Abstract
RNA interference (RNAi) is an important defence against viruses and transposable elements (TEs). RNAi not only protects against viruses by degrading viral RNA, but hosts and viruses can also use RNAi to manipulate each other's gene expression, and hosts can encode microRNAs that target viral sequences. In response, viruses have evolved a myriad of adaptations to suppress and evade RNAi. RNAi can also protect cells against TEs, both by degrading TE transcripts and by preventing TE expression through heterochromatin formation. The aim of our review is to summarize and evaluate the current data on the evolution of these RNAi defence mechanisms. To this end, we also extend a previous analysis of the evolution of genes of the RNAi pathways. Strikingly, we find that antiviral RNAi genes, anti-TE RNAi genes and viral suppressors of RNAi all evolve rapidly, suggestive of an evolutionary arms race between hosts and parasites. Over longer time scales, key RNAi genes are repeatedly duplicated or lost across the metazoan phylogeny, with important implications for RNAi as an immune defence.
Keywords: antiviral RNAi, viral suppressors of RNAi, host–parasite coevolution, miRNA, piRNA, transposable element
1. Introduction
Broadly defined, RNA interference (RNAi) constitutes a class of processes that use short RNAs (approx. 20–30 nucleotides) to recognize and manipulate complementary nucleic acids. RNAi-related pathways have roles in the control of gene expression, epigenetic modification and regulation of heterochromatin, and in the host–parasite interactions (see §2 for references). RNAi-related phenomena are so pervasive that it now seems surprising that the molecular basis of RNAi was first reported little more than 15 years ago (more that 97% of papers using the terms ‘RNAi’ and ‘RNA silencing’ have been published since 2001; for a historical perspective see Matranga & Zamore 2007). From a host–parasite and host–pathogen viewpoint, RNAi-related pathways provide a particularly interesting arena for coevolutionary interaction. For the host, RNAi is a defence against both viruses and ‘genomic parasites’ such as transposable elements (TEs), and can provide fine control over other components of innate immunity by modulating gene expression (Welker et al. 2007; Lodish et al. 2008). For the parasite, host RNAi can be exploited to subvert host cell function by manipulating host gene expression and blocking host resistance mechanisms. Strikingly, components of RNAi are widely distributed across the eukaryotic phylogeny, implying that the common ancestor of all eukaryotes had a functional RNAi pathway (Cerutti & Casas-Mollano 2006), over a billion years or more ago (Roger & Hug 2006).
Immune system genes and the parasite molecules they interact with often evolve rapidly (Schlenke & Begun 2003). It is thought that this results from a coevolutionary arms race, in which there is a reciprocal process of adaptation and counteradaptation between parasites and hosts (Dawkins & Krebs 1979). As RNAi is a primary defence of many organisms against viruses and TEs, it is likely to be a key battlefield on which these arms races are played out. This has the potential to drive the rapid evolution of the proteins involved, and ultimately shape the RNAi pathways themselves (Obbard et al. 2006; Aravin et al. 2007; Marques & Carthew 2007).
The major RNAi mechanisms have been recently reviewed (see §2) and it is not our purpose to duplicate that effort here. Instead, following a brief outline of the pathways most relevant to host–parasite biology, we review RNAi-mediated host–parasite interactions from an evolutionary perspective. We also present new analyses of the distribution of key RNAi-pathway genes across the animal phylogeny, and the rates of evolution in both RNAi genes and viral genes that suppress RNAi.
2. RNAi mechanisms that mediate host–parasite interactions
The RNAi-related pathways are unified by their dependence on sequence-specific binding between short (approx. 20–30 nt) RNAs and target sequences, and their common use of Argonaute family proteins. However, this belies an enormous underlying diversity between pathways and phylogenetic lineages. For example, the short RNA molecules can be derived from exogenous double-stranded RNA (dsRNA) (including viral genomes and replicative intermediates), fold-back RNAs expressed by the host or virus, or overlapping pairs of sense and antisense transcripts. Different pathways also use different complements of Dicer, Argonaute and accessory proteins, and the outcome of RNAi can be the cleavage and degradation of target RNA, the recruitment of additional factors to modify gene expression, or even long-term expression changes through epigenetic modification and heterochromatin formation. (For recent reviews see Aravin et al. 2007; Chapman & Carrington 2007; Ding & Voinnet 2007; Hartig et al. 2007; Matranga & Zamore 2007; Slotkin & Martienssen 2007; Zaratiegui et al. 2007; Klattenhoff & Theurkauf 2008). For a glossary of relevant terms see table 1.
Table 1.
Glossary
| term | meaning | |
|---|---|---|
| Ago | Argonaute protein | the catalytic core of RISC that binds short RNAs and, in many cases, displays RNase H-like mRNA-cleaving activity. Key domains include the PAZ (Piwi–Argonaute–Zwille) and Piwi domains. Ago is named after an Arabidopsis developmental mutant that resembles the tentacles of a paper nautilus (Argonautidae) |
| Dcr | Dicer protein | the RNase-III family ribonuclease that cleaves dsRNA into short RNAs. Key domains include a helicase C-terminal domain, dsRNA-binding domains, a PAZ domain and two RNase-III domains. Named for its ‘dicing’ activity. |
| RNAi | RNA interference | the class of processes that use short single-stranded RNA molecules in complex with an Argonaute protein to bind complementary nuclei acids and modify their action and/or processing |
| siRNA | short interfering RNA | single-stranded RNAs of 20–30 nt involved in RNAi (especially those not classed as microRNAs) |
| miRNA | microRNA | single-stranded RNAs of 21–22 nt, derived from short fold-back hairpins (pre-miRNAs) and involved translational control |
| piRNA | Piwi-interacting RNA | single-stranded RNAs of 24–29 nt that function in complex with Piwi family Argonaute proteins in the animal germ line |
| viRNA | viral RNA | siRNAs derived from viral sequences |
| rasiRNA | repeat-associated siRNA | short RNAs derived from repeat sequences, such as TEs, sometimes considered a subclass of piRNAs in Drosophila |
| RdRp | RNA-dependent RNA polymerase | RNA polymerase directed by RNA, especially eukaryotic polymerases involved in the amplification of RNAi in nematodes and plants |
| RISC | RNA-induced silencing complex | the complex comprising Argonaute, a short RNA, and several other proteins, which mediates RNAi through sequence-specific complementarity |
| TE | transposable element | a stretch of DNA capable of moving around the genome, either by excision (cut-and-paste transposons) or through an RNA intermediate (retro-elements) |
| VSR | viral suppressor of RNAi | any viral gene that inhibits host RNAi function |
| dsRNA | double-stranded RNA | |
| UTR | untranslated region | non-protein-coding regions at the 5′ and 3′ ends of an mRNA |
| endo-siRNA | endogenous siRNA | a short RNA (other than an miRNA) that is derived from the host genome, rather than and exogenous source (e.g. a virus) |
(a) The viRNA pathway
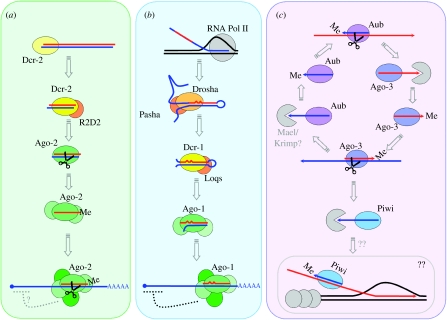
The best-studied role of RNAi in the immune system is as a direct defence against viruses (figure 1a; reviewed by Ding & Voinnet 2007). dsRNA from viruses is recognized by Dicer and cut (‘diced’) into short 21–24 nucleotide fragments called short interfering RNAs (siRNAs, also known as viRNAs when they are derived from viruses; Ding & Voinnet 2007). These are then loaded into an Argonaute-containing effector complex (RISC, the RNA-induced silencing complex; figure 1), and one strand of the viRNA is cleaved and degraded (reviewed by Tolia & Joshua-Tor 2007). The active Argonaute complex then cleaves (‘slices’) viral RNA with the complementary sequence to the viRNA (figure 1; Tolia & Joshua-Tor 2007).
Figure 1.
RNAi pathways in Drosophila. (a) The antiviral siRNA pathway. Dicer-2 cuts dsRNA into siRNAs, which are loaded into an Argonaute-containing RISC that targets RNA for degradation. (b) The miRNA pathway. Primary miRNAs are transcribed from the genome, processed by Drosha and Dicer-1 into mature miRNAs. These are loaded into the Argonaute-containing effector complex (RISC), which binds mRNAs and recruits additional factors to inhibit translation. (c) The ‘ping-pong’ model of TE silencing in the Drosophila germ line. Aubergine and Argonaute-3 (Piwi family Argonautes) alternately cleave sense (red) and antisense (blue) transcripts from TEs, guided by piRNAs generated in the other half of the cycle. Cleavage both inactivates the transcript and generates the 5′ end of a new piRNA. The new piRNA-precursor is bound by the partner Piwi family member and the 3′ end degraded and then modified by the addition of a methyl group (Me). The nuclear localization of Piwi suggests that it might mediate heterochromatin assembly (Klattenhoff & Theurkauf 2008). It is unknown how this occurs, but one possibility is that the active Piwi complex binds nascent TE transcripts to recruit heterochromatin factors (Grewal & Elgin 2007). Recently, a fourth pathway has been identified, which targets TE transcripts in both the soma and the germ line, using Dcr-2 and Ago-2 from the antiviral pathway, but Loqs from the miRNA pathway (Chung et al. 2008; Czech et al. 2008).
In addition to this core siRNA pathway (figure 1a), many eukaryotes are able to amplify the viRNA pool using a host-encoded RNA-dependent RNA polymerase (RdRp), and propagate the antiviral defence signal to other cells (Chapman & Carrington 2007; Jose & Hunter 2007). In Caenorhabditis elegans, a host RdRp binds the siRNA–target duplex and directly synthesizes secondary siRNAs, reportedly independently of additional Argonaute or Dicer activity (Pak & Fire 2007; Sijen et al. 2007). In plants, a host RdRp is also recruited by the siRNA–Argonaute complex, but in this case it synthesizes long dsRNAs which are then processed into siRNAs by a Dicer family protein (Himber et al. 2003; Moissiard et al. 2007). This is thought to amplify viral-derived sequences in both plants (Schwach et al. 2005) and nematodes (Schott et al. 2005; Wilkins et al. 2005), and in plants RdRp knockout mutants are more susceptible to viral infection (Deleris et al. 2006).
In plants and nematodes, RNAi initiated in one cell can be propagated to neighbouring cells, and even to more distant tissues (‘non-cell-autonomous RNAi’; Jose & Hunter 2007). In C. elegans, the movement of siRNAs between cells depends on SID-1 (systemic RNAi defective 1), which is thought to be a passive channel for cell-to-cell RNA movement (Feinberg & Hunter 2003). In Arabidopsis, 21 nt siRNAs are transported from cell to cell for short distances (10–15 cells), and an unknown signal (probably dsRNA) is loaded into the phloem to provide long-distance transport of the silencing signal (Brosnan et al. 2007; Dunoyer et al. 2007; Dunoyer & Voinnet 2008). It has been argued that the short RNAs are likely to need to be amplified by an RdRp for the RNAi signal to be effectively propagated between cells, and in Arabidopsis movement beyond 15 cells is indeed dependent on RdRp function. However, many animals encode SID-1 homologues but not RdRps (see §6), and there is evidence of non-cell-autonomous RNAi in several taxa that lack their own RdRp (Jose & Hunter 2007).
