Abstract
Fission yeast has two kinesin-8s, Klp5 and Klp6, which associate to form a heterocomplex. Here, we show that Klp5 and Klp6 are mutually dependent on each other for nuclear mitotic localization. During interphase, they are exported to the cytoplasm. In sharp contrast, during mitosis, Klp5 and Klp6 remain in the nucleus, which requires the existence of each counterpart. Canonical nuclear localization signal (NLS) is identified in the nonkinesin C-terminal regions. Intriguingly individual NLS mutants (NLSmut) exhibit loss-of-function phenotypes, suggesting that Klp5 and Klp6 enter the nucleus separately. Indeed, although neither Klp5-NLSmut nor Klp6-NLSmut enters the nucleus, wild-type Klp6 or Klp5, respectively, does so with different kinetics. In the absence of Klp5/6, microtubule catastrophe/rescue frequency and dynamicity are suppressed, whereas growth and shrinkage rates are least affected. Remarkably, chimera strains containing only the N-terminal Klp5 kinesin domains cannot disassemble interphase microtubules during mitosis, leading to the coexistence of cytoplasmic microtubules and nuclear spindles with massive chromosome missegregation. In this strain, a marked reduction of microtubule dynamism, even higher than in klp5/6 deletions, is evident. We propose that Klp5 and Klp6 play a vital role in promoting microtubule dynamics, which is essential for the spatiotemporal control of microtubule morphogenesis.
INTRODUCTION
During mitotic cell division, it is vital that chromosomes are segregated equally so that each resulting daughter cell receives a complete copy of all the genetic information (Mitchison and Salmon, 2001). Failure to achieve this can result in aneuploidy, a hallmark of cancer cells. Chromosome segregation is mediated by the mitotic spindle, a structure consisting of microtubule fibers. Microtubules are inherently dynamic, that is, constantly growing and shrinking, and this property is used in the cell to create pushing and pulling forces required for segregating chromosomes (Desai and Mitchison, 1997). Cells contain an array of stabilizing and destabilizing factors to achieve precise spatiotemporal control of microtubule dynamics. During mitosis, this facilitates the capture of kinetochores by spindle microtubules and subsequently enables spindle elongation and shortening to be coordinated with chromosome segregation.
Kinesins were originally identified as motor proteins that use energy gained from ATP hydrolysis to power processive movement along microtubules (Vale et al., 1985). Kinesins consist of a large protein family (Lawrence et al., 2004; Miki et al., 2005); and recently, it has been found that several kinesin subfamilies (namely, kinesin-8, -13, and -14) possess microtubule-depolymerizing activity and are therefore important regulators of microtubule dynamics (Kline-Smith and Walczak, 2004; Wordeman, 2005; Howard and Hyman, 2007; Gardner et al., 2008). Most members characterized so far play pivotal roles in mitotic cell division. Klp5 and Klp6 in fission yeast belong to the kinesin-8 family. Klp5 and Klp6 localize to interphase cytoplasmic microtubule, mitotic kinetochores, the spindle, and the spindle midzone (West et al., 2001, 2002; Garcia et al., 2002a,b; Li and Chang, 2003; Rajagopalan et al., 2006; West and McIntosh, 2008). Deletion mutants are viable but have hyper-stable microtubules and show defects in chromosome congression (Garcia et al., 2002b; West et al., 2002; Sanchez-Perez et al., 2005; Griffiths et al., 2008). Members of this family in Saccharomyces cerevisiae and human cells, Kip3 and Kif18A, respectively, have been shown to function both as plus-end–directed motors and as microtubule depolymerizers (Gupta et al., 2006; Varga et al., 2006; Mayr et al., 2007). Interestingly, it was demonstrated that Kip3 depolymerizes microtubules in a length-dependent manner.
Most kinesins function as homodimers. This structure enables them to step along the microtubule by using a “hand-overhand” mechanism (Asbury, 2005; Carter and Cross, 2005; Yildiz and Selvin, 2005). Klp5 and Klp6 are unusual members of the kinesin-8 family, because they are the only members reported thus far that form a heterocomplex (Garcia et al., 2002b; Li and Chang, 2003), colocalizing during the entire cell cycle. This heterodimerization is essential for Klp5/6 function, because deletion mutants of either Klp5 or Klp6 exhibit identical phenotypes. Furthermore, no additive phenotypes are observed in double deletion mutants; and finally, overexpression of Klp5 or Klp6 is unable to suppress deletion phenotypes of klp6 or klp5, respectively (Garcia et al., 2002a,b; West et al., 2002; Li and Chang, 2003).
In metazoans, the nuclear envelope breaks down at the beginning of mitosis (open mitosis). However, yeast undergo a closed mitosis in which the nuclear envelope is maintained, meaning that components required for mitotic progression need to be imported into the nucleus during mitosis. Many proteins contain nuclear localization signals (NLSs) and/or nuclear export signals (NESs) that are recognized and bound by importin or exportin molecules, respectively (Yoneda, 2000). Classical, canonical NLSs consist of a short sequence rich in three to five basic amino acids, whereas NESs are not as easily defined as NLSs (Dingwall and Laskey, 1991; Kutay and Güttinger, 2005). The first and best characterized exportin is Crm1/exportin-1, which exports proteins containing a leucine-rich NES (Mattaj and Englmeier, 1998; Gorlich and Kutay, 1999). Proteins containing both NLS and NES constantly shuttle between the cytoplasm and the nucleus, and rate of import or export may be modified as necessary to change the subcellular location of the protein during the cell cycle or upon extra- or intracellular cues. Several mechanisms that regulate nuclear transport of cargo proteins are known. For example, the phosphorylation and dephosphorylation of protein substrates that modulate nuclear transport is one of the best-understood mechanisms (Poon and Jans, 2005). In addition, intramolecular masking or chemical modification, such as oxidization, of NLSs/NESs to reduce their accessibility to importins/exportins play important roles in nucleocytoplasmic protein transport (Veal et al., 2007). Alternatively, the intrinsic NLS/NES activity of a protein may also be overcome if it is anchored in the nucleus or cytoplasm by physically associating with specific subcellular structures such as organelles and the actin or microtubule cytoskeleton (Dong et al., 2000; Haller et al., 2004).
