.3 million smoked every day, and 9.0 million smoked some days (1). Economically, smoking is responsible for about 7% of the total US health care costs, or an estimated $167 billion each year (1). It is estimated that smoking causes approximately 438,000 deaths annually in the United States or 18.1% of all deaths nationwide (2).
Smoking is a complex behavior influenced by both genetic and environmental factors. Environmental influences on the development of nicotine dependence (ND) are well documented and include peer and familial influences as the strongest contributors to how and when cigarette experimentation occurs among young people. Moreover, multiple lines of evidence indicate that genetics plays a large part in determining smoking behavior. Over the last decade, many large-sample twin studies in the United States and other countries have yielded results supporting the conclusion that genetics contribute to the risk of becoming a regular smoker (3–7). A meta-analysis of the genetic parameter estimates for ND based on 17 twin studies determined that the weighted mean polygenic heritability for ND is 0.59 in male and 0.46 in female smokers, with an average of 0.56 for all smokers (7). Thus, identifying the genes predisposing to ND and understanding their molecular mechanisms are vital to prevention and treatment.
Recent evidence supports genetic associations with ND of the nicotinic acetylcholine receptor (nAChR) α4 subunit (CHRNA4) (8–10), of the brain-derived neurotrophic factor (BDNF) (11,12), and of the neurotrophic tyrosine kinase receptor 2 (NTRK2, also known as the tyrosine kinase receptor gene, TrkB) (13). Biochemical studies have revealed that the α4β2-containing nAChR subtype makes up the majority of the high-affinity nicotine-binding sites in the brain (14) and that the genes for both subunits are upregulated under chronic nicotine exposure (15,16). Furthermore, it was reported that activation of CHRNA4 is sufficient for nicotine-induced reward, tolerance, and sensitization (17). Moreover, knock-out mice for the α4 or β2 (CHRNB2) subunit show no high-affinity binding sites in their brains and fail to self-administer nicotine, indicating that the α4β2 subtype plays a primary role in the reinforcing effects of nicotine (17,18). However, except for CHRNA4 that has been found to be associated with smoking in three independent samples (8–10), almost all reported studies found no association of CHRNB2 with ND in humans (8,9,19,20).
On the other hand, BDNF must act through its high-affinity receptor NTRK2 in order to support the survival and growth of diverse neuronal populations and influences the form and function of chemical synapse (21). Furthermore, it was reported that nicotine modulates expression of BDNF and NTRK2 at both RNA and protein levels, indicating that nicotine regulates the BDNF/TrkB signaling pathway (22–24). Although the biological interactions of BDNF with NTRK2 and CHRNA4 with CHRNB2 have been established experimentally using in vitro and animal models, no study is reported to demonstrate the presence of gene-gene interactions among the four genes. Thus, the primary objective of this study was to determine whether significant gene-gene interactions exist among the four genes in affecting ND, using a novel algorithm, called generalized multifactor dimensionality method (GMDR), reported recently by this group (25).
MATERIALS AND METHODS
Human study population
The subjects used in this study are of either African-American (AA) or European-American (EA) origin and were selected from a family study on ND, recruited primarily from the Mid-South states including Tennessee, Mississippi, and Arkansas, in the U. S. during 1999–2004 (8,26,27). Smokers were required to be at least 21 years of age, have smoked for at least the last five years, and have consumed an average of 20 cigarettes per day for the last 12 months. Extensive data were collected on each participant, including demographics (e.g., sex, age, race, biological relationships, weight, height, years of education, and marital status), medical history, smoking history and current smoking behavior, ND, and personality traits assessed by various questionnaires, available at NIDA Genetics Consortium Website (http://zork.wustl.edu/nida). All participants provided informed consent. The study protocol and forms/procedures were approved by all participating Institutional Review Boards.
In the present study, ND for smokers was ascertained by the Fagerström Test for ND (FTND: 0–10 scale) (28). All smokers selected for inclusion into the study had a FTND score of 4 or above and controls are defined as those who had exposed to cigarette smoking but smoked less than 100 cigarettes in their lifetime. Given the requirement of the GMDR method for the case-control design, we selected parent(s) or only one child (either smoker or control) from each family.
DNA extraction, SNP selection, and genotyping
The DNA was extracted from peripheral blood samples of each participant using a kit from Qiagen Inc (Valencia, CA). On the basis of the high heterozygosity (minor allele frequency ≥ 0.05) and coverage of the gene of interest that was as uniform as possible, we selected 6 SNPs for CHRNA4 and BDNF, 4 SNPs for CHRNB2, and 9 SNPs for NTRK2 from the National Center for Biotechnology Information (NCBI) database. Table 1 provides detailed information on these SNPs, which include their location within the gene, chromosomal position, allelic variants, the minor allele frequency, and the primer and probe sequences.
Table 1.