(b) The miRNA pathway
The microRNA (miRNA; figure 1b) pathway is a vital component of post-transcriptional control of gene expression in plants and animals (Carthew 2006; Bushati & Cohen 2007; Chapman & Carrington 2007; Pillai et al. 2007). MicroRNAs are encoded by the host genome and transcribed by RNA polymerase II as part of larger fold-back transcripts (primary miRNAs), which are processed in the nucleus by Drosha family members to form short stem-loops (pre-miRNAs), and then exported to the cytoplasm for processing by a Dicer family member to form the mature miRNA (figure 1b, reviewed by Du & Zamore 2005; Chapman & Carrington 2007; Pillai et al. 2007). As with viRNAs, miRNAs are loaded into an Argonaute-containing effector complex (RISC), where the complementary ‘passenger’ strand is lost. This miRNA effector complex can then interact with target messenger RNAs that contain complementary sequence to the miRNA. In plants, miRNAs are usually an exact match to their mRNA targets and cause the cleavage and degradation of the target transcript. In animals, miRNAs usually pair imperfectly with one or more targets in the 3′-UTR and do not induce mRNA endonucleolytic cleavage, but inhibit translation by a variety of mechanisms (Pillai et al. 2007). Although historically considered as a mechanism for the host to control its own gene expression, the miRNA pathway has more recently been shown to be important in interactions with viruses, with both hosts and viruses producing miRNAs that mediate host–virus interactions (see §4).
(c) The piRNA and endogenous siRNA pathways
A third group of RNAi-related pathways target transposable elements (TEs: transposons, retrotransposons and endogenous retroviruses; Aravin et al. 2007; Hartig et al. 2007; Matzke et al. 2007; Mevel-Ninio et al. 2007; Slotkin & Martienssen 2007; Chung et al. 2008; Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008). It was observed in as early as 1999 that several RNAi mutants of C. elegans had elevated rates of transposition (Ketting et al. 1999; Tabara et al. 1999), and it is now apparent that RNAi also plays an important role in determining the activity of TEs in many other eukaryotes (Vastenhouw & Plasterk 2004; Matzke et al. 2007; Zaratiegui et al. 2007; Czech et al. 2008). TE transcripts are processed by Argonaute and/or Dicer family members, both reducing transcript numbers and leading to epigenetic modification of genomic copies of the transposons, heterochromatin formation and reduced expression of TEs.
In Arabidopsis, the pathway that targets transposons involves Dicer-like 3 and Argonaute 4, and a class of 24–26 nucleotide siRNAs derived from the TE transcripts (Matzke et al. 2007; Zaratiegui et al. 2007). In Drosophila and vertebrate germ lines, TE silencing relies on Argonaute proteins in the Piwi family, and a class of short RNAs known as Piwi-interacting short RNAs (piRNAs; Aravin et al. 2007; Hartig et al. 2007; Klattenhoff & Theurkauf 2008). These are longer (24–30 nt) than viRNAs and miRNAs, and originate from a single-stranded RNA (ssRNA) precursor without the involvement of a Dicer protein. In Drosophila, the majority of piRNAs target repeat sequences such as TEs (also known as repeat-associated siRNAs or rasiRNAs), while in vertebrates most target other repetitive sequences and only a minority are complementary to TEs. A model has been proposed in which the suppression of TEs is maintained by a cyclic feedback process that alternately cleaves sense and antisense TE transcripts (‘ping-pong’: figure 1c; see Klattenhoff & Theurkauf (2008) for a review).
Very recently, it has also been shown that Drosophila (Chung et al. 2008; Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008) and mice (Watanabe et al. 2008) have an additional RNAi pathway that targets endogenous dsRNA such as that produced by overlapping 3′-UTRs from genes on opposite strands, long hair-pin fold-back sequences and TEs (Chung et al. 2008; Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008). In Drosophila, this pathway is similar to the antiviral pathway (figure 1a)—requiring both Ago-2 and Dcr-2—but it differs in that Loqs (from the miRNA pathway; figure 1b) takes the role of R2D2, at least for some transcripts (Chung et al. 2008; Czech et al. 2008). The resulting ‘endogenous siRNAs’ have similar size and properties to viral-derived siRNAs (viRNAs), and have a role in regulating the expression of several host genes in addition to affecting TE transcript levels.
3. The viRNA pathway and host–virus coevolution
Most organisms lack the adaptive immune system of vertebrates, and must rely on innate immunity to protect themselves from infection. The innate immune system often detects and destroys pathogens by recognizing families of molecules that are conserved across broad classes of pathogens (Medzhitov & Janeway 1997). In the case of bacteria and fungi, these molecules are primarily cell-surface polysaccharides, while viruses can be recognized if they produce dsRNA. dsRNA can be used to distinguish self from non-self because eukaryotes typically do not produce long stretches of dsRNA (other than that destined for processing by RNAi). However, many RNA viruses have double-stranded genomes, and even viruses with single-stranded genomes can produce dsRNA during replication, when viral RNA forms secondary structures, or when there are complementary sense and antisense replicative intermediates. DNA viruses can potentially also produce the dsRNA necessary for RNAi through secondary structure in their transcripts or sense–antisense transcript pairs (Ding & Voinnet 2007). It is now clear that the viRNA pathway (figure 1a) is an important component of innate antiviral immunity in plants, fungi, arthropods, nematodes and many animals.
(a) RNAi as innate antiviral immunity
The possibility that RNAi might have an antiviral function was first raised in plants when experimentally induced ‘gene silencing’ was found to provide resistance to viruses carrying an identical sequence (Lindbo et al. 1993). It was then identified as a natural component of innate antiviral immunity when viruses were found to naturally induce a similar response (Covey et al. 1997; Ratcliff et al. 1997). For animals, antiviral RNAi was first identified in Drosophila cell culture, where the beetle Nodamura virus FHV (flock house virus) acts as both an initiator and a target of the viRNA pathway (Li et al. 2002). It was later confirmed as biologically relevant defence outside of cell culture using the natural host–virus combination of O'nyong-nyong virus (alphavirus; Togaviridae) and Anopheles gambiae mosquitoes (Keene et al. 2004). RNAi has subsequently been identified as a component of antiviral immunity in adult Drosophila (Galiana-Arnoux et al. 2006; van Rij et al. 2006; Wang et al. 2006; Zambon et al. 2006) and in nematode worms (Lu et al. 2005; Schott et al. 2005; Wilkins et al. 2005). Very recently, RNAi has also been identified as an important antiviral defence in fungi (Segers et al. 2007; Hammond et al. 2008). Therefore, although several isolated lineages lack components of RNAi (e.g. Saccharomyces), it is likely that the viRNA antiviral pathway is a shared character of most eukaryotes.
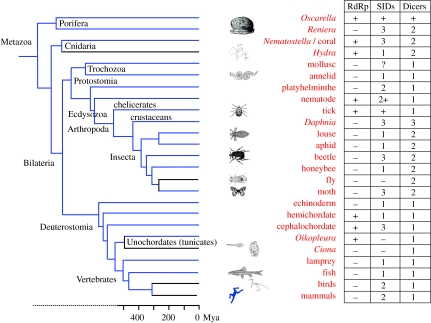
It remains an open question as to whether this pathway might be a component of antiviral immunity in vertebrates (Cullen 2006; Ding & Voinnet 2007; Haasnoot et al. 2007). Although vertebrates encode relevant genes (including Argonaute, Dicer and SID family members; figure 5), there is no strong evidence of a viRNA antiviral pathway, and these RNAi genes are used in other RNAi pathways (e.g. miRNA function and anti-TE function) that mediate host cellular processes.
Figure 5.
Evolutionary relationships of metazoans and their complements of selected RNAi genes. The presence of an RdRp and the number (where known) of SID and Dicer family members (Weinstock et al. 2006; Gordon & Waterhouse 2007) are indicated in table 1. The metazoan phylogeny is compiled from Bourlat et al. (2006), Delsuc et al. (2006) and Simionato et al. (2007). See figure 1 in the electronic supplementary material for sources.
(b) Viral suppression and evasion of the viRNA pathway
Viral suppression of the immune system is a widespread phenomenon (e.g. Katze et al. 2002; Marques & Carthew 2007). It is therefore not surprising that many viruses inhibit the viRNA pathway by expressing viral suppressors of RNAi (VSRs; Li & Ding 2006; Ding & Voinnet 2007). Very soon after the natural antiviral role of RNAi was revealed, the Hc-Pro protein of plant Potyviruses (Anandalakshmi et al. 1998; Brigneti et al. 1998; Kasschau & Carrington 1998) and the 2b protein of plant cucumoviruses (Li et al. 1999) were identified as VSRs. The first VSR identified in an animal virus was the B2 protein of the beetle virus FHV, which was initially found to suppress RNAi in plants, before being shown to play the same role in insects (Li et al. 2002). As of 2006, Li and Ding were able to review the function of more than 50 VSRs from over 30 viral genera. This list includes all categories of RNA viruses (negative sense, positive sense and double stranded) and some DNA viruses, in both plants and animals. Although the VSR function of some of these genes has been questioned (especially those of negative-sense ssRNA animal viruses, e.g. Blakqori et al. 2007), it now appears that active suppression of the host viRNA pathways is often an important component of infection, and nearly universal in plant–RNA virus interactions (Li & Ding 2006; Ding & Voinnet 2007). VSRs appear so important for successful viral infection that several viruses encode multiple VSRs, e.g. Citrus Tristeza Virus (a Closterovirus) encodes three (Lu et al. 2004) and several potyviruses encode two (Valli et al. 2008).
Many VSRs are dsRNA-binding proteins. However, while sequestering viRNA molecules away from the RNAi pathway is apparently the primary mechanism for some—such as P19 of tombusviruses, which shows a strong specificity for 21 nt dsRNA duplexes—it is now clear that there are a diverse range of other suppression mechanisms (Li & Ding 2006; Ding & Voinnet 2007). Indeed, not all VSRs are even proteins—at least two are thought to be RNAs, which may function by binding RNAi components in place of viRNAs. Different VSRs also target different components of RNAi (Ding & Voinnet 2007). For example, both Hc-Pro and B2 appear to inhibit viRNA processing from long dsRNA precursors by Dicer. Conversely, the 2b protein of cucumoviruses and the P0 protein of poleroviruses both target Argonaute family members; 2b by binding Arabidopsis Ago-1 directly to prevent the RISC complex from cleaving its target RNA (Zhang et al. 2006b), and P0 by targeting Argonaute family proteins for degradation (Baumberger et al. 2007).
In plants, where cell-to-cell propagation of RNAi is a key feature of the viRNA pathway (and where assays for non-cell-autonomous silencing are best developed), VSRs differentially affect cell-autonomous RNAi, non-cell-autonomous RNAi and the vascular transport of RNAi (Ding & Voinnet 2007; Jose & Hunter 2007). For example, if the P38 VSR is deleted from Turnip Crinkle Virus (Tombusviridae), the virus is still able to move locally from cell to cell, but cannot spread through the entire plant (Deleris et al. 2006). Similarly, the Potexvirus VSR P25 does not effectively inhibit cell-autonomous RNAi, but does block long distance movement (Guo & Ding 2002). By contrast, Hc-Pro from potyviruses and AC2 from African cassava mosaic virus (a DNA geminivirus) are VSRs that suppress cell-autonomous and short-distance RNAi, respectively, but not long-distance export (Mallory et al. 2003; Vanitharani et al. 2004; Trinks et al. 2005). A second strategy may be for a virus to manipulate the host's own control of the pathway rather than suppressing RNAi directly. For example, the Begomovirus VSR (AC2) appears to act as a transcriptional regulator that alters the expression of approximately 50 host genes, including some that might modify RNAi activity (Trinks et al. 2005).