Although fission yeast Klp5 and Klp6 belong to the kinesin 8 family and their deletions lead to longer, hyperstabilized microtubules, the contribution of these two molecules to in vivo microtubule dynamics has never been addressed before. Furthermore a physical interaction between Klp5 and Klp6 gives us a unique opportunity to explore the importance of kinesin-8 dimerization, because it is possible to examine the behavior of one “half” of the dimer in the absence of the partner. It is also of interest to distinguish whether Klp5 and Klp6 have different biochemical and cell biological properties; and if so, what the physiological significance of these is for the function of the Klp5/6 complex. In this study, we have pursued these issues and uncover novel regulatory mechanisms.
MATERIALS AND METHODS
Schizosaccharomyces pombe Strains, Media, and Genetic Methods
The strains used in this study are listed in Table 1 and Supplemental Table S1. The growth and the maintenance of the strains and strain constructions were carried out according to standard procedures (Moreno et al., 1991; Bähler et al., 1998; Sato et al., 2005). Most of the experiments were performed at 30°C otherwise stated. For live analysis, the room temperature (26°C) was used.
Table 1.
Strain list
| Strain | Genotype | Derivation |
|---|---|---|
| 513 | h− | Lab stock |
| AR017 | h−klp5::ura4+ | This study |
| AR065 | h−klp5+-GFP-kan pREP1(mRFP-atb2+) | This study |
| AR066 | h−klp5+-GFP-kan klp6::ura4+pREP1(mRFP-atb2+) | This study |
| AR067 | h+ade6–216 klp5::ura4+klp6+-GFP-kanR pREP1(mRFP-atb2+) | This study |
| AR085 | h−klp6+-GFP-kan pREP1(mRFP-atb2+) | This study |
| AR214 | h−klp5::ura4+klp6::hph pREP41(GFP-klp5+) | This study |
| AR216 | h−klp5::ura4+klp6::hph pREP41(GFP-klp5-NLSmut) | This study |
| AR217 | h−klp5::ura4+klp6::hph pREP41(GFP-klp6-NLSmut) | This study |
| AR218 | h−klp5::ura4+klp6::hph pREP41(GFP-klp6+) | This study |
| AR231 | h+klp5::nat klp6::ura4+ | This study |
| AR245 | h−his7 klp5-NLSmut | This study |
| AR247 | h−his2/7 klp6-NLSmut | This study |
| AR285 | h−kan-nmtP3-GFP-atb2+klp5::ura4+klp6::hph | This study |
| AR287 | h−kan-nmtP3-GFP-atb2+klp5::ura4+ | This study |
| AR289 | h−kan-nmtP3-GFP-atb2+klp6::ura4+ | This study |
| AR374 | h+his2 klp6N(1–410)::ura4+::klp5N(1–404) | This study |
| AR399 | h−klp6N(1–410)::ura4+::klp5N(1–404)-GFP-nat | This study |
| AR402 | h−klp5N(1–404)::ura4+::klp6N( 1–410) | This study |
| AR445 | h+his2 klp5N(1–404)::ura4+::klp6(1–403)-GFP-nat | This study |
| AR449 | h? kan-nmtP3-GFP-atb2+klp6N(1–410)::ura4+::klp5N(1–404) | This study |
| AR450 | h−kan-nmtP3-GFP-atb2+klp5N(1–404)::ura4+::klp6N(1–403) | This study |
| AR614 | h−klp5+-GFP-kan aur1-mCherry-atb2+ | This study |
| AR615 | h−klp5+-GFP-kan klp6::ura4+aur1-mCherry-atb2+ | This study |
| AR616 | h−ade6–216 klp6+-GFP-kan aur1-mCherry-atb2+ | This study |
| AR617 | h+his7 klp5::ura4+klp6+-GFP-kan aur1-mCherry-atb2+ | This study |
| AR640 | h+his2 klp5::ura4+klp6N(1–410)::ura4+::klp5N(1–404)-GFP-nat aur1-mCherry-atb2+pREP41(GFP-klp5+) | This study |
| AR644 | h−his7 klp5::ura4+aur1-mCherry-atb2+pREP41(GFP-klp5+) | This study |
| AR645 | h−his7 klp5::ura4+aur1-mCherry-atb2+pREP41(GFP-klp5-NLSmut) | This study |
| AR646 | h+his2 klp6::ura4+aur1-mCherry-atb2+pREP41(GFP-klp6+) | This study |
| AR647 | h+his2 klp6::ura4+aur1-mCherry-atb2+pREP41(GFP-klp6-NLSmut) | This study |
| AR648 | h+his2 klp5::ura4+klp6-NLSmut aur1-mCherry-atb2+pREP41(GFP-klp5+) | This study |
| AR650 | h−his7 klp5-NLSmut klp6-hph aur1-mCherry-atb2+pREP41(GFP-klp6+) | This study |
| NK28–5A | h−his2/7 klp6::ura4+ | Lab stock |
| MA201 | h−kan-nmtP3-GFP-atb2+ | Lab stock |
All strains listed in this table contain leu1–32 ura4-D18.
Nucleic Acid Preparation and Manipulation
Enzymes were used as recommended by the suppliers (New England Biolabs, Beverly, MA; Takara Shuzo, Kyoto, Japan; and Stratagene, La Jolla, CA.). Site-directed mutants of klp5 and klp6 were generated using QuikChange II kit (Stratagene) according to the manufacturer's protocol. The plasmids pREP41-EGFP-klp5 or pREP41-EGFP-klp6 (kind gift from Dr. Eric Chang, Baylor College of Medicine, Houston, TX) were used as templates. Mutated plasmids were verified by nucleotide sequencing. pREP41-EGFP-klp5-NLSmut contains two substitutions from arginines to alanines in the NLS sequence (R693A and R695A), whereas pREP41-EGFP-klp6-NLSmut contains R673A and R675A.
Strain Constructions
Chimeric molecules were generated by a two-step process as follows. First, the region corresponding to the N terminus of Klp5 (1–404 amino acids) or Klp6 (1–403 or 410 amino acids) was replaced with ura4+ (klp5 ΔN or klp6 ΔN, respectively). Next, the gene sequence coding for the N terminus of the other klp5/6 molecule (Klp6 or Klp5, respectively) was integrated into the ura4+ marker, and transformants were selected on plates containing 5-fluoroorotic acid (5-FOA). This second step was carried out in a strain deleted for the other klp5/6 (Klp5 or Klp6, respectively) to prevent unwanted homologous recombination between the N terminus PCR product and the endogenous klp+ locus. Correct replacement was verified by colony PCR and furthermore by sequencing across the Klp5/Klp6 boundaries. Once chimera strains had been generated, they were crossed back to a wild-type strain to create strains containing the N-terminal kinesin domains derived from only Klp5 or Klp6 (Klp5N+Klp5N or Klp6N+Klp6N), respectively. Klp5- and Klp6-NLS mutants (NLSmut) were constructed by integrating polymerase chain reaction (PCR)-amplified fragments containing klp5-NLSmut or klp6-NLSmut into a klp5::ura4+ or klp6::ura4+ strain, followed by selection on plates containing 5-FOA. Correct replacement was confirmed by PCR.