Position, nucleotide variation, minor allele frequency and primer/probe sequences of 25 SNPs within CHRNA4, CHRNB2, BDNF, and NTRK2
| Gene | SNP | Domain | Physical position | Alleles a | Reported MAF b | Forward (F) and Reverse ® Primer and Probe Sequences (5’-3”) |
|---|---|---|---|---|---|---|
| CHRNA4 | rs2273505 | Intron 2 | 61461322 | C/T | 0.119 | F: CCCCTGTGCTCCTTGCA R: CAGCTCATTGACGTGGTAGGT P: CCATCG/ATGCACTGTGA |
| rs2273504 | Intron 2 | 61458505 | A/G | 0.470 | F: AGGCTTCCCCAGCTAAGGA R: GCCGGGAGGGAGCAA P: AAGGAGGCTCT/CCCATGC |
|
| rs2229959 | Exon 5 | 61451998 | G/T | 0.179 | F: AGCGGCACCCAGAGC R: CTCAGCCGGCACATCCA P: TGCACCCT/GCCCTCAC |
|
| rs1044396 | Exon 5 | 61451578 | C/T | 0.389 | F: GCAAATGCACATGCAAGAAGGA R: GTGCTGCGGGTCTTGAC P: TCCCCGAGC/TGCCACG |
|
| rs3787137 | Intron 5 | 61449544 | A/G | 0.261 | F: ACAACCCCCAAGCTGATGAG R: GGGACCCAGGACACCCT P: CTGGGCCCA/GGCCGT |
|
| rs2236196 | Intron 6 | 61448000 | A/G | 0.266 | F: ACCCTCTCCTAGCGAAGCA R: CTCTCGGGCCCCATGAG P: ATTGGAGCA/GCTGCTGG |
|
| CHRNB2 | rs2072658 | 5’-flanking | 152806849 | A/G | 0.074 | F: CCCCGGAGGCGGAAAC R: GGCAGAACCAATCGAAGACTATCC P: CTTTTTTTTCCTG/AGGACCC |
| rs2072660 | Exon 6 | 152815345 | C/T | 0.258 | F:GCTGCTAAGTGGAAGACAGAGATG R: GGAGGCAGCAGACAATCCT P: CCTTGCCCG/ATCACTC |
|
| rs2072661 | Exon 6 | 152815504 | A/G | 0.261 | F:CCTGACACAATGGTAGCTCTGAAG R: CCAGCTGCTGTCCACTCAAG P: CCTGGGTA/GTGACCTG |
|
| rs3811450 | 3’-flanking | 152817656 | C/T | 0.132 | F: GCCCTCACCTCTTCCTTATTGTG R: GCAGTGTCATTCCCTCCATCT P: CAAAGGGCTGCG/ATACAG |
|
| BDNF | rs6265 | Exon 2 | 27636492 | A/G | 0.269 | F: CTTGACATCATTGGCTGACACTTT R: TTCTTCATTGGGCCGAACTTT P: CGAACACG/ATGATAGAA |
| rs2049045 | Intron 1 | 27650817 | C/G | 0.053 | F: CCCTCTCCAACCAAAATCTCTCTT R: CACCAACCTAGAAATTGGGTTACCT P: CTTCGATAAAC/GTTCC |
|
| rs6484320 | Intron 1 | 27659764 | A/T | 0.310 | F: AACACATGAGACTCAGAGAATTACAACAA R: GCATGCTTCATACCTAAATATGCTTCAC P: AACAGTTTTAGTCA/TTGTAAAC |
|
| rs988748 | Intron 1 | 27681321 | C/G | 0.340 | F: ATTCATCTTACAACCTGGGAACCAA R: GAGGGCATGAAGCTGGATACC P: TACCCCAG/CAGACCCT |
|
| rs2030324 | Intron 1 | 27683491 | C/T | 0.431 | F: CACAGCCTAAATAGGTGAGTCTCAA R: ACCAAAGGGTTTCAGGACATTGA P: ATGAAAGATGAACG/ATTAGCT |
|
| rs7934165 | Intron 1 | 27688559 | A/G | 0.438 | F: CCCTTACCCAGAATATATCCCAGCTT R: CTGGGTCCTTTGTGTCTTTGC P: CAGAGTTCTGAA/GTTGGT |
|
| NTRK2 | rs993315 | Intron 2 | 86477541 | C/T | 0.495 | F: TCCTGATCTTAGCTGCCTGATGTAT R: GCAACAACAATTGTGAGGACAGAT P: CTGCTGAAATCAG/ATTGATT |
| rs1659400 | Intron 6 | 86515814 | C/T | 0.353 | F: TGTACCGTGGACTGGCTACT R: TGCCATACTCACCTTTACTTTGTTCT P: CACTTTCGGTAATTC/TTTGAC |
|
| rs1187272 | Intron 12 | 86593906 | C/T | 0.443 | F: GTCCCTGTTAGCCTCACTGTT R: TTTGCCAAGAGCTGAGGTATCTG P: CTTCTCTCCCC/TCTGCGTG |
|
| rs1122530 | Intron 12 | 86654172 | C/T | 0.218 | F: GTCCAATTATCTTCTGGGCAAGGAA R: GCACGGATCCATGATTACGTTAGAT P: CTCTTCTT/CGCCTCTCC |
|
| rs736744 | Intron 14 | 86704227 | A/G | 0.221 | F: CCAGACTGGGAAGTAGGTCATG R: TCTCTCTCTCTGGAAAGTCAGGAG P: TGGGAGGGCAA/GGTG |
|
| rs920776 | Intron 14 | 86728156 | C/T | 0.217 | F: CTGACCTGTATCTTAATGAATGAACATTTTATTTTTACAT R: CCACATTCTGATTGTGTTATTGTCATTGATATATT P: CCATCATTAGAGTATG/ATGCACA |
|
| rs1078947 | Intron 15 | 86753072 | C/T | 0.262 | F: GCAAAAGATGAGAAAACAGCCTAGT R: AGCTTTGCATATGCCTAAGGAGTT P: CTAAGGAAATAACA/GTTTGTG |
|
| rs4075274 | Intron 17 | 86786382 | A/G | 0.332 | F: TGTGGACATAATAGCCAAAGTAAAATGCT R: AAAGCTGATGTGTTTTTTCATTGTCTTCT P: AACATGTAAGTATCA/GTACCTT |
|
| rs729560 | Intron 17 | 86824125 | A/G | 0.387 | F: GGACTACTGTGCAGAAATCTGTTCA R: CATGGAGGAAAGTTAGGAAGCTCAT P: TTTGAAATCA/GGGCATCC |
The nucleotide of each SNP shown in bold font represents the minor allele as given in NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/).