Viruses may also reduce the effects of RNAi by preventing dsRNA from coming into contact with the RNAi pathway, or avoiding degradation once viRNAs have been produced. Viruses have several adaptations to reduce the production of dsRNA and shield the viral genome from degradation (Ahlquist 2002). For example, when single-stranded viruses replicate they must produce the complementary replication intermediate, and the nucleocapsid protein of negatively stranded viruses prevents the two strands from annealing and producing dsRNA. Because viRNAs are rarely derived uniformly from across the viral genome (e.g. Molnar et al. 2005; Deleris et al. 2006), it seems likely that certain regions of the genome are more vulnerable than others. The secondary structure of RNA can determine which fragments are processed to form viRNAs, and whether the active silencing complex has access to degrade the target RNA. For example, although viroids (subviral RNA pathogens that do not encode proteins) are a source of functional viRNAs, they are not cleaved by the silencing complex owing to their secondary structure (Itaya et al. 2007). When HIV is cultured with an artificial RNAi construct, its secondary structure evolves rapidly to block access by RNAi components, supporting the idea that this could be a viable mechanism of viral evasion, at least in principle (Westerhout et al. 2005).
(c) Viral evolution in response to host RNAi
There is clearly an evolutionary conflict between the viRNA pathway and its suppression by viruses. This has the potential to result in a never-ending arms race, where the RNAi pathway continually evolves new ways to escape suppression by VSRs, which leads to counteradaptations by the virus that restore suppression (Carthew 2006; Obbard et al. 2006). If this is the case, then VSRs might be expected to evolve much faster than other viral genes. This is certainly true when comparing VSRs across different viral families (Li & Ding 2006; Ding & Voinnet 2007). Li & Ding (2006) observed that VSRs from different viral families have essentially no structural similarity, even where they have similar functions. This is particularly striking when the dsRNA-binding motifs are compared across different VSRs: the dsRNA-binding domains from P19 (tombusviruses), P21 (Beet Yellows Virus) and B2 (FHV) each appear to have evolved different and entirely novel dsRNA-binding motifs (reviewed by Li & Ding 2006). Many VSRs also appear to have arisen relatively recently, and it has been argued that this is an indication of frequent viral adaptation to host RNAi (Li & Ding 2006).
If VSRs are engaged in an evolutionary arms race, then we might also expect to see the rapid sequence evolution of homologous VSRs within the same virus family. There is circumstantial evidence that this is the case (Li & Ding 2006). For example, the P1 VSR of potyviruses (Valli et al. 2006, 2008) is highly variable both in length and sequence, and shows evidence of recombination between potyviral lineages (Valli et al. 2007). Potyviruses typically have a second VSR called Hc-Pro, but in cucumber vein yellowing virus from the related genus Ipomovirus, Hc-Pro has been lost and its function appears to have been taken on by a tandemly duplicated copy of P1 (Valli et al. 2006). Similarly, in the Closteroviridae there is a considerable sequence diversity in the VSR P22 (Cuellar et al. 2008), and in one species (Sweet Potato Chlorotic Stunt Virus) some viral isolates have lost this suppressor altogether but still maintain VSR activity, presumably from a second locus (Cuellar et al. 2008).
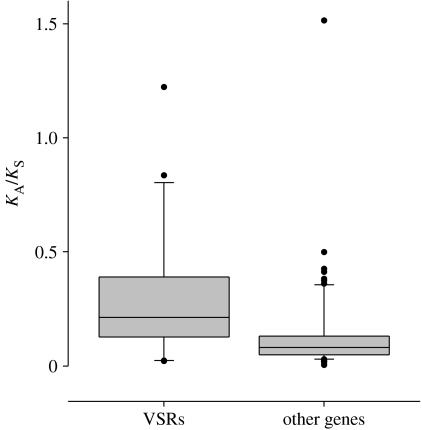
To test whether VSRs evolve faster than other viral genes, we analysed the rate of protein divergence for all genes in pairs of isolates from 17 ssRNA plant viruses with experimentally verified VSRs (figure 2). We found that protein divergence (relative to silent site divergence) between pairs of isolates was significantly higher for VSRs than for other genes, which is consistent with strong selection acting on VSRs. However, we would caution that the rapid evolution of VSRs could also be explained by low selective constraints (if most of the amino acid changing mutations have little effect on the function of the protein, they may be fixed at a high rate by random genetic drift), and more sophisticated analyses are needed to reject this possibility.
Figure 2.
The evolutionary rate of VSRs in positive sense ssRNA plant viruses. The rate of protein evolution (KA, non-synonymous sites) relative to the rate of neutral evolution (KS, synonymous sites) for genes taken from 17 closely related pairs of ssRNA plant viruses. For viral suppressors of RNAi (VSRs) mean KA/KS=0.29 (n=20), and for other genes mean KA/KS=0.14 (n=72). In 14 of the 17 viruses, the rate of VSR evolution was higher than the average rate of the other genes (p<0.01, sign test). Where possible, genome pairs comprised two isolates of the same viral taxon (see the electronic supplementary material for accession details).
(d) Subversion of the viRNA pathway
There is evidence that some viruses might have evolved to subvert the viRNA pathway for their own benefit (reviewed by Ding & Voinnet 2007). Several short RNAs derived from Cauliflower Mosaic Virus are complementary to Arabidopsis messenger RNAs, and downregulate those genes (Moissiard & Voinnet 2006). This may be coincidental rather than adaptive—since Arabidopsis is unlikely to be a natural host for cauliflower mosaic virus, this would require the short RNA targets to be very highly conserved between members of the Brassicaceae—but again illustrates the potential for viral evolution in response to host RNAi. It opens up interesting opportunities for the virus, as these host-manipulating viRNAs could in principle be exported from the site of infection and ‘prime’ the rest of the plant for viral invasion (Ding & Voinnet 2007).
(e) The evolution of host viRNA-pathway genes
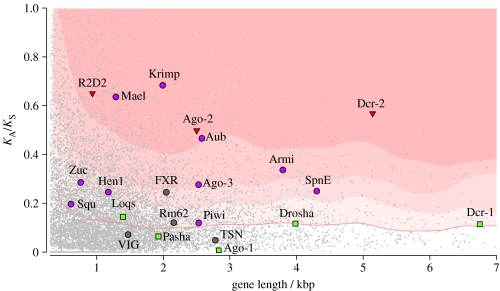
Because many viruses rely on suppressing the viRNAi pathway to infect their hosts, there will be a considerable advantage to mutations in viRNAi genes that escape this suppression. Furthermore, as the VSRs themselves are evolving quickly, the viRNAi pathway faces a continually changing array of suppressors to adapt to, and this could result in an evolutionary arms race driving the rapid evolution of both host and viral proteins. As predicted by this hypothesis, three key proteins in the viRNAi pathway of Drosophila (Dcr-2, Ago-2 and R2D2) are among the top 3 per cent of the most rapidly evolving in the entire genome (Obbard et al. 2006; figure 3). Proteins involved in the immune system often have a higher rate of evolution than the genome average (Hurst & Smith 1999; Schlenke & Begun 2003), but even compared with other immunity genes the viRNAi genes evolve exceptionally fast (Obbard et al. 2006). Indeed, no other functional class of immunity genes shows such consistently rapid evolution. These three viRNA genes have paralogs in the miRNA pathway (Dcr-1, Ago-1 and R3D1/Loqs; figure 1), which have similar molecular functions but no direct antiviral function. The miRNA-pathway genes evolve slowly, suggesting that it is the antiviral role of viRNAi genes that is causing the high rate of evolution.
Figure 3.
Evolutionary rate of RNAi-related genes in Drosophila. The rate of protein evolution (KA, non-synonymous sites) relative to the rate of neutral evolution (KS, synonymous sites) between D. melanogaster and D. simulans, plotted against aligned gene length. All 10 581 orthologous genes for which sequences are available are shown in grey, and 22 RNAi-pathway genes are shown as filled circles, squares and triangles. The smoothed median (red line) and the 75th, 85th and 95th percentiles (shaded background) are plotted for a sliding window (average 380 genes wide). RNAi-related genes evolve significantly more rapidly than the genome average (Wilcoxon signed-rank test, p<0.01). Six are in the fastest 5 per cent of genes (R2D2, Maelstrom, Krimper, Ago-2, Aubergine and Dcr-2), and a further 12 are in the upper 50 per cent. The piRNA pathway (purple circles) and viRNA pathway (red triangles) each evolve more rapidly than other genes (p<0.001 and p<0.01, respectively) but the miRNA pathway (green squares) does not (p=0.34). KA/KS was calculated by the method of Li et al. (1985). This analysis partially duplicates that of Obbard et al. (2006), but uses newer genome releases and an alternative estimator of KA/KS.
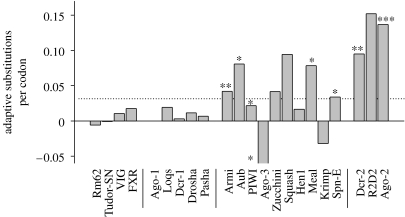
However, rapid protein evolution need not be caused by positive natural selection, but can also result from low selective constraints. If a gene is evolving neutrally, the ratio of non-synonymous (amino acid changing) to synonymous differences between species will be the same as the ratio for polymorphism within species (McDonald & Kreitman 1991). In the case of the viRNAi genes, the high amino acid divergence between species is not matched by the high levels of amino acid polymorphism, providing compelling evidence that selection has driven the rapid evolution of these genes (Obbard et al. 2006). Using this excess of amino acid divergence relative to polymorphism, we have estimated the number of amino acid differences between Drosophila melanogaster and its close relative Drosophila simulans that were fixed because they had a selective advantage (figure 4). The rate at which natural selection has fixed selectively advantageous amino acid substitutions in the viRNAi genes is 24-fold greater than their miRNA paralogs. For R2D2 and Dicer-2 these results have also been confirmed in a second study, using a different approach that compared the genome sequences of 12 species of Drosophila (Heger & Ponting 2007).
Figure 4.
The rate of adaptive evolution in Drosophila RNAi genes. The estimated number of adaptive substitutions per codon (as opposed to neutral substitutions fixed by random genetic drift) for RNAi-pathway genes was estimated using the method of Smith & Eyre-Walker (2002). The analysis uses the D. simulans polymorphism data from Obbard et al. (2006; Ago-1, Ago-2, Dcr-1, Dcr-2, Loqs, R2D2) and Begun et al. (2007; all other genes), together with the fixed differences between these datasets and the D. melanogaster genome. Negative estimates (Ago-3 and Krimper) arise due to the relatively large number of amino acid polymorphisms in these genes. Genes with individually significant McDonald–Kreitman tests (McDonald & Kreitman 1991) are indicated by asterisks (*p<0.05, **p<0.01, ***p<0.001, no correction for multiple tests). Note that the differences in sample size mean that the power of the test varies greatly between genes (details available in the electronic supplementary material).
4. The miRNA pathway and host–virus coevolution
It has recently become clear that the miRNA pathway can also play a role in host–virus interactions (Sarnow et al. 2006). Unlike the viRNA pathway, which primarily uses short RNAs derived from viruses as an immune defence, both host and virus sequences can be processed into miRNAs, and the miRNA pathway is not only antiviral but can also be exploited by viruses for their own benefit.
(a) Virus-encoded miRNAs
Virus-encoded miRNAs were first identified in the Epstein–Barr virus (EBV; Pfeffer et al. 2004) and have now been found in at least 12 mammalian viruses, spanning the three herpesvirus subfamilies, polyomaviruses and retroviruses (for a current listing see the miRBase Sequence Database; Griffiths-Jones et al. 2008) (for a review see Sarnow et al. 2006). The miRNA pathway (figure 1b) provides a powerful mechanism for fine tuning gene expression in the host, and viruses make use of this pathway to modulate their own gene expression. For example, in Simian Virus 40 (SV40), the viral miRNA miR-S1 is expressed late in infection and downregulates an early viral gene that is encoded on the opposite strand of the miRNA. This autoregulation of the viral gene (the viral T-antigen) by an miRNA is proposed to be important for enabling the virus to evade the host immune response (Sullivan et al. 2005).