For the construction of a strain containing mCherry-atb2+ (encoding α2-tubulin), the following procedures were undertaken. The nda3p-mCherry-atb2+ fragment consisting of the promoter from the nda3+ gene (encoding β-tubulin), mCherry, and the atb2+ open reading frame (ORF) was created by replacing green fluorescent protein (GFP) with mCherry (Shaner et al., 2004) in the nda3p-GFP-atb2+ DNA fragment (Masuda et al., 2006), and this fusion gene was inserted into pYC19 that contains the aur1r gene as a selectable marker (aureobasidin A-resistance; Takara Shuzo). The pYC19 harboring nda3p-mCherry-atb2+ was linearized by cutting a single site in aur1r and subsequently integrated into the aur1+ locus, by which mCherry-Atb2 could be marked by aureobasidin A-resistance phenotypes. This construct seems to be least detrimental to microtubule function, because no adverse effect is observed when combined with an alp14 deletion (defective in a member of the Dis1/TOG microtubule-associated protein family), which is known to be sensitive to compromised tubulin function (Garcia et al., 2001; H. M. and T. T., unpublished data).
Live Cell Analysis
For live cell analysis, 200 μl of a log-phase culture in minimal media-NH4Cl2 supplemented with glutamate and appropriate amino acids was mounted on a glass-bottomed culture dish (MatTek, Ashland, MA) coated with lectin and incubated for 30 min or more for cell adhesion. In some experiments, 2 ml of media was added to the dish after cell adhesion. Cells were observed with an Olympus IX-70 inverted fluorescence microscope equipped with a Roper CoolSNAP HQ (Photometrix, Tucson, AZ) charge-coupled device camera and a UPlanApo ×60/1.40 numerical aperture (NA) oil or UPlanSApo ×100/1.40 NA oil objective lens (Olympus, Tokyo, Japan). Images were collected using DeltaVision software (Applied Precision, Issaquah, WA). Ten to 14 Z-sections (0.3 μm apart) were acquired and subsequently deconvolved and projected to two-dimensional images by using SoftWoRx software (Applied Precision).
Drug Treatment
Leptomycin B (LMB; provided by M. Yoshida, RIKEN, Saitama, Japan) was used to inhibit Crm1-dependent nuclear export. Fifty to 140 μl of 250 ng/ml LMB diluted in media from 100 μg/ml stock solution (in ethanol) was added to 200 μl of cells mounted on a glass-bottomed dish (final concentration, 50–140 ng/ml). Carbendazim (MBC/CBZ; Sigma-Aldrich, St. Louis, MO) was used to depolymerize microtubules in liquid culture. Thirty microliters of 5 mg/ml stock (dissolved in dimethyl sulfoxide) was added to 970 μl of media, mixed well, and centrifuged at 13,000 rpm for 5 min to precipitate crystals. The supernatant was then added in a culture dish already containing cells and 2 ml of media. This gave a final concentration of MBC/CBZ of 50 μg/ml.
Fluorescence Intensity Profiling
To quantify nuclear accumulation of GFP-tagged Klp5 or Klp6, projection images of summed fluorescence intensity were reconstituted, and the intensity of the nuclear region was measured using SoftWoRx software. As a control, fluorescence intensity of the nuclear region in wild-type cells expressing no GFP was measured, and the average intensity was subtracted as a background from the data obtained from GFP-tagged Klp5/6. For line-scan analysis, the projections of maximum intensity were imported into ImageJ software, version 1.37 (National Institutes of Health, Bethesda, MD). Lines were drawn from one end of a cell to the other, along the longitudinal axis, and fluorescence intensity across each line was measured using the plot profile function. Fluorescence data were exported in numerical format and graphs drawn using Excel (Microsoft, Redmond, WA).
Measurement of Microtubule Dynamics
Time-lapse images of 10 Z-sections (0.3 μm apart) were taken every 10 s for 10 min, deconvolved, and projected as described above. Parameters of microtubule dynamics (growth and shrinkage rates, catastrophe frequency, rescue frequency, dynamicity, and dwell time) were then quantified for individual microtubules. Dynamicity, the mean rate of total tubulin exchange, was calculated using a conversion factor of 1 μm of microtubule length = 1690 tubulin dimers (Toso et al., 1993). For the calculation of this parameter, we considered the time spent in microtubule growth and shortening and pause states, as reported previously (Gupta et al., 2006).
RESULTS
Klp5 and Klp6 Are Mutually Dependent for Their Mitotic Localization
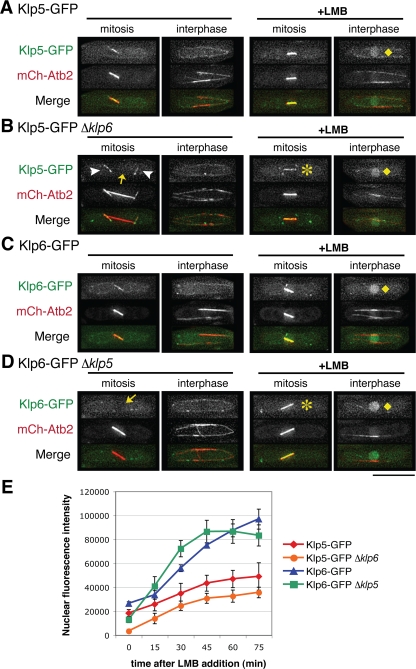
As an initial step in addressing the role of Klp5/6 association, the localization of Klp5-GFP was examined in a Δklp6 background and vice versa, i.e., the localization of Klp6-GFP in a Δklp5 background. Figure 1 compares the localization of Klp5-GFP between wild-type (Figure 1A) and Δklp6 cells (Figure 1B). To assign precisely cell cycle stages of individual cells, microtubules are visualized by mCherry-Atb2 (α2-tubulin fused to monomeric Cherry). Although Klp5-GFP in the absence of Klp6 localizes correctly to cytoplasmic microtubules during interphase, as in wild-type cells (second panels), during mitosis its localization to the nuclear spindle was abolished (arrow, far left in B). It is of note that under this condition, Klp5-GFP can be seen localizing to cytoplasmic astral microtubules (arrowheads), indicating that Klp5 alone is capable of localizing to microtubules independently of Klp6. This result suggested that Klp5-GFP might well be excluded from the nucleus in the absence of Klp6.