Based on the allele frequency presented in the NCBI dbSNP database (Build 123).
All SNPs were genotyped using the TaqMan SNP Genotyping Assay in a 384-well microplate format (Applied Biosystems, Foster, CA). Briefly, 15 ng of DNA was amplified in a total volume of 7 µl containing an MGB probe and 2.5 µl of TaqMan universal PCR master mix. Allelic discrimination analysis was performed on the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster CA). To ensure the quality of the genotyping, SNP-specific control samples were added to each 384-well plate.
Statistical analysis for gene-gene interaction
Prior to performing the statistical analysis for detecting gene-gene interaction reported here, we used the PedCheck program (29) to determine genotyping consistency for Mendelian inheritance on the basis of the genotype data from other family members. Also, we checked the SNP data for any significant departure from Hardy-Weinberg equilibrium (HWE). The HWE at each locus was assessed by the χ2 test. The allele frequencies for each genetic marker were calculated using the FREQ program of S.A.G.E. (v. 5.0).
Statistical analyses of all SNPs between all possible pairs of the four genes were performed using a newly developed GMDR (25), as an extension of the MDR methods developed previously by other research groups (30–32) for allowing covariate adjustment. Given that both human and animal studies of nicotine administration or smoking behavior have documented that age, gender and ethnicity play significant roles in ND (7,33,34), we included age, gender and ethnicity as covariates in all our gene-gene interaction analyses. Residual scores of all individuals were computed under a null logistic model for smoking status.
Ten-fold cross-validation was used in our GMDR analysis. That is, data were randomly split into 10 approximately equal parts; one subdivision was used as the testing set and the rest as the training set. The genotypes formed by a subset of n SNPs (called n-locus model) were classified into high-risk, low-risk, or empty cells according to whether the average score in the training set was ≥ 0, < 0, or, empty (no data). Two contrasting (i.e, high-risk and low-risk) groups were formed by pooling all high-risk and low-risk cells, respectively, and the difference between two group scores was used to measure the classification accuracy. All potential n-locus models were evaluated sequentially and the model with the maximum classification accuracy was selected as the best model from those with the same dimensionality. We then used the independent testing set to estimate the prediction accuracy, i.e., to calculate the score difference in the testing set between two groups formed by the same pooling pattern as the training set. Finally, the model with the maximum prediction accuracy was chose as the best model among this set of best models from different locus numbers. To reduce the fluctuations due to chance divisions of the data, each possible training set and its corresponding testing set were successively used and the results were averaged. The consistency of the model across cross-validation training sets, i.e., how many times the same MDR model is identified in all the possible training sets, is also evaluated.
To identify the best interaction model, we did an exhaustive search for all possible two- to seven-SNP models for the SNPs between all six gene pairs. We also performed 10,000 permutations to generate the null distribution of maximum prediction accuracy for each dimensionality of the multilocus models evaluated so that the obtained empirical P values can be used as a yardstick to rank the models. Specifically, we randomly permuted the affection status for all participants to generate a set of pseudo-samples in which any potential relationship between the SNPs and the phenotype was disrupted. And then, as conducted in the above GMDR analysis for the real dataset, we used cross-validation to identify and evaluate the best interaction model. That is, we divided the pseudo-sample into two subsets of either training or testing, and then used training data to determine the best model based on the classification accuracy of the model under investigation, and testing data to evaluate the prediction accuracy for the identified model. Because of the independence of two subsets and no genetic association between the SNPs and the phenotype, the prediction accuracy from each permutation represented a sample drawn from the null distribution. The P-value was estimated by the proportion of the pseudo-samples resulting in larger prediction accuracy than the observed one in the real dataset. Based on these P values, along with the corresponding prediction accuracy and cross-validation consistency, we identified the best statistical gene-gene interaction model for all possible pairs among the four genes.
Results
To determine whether gene-gene interaction exist among the four genes in affecting ND, we studied 275 unrelated smokers with a FTND score ≥ 4.0 and 348 unrelated nonsmokers selected from the U. S. Mid-South Tobacco Family cohort (26). Of the subjects included, the average age was 46.6 ± 13.0 for smokers and 41.6 ± 17.8 for controls; 70% and 78% were females for smokers and controls; and 65% and 66% were African Americans for smokers and controls, respectively. For the smoker group, the average FTND score was 8.4 ± 1.6. Table 2 provides a summary of clinical characteristics for the cases and controls used in the current study.
Table 2.