It has been estimated that more than 50 per cent of the genes in herpesviruses and poxviruses could be devoted to manipulating the host immune system (Alcami & Koszinowski 2000) and viral miRNAs are an efficient way to do this as they only require a small amount of coding sequence in the genome, yet can have a large capacity for gene regulation as they can have multiple targets. This field is in its infancy and the actual number of targets and in vivo functions of virus-encoded miRNAs remains to be elucidated. However, a recent report demonstrates that a miRNA encoded by Human Cytomegalovirus (HCMV) can target the MHC class I-related chain B gene (Stern-Ginossar et al. 2007) and thereby protect HCMV-infected cells from lysis by natural killer cells. Kaposi's sarcoma-associated herpesvirus appears to use a different strategy to manipulate host function: it encodes a miRNA (termed mir-k12-11) that mimics the host-encoded miRNA mir-155, both having the same sequence in the seed region and sharing several targets (Gottwein et al. 2007; Skalsky et al. 2007). Some of these genes regulate cell growth and B-cell transformation, so the miRNA may be partly responsible for B-cell tumours in infected patients.
Given the apparent benefits to the virus, it might be expected that many viral lineages will be found to encode miRNAs. Of the 12 viruses reported to encode miRNAs, 10 belong to the herpesvirus family, one belongs to the polyomavirus family and one to the retrovirus family. This bias is at least partly attributable to the large volume of research aimed at herpesviral miRNAs (Grey et al. 2008) and additional sequencing efforts will be required to understand the generality of viral-miRNA existence. However, computational analyses reveal a low probability that pre-miRNA structures occur in human and mouse ssDNA and ssRNA viruses, and cloning efforts failed to identify miRNAs in RNA viruses including Yellow Fever Virus and Hepatitis C Virus (Pfeffer et al. 2005). Indeed, with the exception of miRNAs recently reported from HIV, viral miRNAs have only been found in dsDNA viruses. Why might this be? First, the primary miRNA-processing machinery (e.g. Drosha) is found in the nucleus, and thus is unlikely to be available to cytoplasmic viruses. Second, host-targeted viral miRNAs must match the host sequences, and may only have time to evolve where there is a long-term host–parasite association. Third, some viruses produce VSRs that can inhibit the miRNA pathway. These factors may explain the apparent bias for miRNA representation in herpesviruses, which replicate in the nucleus, tend to remain associated with the same host lineage for millions of years (Sharp 2002), and are not known to inhibit the miRNA pathway.
(b) Host-encoded miRNAs
Hosts can also use the miRNAs pathway as a defence against viruses by expressing miRNAs that target viral RNA (Lecellier et al. 2005; Pedersen et al. 2007). For example, Primate Foamy Virus 1 (PFV-1) mRNA is targeted by the human miRNA mir-32, which results in decreased PFV-1 replication (Lecellier et al. 2005). However, the physiological relevance of this might be questioned, since mir-32 may not be expressed in cell types relevant to PFV-1 replication (Cullen 2006), foamy viruses currently only occur as rare zoonoses in humans (Calattini et al. 2007) and it is not known whether the target sequence is conserved in related viruses that regularly infect humans. In another recent report, mice deficient in Dicer-1 were shown to be more susceptible to infection by Vesicular Stomatitis Virus (VSV) (Otsuka et al. 2007): two cellular miRNAs, mir-24 and mir-93, are predicted to target VSV genes encoding the viral RdRp and a polymerase cofactor, and mutation of the miRNA-binding sites in these genes rescues the growth defect in Dicer-1 deficient cells (Otsuka et al. 2007). It has also been suggested that miRNAs may be an effector in the classical vertebrate innate immune response (Pedersen et al. 2007). Activation of the interferon beta pathway in human cells infected by Hepatitis C Virus induces the expression of several host-encoded miRNAs, some of which have predicted targets in the virus (Pedersen et al. 2007). In support of their antiviral role, when these miRNAs are experimentally introduced they reproduce the antiviral effects of interferon beta on HCV, and the interferon defence is lost when they are experimentally removed (Pedersen et al. 2007). It appears that host miRNAs can also modulate cellular genes involved in the interferon response, as demonstrated for mir-146 (Taganov et al. 2006), and the expression of mir-146 is induced by the EBV-encoded latent membrane protein (LMP1) (Cameron et al. 2008), which suggests an intricate role of miRNAs in viral–host interactions.
Viruses may also exploit host-encoded miRNAs for their own advantage. For example, Hepatitis C Virus requires a host miRNA expressed in the liver, mir-122, which interacts with the 5′-non-coding region of the viral genome to increase replication (Jopling et al. 2005). Moreover, the host innate immune response downregulates mir-122, presumably reducing the viral replication (Pedersen et al. 2007).
(c) Host–virus coevolution in the miRNA pathway
In the viRNA pathway, short RNAs are derived from the virus itself and will thus always match the target. This means that there is no possibility of the viral genome evolving to avoid sequence-based recognition. This is clearly important, as when a ‘fixed’ siRNA sequence is experimentally applied (i.e. one that is exogenous, rather than derived from the virus), the targeted viral sequence rapidly evolves to avoid being targeted (Westerhout et al. 2005). By contrast, in the miRNA pathway both host- and virus-encoded miRNAs may have an opportunity to coevolve with the target sequences in their opponent. As viruses generally evolve faster, thanks to their larger population sizes, shorter generation times and higher mutation rates, they may often have the upper hand.
Host-encoded miRNAs involved in the control of gene expression are often conserved over long periods of evolutionary time in both plants (Zhang et al. 2006a; Willmann & Poethig 2007) and animals (Bartel 2004; Ruby et al. 2007). Superficially, this might suggest that miRNA sequences are not engaged in rapid arms race evolution. However, recent sequencing of Drosophila miRNAs, in conjunction with analysis of 12 Drosophila genomes, suggests that some miRNAs are evolutionarily young and can be frequently gained and lost (Lu et al. 2008), opening up the possibility that there may be a class of rapidly evolving miRNAs that could target and coevolve with viruses.
The VSRs of plant viruses typically suppress the miRNA pathway as well as the viRNA pathways, and viral suppression of the host miRNA functions can be a major cost of infection (Kasschau et al. 2003; Chapman et al. 2004). Similarly, many viruses of vertebrates encode genes that suppress both RNAi pathways, possibly as a by-product of suppressing the interferon response (Cullen 2006). Given the possibility of antiviral miRNAs, it now seems plausible that this is an adaptive strategy. However, because many viruses use the miRNA pathway for their own advantage, a trade-off may occur between a virus' ability to suppress RNAi, and its ability to use miRNAs to its own advantage. Such a trade-off might not occur in Drosophila, where there is a separation of the core miRNA and viRNA pathways (Kavi et al. 2005), and VSRs do not impede the miRNA pathway in vivo (Chou et al. 2007).
5. Heterochromatin, the piRNA pathway, and intragenomic conflict
(a) RNAi as a defence against TEs
TEs copy themselves throughout the genome, and can harm the host when they cause mutations or ectopic recombination (reviewed by Charlesworth et al. 1994). RNAi can act as a defence against TEs, both by degrading transcripts, and by preventing expression by heterochromatin formation (Grewal & Elgin 2007; Matzke et al. 2007; Slotkin & Martienssen 2007; Zaratiegui et al. 2007). Heterochromatin is usually strongly enriched for TEs (Zaratiegui et al. 2007), and although it is required for cellular functions such as the control of gene expression and successful chromosome segregation, it has been widely suggested that it originally evolved as an anti-TE defence (Grewal & Elgin 2007; Slotkin & Martienssen 2007; Zaratiegui et al. 2007).
The de novo initiation of heterochromatic silencing, including that targeted against TEs, seems almost universally mediated by RNAi-related pathways (Grewal & Elgin 2007; Zaratiegui et al. 2007), and in many organisms RNAi also plays a central role in its ongoing maintenance (Grewal & Elgin 2007). In Arabidopsis, more than half the endogenous siRNAs are derived from heterochromatic sequence, and a third of these target TEs and other repeats (Matzke et al. 2007). Here, the 24–26 nt siRNAs that target TEs are produced by a Dicer family member (DCL3), and then bound by an Ago-4-containing complex that targets the genomic copies of the TE for methylation and heterochromatin formation. A host RdRp (RDR2) is also required, suggesting that there may be an amplification step. Although it is not yet certain how this anti-TE RNAi is initiated, it has been suggested that dsRNA may be generated both by antisense TE transcripts expressed by host promoters, and by fold-back structures such as those resulting from inverted terminal repeats produced by some classes of TE (Slotkin & Martienssen 2007).
In vertebrates and Drosophila the piRNA pathway (figure 1c) appears to be a major regulator of TE activity in germ line-related cells (Aravin et al. 2007; Hartig et al. 2007). In Drosophila there are three Piwi-class Argonaute family proteins (Argonaute-3, Piwi and Aubergine) that are strongly expressed in the germ line. All three of these genes bind piRNAs that are primarily derived from TEs, and mutants show an elevated rate of transposition. Similarly, mutants in two of the three mouse Piwi homologues lead to increased expression of some TEs (Aravin et al. 2007; Carmell et al. 2007). Several more RNAi-related genes that are required for germ line TE silencing have been identified in the piRNA pathway of Drosophila (reviewed by Klattenhoff & Theurkauf 2008). The putative nucleases Zucchini and Squash are thought to be required for 3′-processing in the generation of mature piRNAs (Pane et al. 2007), and 3′-methylation by Hen1 stabilizes mature piRNAs (Horwich et al. 2007). Additionally, mutations in Spindle-E/Homeless and Armitage (which are thought to be helicases), Krimper (a tudor domain protein), and Maelstrom all disrupt piRNA formation and increase the activity of retrotransposons (Aravin et al. 2004; Vagin et al. 2006; Lim & Kai 2007).
In C. elegans, several RNAi-pathway genes are necessary for silencing DNA transposons in the germ line (Tabara et al. 1999; Sijen & Plasterk 2003), and numerous siRNAs are derived from TE sequences (Ruby et al. 2006). However, although Piwi family Argonautes are found in C. elegans and are required for germ line function, with the exception of the TE Tc3 (Das et al. 2008), the C. elegans Piwi-related genes (PRG-1 and PRG-2) do not appear to be required for the suppression of TE transposition (Batista et al. 2008).
Very recently, a second anti-TE RNAi pathway has been identified in Drosophila (Chung et al. 2008; Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008). This pathway uses Ago-2 and Dicer-2 from the viRNA pathway, and processes the TE transcripts into endogenous siRNAs in both somatic (Chung et al. 2008) and germ line (Czech et al. 2008) tissue. Although it has not yet been shown to affect rates of transposition, both Ago-2 and Dcr-2 knockouts display increased levels of TE transcript, strongly suggesting that this may be a second regulatory mechanism. Why two different RNAi pathways should be required to suppress TEs is unclear: it is possible that there is crosstalk between the pathways, and it has been hypothesized that the second pathway may be involved in the maintenance of heterochromatin in somatic tissue (Kawamura et al. 2008; Obbard & Finnegan 2008).
(b) Evolution of the piRNA pathway in Drosophila
In the germ line, the conflict between the host genome and TEs could drive an evolutionary arms race (Charlesworth & Langley 1989), in the same way as has been hypothesized for viruses (Obbard et al. 2006; Marques & Carthew 2007). In support of this, a recent analysis of the genome sequences of 12 species of Drosophila found that natural selection has driven the rapid evolution of both Krimper and Spindle-E (Heger & Ponting 2007). To examine the other piRNA-pathway genes, we have summarized the rate of protein evolution in several components of the piRNA pathway (figure 3), and have tested these genes for effects of positive selection using the DNA sequence polymorphism data from the genome sequences of several D. simulans strains (Begun et al. 2007) (figure 4). We found that Maelstrom, Krimper and Aubergine are all among the top 5 per cent of the most rapidly evolving genes in the genome, and Armitage, Aubergine, Piwi, Maelstrom and Spindle-E show evidence of positive selection (natural selection fixing advantageous amino acid substitutions; Krimper is not significant in this test, but does appear to have an extremely high level of amino acid polymorphism within D. simulans). This mirrors what is seen for the viRNA-pathway genes Ago-2, Dcr-2 and R2D2, and is in contrast to the miRNA-pathways genes that evolve very slowly (see §4).