Figure 1.
Klp5 and Klp6 are interdependent for nuclear mitotic localization. Wild-type (A and C), Δklp6 (B), or Δklp5 (D) strains containing mCherry-Atb2 and either Klp5-GFP (A and B) or Klp6-GFP (C and D) were used to visualize microtubules and Klp5/6 localization. (A–D) Representative images before (first and second panels) or after LMB addition (45 min; third and fourth panels) are shown during mitosis (first and third panels) or interphase cells (second and fourth panels). Cytoplasmic astral microtubules and dim nuclei are marked with arrowheads and arrows, respectively. After LMB addition, both Klp5 and Klp6 accumulate in the nucleoplasm (B and D; diamonds) or localize to nuclear spindles (asterisks) in the absence of each counterpart. In merged images, GFP-Klp5/6 and mCherry-Atb2 are shown in green and red, respectively. Bar, 10 μm. (E) Kinetics of nuclear accumulation during interphase after addition of LMB (0–75 min) are plotted in individual strains shown in A–D. n ≥5 for each strain.
The reciprocal experiment using Klp6-GFP showed a similar result (wild type in Figure 1C and Δklp5 in Figure 1D). Klp6-GFP could localize to cytoplasmic microtubules during interphase in the absence of Klp5 (second panels). However, no localization of Klp6-GFP to the nuclear mitotic spindle was observed in the Δklp5 strain (arrow, far left in D). Klp5 and Klp6 are therefore interdependent for localization to the nuclear spindle during mitosis.
Klp5 and Klp6 Shuttle between the Cytoplasm and the Nucleus
The interdependency of Klp5 and Klp6 for localization to the nucleus during mitosis could be for one of the following reasons: 1) for activation of nuclear import or 2) for inhibition of nuclear export. To distinguish between these two possibilities, nuclear export was inhibited in each strain observed in the previous experiment. This was achieved with LMB, which specifically inhibits leucine rich NES-dependent nuclear export by directly binding Crm1/exportin-1 (Fornerod et al., 1997; Fukuda et al., 1997; Ossareh-Nazari et al., 1997). If a Klp5/6-GFP protein cannot enter the nucleus, incubation with LMB will not cause any GFP accumulation in the nucleus; however, if a Klp5/6-GFP protein is able to enter the nucleus, then nuclear accumulation of the GFP signal should be observed. LMB addition resulted in mitotic spindle localization of either Klp5-GFP or Klp6-GFP in Δklp6 or Δklp5 cells, respectively (asterisks in third panels, Figure 1, B and D). Furthermore, even during interphase, Klp5-GFP and Klp6-GFP accumulated in the nucleoplasm in either wild-type or klp5/6-deleted cells (diamonds, far right in Figure 1, A–D). LMB-sensitive nuclear localization, at least for Klp6, has been shown recently in an independent genome-wide analysis (Matsuyama et al., 2006). This result indicates that it is the Crm1-dependent nuclear export that is inhibited during mitosis by coexistence of Klp5 and Klp6, presumably via the heterodimerization or oligomerization.
During the course of experiments with LMB, we realized that intensities of nuclear Klp5-GFP and Klp6-GFP might not be identical; instead, those of Klp6-GFP are stronger than those of Klp5-GFP. To inspect this notion and quantify the kinetics of nuclear import of these two proteins, nuclear GFP signals were quantified every 15 min upon addition of LMB. As posited, Klp6-GFP nuclear signals indeed accumulated faster and more intensely than those of Klp5-GFP (red diamonds and blue triangles in Figure 1E; see Supplemental Figure S1). This result indicates that NLS activities of Klp6 are more efficient than those of Klp5 during interphase. Furthermore it might imply that Klp5 and Klp6 are imported into the nucleus separately, rather than together as a heterocomplex (see below).
To test the idea concerning independent nuclear transport of Klp5 and Klp6, the kinetics of nuclear import was examined in the absence of each partner (Klp5-GFP Δklp6 and Δklp5 Klp6-GFP). Consistent with the notion of separate transport, import profiles of Klp6-GFP were mostly indistinguishable, if not identical, in either presence or absence of Klp5 (green squares and blue triangles in Figure 1E). However in Klp5-GFP, in the absence of Klp6, we did reproducibly observe the reduction of nuclear import by 30–40% (compare red diamonds and orange circles). The decrease in nuclear import of Klp5-GFP suggests that there are two populations for Klp5 nuclear import, one population on its own and the other population concomitant with Klp6. It is possible that the cellular levels of Klp6 are higher than those of Klp5, by which more Klp6 might exist independently of Klp5. We have examined the native size of Klp5 and Klp6 by gel filtration to distinguish physical forms between a Klp5/6 heterodimer and each monomer. However irrespective of the presence or absence of each counterpart, Klp5 and Klp6 were fractionated in a large size (∼2000 kDa; see Supplemental Figure S2); therefore, we could not deduce a physical state of Klp5 and Klp6 with this method.
Identification of NLSs
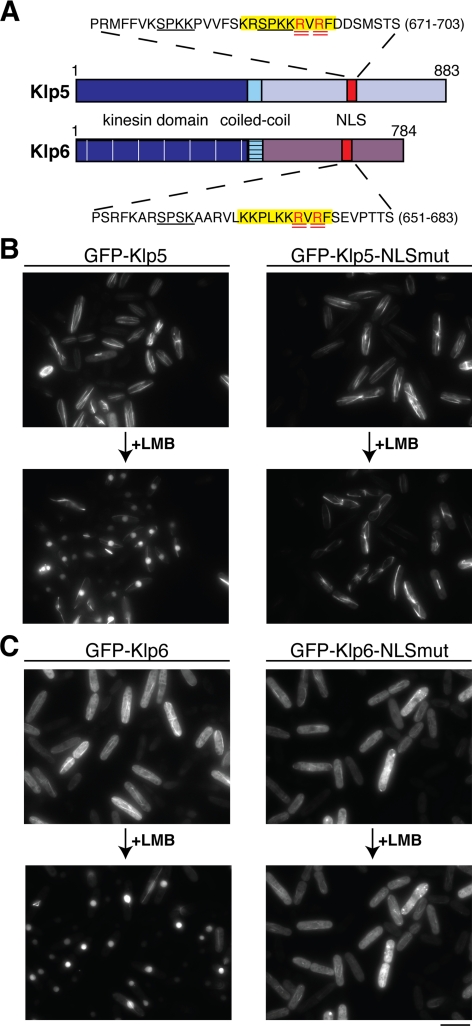
Having established that Klp5 and Klp6 are both able to enter the nucleus independently of each other, the mechanism of their nuclear transport was explored. A search for possible NLSs within Klp5 and Klp6 was performed by submitting the protein sequences to PSORT (http://psort.nibb.ac.jp/). This revealed that both Klp5 and Klp6 contain a classical canonical NLS, rich in basic amino acid residues, within their C-terminal tails (highlighted in yellow in Figure 2A).