Clinical characteristics for the cases and controls used in the study
| Characteristics | Cases | Controls |
|---|---|---|
| Sample size | 275 | 348 |
| Age (± SD; years) | 46.6 ± 13.0 | 41.6 ± 17.8 |
| Females (%) | 70% | 78% |
| African Americans (%) | 65% | 66% |
| FTND (± SD) | 8.4 ± 1.6 | NA |
As shown in Table 3, we detected highly significant interactive genetic effects on ND for the gene pairs CHRNA4 and CHRNB2, CHRNA4 and NTRK2, CHRNB2 and NTRK2, and BDNF and NTRK2 (with prediction accuracy ranging from 0.565 to 0.593; empirical P values < 0.01 for all these pairs). In addition, we found significant statistical interaction on ND between the genes CHRNA4 and BDNF, with prediction accuracy 0.552 (empirical P = 0.031). For the case of BDNF and CHRNB2, we did not find any interaction model better than we show in Table 3 (empirical P = 0.068) even when we tested more than seven SNPs for the gene pair. We also examined these interactions for each ethnic sample separately. Although the detected interactions in each ethnic sample appeared to be less significant (data not shown), we did obtain similar results as reported here for the combined sample.
Table 3.
The best interaction model detected for each gene pair on the basis of prediction accuracy, cross-validation consistency, and empirical P value from 10,000 permutations (Columns 5–7), and the prediction ability of the SNPs included in the identified interaction model for each corresponding gene (Columns 3 and 4)
| Gene pair | SNP(s) included in each interaction model | Prediction by the SNP(s) of each gene in the identified model | Prediction by interaction model | |||
|---|---|---|---|---|---|---|
| Prediction accuracy | Empirical P value | Prediction accuracy | Cross-validation consistency | Empirical P value | ||
| CHRNA4 | rs2273504, rs2229959, rs2236196 | 0.508 | 0.348 | 0.565 | 6 | 0.007 |
| CHRNB2 | rs2072661, rs2072660 | 0.507 | 0.345 | |||
| CHRNA4 | rs2229959, rs1044396 | 0.552 | 0.024 | 0.552 | 4 | 0.031 |
| BDNF | rs2030324 | 0.535 | 0.094 | |||
| CHRNA4 | rs2273505 | 0.534 | 0.076 | 0.578 | 9 | <0.0001 |
| NTRK2 | rs4075274 | 0.533 | 0.107 | |||
| CHRNB2 | rs3811450, rs2072661 | 0.508 | 0.322 | 0.541 | 6 | 0.068 |
| BDNF | rs2030324 | 0.535 | 0.079 | |||
| CHRNB2 | rs2072661 | 0.494 | 0.454 | 0.593 | 6 | 0.002 |
| NTRK2 | rs993315, rs729560, rs1187272, rs1122530, rs1078947, rs4075274 | 0.564 | 0.020 | |||
| BDNF | rs2030324 | 0.534 | 0.090 | 0.578 | 9 | 0.002 |
| NTRK2 | rs4075274 | 0.533 | 0.106 | |||
To determine whether interaction analysis between each gene pair yield a better model than a single gene approach, we also performed interaction analysis on the SNP(s) included in the best interaction model for each gene pair (see Table 3). A comparison of the prediction accuracy and empirical P value of each gene pair and the corresponding individual gene (with prediction accuracy from 0.494 to 0.564; empirical P value from 0.020 to 0.454; Table 3) further confirmed our finding that significant gene-gene interaction exists among these gene pairs in affecting ND.
By examining the SNPs included in the best prediction model for each gene pair, we found that SNP rs2072661 was included in all the CHRNB2-related gene pairs, as were SNPs rs2030324 for BDNF and rs4075274 for NTRK2. This suggests that the gene region where each SNP is located more likely harbors causative SNP(s) involved in the etiology of ND. As for CHRNA4, SNPs included in the three gene pairs with significant interaction are almost evenly distributed across the whole gene, with no single SNP detected in all the gene pairs, suggesting multiple causative or functional SNPs exist within the gene. We also analyzed ND as a quantitative trait and obtained almost identical results as when it was treated as a binary trait (data not shown).
Discussion
Mounting studies have pointed to the view that genes act in concert, rather than isolatedly, to affect ND. Statistical gene-gene interactions have been reported for smoking-related phenotypes for DRD2 with SLC6A3, DBH, and CYP2B6 (35–37). It has been long known that subunits α4 and β2 must assemble together in order to form a functional α4β2-containing heteromeric nAChR, a major highly expressed receptor type in the central nervous system. Similarly, in order for BDNF to exert its biological functions in the regulation of dopamine and serotonin neurotransmission in the brain, BDNF has to bind to its receptor, NTRK2, leading to activation of BDNF/NTRK2 signaling pathway. However, to date, there is no human genetic epidemiological study showing evidence that CHRNA4 indeed interacts with CHRNB2 and so does for BDNF and NTRK2 in affecting ND. Thus, this study provides first evidence that gene-gene interactions exist among the two gene pairs in influencing ND.