6. The phylogenetic distribution of RNAi components
While the core machinery of the RNAi pathways (figure 1) is conserved in plants and animals (Cerutti & Casas-Mollano 2006; Murphy et al. 2008), a fundamental difference between the kingdoms has emerged from studies of signal amplification and non-cell-autonomous RNAi. In particular, not all animal lineages carry key genes implicated in signal amplification and non-cell-autonomous RNAi (figure 5), and these differences are reflected in the diversity of RNAi phenotypes among animals (reviewed by Jose & Hunter 2007). Given the importance of signal amplification for antiviral RNAi in plants, this might be expected to have significant implications for the contribution RNAi is able to make to innate immunity in animals.
The distribution of RdRps, which are thought to be required for signal amplification, is a striking example. With extensive genome data now available from all major metazoan groups, we have been able to identify this gene in lineages on each major branch of the bilateria, consistent with its presence in the metazoan ancestor (figure 5). Although the gene has been retained in some Porifera, the Cnidaria and some of the Protostomia (e.g. nematodes and the tick) and Deuterostomia (hemichordates, cephalochordate and a tunicate), in most of these groups, it has been independently lost from at least one representative. Among the lineages known to have lost the gene are the vertebrates and insects (Wassenegger & Krczal 2006; Gordon & Waterhouse 2007; Tomoyasu et al. 2008), as well as the others shown in figure 5. The RdRp genes found in all animal, fungal and plant genomes are characterized by a highly conserved domain (see figure 1 in the electronic supplementary material).
SID-1, which was first identified in nematodes and forms a channel for cell-to-cell movement of dsRNA, has no orthologue in plants, but it does in many animal lineages—with the notable exception of the dipterans (figure 5). Compared to RdRp, the gene is not only more widespread, but the number of genes in each metazoan group is more variable. In the arthropods, for example, a single orthologue of SID-1 is found in each of the bee (Weinstock et al. 2006), louse and aphid genomes (figure 5), while multiple (three) orthologues are found in the genomes of the moth Bombyx and the beetle Tribolium (Tomoyasu et al. 2008). Furthermore, the relationships of the insect SIDs do not match the phylogeny of the animals they are found in. The bee SID-1 protein clusters with one of three beetle and two of three moth SID homologues (fig. 12 in Weinstock et al. 2006), as well as the two vertebrate SID paralogues, one of which was lost in fish, but preserved in birds and mammals. This suggests that there were ancient duplications of this gene, and some of the resulting copies have subsequently been lost in some lineages. The variation in the number of SID-1 genes may explain why the effects of RNAi seem so variable in different species. For example, if RNAi is used experimentally to knock down gene expression, it can result in systemic silencing in moths and beetles (Baum et al. 2007; Mao et al. 2007), while in flies, which have no SID-1-like genes, transgene silencing has only a localized effect. A more recent study using multiple RNAi of all SID-1 orthologues in Tribolium has cast a doubt on whether these genes function in systemic RNAi at all (Tomoyasu et al. 2008). The basis for the varying, albeit limited, extent of systemic RNAi in arthropods therefore remains unexplained. Understanding the function of the different arthropod SID genes will be of great interest, as even in C. elegans the function of only two of the several SID genes is known (Jose & Hunter 2007; Winston et al. 2007).
Dicer genes also show considerable variation among eukaryotic lineages. In plants, for example, there are larger numbers—Arabidopsis has four Dicer-like genes—and these show some functional redundancy (reviewed by Ding & Voinnet 2007). For example, resistance phenotypes against some viral infections are seen in Arabidopsis only when multiple Dicers are knocked out, suggesting that they can substitute for each other (Deleris et al. 2006). In Arabidopsis, it appears that the Dicer DCL4 is a primary effector against ssRNA viruses, but that DCL2 substitutes when DCL4 is knocked out or suppressed, and DCL3 (which normally mediates chromatin modification) plays an important role in some viral infections, and in DCL2/DCL4 double mutants (Moissiard & Voinnet 2006; Ding & Voinnet 2007). This may suggest that, in plants at least, multiple paralogous viRNA-pathway genes have specialized functions, perhaps evolving in response to viral suppression of RNAi. In animals, one or two Dicers have been found in all genomes explored to date, with the exception of Daphnia (figure 5; McTaggart et al. in press). The vertebrates and nematodes have a single Dicer responsible for both siRNA and miRNA production. Genes for two Dicers have been found in first Drosophila, and now in the honeybee and Tribolium genomes (Weinstock et al. 2006; Tomoyasu et al. 2008).
Even more striking is the variability in the components of the RISC complexes. Although it is not always clear that they function within RNAi-related pathways, Argonaute family members, in particular, seem to be very variable in number (Hock & Meister 2008; Hutvagner & Simard 2008; Murphy et al. 2008)—Arabidopsis has 10, fission yeast (Schizosaccharomyces pombe) 1, Drosophila 5, and C. elegans in excess of 20. Why this should be is not clear. Although there is sometimes pathway specialization (Piwi-like Argonautes function in the piRNA pathway), there is also overlap between the pathways, and genes can be functionally redundant. In the nematode, there is discrimination in loading siRNAs or miRNAs onto RISCs after processing by its single Dicer (Jannot et al. 2008), so that short RNAs with different functional roles are associated with different members of the large family of Argonautes. The siRNAs are found bound to RDE-1, which has RNase H activity (Yigit et al. 2006); by contrast, miRNAs are bound to the Argonaute proteins ALG-1 and ALG-2 (Gu et al. 2007). Insects and vertebrates appear to show less discrimination in this respect. In Drosophila, miRNAs and siRNAs are generated by distinct Dicers but the duplex intermediates undergo independent sorting processes, one involving Dcr-2/R2D2 that favours loading of siRNAs into Ago-2 (Tomari et al. 2007), and may be loaded into Ago-1 or into Ago-2 (Okamura et al. 2004; Forstemann et al. 2007). Ago-1 has inefficient RNase H activity (Valencia-Sanchez et al. 2006), so that only Ago-2 is capable of cleaving target RNAs, where there is sufficient complementarity (Zeng et al. 2003). In humans, which also have only one Dicer, miRNAs may be loaded onto any of the four proteins Ago-1–4; only Ago-2 has RNase H activity (Meister & Tuschl 2004).
7. Outlook and prospects
Despite evolution and arms races being frequently mentioned in the RNAi literature, there are still few explicitly evolutionary studies of RNAi. In this final section, we highlight three interesting topics for future work.
(a) Specific immune responses
In this review, we have discussed how coevolution between hosts and parasites can take the form of an arms race, in which novel mutations that either increase resistance or overcome this resistance sweep through the host and parasite populations. However, coevolution can also act to maintain genetic variation in the resistance of individuals in host populations and infectivity of strains in populations (Woolhouse et al. 2002). This occurs because as the frequency of a resistance allele increases in the host population, the frequency of the pathogens it targets and hence its selective advantage can decrease. If the resistant allele confers susceptibility to other pathogen genotypes, or has some other fitness cost, there comes a point where it is at a selective disadvantage. An analogous process can occur within the parasite population. This results in negative frequency-dependent selection, and can maintain genetic variation in populations and promote complex dynamics, such as cycles in allele frequency.
It is possible that this form of coevolution could maintain variation in RNAi genes and viral populations. For example, because host-encoded miRNAs that target viral sequences are a form of specific immunity that is germ line encoded (as opposed to viRNAs, where immunity is acquired after infection), antiviral miRNAs will select for escape mutants in the viral population, and these escape mutants could in turn select for matching miRNAs. This could lead to specific ‘matching alleles’ in the host and viral population, in which each host allele confers resistance to some viral genotypes but susceptibility to others, resulting in the coevolutionary dynamics described above (Hamilton 1980) This could both maintain genetic variation in miRNAs and their viral targets, and lead to dynamic changes in the effectiveness of miRNA defences through time and space.
(b) Acquired immunity
Unlike the miRNA pathway, where immunity is germ line encoded and may take many generations to evolve, highly specific viRNA-based immunity can develop rapidly after exposure to the virus. In this respect, the viRNAi pathway is reminiscent of the acquired immune response of vertebrates, which is also highly specific and develops after infection. In vertebrates, acquired immunity can maintain a high diversity of antigens in the parasite population—hosts are most likely to be immune to common antigenic types, so there is an advantage to rare antigen alleles (Hughes 1991). A similar process can occur within an infected host, where mutations in antigens that escape the immune response may be favoured, leading to a turnover of antigens through time (Allen et al. 2000). Could such processes occur in response to the viRNAi pathway? Plants that have been infected by a virus can develop new growth that is free from viral infection and resistant to secondary infection due to RNAi targeted against the virus (Baulcombe 2004). If these hosts are exposed to a second viral strain, then this incoming strain may have an advantage if it is not recognized by viRNAs derived from the resident strain. This could potentially maintain genetic polymorphisms in the viral population. Similarly, there may be selection for mutants that escape recognition by the viRNA pathway within an infected host, by evolving new (and thus rare) sequences (Ding & Voinnet 2007). However, the acquired immune response of vertebrates and viRNA differ in the speed of the response, whether or not it is systemic, and the number of different epitopes or viRNAs produced from a pathogen, and these factors may all affect the outcome. These ideas could be tested both theoretically and experimentally.
(c) Why do RNAi-related genes evolve so fast?
In Drosophila, antiviral and anti-TE RNAi genes are among the most rapidly evolving genes in the genome (figures 4 and 5), and both viruses (Marques & Carthew 2007) and TEs (Charlesworth & Langley 1989) select for resistance in their hosts. However, while it is easy to see how the rapid evolution of antiviral genes might be driven by VSRs, it is less clear how TEs could impose such strong selection, as they are not known to suppress RNAi (although this possibility has been proposed, see Blumenstiel & Hartl 2005).
An alternative explanation for rapid evolution in the Drosophila piRNA pathway might be that some piRNA components also have antiviral function in addition to their anti-TE role. In support of this, some piRNA-pathway mutants appear to be more susceptible to a dsRNA virus (although these results await further experimental confirmation; Zambon et al. 2006), and there is an extensive overlap between the antiviral and anti-TE pathways in other organisms (e.g. Moissiard & Voinnet 2006; Brosnan et al. 2007). Moreover, components of the piRNA pathway might also interact with DNA viruses and retroviruses. First, DNA virus function often depends upon chromatin status (e.g. Jenkins et al. 2000) and in plants this can be mediated by RNAi (Bian et al. 2006). Second, the distinction between retroviruses and retrotransposons is a subtle one (retrotransposons are often considered ‘endogenous retroviruses’; e.g. Kim et al. 1994; Lecher et al. 1997) and in Drosophila retrotransposons are silenced by the piRNA pathway in the germ line (Mevel-Ninio et al. 2007; Pelisson et al. 2007), opening up the possibility that the piRNA pathway could also suppress ‘true’ retroviruses.