Figure 2.
Identification and characterization of Klp5 and Klp6 NLSs. (A) Cartoon depicting Klp5 and Klp6 domains. Dark blue, kinesin domain (note that for simplicity, the N-terminal nonkinesin extensions consisting of −100 amino acid residues are not shown); light blue, coiled-coil; and red, NLS. Amino acid sequences surrounding consensus NLS sequences in Klp5 and Klp6 are highlighted in yellow, and arginine residues mutated to alanines are doubly underlined in red. Underlined sequences in black denote CDK consensus phosphorylation sites. (B and C) Cellular localization of NLS mutants. Δklp5 Δklp6 cells were transformed with plasmids containing GFP-klp5+ (B; left), GFP-klp5-NLSmut (B; right), GFP-klp6+(C; left), GFP-klp6-NLSmut (C; right) and observed by fluorescence microscopy. The same field of cells is shown before and after (60 min) LMB addition. Bar, 10 μm.
To test whether these were genuine localization signals, arginine residues (doubly underlined in red) within each putative NLS were mutated to alanines to create the Klp5-R693A R695A (Klp5-NLSmut) and Klp6-R673A R675A (Klp6-NLSmut). Δklp5 Δklp6 double mutant cells were then transformed with plasmids containing either wild-type GFP-Klp5, GFP-Klp5-NLSmut, wild type GFP-Klp6 or GFP-Klp6-NLSmut. This host strain was chosen so that the nuclear import competency of the Klp5 and Klp5-NLSmut proteins could be assessed independently of their interaction with Klp6, and vice versa. As shown in Figure 2, B and C, live observation of GFP-Klp5/6 signals before and after LMB addition (the same field of cells) unequivocally showed that NLSmut constructs fail to enter the nucleus (right). In contrast both wild-type proteins accumulated in the nucleoplasm in interphase cells upon drug addition (bottom left). We noticed that Klp6 showed diffuse cytoplasmic staining in addition to microtubule localization, whereas both wild-type and Klp5-NLSmut constructs show much stronger colocalization with cytoplasmic microtubules (compare top two panels in Figure 2, B and C). This raises the interesting possibility that Klp5 and Klp6 may have different microtubule binding affinities, which is consistent with a recent report on microtubule localization of klp5 and Klp6 (West and McIntosh, 2008). Together, we have identified the NLSs of Klp5 and Klp6 in analogous positions within the nonkinesin C-terminal tails, and these NLSs are critical to nuclear import at least in the absence of individual counterparts.
Klp5 Is Not Anchored in the Cytoplasm by Microtubule Binding during Interphase
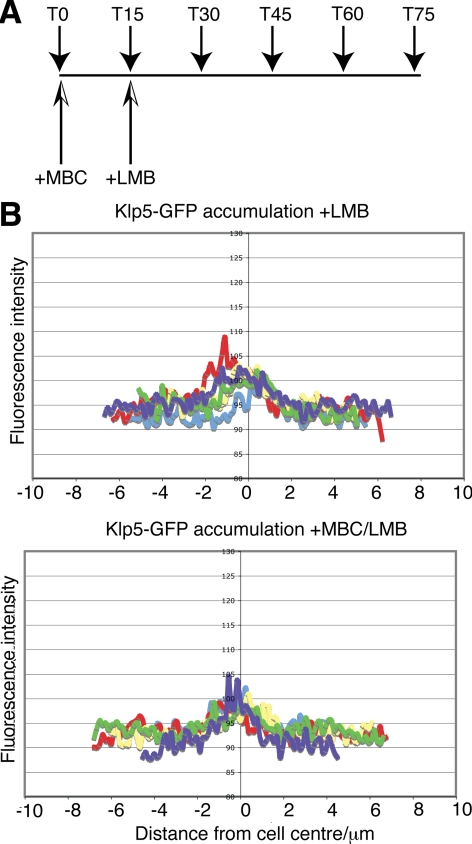
Our previous results showed that lower levels of Klp5 enter the nucleus during interphase compared with Klp6 and they hinted that Klp5 might possess robust affinities for microtubules. We hypothesized that Klp5 might be sequestered away from the nucleus and/or retained in the cytoplasm during interphase by binding the cytoplasmic interphase microtubules. If this were the case, we reasoned that induced depolymerization of cytoplasmic microtubules would release Klp5-GFP to enter the nuclei of interphase cells. To test this possibility, cytoplasmic microtubules were depolymerized using the drug methyl 2-benzimidazolecarbamate (MBC). The extent of microtubule depolymerization was monitored by visualizing microtubules with monomeric red fluorescent protein (mRFP)-Atb2. After MBC treatment (15 min), the efficiency of Klp5-GFP nuclear import was assessed by inhibiting nuclear export by using LMB (see experimental plan, Figure 3A). The nuclear accumulation of Klp5-GFP after both MBC and LMB treatment was compared with the accumulation after LMB treatment alone. The line profiles of fluorescence intensity in Figure 3B show that nuclear accumulation of Klp5-GFP in interphase cells is not increased when microtubules are depolymerized by MBC treatment. This result shows that the cell cycle-dependent localization of Klp5, at least interphase microtubule localization, is not achieved by microtubule anchoring in the cytoplasm.
Figure 3.
Depolymerization of cytoplasmic microtubules does not result in Klp5 nuclear entry during interphase. (A) Experimental plan. At time 0, cultures were split into two parts and MBC (50 μg/ml) was added in one of them. After 15 min, LMB (50 ng/ml) was further added in both cultures. Time points (T) refer to minutes. (B) Line profiles of fluorescence intensity showing nuclear accumulation of Klp5-GFP during interphase 60 min after LMB addition (T75 of experiment plan). Top graph, LMB; bottom graph, MBC + LMB. n = 5.