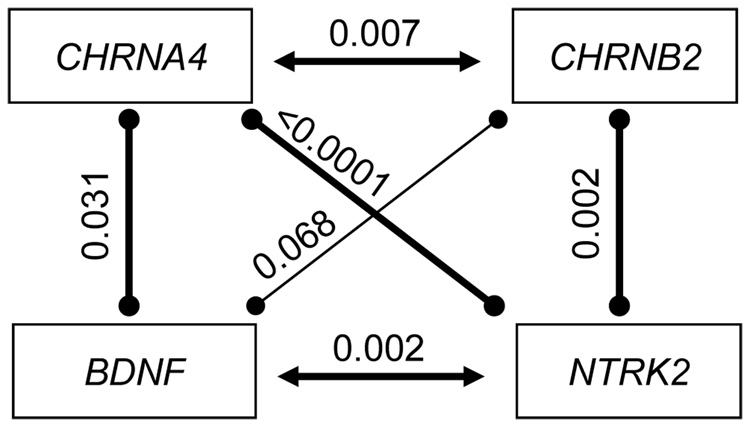
To gain further insight into the contributions of the four genes to ND, we propose a genetic model to explain our findings (Figure 1). Because it has been demonstrated experimentally that CHRNA4 interacts with CHRNB2 and BDNF with NTRK2, we conclude that these two gene pairs contribute to ND by a known biological interactive mechanism. On the other hand, since α4β2-containing nAChR and BDNF/NTRK2 represent two independent signaling systems, we conclude that CHRNA4 and BDNF or NTRK jointly contribute to ND in an as yet unknown indirect manner, as is also the case for CHRNB2 and NTRK2.
Figure 1.
Illustration of detected genetic effects of CHRNA4, CHRNB2, BDNF, and NTRK2 on ND. Two types of genetics effects are shown in the figure: known biological interaction (shown in horizontal line) and joint action (shown in vertical line). Value shown on the top of each line represents the empirical p-value from 10,000 replicates.
Compared with our earlier family-based individual SNP and haplotype analyses for each individual gene, we found most of the reported significant SNPs in the earlier studies yielded a stronger signal (significance) in the present study. For example, SNP rs2030324 in BDNF that was significant in the EA and the pooled samples (11) was identified to interact with NTRK2 (P = 0.002) and CHRNA4 (P = 0.031). Significant SNPs in NTRK2, rs1187272 and rs1122530 in EAs, and rs993315, rs4075274, rs729560 in AAs (13), were included in the lists of the best interaction models of the current report. Significant SNPs in CHRNA4, rs2273504 and rs1044396 in EAs, and rs2273504, rs2273505, and rs2236196 in AAs (8) also interacted with at least one SNP of CHRNB2, BDNF, and NTRK2. On the other hand, we also noticed that few significant SNPs, e.g., rs1659400 of NTRK2 significant in EAs (13) and rs3787137 of CHRNA4 significant in AAs (8) were not included in our detected interaction models. This is more likely due to that those SNPs were in very strong LD with some SNPs included in the identified interaction models, e.g., rs1659400 with rs1187272 and rs3787137 with rs2236196, and inclusion of them in the final interaction model would be redundant and contribute no or less extra information to account for ND variation in our samples.
Also, it is worthy of mention that, although CHRNB2 alone was not found to be associated with ND in several reported studies (8,9,19,20), it does appear to play a role in ND when analyzed jointly with CHRNA4 or NTRK2. In other words, although no significant contribution of CHRNB2 to ND could be detected in the four independent human genetics studies (8,9,19,20), it does not exclude the possibility of involvement of the gene to ND. The reason of failing to detect significant association of CHRNB2 with ND might be due to the strong dependency of CHRNB2 effects on specific CHRNA4 variants and/or small sample size for relatively small marginal effects of CHRNB2 used in those studies. With the GMDR approach that can detect genetic contribution of multiple genes on ND, we detected significant gene-gene interaction between CHRNB2 and CHRNA4 in influencing ND. This indicates that it is critical to develop more powerful and sensitive methodology of detecting gene-gene and gene-environment interactions and apply them to our genetics research, which will help us to identify more genetic contributors to ND that cannot be detected at single gene level based on conventional association analysis. More importantly, detection of significant joint contribution of CHRNB2 with CHRNA4 in human provide a plausible explanation to well-documented experimental evidence observed in animals where knockout mice for the α4- or β2-subunit of nAChRs show no high-affinity binding site in their brains and fail to self-administer nicotine (18,38,39).
It is also interesting to notice that we detected significant interaction between CHRNA4/ CHRNB2 and BDNF/NTRK2. From our current knowledge on the biological function and signaling pathways involved by the four genes, it seems that these four genes function independently as two separate functional groups. The first group consists of CHRNA4 and CHRNB2 whose gene products must assemble together to form functional α4β2-containing nAChRs involved in modulation the release of dopamine and gamma-aminobutyric acid (40). Another group contains BDNF and NTRK2 genes that make up BDNF/NTRK2 signaling pathway with a critical role in regulating the survival and differentiation of neuronal populations during development and synaptic transmission and plasticity at adult synapses in many brain regions (21). Thus, to some extent, our detected interactions of CHRNA4 and CHRNB2 with BDNF and NTRK2 is considered to be novel and intriguing from biological point of view, and such predictions deserve to be tested experimentally using in vitro and animal models.