It is also possible that a factor other than viruses or TEs drives rapid adaptive evolution in both RNAi pathways. For example, both Ago-2 and Dcr-2 have other germ line functions in Drosophila (Deshpande et al. 2005; Jin & Xie 2007), and the Piwi family genes of both C. elegans (Batista et al. 2008) and Drosophila have non-TE-related germ line functions, such as mediating the silencing of other potentially ‘selfish’ sequences such as Stellate in Drosophila (Vagin et al. 2006; Aravin et al. 2007). Indeed, in general, the most rapidly evolving genes in Drosophila are those that regulate chromatin, mediate nuclear import and export, and are involved in meiosis and reproduction (Begun et al. 2007), and it may be that rapidly evolving RNAi genes are a part of this phenomenon. Thus, although viruses remain a prime candidate, it remains an open question as to whether viruses, TEs, other parasites, meiotic drive, sexual conflict or something else entirely has driven the adaptive evolution seen in RNAi-pathway genes.
Acknowledgements
We thank Andreas Heger for alignments used in figure 3, and Elizabeth Bayne, Alain Kohl, Mike Siva-Jothy and two anonymous referees for their helpful comments on an earlier version of the manuscript. D.J.O. was funded by the Wellcome Trust; F.M.J. was funded by the Wellcome Trust and the Royal Society; A.H.B. was funded by a Marie Curie Fellowship (EU) and a Thomas Work fellowship (CID, University of Edinburgh); K.H.J.G. received no specific funding for this work.
Footnotes
One contribution of 11 to a Theme Issue ‘Ecological immunology’.
Supplementary Material
Genbank accession numbers for the sequences used in figure 2
Alignment of sequences of RdRps from selected animal and other genomes
Numbers used for the MK tests shown in figure 4 of the main test
References
- Ahlquist P. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science. 2002;296:1270–1273. doi: 10.1126/science.1069132. doi:10.1126/science.1069132 [DOI] [PubMed] [Google Scholar]
- Alcami A., Koszinowski U.H. Viral mechanisms of immune evasion. Trends Microbiol. 2000;8:410–418. doi: 10.1016/S0966-842X(00)01830-8. doi:10.1016/S0966-842X(00)01830-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T.M., O'Connor D.H., Jing P.C., Dzuris J.L., Mothe B.R., Vogel T.U. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. doi:10.1038/35036559 [DOI] [PubMed] [Google Scholar]
- Anandalakshmi R., Pruss G.J., Ge X., Marathe R., Mallory A.C., Smith T.H. A viral suppressor of gene silencing in plants. Proc. Natl Acad. Sci. USA. 1998;95:13 079–13 084. doi: 10.1073/pnas.95.22.13079. doi:10.1073/pnas.95.22.13079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A.A., Klenov M.S., Vagin V.V., Bantignies F., Cavalli G., Gvozdev V.A. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol. Cell. Biol. 2004;24:6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. doi:10.1128/MCB.24.15.6742-6750.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A.A., Hannon G.J., Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. doi:10.1126/science.1146484 [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. doi:10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Batista P.J., et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. doi:10.1016/j.molcel.2008.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. doi:10.1038/nature02874 [DOI] [PubMed] [Google Scholar]
- Baum J.A., Bogaert T., Clinton W., Heck G.R., Feldmann P., Ilagan O. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007;25:1322–1326. doi: 10.1038/nbt1359. doi:10.1038/nbt1359 [DOI] [PubMed] [Google Scholar]
- Baumberger N., Tsai C.-H., Lie M., Havecker E., Baulcombe D.C. The polerovirus silencing suppressor P0 targets Argonaute proteins for degradation. Curr. Biol. 2007;17:1609. doi: 10.1016/j.cub.2007.08.039. doi:10.1016/j.cub.2007.08.039 [DOI] [PubMed] [Google Scholar]
- Begun D.J., Holloway A.K., Stevens K., Hillier L.W., Poh Y.P., Hahn M.W. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:2534–2559. doi: 10.1371/journal.pbio.0050310. doi:10.1371/journal.pbio.0050310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian X.Y., Rasheed M.S., Seemanpillai M.J., Rezaian M.A. Analysis of silencing escape of tomato leaf curl virus: an evaluation of the role of DNA methylation. Mol. Plant Microbe Interact. 2006;19:614–624. doi: 10.1094/MPMI-19-0614. doi:10.1094/MPMI-19-0614 [DOI] [PubMed] [Google Scholar]
- Blakqori G., Delhaye S., Habjan M., Blair C.D., Sanchez-Vargas I., Olson K.E. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J. Virol. 2007;81:4991–4999. doi: 10.1128/JVI.01933-06. doi:10.1128/JVI.01933-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstiel J.P., Hartl D.L. Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proc. Natl Acad. Sci. USA. 2005;102:15 965–15 970. doi: 10.1073/pnas.0508192102. doi:10.1073/pnas.0508192102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourlat S.J., Juliusdottir T., Lowe C.J., Freeman R., Aronowicz J., Kirschner M. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–88. doi: 10.1038/nature05241. doi:10.1038/nature05241 [DOI] [PubMed] [Google Scholar]
- Brigneti G., Voinnet O., Li W.X., Ji L.H., Ding S.W., Baulcombe D.C. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. doi:10.1093/emboj/17.22.6739 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Brosnan C.A., Mitter N., Christie M., Smith N.A., Waterhouse P.M., Carroll B.J. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc. Natl Acad. Sci. USA. 2007;104:14 741–14 746. doi: 10.1073/pnas.0706701104. doi:10.1073/pnas.0706701104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N., Cohen S.M. MicroRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. doi:10.1146/annurev.cellbio.23.090506.123406 [DOI] [PubMed] [Google Scholar]
- Calattini S., Betsem E.B.A., Froment A., Mauclere P., Tortevoye P., Schmitt C. Simian foamy virus transmission from apes to humans, rural Cameroon. Emerg. Infect. Dis. 2007;13:1314–1320. doi: 10.3201/eid1309.061162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron J.E., Yin Q.Y., Fewell C., Lacey M., McBride J., Wang X. Epstein–Barr virus latent membrane protein 1 induces cellular microRNA miR-146a, a modulator of lymphocyte signaling pathways. J. Virol. 2008;83:1946–1958. doi: 10.1128/JVI.02136-07. doi:10.1128/JVI.02136-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell M.A., Girard A., van de Kant H.J.G., Bourc'his D., Bestor T.H., de Rooij D.G., Hannon G.J. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. doi:10.1016/j.devcel.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Carthew R.W. Gene regulation by microRNAs. Curr. Opin. Genet. Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. doi:10.1016/j.gde.2006.02.012 [DOI] [PubMed] [Google Scholar]
- Cerutti H., Casas-Mollano J.A. On the origin and functions of RNA-mediated silencing: from protists to man. Curr. Genet. 2006;50:81–99. doi: 10.1007/s00294-006-0078-x. doi:10.1007/s00294-006-0078-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.J., Carrington J.C. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. doi:10.1038/nrg2179 [DOI] [PubMed] [Google Scholar]
- Chapman E.J., Prokhnevsky A.I., Gopinath K., Dolja V.V., Carrington J.C. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 2004;18:1179–1186. doi: 10.1101/gad.1201204. doi:10.1101/gad.1201204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Langley C.H. The population-genetics of Drosophila transposable elements. Annu. Rev. Genet. 1989;23:251–287. doi: 10.1146/annurev.ge.23.120189.001343. doi:10.1146/annurev.ge.23.120189.001343 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Sniegowski P., Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. doi:10.1038/371215a0 [DOI] [PubMed] [Google Scholar]
- Chou Y.-t., Bergin T., Fabien L., Lai E.C. Transgenic inhibitors of RNA interference in Drosophila. Fly. 2007;1:311–316. doi: 10.4161/fly.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W.-J., Okamura K., Martin R., Lai E.C. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. doi:10.1016/j.cub.2008.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey S.N., AlKaff N.S., Langara A., Turner D.S. Plants combat infection by gene silencing. Nature. 1997;385:781–782. doi:10.1038/385781a0 [Google Scholar]
- Cuellar W.J., Tairo F., Kreuze J.F., Valkonen J.P.T. Analysis of gene content in sweet potato chlorotic stunt virus RNA1 reveals the presence of the p22 RNA silencing suppressor in only a few isolates: implications for viral evolution and synergism. J. Gen. Virol. 2008;89:573–582. doi: 10.1099/vir.0.83471-0. doi:10.1099/vir.0.83471-0 [DOI] [PubMed] [Google Scholar]
- Cullen B.R. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat. Immunol. 2006;7:563–567. doi: 10.1038/ni1352. doi:10.1038/ni1352 [DOI] [PubMed] [Google Scholar]
- Czech B., et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:729–731. doi: 10.1038/nature07007. doi:10.1038/nature07007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P.P., et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. doi:10.1016/j.molcel.2008.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins R., Krebs J.R. Arms races between and within species. Proc. R. Soc. B. 1979;205:489–511. doi: 10.1098/rspb.1979.0081. doi:10.1098/rspb.1979.0081 [DOI] [PubMed] [Google Scholar]
- Deleris A., Gallego-Bartolome J., Bao J.S., Kasschau K.D., Carrington J.C., Voinnet O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. doi:10.1126/science.1128214 [DOI] [PubMed] [Google Scholar]
- Delsuc F., Brinkmann H., Chourrout D., Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. doi:10.1038/nature04336 [DOI] [PubMed] [Google Scholar]
- Deshpande G., Calhoun G., Schedl P. Drosophila Argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Gene Dev. 2005;19:1680–1685. doi: 10.1101/gad.1316805. doi:10.1101/gad.1316805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.-W., Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413. doi: 10.1016/j.cell.2007.07.039. doi:10.1016/j.cell.2007.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T.T., Zamore P.D. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. doi:10.1242/dev.02070 [DOI] [PubMed] [Google Scholar]
- Dunoyer P., Himber C., Ruiz-Ferrer V., Alioua A., Voinnet O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat. Genet. 2007;39:848–856. doi: 10.1038/ng2081. doi:10.1038/ng2081 [DOI] [PubMed] [Google Scholar]
- Dunoyer, P. & Voinnet, O. 2008 Mixing and matching: the essence of plant systemic silencing? Trends Genet. 24, 151–154. (doi:10.1016/j.tig.2008.01.005) [DOI] [PubMed]
- Feinberg E.H., Hunter C.P. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–1547. doi: 10.1126/science.1087117. doi:10.1126/science.1087117 [DOI] [PubMed] [Google Scholar]
- Forstemann K., Horwich M.D., Wee L., Tomari Y., Zamore P.D. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by Dicer-1. Cell. 2007;130:287. doi: 10.1016/j.cell.2007.05.056. doi:10.1016/j.cell.2007.05.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiana-Arnoux D., Dostert C., Schneemann A., Hoffmann J.A., Imler J.L. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat. Immunol. 2006;7:590–597. doi: 10.1038/ni1335. doi:10.1038/ni1335 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M., et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. doi:10.1126/science.1157396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon K.H.J., Waterhouse P.M. RNAi for insect-proof plants. Nat. Biotechnol. 2007;25:1231–1232. doi: 10.1038/nbt1107-1231. doi:10.1038/nbt1107-1231 [DOI] [PubMed] [Google Scholar]