Each NLS Is Essential for Klp5 and Klp6 Function
If Klp5 and Klp6 are imported into the nucleus independently of each other, one prediction derived from this notion is that each NLS mutant of Klp5 and Klp6 would exhibit phenotypes identical to deletion mutants, because either Klp5-NLSmut or Klp6-NLSmut should be unable to enter the nucleus on its own. To test this hypothesis, we replaced the wild-type genomic klp5+ or klp6+ gene with klp5-NLSmut or klp6-NLSmut, respectively, and examined sensitivity/resistance of these strains to a microtubule-depolymerizing drug thiabendazole (TBZ; a derivative of MBC). Previous work showed that TBZ resistance is a reliable assay for Klp5/6 functionality, because loss-of-function mutants display drug resistance due to microtubule hyperstabilization (West et al., 2001; Garcia et al., 2002b). As shown in Figure 4A, either Klp5/6-NLSmut strain displayed hyperresistance to this drug, which is indistinguishable from each deletion mutant. Thus, mutations in individual NLSs are sufficient to impair Klp5/6 functions, consistent with the idea that Klp5 and Klp6 are imported into the nucleus separately.
Figure 4.
Each NLS mutant is nonfunctional and fails to accumulate in the nucleus upon LMB addition even in the presence of its wild-type counterpart. (A) Functional test of NLS mutants. Ten-fold serial dilutions of various mutants plus a wild-type control strain as shown were spotted onto rich YE medium containing 0, 10, or 20 μg/ml TBZ as indicated (5 × 103 cells were applied in the first spot). Plates incubated for 2 d at 30°C. (B–G) Localization of Klp5/6 wild-type and NLS mutants in the presence of each counterpart. Plasmids containing GFP-klp5+ (B and C), GFP-klp5-NLSmut (D), GFP-klp6+ (E and F), or GFP-klp6-NLSmut (G) were transformed into Δklp5 cells (B and D), Δklp5 klp6-NLSmut (C), Δklp6 (E and G), or klp5-NLSmut Δklp6 (F) strains each containing mCherry-Atb2. Representative images of interphase cells before (left three panels) or after LMB addition (75–90 min; right three panels) are shown. In merged images, GFP-Klp5/6 and mCherry-Atb2 are shown in green and red, respectively. After LMB addition, both Klp5 and Klp6 wild-type molecules accumulate in the nucleoplasm (B, C, E, and F), whereas no nuclear signals of NLS mutants are observed (D and G). Bar, 10 μm.
Next, to examine the cellular localization of NLS mutants in the presence of the wild-type counterpart (note that earlier experiments shown in Figure 2 were performed in the absence of each counterpart), plasmids containing GFP-tagged Klp5-NLSmut or Klp6-NLSmut were introduced into Δklp5 klp6+ or klp5+ Δklp6 cells, respectively, and GFP signals were observed upon LMB addition. As predicted, unlike wild-type Klp5 or Klp6 (Figure 4, B and E), neither Klp5-NLSmut nor Klp6-NLSmut was capable of accumulating in the nucleus upon drug treatment (Figure 4, D and G). In sharp contrast, reciprocal experiments, namely, localization of wild-type Klp5 or Klp6 in a genetic background of Δklp5 klp6-NLSmut or klp5-NLSmut Δklp6, respectively, showed that Klp5 or Klp6 molecules accumulated normally upon LMB addition irrespective of NLS malfunction in its counterpart (Figure 4, C and F). Collectively, both genetic and localization data are fully consistent with the idea that Klp5 and Klp6 enter the nucleus separately, although some population might be imported together.
Individual Kinesin Domains of Klp5 and Klp6 Are Required for Proper Function
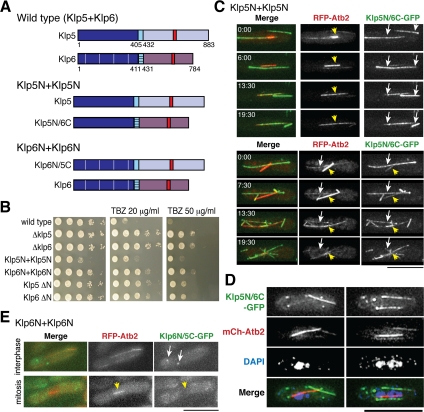
We next addressed the roles of the N-terminal kinesin domains. To do this, we created two chimera strains, one containing only N-terminal Klp5 domains and the other containing only Klp6 kinesin domains. These are designated Klp5N+Klp5N and Klp6N+Klp6N accordingly (see schematic diagrams in Figure 5A). The Klp5N+Klp5N strain contains a wild-type Klp5 molecule and a chimeric Klp5N/6C molecule, consisting of the N-terminal Klp5 kinesin domain and the C-terminal Klp6 tail region. Conversely, the Klp6N+Klp6N strain contains wild-type Klp6 and a chimeric Klp6N/5C molecule consisting of the N-terminal Klp6 kinesin domain and the C-terminal Klp5 tail region.
Figure 5.
Construction and characterization of strains containing two identical Klp5 or Klp6 kinesin domains. (A) Schematic drawings showing domain structures in wild-type, Klp5N+Klp5N, and Klp6N+Klp6N strains. As in Figure 2A, the kinesin domain (dark blue) and coiled-coil domain (light blue) of Klp6 are cross-hatched to distinguish them from those of Klp5. (B) Ten-fold serial dilutions of Klp5N+Klp5N and Klp6N+Klp6N strains, plus controls as shown, were spotted onto rich YE medium containing 0, 20, or 50 μg/ml TBZ as indicated (5 × 104 cells were applied in the first spot). Plates incubated for 2 d at 30°C. (C) Time-lapse microscopy of mitotic Klp5N+Klp5N cells showing localization of Klp5N/6C-GFP and RFP-tubulin (pREP1-mRFP-atb2+; Yamashita et al., 2005). Images from two different movies are shown, the top cell started from early mitosis, whereas the bottom cell was filmed from mid-mitosis. Time in minutes is shown in the top left corner in the first panels. (D) Chromosome segregation defects of a Klp5N+Klp5N strain. Mitotic Klp5N+Klp5N cells that contain mCherry-Atb2 and Klp5N/6C-GFP were fixed and stained with DAPI. (E) Still images of Klp6N+Klp6N interphase (top) and mitotic live cells (bottom) showing localization of Klp6N/5C-GFP and RFP-tubulin. In merged images, GFP-Klp5/6, RFP-Atb2 (for C and E) or mCherry-Atb2 (for D) and DAPI (for D) are shown in green, red, and blue respectively. Bars, 10 μm.