It is agreed that ND, like any other complex trait, is controlled by multiple genetic factors, with each having a relatively small effect, as well as by environmental factors and interactions between genes or between genes and the environment. During the past decade, significant progress has been made in searching for susceptibility genes for ND through linkage and association analyses (41–44). However, these approaches are effective only for genes with moderate to major effects. The ability to identify susceptibility genes for ND and other complex traits has been improving but is still limited because of various factors such as gene interaction, modest marginal contribution, variable expressivity, small sample size, and heterogeneities (41,45,46). Of these factors, gene-gene and gene-environment interactions are of the greatest importance. To search for determinants of gene-gene and gene-environment interactions, considerable effort has been made in the past. Several combinatorial approaches, such as the multifactor dimensionality reduction (MDR) method (30–32,47), the combinatorial partitioning method (CPM) (48), and the restricted partition method (RPM) (49), have been developed as promising tools for detecting gene-gene and gene-environment interactions (50–52). Since the original report, MDR has been applied by many research groups to detect interactions for a number of complex disorders, and the list of publications is expanding rapidly (for details, please see the Epistasis Blog Website: http://compgen.blogspot.com/2006/05/mdr-applications.html). However, these established methods have limitations that restrict their practical use. For example, MDR, CPM, and RPM do not allow adjustment for covariates; MDR is applicable only to dichotomous phenotypes, and CPM and RPM cannot handle categorical phenotypes. To overcome the limitations of these existing combinatorial approaches and meet our research needs in determining gene-gene and gene-environment interactions for ND, we recently developed a generalized approach, called generalized MDR (25) and applied this new methodology to our genetic analysis of SNP data on ND. The results reported in this communication represent a new research dimension for our ongoing efforts in searching for susceptibility genes for ND.
In summary, we have determined not only that CHRNA4 interacts with CHRNB2 and BDNF with NTRK2 to contribute to ND by known biological interactive mechanisms that have been demonstrated experimentally, but also that CHRNA4 interacts with BDNF or NTRK, and CHRNB2 with NTRK2, contributing to ND in an as yet unknown manner, as the gene pairs represent two independent signaling systems. Further replication of these findings in independent samples is thus needed in future study. Moreover, our interaction investigation suggests that CHRNB2 is involved in the etiology of ND when analyzed jointly with CHRNA4 or NTRK2. This provides an example of how traditional analysis may fail to identify important risk genes and thus that the use of a validated detection strategy for interactions is warranted.
Acknowledgements
We acknowledge the invaluable contributions of personal information and blood samples by all participants in the study. This project was funded by National Institutes of Health Grant DA-12844 to MDL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures
Dr. Lou, Mr. Chen, Dr. Ma, and Dr. Elston reported no biomedical financial interests or potential conflicts of interest. Dr. Li disclosed consulting fees from Information Management Consultants, Inc. and having received lecture fees from CME Enterprise.
References
- 1.CDC. Cigarette smoking among adults--United States, 2006. MMWR Morb Mortal Wkly Rep. 2007;56:1157–1161. [PubMed] [Google Scholar]
- 2.CDC. Annual smoking-attributable mortality, years of potential life lost, and productivity losses--United States, 1997–2001. MMWR Morb Mortal Wkly Rep. 2005;54:625–628. [PubMed] [Google Scholar]
- 3.Carmelli D, Swan GE, Robinette D, Fabsitz R. Genetic influence on smoking--a study of male twins. N Engl J Med. 1992;327:829–833. doi: 10.1056/NEJM199209173271201. [DOI] [PubMed] [Google Scholar]
- 4.Heath AC, Madden PA, Slutske WS, Martin NG. Personality and the inheritance of smoking behavior: a genetic perspective. Behav Genet. 1995;25:103–117. doi: 10.1007/BF02196921. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan PF, Kendler KS. The genetic epidemiology of smoking. Nicotine Tob. Res. 1999;1 Suppl 2:S51–S57. doi: 10.1080/14622299050011811. discussion S69–70. [DOI] [PubMed] [Google Scholar]
- 6.Tyndale RF. Genetics of alcohol and tobacco use in humans. Ann. Med. 2003;35:94–121. doi: 10.1080/07853890310010014. [DOI] [PubMed] [Google Scholar]
- 7.Li MD, Cheng R, Ma JZ, Swan GE. A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction. 2003;98:23–31. doi: 10.1046/j.1360-0443.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- 8.Li MD, Beuten J, Ma JZ, Payne TJ, Lou XY, Garcia V, et al. Ethnic- and gender-specific association of the nicotinic acetylcholine receptor alpha4 subunit gene (CHRNA4) with nicotine dependence. Hum Mol Genet. 2005;14:1211–1219. doi: 10.1093/hmg/ddi132. [DOI] [PubMed] [Google Scholar]