- Gottwein E., Mukherjee N., Sachse C., Frenzel C., Majoros W.H., Chi J.T.A. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–U1017. doi: 10.1038/nature05992. doi:10.1038/nature05992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S.I.S., Elgin S.C.R. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. doi:10.1038/nature05914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F., Hook L., Nelson J. The functions of herpesvirus-encoded microRNAs. Med. Microbiol. Immunol. 2008;197:261–267. doi: 10.1007/s00430-007-0070-1. doi:10.1007/s00430-007-0070-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. doi:10.1093/nar/gkm952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S.G., Pak J., Barberan-Soler S., Ali M., Fire A., Zahler A.M. Distinct ribonucleoprotein reservoirs for microRNA and siRNA populations in C. elegans. RNA. 2007;13:1492–1504. doi: 10.1261/rna.581907. doi:10.1261/rna.581907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H.S., Ding S.W. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 2002;21:398–407. doi: 10.1093/emboj/21.3.398. doi:10.1093/emboj/21.3.398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot J., Westerhout E.M., Berkhout B. RNA interference against viruses: strike and counterstrike. Nat. Biotechnol. 2007;25:1435–1443. doi: 10.1038/nbt1369. doi:10.1038/nbt1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton W.D. Sex versus non-sex versus parasite. Oikos. 1980;35:282–290. doi:10.2307/3544435 [Google Scholar]
- Hammond T.M., Andrewski M.D., Roossinck M.J., Keller N.P. Aspergillus mycoviruses are targets and suppressors of RNA silencing. Eukaryot. Cell. 2008;7:350–357. doi: 10.1128/EC.00356-07. doi:10.1128/EC.00356-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig J.V.a., Tomari Y., Forstemann K. piRNAs the ancient hunters of genome invaders. Gene Dev. 2007;21:1707–1713. doi: 10.1101/gad.1567007. doi:10.1101/gad.1567007 [DOI] [PubMed] [Google Scholar]
- Heger A., Ponting C.P. Evolutionary rate analyses of orthologs and paralogs from 12 Drosophila genomes. Genome Res. 2007;17:1837–1849. doi: 10.1101/gr.6249707. doi:10.1101/gr.6249707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himber C., Dunoyer P., Moissiard G., Ritzenthaler C., Voinnet O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. doi:10.1093/emboj/cdg431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock J., Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. doi:10.1186/gb-2008-9-2-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich M.D., Li C.J., Matranga C., Vagin V., Farley G., Wang P., Zamore P.D. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. doi:10.1016/j.cub.2007.06.030 [DOI] [PubMed] [Google Scholar]
- Hughes A.L. Circumsporozoite protein genes of malaria parasites (Plasmodium spp.)—evidence for positive selection on immunogenic regions. Genetics. 1991;127:345–353. doi: 10.1093/genetics/127.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst L.D., Smith N.G.C. Do essential genes evolve slowly? Curr. Biol. 1999;9:747–750. doi: 10.1016/s0960-9822(99)80334-0. doi:10.1016/S0960-9822(99)80334-0 [DOI] [PubMed] [Google Scholar]
- Hutvagner G., Simard M.J. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22. doi: 10.1038/nrm2321. doi:10.1038/nrm2321 [DOI] [PubMed] [Google Scholar]
- Itaya A., et al. A structured viroid RNA serves as a substrate for Dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J. Virol. 2007;81:2980–2994. doi: 10.1128/JVI.02339-06. doi:10.1128/JVI.02339-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannot G., Boisvert M.E.L., Banville I.H., Simard M.J. Two molecular features contribute to the Argonaute specificity for the microRNA and RNAi pathways in C. elegans. RNA. 2008;14:829–835. doi: 10.1261/rna.901908. doi:10.1261/rna.901908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins P.J., Binne U.K., Farrell P.J. Histone acetylation and reactivation of Epstein–Barr virus from latency. J. Virol. 2000;74:710–720. doi: 10.1128/jvi.74.2.710-720.2000. doi:10.1128/JVI.74.2.710-720.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z.G., Xie T. Dcr-1 maintains Drosophila ovarian stem cells. Curr. Biol. 2007;17:539–544. doi: 10.1016/j.cub.2007.01.050. doi:10.1016/j.cub.2007.01.050 [DOI] [PubMed] [Google Scholar]
- Jopling C.L., Yi M.K., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. doi:10.1126/science.1113329 [DOI] [PubMed] [Google Scholar]
- Jose A.M., Hunter C.P. Transport of sequence-specific RNA interference information between cells. Annu. Rev. Genet. 2007;41:305–330. doi: 10.1146/annurev.genet.41.110306.130216. doi:10.1146/annurev.genet.41.110306.130216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau K.D., Carrington J.C. A counter defensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. doi:10.1016/S0092-8674(00)81614-1 [DOI] [PubMed] [Google Scholar]
- Kasschau K.D., Xie Z.X., Allen E., Llave C., Chapman E.J., Krizan K.A. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. doi:10.1016/S1534-5807(03)00025-X [DOI] [PubMed] [Google Scholar]
- Katze M.G., He Y.P., Gale M. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2002;2:675–687. doi: 10.1038/nri888. doi:10.1038/nri888 [DOI] [PubMed] [Google Scholar]
- Kavi H.H., Fernandez H.R., Xie W., Birchler J.A. RNA silencing in Drosophila. FEBS Lett. 2005;579:5940. doi: 10.1016/j.febslet.2005.08.069. doi:10.1016/j.febslet.2005.08.069 [DOI] [PubMed] [Google Scholar]
- Kawamura Y., Saito K., Kin T., Ono Y., Asai K., Sunohara T., Okada T.N., Siomi M.C., Siomi H. Drosophila endogenous small RNAs bind to Argonaute-2 in somatic cells. Nature. 2008;453:753–797. doi: 10.1038/nature06938. doi:10.1038/nature06938 [DOI] [PubMed] [Google Scholar]
- Keene K.M., Foy B.D., Sanchez-Vargas I., Beaty B.J., Blair C.D., Olson K.E. RNA interference acts as a natural antiviral response to O'nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc. Natl Acad. Sci. USA. 2004;101:17 240–17 245. doi: 10.1073/pnas.0406983101. doi:10.1073/pnas.0406983101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting R.F., Haverkamp T.H.A. Plasterk RHA mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. doi:10.1016/S0092-8674(00)81645-1 [DOI] [PubMed] [Google Scholar]
- Kim A., Terzian C., Santamaria P., Pelisson A., Prudhomme N., Bucheton A. Retroviruses in invertebrates—the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 1994;91:1285–1289. doi: 10.1073/pnas.91.4.1285. doi:10.1073/pnas.91.4.1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C., Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. doi:10.1242/dev.006486 [DOI] [PubMed] [Google Scholar]
- Lecellier C.H., Dunoyer P., Arar K., Lehmann-Che J., Eyquem S., Himber C., Saib A., Voinnet O. A cellular MicroRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. doi:10.1126/science.1108784 [DOI] [PubMed] [Google Scholar]
- Lecher P., Bucheton A., Pelisson A. Expression of the Drosophila retrovirus gypsy as ultrastructurally detectable particles in the ovaries of flies carrying a permissive flamenco allele. J. Gen. Virol. 1997;78:2379–2388. doi: 10.1099/0022-1317-78-9-2379. [DOI] [PubMed] [Google Scholar]
- Li F., Ding S.-W. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. doi:10.1146/annurev.micro.60.080805.142205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.H., Wu C.I., Luo C.C.A. New method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol. Biol. Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- Li H.W., Lucy A.P., Guo H.S., Li W.X., Ji L.H., Wong S.M. Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J. 1999;18:2683–2691. doi: 10.1093/emboj/18.10.2683. doi:10.1093/emboj/18.10.2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.W., Li W.X., Ding S.W. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. doi:10.1126/science.1070948 [DOI] [PubMed] [Google Scholar]
- Lim A.K., Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl Acad. Sci. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. doi:10.1073/pnas.0701920104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbo J.A., Silvarosales L., Proebsting W.M., Dougherty W.G. Induction of a highly specific antiviral state in transgenic plants—implications for regulation of gene-expression and virus-resistance. Plant Cell. 1993;5:1749–1759. doi: 10.1105/tpc.5.12.1749. doi:10.1105/tpc.5.12.1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H.F., Zhou B., Liu G., Chen C.Z. Micromanagement of the immune system by microRNAs. Nat. Rev. Immunol. 2008;8:120–130. doi: 10.1038/nri2252. doi:10.1038/nri2252 [DOI] [PubMed] [Google Scholar]
- Lu J., Shen Y., Wu Q., Kumar S., He B., Shi S. The birth and death of microRNA genes in Drosophila. Nat. Genet. 2008;40:351. doi: 10.1038/ng.73. doi:10.1038/ng.73 [DOI] [PubMed] [Google Scholar]
- Lu R., Folimonov A., Shintaku M., Li W.X., Falk B.W., Dawson W.O. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl Acad. Sci. USA. 2004;101:15 742–15 747. doi: 10.1073/pnas.0404940101. doi:10.1073/pnas.0404940101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Maduro M., Li F., Li H.W., Broitman-Maduro G., Li W.X. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436:1040–1043. doi: 10.1038/nature03870. doi:10.1038/nature03870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Mlotshwa S., Bowman L.H., Vance V.B. The capacity of transgenic tobacco to send a systemic RNA silencing signal depends on the nature of the inducing transgene locus. Plant J. 2003;35:82–92. doi: 10.1046/j.1365-313x.2003.01785.x. doi:10.1046/j.1365-313X.2003.01785.x [DOI] [PubMed] [Google Scholar]
- Mao Y.B., Cai W.J., Wang J.W., Hong G.J., Tao X.Y., Wang L.J., Huang Y-P., Chen X-Y. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 2007;25:1307–1313. doi: 10.1038/nbt1352. doi:10.1038/nbt1352 [DOI] [PubMed] [Google Scholar]
- Marques J.T., Carthew R.W. A call to arms: coevolution of animal viruses and host innate immune responses. Trends Genet. 2007;23:359. doi: 10.1016/j.tig.2007.04.004. doi:10.1016/j.tig.2007.04.004 [DOI] [PubMed] [Google Scholar]
- Matranga C., Zamore P.D. Small silencing RNAs. Curr. Biol. 2007;17:R789–R793. doi: 10.1016/j.cub.2007.07.014. doi:10.1016/j.cub.2007.07.014 [DOI] [PubMed] [Google Scholar]
- Matzke M., Kanno T., Huettel B., Daxinger L., Matzke A.J.M. Targets of RNA-directed DNA methylation. Curr. Opin. Plant Biol. 2007;10:512–519. doi: 10.1016/j.pbi.2007.06.007. doi:10.1016/j.pbi.2007.06.007 [DOI] [PubMed] [Google Scholar]
- McDonald J.H., Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. doi:10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- McTaggart S.J., Conlon C., Colbourne J.K., Blaxter M.L., Little T.J. The components of the Daphnia pulex immune system as revealed by complete genome sequencing. BMC Genomics. In press. doi: 10.1186/1471-2164-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R., Janeway C.A. Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. doi:10.1016/S0092-8674(00)80412-2 [DOI] [PubMed] [Google Scholar]
- Meister G., Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343. doi: 10.1038/nature02873. doi:10.1038/nature02873 [DOI] [PubMed] [Google Scholar]
- Mevel-Ninio M., Pelisson A., Kinder J., Campos A.R., Bucheton A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics. 2007;175:1615–1624. doi: 10.1534/genetics.106.068106. doi:10.1534/genetics.106.068106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissiard G., Parizotto E.A., Himber C., Voinnet O. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA. 2007;13:1268–1278. doi: 10.1261/rna.541307. doi:10.1261/rna.541307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissiard G., Voinnet O. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc. Natl Acad. Sci. USA. 2006;103:19 593–19 598. doi: 10.1073/pnas.0604627103. doi:10.1073/pnas.0604627103 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Molnar A., Csorba T., Lakatos L., Varallyay E., Lacomme C., Burgyan J. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 2005;79:7812–7818. doi: 10.1128/JVI.79.12.7812-7818.2005. doi:10.1128/JVI.79.12.7812-7818.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D., Dancis B., Brown J.R. The evolution of core proteins involved in microRNA biogenesis. BMC Evol. Biol. 2008;8:92. doi: 10.1186/1471-2148-8-92. doi:10.1186/1471-2148-8-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard D.J., Finnegan D.J. RNA interference: endogenous siRNAs derived from transposable elements. Curr. Biol. 2008;18:R561–R563. doi: 10.1016/j.cub.2008.05.035. doi:10.1016/j.cub.2008.05.035 [DOI] [PubMed] [Google Scholar]
- Obbard D.J., Jiggins F.M., Halligan D.L., Little T.J. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 2006;16:580–585. doi: 10.1016/j.cub.2006.01.065. doi:10.1016/j.cub.2006.01.065 [DOI] [PubMed] [Google Scholar]