We first examined the growth and resistance to TBZ of these chimera strains. Neither the Klp5N+Klp5N nor the Klp6N+Klp6N strains behaved like wild-type cells; and intriguingly, they exhibited very different phenotypes (Figure 5B). The Klp5N+Klp5N strain showed substantial growth retardation and was hyperresistant to TBZ (fourth row, compare this with wild type, top row). It is important to note that the appearance of this phenotype requires wild-type Klp5, because a strain containing Klp5N/6C in a klp5Δ background behaves like a klp5Δ strain (the second panel; data not shown). In addition, the phenotype is not due to loss of the N-terminal Klp6 kinesin domain either, because the klp6 ΔN strain (deleted for only the N-terminal Klp6 domain in the presence of wild-type Klp5) exhibited apparently normal growth (bottom row), like a complete deletion of klp6 (Δklp6, third row), although all these deletion stains displayed TBZ-resistance phenotypes. It therefore seemed that the existence of the Klp5 kinesin domains in a trans configuration leads to dominant negative effects on mitotic cell division. In contrast, the phenotype of a Klp6N+Klp6N strain was similar, if not identical, to the deletion phenotype, N-terminal (klp5 ΔN, sixth row) or complete (Δklp5, second row), i.e., it exhibited normal growth and resistance to TBZ. This result raised the possibility that the N-terminal kinesin domains of Klp5 and Klp6 might play distinct roles in microtubule dynamics and morphogenesis.
Coexistence of Cytoplasmic Microtubules and Mitotic Nuclear Spindles in Cells Containing Two Klp5-Kinesin Domains
We determined whether chimera molecules (Klp5N/6C or Klp6N/5C) were capable of localizing correctly during the cell cycle. In each case, its C terminus was tagged with GFP, and colocalization with microtubules was determined by simultaneous observation of mRFP-Atb2. Figure 5C shows time-lapse microscopy of two representative mitotic Klp5N+Klp5N cells (time in minutes is shown in the top left corner of left panels). What is remarkable in these images is that cytoplasmic microtubules persist long into mitosis; and as a result, cytoplasmic and spindle microtubules coexist in a single mitotic cell. It is worth noting that chimeric Klp5N/6C-GFP does not localize noticeably to the spindle during early mitosis (top row, 0 min in the upper part, spindle microtubules shown with mRFP-Atb2 [arrowheads in middle panels]), instead showing predominant localization to two long, straight cytoplasmic microtubules that are still present in this “mitotic” cells (arrows in right panels). As mitosis progresses, these cytoplasmic microtubules break down very slowly, and Klp5N/6C-GFP gradually accumulates on the mitotic spindle (13.5 and 19.5 min; see merged images in left panels). The bottom part of Figure 5C displays progression from mid-mitosis (top row, 0 min) to telophase (bottom row, 19.5 min). Notably, Klp5N/6C-localizing cytoplasmic microtubules were still visible even in the later stage (13.5 min), during which breakdown of the mitotic spindle was initiated.
Cytoplasmic microtubules usually disappear at the transition from interphase to mitosis, growth of new microtubules is effectively inhibited because the rate of microtubule shrinkage and/or the frequency of catastrophe exceeds that of microtubule assembly (Sagolla et al., 2003). The time-lapse images in Figure 5C do not show de novo microtubule nucleation but instead show that existing cytoplasmic microtubules are extremely slow to depolymerize. This suggests that microtubule dynamics are perturbed in the Klp5N+Klp5N strain, such that disassembly of cytoplasmic microtubules is substantially suppressed even during mitosis. Consistent with low viability phenotypes (Figure 5B), 4,6-diamidino-2-phenylindole (DAPI) staining of these Klp5N+Klp5N cells showed massive chromosome segregation defects, in which both cytoplasmic and nuclear spindle microtubules coexisted (Figure 5D), which were not observed in klp5/6 deletion strains (West et al., 2001; Garcia et al., 2002b).
We next observed the localization of Klp6N/5C-GFP (Klp6N+Klp6N). In sharp contrast to Klp5N/6C-GFP, no visible localization of Klp6N/5C-GFP along the microtubules was detected (Figure 5E). During interphase Klp6N/5C-GFP concentrated in one or two bright foci that seemed to be located on cytoplasmic microtubules (arrows in top panels). During mitosis, in contrast, Klp6N/5C-GFP was excluded from the nucleus and neither spindle nor nucleoplasm localization was observed (arrowheads in bottom panels). The lack of nuclear mitotic localization mimics the phenotype seen in a single klp5/6 deletion strain (Figure 1, B and D) and is consistent with the TBZ resistance observed for this Klp6N+Klp6N strain (fifth panel in Figure 5B). This result implies that both microtubule binding and NLS activity may be perturbed in this chimera configuration (see Supplemental Figure S3). However, because we could not rule out the possibility that Klp6N+Klp6N is defective in folding into the proper three-dimensional structure as a result of chimera construction, we have not pursued the physiology of this strain any further.
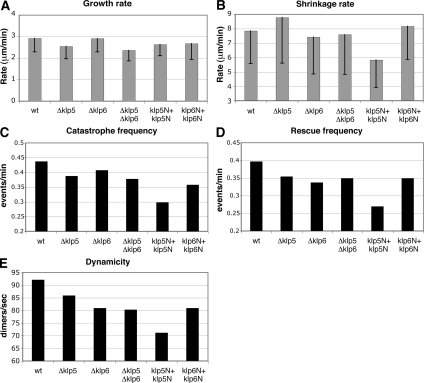
Dynamics of Cytoplasmic Microtubules in Various klp5/6 Mutants: Growth and Shrinkage Rates
Deletion mutants of Klp5 or Klp6 display longer, stabilized microtubules (West et al., 2001; Garcia et al., 2002b), and the live images of the Klp5N+Klp5N strain indicated that cytoplasmic microtubules disassemble very slowly, suggesting that microtubule dynamics is impaired. To investigate this quantitatively, the rate of microtubule growth and shrinkage, frequency of catastrophe/rescue, and microtubule dynamicity (length change of microtubules in any direction over time) were measured during interphase in wild-type, Δklp5, Δklp6, Δklp5 Δklp6, Klp5N+Klp5N, and Klp6N+Klp6N strains (integrated nmtP3-GFP-atb2+ under repressed conditions; Garcia et al., 2001). The average rate of microtubule growth in wild-type cells was 2.93 μm/min (wt in Figure 6A and Table 2), similar to previously reported values, including our own published data (Drummond and Cross, 2000; Sagolla et al., 2003; Busch and Brunner, 2004; Zimmerman and Chang, 2005; Asakawa et al., 2006; Masuda et al., 2006). Although several groups have reported different rates of microtubule growth and shrinkage under different experimental conditions (e.g., media, temperatures, and Atb2 constructs), a comparison of these parameters in our study between wild type and klp5/6 mutants under identical conditions is still valid. The measurement of growth rates in various klp5/6 mutant strains showed that values were similar (2.37–2.92 μm/min; Figure 6A and Table 2; see Supplemental Figure S4A for more quantitative data set), indicating that Klp5 and Klp6 do not influence microtubule growth rate significantly during interphase. Thus fission yeast kinesin 8 molecules play no major roles in microtubule growth rate per se, consistent with a report on budding yeast Kip3 (Gupta et al., 2006).