- 9.Feng Y, Niu T, Xing H, Xu X, Chen C, Peng S, et al. A common haplotype of the nicotine acetylcholine receptor alpha 4 subunit gene is associated with vulnerability to nicotine addiction in men. Am J Hum Genet. 2004;75:112–121. doi: 10.1086/422194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutchison KE, Allen DL, Filbey FM, Jepson C, Lerman C, Benowitz NL, et al. CHRNA4 and tobacco dependence: from gene regulation to treatment outcome. Arch Gen Psychiatry. 2007;64:1078–1086. doi: 10.1001/archpsyc.64.9.1078. [DOI] [PubMed] [Google Scholar]
- 11.Beuten J, Ma JZ, Payne TJ, Dupont RT, Quezada P, Huang W, et al. Significant association of BDNF haplotypes in European-American male smokers but not in European-American female or African-American smokers. Am J Med Genet B Neuropsychiatr Genet. 2005;139:73–80. doi: 10.1002/ajmg.b.30231. [DOI] [PubMed] [Google Scholar]
- 12.Lang UE, Sander T, Lohoff FW, Hellweg R, Bajbouj M, Winterer G, Gallinat J. Association of the met66 allele of brain-derived neurotrophic factor (BDNF) with smoking. Psychopharmacology (Berl) 2007;190:433–439. doi: 10.1007/s00213-006-0647-1. [DOI] [PubMed] [Google Scholar]
- 13.Beuten J, Ma JZ, Payne TJ, Dupont RT, Lou XY, Crews KM, et al. Association of Specific Haplotypes of Neurotrophic Tyrosine Kinase Receptor 2 Gene (NTRK2) with Vulnerability to Nicotine Dependence in African-Americans and European-Americans. Biol Psychiatry. 2007;61:48–55. doi: 10.1016/j.biopsych.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 14.Flores CM, Rogers SW, Pabreza LA, Wolfe BB, Kellar KJ. A subtype of nicotinic cholinergic receptor in rat brain is composed of alpha 4 and beta 2 subunits and is up-regulated by chronic nicotine treatment. Mol Pharmacol. 1992;41:31–37. [PubMed] [Google Scholar]
- 15.Marks MJ, Pauly JR, Gross SD, Deneris ES, Hermans-Borgmeyer I, Heinemann SF, Collins AC. Nicotine binding and nicotinic receptor subunit RNA after chronic nicotine treatment. J Neurosci. 1992;12:2765–2784. doi: 10.1523/JNEUROSCI.12-07-02765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whiteaker P, Sharples CG, Wonnacott S. Agonist-induced up-regulation of alpha4beta2 nicotinic acetylcholine receptors in M10 cells: pharmacological and spatial definition. Mol Pharmacol. 1998;53:950–962. [PubMed] [Google Scholar]
- 17.Tapper AR, McKinney SL, Nashmi R, Schwarz J, Deshpande P, Labarca C, et al. Nicotine activation of alpha4* receptors: sufficient for reward, tolerance, and sensitization. Science. 2004;306:1029–1032. doi: 10.1126/science.1099420. [DOI] [PubMed] [Google Scholar]
- 18.Picciotto MR, Zoli M, Rimondini R, Lena C, Marubio LM, Pich EM, et al. Acetylcholine receptors containing the beta2 subunit are involved in the reinforcing properties of nicotine. Nature. 1998;391:173–177. doi: 10.1038/34413. [DOI] [PubMed] [Google Scholar]
- 19.Silverman MA, Neale MC, Sullivan PF, Harris-Kerr C, Wormley B, Sadek H, et al. Haplotypes of four novel single nucleotide polymorphisms in the nicotinic acetylcholine receptor beta2-subunit (CHRNB2) gene show no association with smoking initiation or nicotine dependence. Am J Med Genet. 2000;96:646–653. [PubMed] [Google Scholar]
- 20.Lueders KK, Hu S, McHugh L, Myakishev MV, Sirota LA, Hamer DH. Genetic and functional analysis of single nucleotide polymorphisms in the beta2-neuronal nicotinic acetylcholine receptor gene (CHRNB2) Nicotine Tob Res. 2002;4:115–125. doi: 10.1080/14622200110098419. [DOI] [PubMed] [Google Scholar]
- 21.Bramham CR, Messaoudi E. BDNF function in adult synaptic plasticity: the synaptic consolidation hypothesis. Prog Neurobiol. 2005;76:99–125. doi: 10.1016/j.pneurobio.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Yamada K, Nabeshima T. Brain-derived neurotrophic factor/TrkB signaling in memory processes. J Pharmacol Sci. 2003;91:267–270. doi: 10.1254/jphs.91.267. [DOI] [PubMed] [Google Scholar]
- 23.Sun D, Huang W, Hwang YY, Zhang Y, Zhang Q, Li MD. Regulation by nicotine of Gpr51 and Ntrk2 expression in various rat brain regions. Neuropsychopharmacology. 2007;32:110–116. doi: 10.1038/sj.npp.1301134. [DOI] [PubMed] [Google Scholar]
- 24.Serres F, Carney SL. Nicotine regulates SH-SY5Y neuroblastoma cell proliferation through the release of brain-derived neurotrophic factor. Brain Res. 2006;1101:36–42. doi: 10.1016/j.brainres.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 25.Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, Li MD. A Generalized Combinatorial Approach for Detecting Gene-by-Gene and Gene-by-Environment Interactions with Application to Nicotine Dependence. Am J Hum Genet. 2007;80:1125–1137. doi: 10.1086/518312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li MD, Payne TJ, Ma JZ, Lou XY, Zhang D, Dupont RT, et al. A genomewide search finds major susceptibility Loci for nicotine dependence on chromosome 10 in african americans. Am J Hum Genet. 2006;79:745–751. doi: 10.1086/508208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li MD, Ma JZ, Payne TJ, Lou XY, Zhang D, Dupont RT, Elston RC. Genome-wide linkage scan for nicotine dependence in European Americans and its converging results with African Americans in the Mid-South Tobacco Family sample. Mol Psychiatry. 2007 doi: 10.1038/sj.mp.4002038. [DOI] [PubMed] [Google Scholar]