- Okamura K., Ishizuka A., Siomi H., Siomi M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. doi:10.1101/gad.1210204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Chung W.-J., Ruby J.G., Guo H., Bartel D.P., Lai E.C. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. doi:10.1038/nature07015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M., Jing Q., Georgel P., New L., Chen J.M., Mols J. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. doi:10.1016/j.immuni.2007.05.014 [DOI] [PubMed] [Google Scholar]
- Pak J., Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. doi:10.1126/science.1132839 [DOI] [PubMed] [Google Scholar]
- Pane A., Wehr K., Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. doi:10.1016/j.devcel.2007.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen I.M., Cheng G., Wieland S., Volinia S., Croce C.M., Chisari F.V., David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–U913. doi: 10.1038/nature06205. doi:10.1038/nature06205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A., Sarot E., Payen-Groschene G., Bucheton A.A. Novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J. Virol. 2007;81:1951–1960. doi: 10.1128/JVI.01980-06. doi:10.1128/JVI.01980-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S., et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. doi:10.1126/science.1096781 [DOI] [PubMed] [Google Scholar]
- Pfeffer S., et al. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. doi:10.1038/nmeth746 [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. doi:10.1016/j.tcb.2006.12.007 [DOI] [PubMed] [Google Scholar]
- Ratcliff F., Harrison B.D., Baulcombe D.C. A similarity between viral defense and gene silencing in plants. Science. 1997;276:1558–1560. doi: 10.1126/science.276.5318.1558. doi:10.1126/science.276.5318.1558 [DOI] [PubMed] [Google Scholar]
- Roger A.J., Hug L.A. The origin and diversification of eukaryotes: problems with molecular phylogenetics and molecular clock estimation. Phil. Trans. R. Soc. B. 2006;361:1039–1054. doi: 10.1098/rstb.2006.1845. doi:10.1098/rstb.2006.1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Jan C., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. doi:10.1016/j.cell.2006.10.040 [DOI] [PubMed] [Google Scholar]
- Ruby J.G., Stark A., Johnston W.K., Kellis M., Bartel D.P., Lai E.C. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. doi:10.1101/gr.6597907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow P., Jopling C.L., Norman K.L., Schutz S., Wehner K.A. MicroRNAs: expression, avoidance and subversion by vertebrate viruses. Nat. Rev. Microbiol. 2006;4:651–659. doi: 10.1038/nrmicro1473. doi:10.1038/nrmicro1473 [DOI] [PubMed] [Google Scholar]
- Schlenke T.A., Begun D.J. Natural selection drives Drosophila immune system evolution. Genetics. 2003;164:1471–1480. doi: 10.1093/genetics/164.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott D.H., Cureton D.K., Whelan S.P., Hunter C.P. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 2005;102:18 420–18 424. doi: 10.1073/pnas.0507123102. doi:10.1073/pnas.0507123102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach F., Vaistij F.E., Jones L., Baulcombe D.C. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005;138:1842–1852. doi: 10.1104/pp.105.063537. doi:10.1104/pp.105.063537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers G.C., Zhang X.M., Deng F.Y., Sun Q.H., Nuss D.L. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl Acad. Sci. USA. 2007;104:12 902–12 906. doi: 10.1073/pnas.0702500104. doi:10.1073/pnas.0702500104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M. Origins of human virus diversity. Cell. 2002;108:305. doi: 10.1016/s0092-8674(02)00639-6. doi:10.1016/S0092-8674(02)00639-6 [DOI] [PubMed] [Google Scholar]
- Sijen T., Plasterk R.H.A. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–314. doi: 10.1038/nature02107. doi:10.1038/nature02107 [DOI] [PubMed] [Google Scholar]
- Sijen T., Steiner F.A., Thijssen K.L., Plasterk R.H.A. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. doi:10.1126/science.1136699 [DOI] [PubMed] [Google Scholar]
- Simionato E., Ledent V., Richards G., Thomas-Chollier M., Kerner P., Coornaert D., Degnan B.M., Vervoort M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol. Biol. 2007;7:1–18. doi: 10.1186/1471-2148-7-33. doi:10.1186/1471-2148-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky R.L., Samols M.A., Plaisance K.B., Boss I.W., Riva A., Lopez M.C., Baker H.V., Renne R. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 2007;81:12 836–12 845. doi: 10.1128/JVI.01804-07. doi:10.1128/JVI.01804-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R.K., Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007;8:272. doi: 10.1038/nrg2072. doi:10.1038/nrg2072 [DOI] [PubMed] [Google Scholar]
- Smith N.G.C., Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. doi:10.1038/4151022a [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N., et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. doi:10.1126/science.1140956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan C.S., Grundhoff A.T., Tevethia S., Pipas J.M. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. doi:10.1038/nature03576 [DOI] [PubMed] [Google Scholar]
- Tabara H., Sarkissian M., Kelly W.G., Fleenor J., Grishok A., Timmons L., Fire A., Mello C.C. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. doi:10.1016/S0092-8674(00)81644-X [DOI] [PubMed] [Google Scholar]
- Taganov K.D., Boldin M.P., Chang K.J., Baltimore D. NF kappa B-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl Acad. Sci. USA. 2006;103:12 481–12 486. doi: 10.1073/pnas.0605298103. doi:10.1073/pnas.0605298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolia N.H., Joshua-Tor L. Slicer and the argonautes. Nat. Chem. Biol. 2007;3:36–43. doi: 10.1038/nchembio848. doi:10.1038/nchembio848 [DOI] [PubMed] [Google Scholar]
- Tomari Y., Du T., Zamore P.D. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. doi:10.1016/j.cell.2007.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu Y., Miller S.C., Tomita S., Schoppmeier M., Grossmann D., Bucher G. Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol. 2008;9:R10. doi: 10.1186/gb-2008-9-1-r10. doi:10.1186/gb-2008-9-1-r10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinks D., Rajeswaran R., Shivaprasad P.V., Akbergenov R., Oakeley E.J., Veluthambi K. Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J. Virol. 2005;79:2517–2527. doi: 10.1128/JVI.79.4.2517-2527.2005. doi:10.1128/JVI.79.4.2517-2527.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin V.V., Sigova A., Li C.J., Seitz H., Gvozdev V., Zamore P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. doi:10.1126/science.1129333 [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez M.A., Liu J., Hannon G.J., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. doi:10.1101/gad.1399806 [DOI] [PubMed] [Google Scholar]
- Valli A., Martin-Hernandez A.M., Lopez-Moya J.J., Garcia J.A. RNA silencing suppression by a second copy of the P1 serine protease of cucumber vein yellowing Ipomovirus, a member of the family Potyviridae that lacks the cysteine protease HCPro. J. Virol. 2006;80:10 055–10 063. doi: 10.1128/JVI.00985-06. doi:10.1128/JVI.00985-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli A., Lopez-Moya J.J., Garcia J.A. Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae. J. Gen. Virol. 2007;88:1016–1028. doi: 10.1099/vir.0.82402-0. doi:10.1099/vir.0.82402-0 [DOI] [PubMed] [Google Scholar]
- Valli A., Dujovny G., Garcia J.A. Protease activity, self interaction, and small interfering RNA binding of the silencing suppressor P1b from cucumber vein yellowing ipomovirus. J. Virol. 2008;82:974–986. doi: 10.1128/JVI.01664-07. doi:10.1128/JVI.01664-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij R.P., Saleh M-C., Berry B., Foo C., Houk A., Antoniewski C., Andino R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. doi:10.1101/gad.1482006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanitharani R., Chellappan P., Pita J.S., Fauquet C.M. Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 2004;78:9487–9498. doi: 10.1128/JVI.78.17.9487-9498.2004. doi:10.1128/JVI.78.17.9487-9498.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw N.L., Plasterk R.H.A. RNAi protects the Caenorhabditis elegans germline against transposition. Trends Genet. 2004;20:314–319. doi: 10.1016/j.tig.2004.04.011. doi:10.1016/j.tig.2004.04.011 [DOI] [PubMed] [Google Scholar]
- Wang X.H., Aliyari R., Li W.X., Li H.W., Kim K., Carthew R., Atkinson P., Ding S.W. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. doi:10.1126/science.1125694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M., Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006;11:142–151. doi: 10.1016/j.tplants.2006.01.003. doi:10.1016/j.tplants.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Watanabe T., et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. doi:10.1038/nature06908 [DOI] [PubMed] [Google Scholar]
- Weinstock G.M., et al. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. doi:10.1038/nature05260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker N.C., Habig J.W., Bass B.L. Genes misregulated in C. elegans deficient in Dicer, RDE-4, or RDE-1 are enriched for innate immunity genes. RNA. 2007;13:1090–1102. doi: 10.1261/rna.542107. doi:10.1261/rna.542107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerhout E.M., Ooms M., Vink M., Das A.T., Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. doi:10.1093/nar/gki220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins C., Dishongh R., Moore S.C., Whitt M.A., Chow M., Machaca K. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. doi:10.1038/nature03957 [DOI] [PubMed] [Google Scholar]
- Willmann M.R., Poethig R.S. Conservation and evolution of miRNA regulatory programs in plant development. Curr. Opin. Plant Biol. 2007;10:503–511. doi: 10.1016/j.pbi.2007.07.004. doi:10.1016/j.pbi.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston W.M., Sutherlin M., Wright A.J., Feinberg E.H., Hunter C.P. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl Acad. Sci. USA. 2007;104:10 565–10 570. doi: 10.1073/pnas.0611282104. doi:10.1073/pnas.0611282104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M.E.J., Webster J.P., Domingo E., Charlesworth B., Levin B.R. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 2002;32:569–577. doi: 10.1038/ng1202-569. doi:10.1038/ng1202-569 [DOI] [PubMed] [Google Scholar]
- Yigit E., Batista P.J., Bei Y.X., Pang K.M., Chen C.C.G., Tolia N.H. Analysis of the C. elegans argonaute family reveals that distinct argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. doi:10.1016/j.cell.2006.09.033 [DOI] [PubMed] [Google Scholar]
- Zambon R.A., Vakharia V.N., Wu L.P. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell. Microbiol. 2006;8:880–889. doi: 10.1111/j.1462-5822.2006.00688.x. doi:10.1111/j.1462-5822.2006.00688.x [DOI] [PubMed] [Google Scholar]
- Zaratiegui M., Irvine D.V., Martienssen R.A. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. doi:10.1016/j.cell.2007.02.016 [DOI] [PubMed] [Google Scholar]
- Zeng Y., Yi R., Cullen B.R. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl Acad. Sci. USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. doi:10.1073/pnas.1630797100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.H., Pan X.P., Cannon C.H., Cobb G.P., Anderson T.A. Conservation and divergence of plant microRNA genes. Plant J. 2006;46:243–259. doi: 10.1111/j.1365-313X.2006.02697.x. doi:10.1111/j.1365-313X.2006.02697.x [DOI] [PubMed] [Google Scholar]
- Zhang X.R., Yuan Y.R., Pei Y., Lin S.S., Tuschl T., Patel D.J. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Gene Dev. 2006b;20:3255–3268. doi: 10.1101/gad.1495506. doi:10.1101/gad.1495506 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genbank accession numbers for the sequences used in figure 2
Alignment of sequences of RdRps from selected animal and other genomes
Numbers used for the MK tests shown in figure 4 of the main test