- 28.Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br. J. Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 29.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogenmetabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–382. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- 32.Moore JH, Gilbert JC, Tsai CT, Chiang FT, Holden T, Barney N, White BC. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol. 2006;241:252–261. doi: 10.1016/j.jtbi.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 33.Picciotto MR. Nicotine as a modulator of behavior: beyond the inverted U. Trends Pharmacol. Sci. 2003;24:493–499. doi: 10.1016/S0165-6147(03)00230-X. [DOI] [PubMed] [Google Scholar]
- 34.Madden PA, Heath AC, Pedersen NL, Kaprio J, Koskenvuo MJ, Martin NG. The genetics of smoking persistence in men and women: a multicultural study. Behav Genet. 1999;29:423–431. doi: 10.1023/a:1021674804714. [DOI] [PubMed] [Google Scholar]
- 35.Lerman C, Caporaso NE, Audrain J, Main D, Bowman ED, Lockshin B, et al. Evidence suggesting the role of specific genetic factors in cigarette smoking. Health Psychol. 1999;18:14–20. doi: 10.1037//0278-6133.18.1.14. [DOI] [PubMed] [Google Scholar]
- 36.Johnstone EC, Yudkin PL, Hey K, Roberts SJ, Welch SJ, Murphy MF, et al. Genetic variation in dopaminergic pathways and short-term effectiveness of the nicotine patch. Pharmacogenetics. 2004;14:83–90. doi: 10.1097/00008571-200402000-00002. [DOI] [PubMed] [Google Scholar]
- 37.David SP, Brown RA, Papandonatos GD, Kahler CW, Lloyd-Richardson EE, Munafo MR, et al. Pharmacogenetic clinical trial of sustained-release bupropion for smoking cessation. Nicotine Tob Res. 2007;9:821–833. doi: 10.1080/14622200701382033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Picciotto MR, Zoli M, Lena C, Bessis A, Lallemand Y, Le Novere N, et al. Abnormal avoidance learning in mice lacking functional high-affinity nicotine receptor in the brain. Nature. 1995;374:65–67. doi: 10.1038/374065a0. [DOI] [PubMed] [Google Scholar]
- 39.Marubio LM, del Mar Arroyo-Jimenez M, Cordero-Erausquin M, Lena C, Le Novere N, de Kerchove d'Exaerde A, et al. Reduced antinociception in mice lacking neuronal nicotinic receptor subunits. Nature. 1999;398:805–810. doi: 10.1038/19756. [DOI] [PubMed] [Google Scholar]
- 40.Lindstrom JM. Nicotinic acetylcholine receptors of muscles and nerves: comparison of their structures, functional roles, and vulnerability to pathology. Ann N Y Acad Sci. 2003;998:41–52. doi: 10.1196/annals.1254.007. [DOI] [PubMed] [Google Scholar]
- 41.Li MD. Identifying susceptibility loci for nicotine dependence: 2008 update based on recent genome-wide linkage analyses. Hum Genet. 2008;123:119–131. doi: 10.1007/s00439-008-0473-0. [DOI] [PubMed] [Google Scholar]
- 42.Li MD. The genetics of nicotine dependence. Curr Psychiatry Rep. 2006;8:158–164. doi: 10.1007/s11920-006-0016-0. [DOI] [PubMed] [Google Scholar]
- 43.Lessov-Schlaggar CN, Pergadia ML, Khroyan TV, Swan GE. Genetics of nicotine dependence and pharmacotherapy. Biochem Pharmacol. 2008;75:178–195. doi: 10.1016/j.bcp.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ho MK, Tyndale RF. Overview of the pharmacogenomics of cigarette smoking. Pharmacogenomics J. 2007;7:81–98. doi: 10.1038/sj.tpj.6500436. [DOI] [PubMed] [Google Scholar]
- 45.Schnoll RA, Johnson TA, Lerman C. Genetics and smoking behavior. Curr Psychiatry Rep. 2007;9:349–357. doi: 10.1007/s11920-007-0045-3. [DOI] [PubMed] [Google Scholar]
- 46.Swan GE, Lessov CN. Gene-environment interaction in nicotine addiction: the need for a large-scale, collaborative effort. Subst Use Misuse. 2004;39:2083–2085. [PubMed] [Google Scholar]
- 47.Martin ER, Ritchie MD, Hahn L, Kang S, Moore JH. A novel method to identify gene-gene effects in nuclear families: the MDR-PDT. Genet Epidemiol. 2006;30:111–123. doi: 10.1002/gepi.20128. [DOI] [PubMed] [Google Scholar]
- 48.Nelson MR, Kardia SL, Ferrell RE, Sing CF. A combinatorial partitioning method to identify multilocus genotypic partitions that predict quantitative trait variation. Genome Res. 2001;11:458–470. doi: 10.1101/gr.172901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Culverhouse R, Klein T, Shannon W. Detecting epistatic interactions contributing to quantitative traits. Genet Epidemiol. 2004;27:141–152. doi: 10.1002/gepi.20006. [DOI] [PubMed] [Google Scholar]
- 50.Hoh J, Ott J. Mathematical multi-locus approaches to localizing complex human trait genes. Nat Rev Genet. 2003;4:701–709. doi: 10.1038/nrg1155. [DOI] [PubMed] [Google Scholar]
- 51.Thornton-Wells TA, Moore JH, Haines JL. Genetics, statistics and human disease: analytical retooling for complexity. Trends Genet. 2004;20:640–647. doi: 10.1016/j.tig.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 52.Williams SM, Haines JL, Moore JH. The use of animal models in the study of complex disease: all else is never equal or why do so many human studies fail to replicate animal findings? Bioessays. 2004;26:170–179. doi: 10.1002/bies.10401. [DOI] [PubMed] [Google Scholar]