Figure 6.
Microtubule dynamics in various Klp5/Klp6 mutants. Various parameters representing microtubule dynamics (A, growth rate; B, shrinkage rate; C, catastrophe frequency; D, rescue frequency; and E, dynamicity) were quantified in indicated strains: wild type, Δklp5, Δklp6, Δklp5 Δklp6, Klp5N+Klp5N, and Klp6N+Klp6N (see Table 2 for detailed experimental values). Rates of microtubule growth (A) and shrinkage (B) are shown with box-and-whisker plots.
Table 2.
Dynamisms of cytoplasmic microtubules in various klp5/6 mutants
| Strain | Growth rate (μm/min) | Shrinkage rate (μm/min) | Catastrophe frequency (events/min) | Rescue frequency (events/min) | Dwell timea (min) | Dynamicityb (dimers/s) | Total observation time (s) |
|---|---|---|---|---|---|---|---|
| Wt | 2.93 ± 0.64 (n = 18) | 7.86 ± 2.27 (n = 33) | 0.44 | 0.40 | 1.26 ± 0.76 (n = 14) | 92.1 | 4620 |
| Δklp5 | 2.55 ± 0.58 (n = 25) | 8.79 ± 3.18 (n = 38) | 0.39 | 0.36 | 1.15 ± 1.00 (n = 19) | 85.9 | 5810 |
| Δklp6 | 2.92 ± 0.63 (n = 14) | 7.43 ± 2.57 (n = 32) | 0.41 | 0.34 | 1.91 ± 1.27 (n = 11) | 80.9 | 4740 |
| Δklp5 Δklp6 | 2.37 ± 0.50 (n = 11) | 7.60 ± 2.77 (n = 30) | 0.38 | 0.35 | 1.64 ± 1.60 (n = 22) | 80.3 | 4800 |
| klp5N + klp5N | 2.64 ± 0.53 (n = 17) | 5.84 ± 1.91 (n = 30) | 0.30 | 0.27 | 2.14 ± 1.38 (n = 14) | 71.1 | 6000 |
| klp6N + klp6N | 2.68 ± 0.74 (n = 20) | 8.18 ± 2.31 (n = 36) | 0.36 | 0.35 | 1.65 ± 1.29 (n = 19) | 80.9 | 6000 |
a Duration of contact time (minutes) between microtubule end and the cell tip before catastrophe. In our measurements, microtubules in all the strains examined continue to grow at the cell end.
b Mean rate of total tubulin exchange calculated using a conversion factor of 1 μm of microtubule length = 1690 tubulin dimers.
The average rate of microtubule shrinkage in wild-type cells was 7.86 μm/min (Figure 6B and Table 2). Unlike growth rate, we observed modest differences in this parameter between Δklp5 and Δklp6. In the absence of Klp5, the value was increased by 12% (8.76 μm/min). In contrast Δklp6 and Δklp5 Δklp6 cells displayed small reductions instead by 3–6% (7.43 and 7.6 μm/min, respectively; see Supplemental Figure S4B for more quantitative data set). It is thus formally possible that Klp5 and Klp6 contribute to the rate of microtubule shrinkage in an opposing manner, negative and positive, respectively, and when both genes are deleted, klp6 deletion phenotypes become epistatic. The measurement of dwell time in various mutants showed a marginal reduction in Δklp5 deletions (5%), whereas it was increased in the Δklp6 or Δklp5 Δklp6 mutant (56 or 34% respectively; Table 2). However, because the standard deviations were relatively high in all these measurements, it would be, at the moment, fair to say that we could not make solid conclusions as to whether Klp5 and Klp6 regulate shrinkage rate or dwell time (see Discussion). It should be noted however that consistent with the persistent existence of cytoplasmic microtubules upon mitotic entry, Klp5N+Klp5N cells exhibited a significant decrease in shrinkage rate (35%) and increase in dwell time (70%; Table 2).
All the klp5/6 Mutants Display Suppression of Catastrophe/Rescue Frequency and Dynamicity
In clear contrast to growth and shrinkage rates, we did observe consistent reductions in other three parameters, i.e., catastrophe and rescue frequency and microtubule dynamicity. In these cases, the quantified values were very similar, if not identical, between each single and double deletion mutants (7–15% reduction; Figure 6, C–E, and Table 2; see Supplemental Figure S5 for more quantitative data set). It would be important to point out that these values were further dropped in Klp5N+Klp5N cells (30–47% reduction), again consistent with the notion that cytoplasmic microtubules in cells containing two identical Klp5-kinesin domains acquire more static properties, thereby being hyperstabilized. Together, the measurement of microtubule dynamics in various klp5/6 mutants has highlighted the following three points on the roles for fission yeast kinesin 8 in microtubule dynamics. First, Klp5 and Klp6 promote catastrophe/rescue frequency and microtubule dynamicity without affecting significantly rates of growth and shrinkage. Second, consistent with previous genetic data (Garcia et al., 2002a; b), these two molecules mostly play noncomplementary roles, single and double mutants show similar, if not identical, defects in microtubule dynamics. Finally, when two kinesin domains derived from Klp5 exist in trans in a single cell, microtubules become extremely stabilized and tend to lose dynamic natures, leading to deleterious chromosome missegregation. We, therefore, propose that Klp5 and Klp6 act as crucial regulatory molecules that confer microtubule dynamisms during both interphase and mitosis.
DISCUSSION
Fission yeast kinesin-8 Klp5 and Klp6 are required for length control of both cytoplasmic and nuclear spindle microtubules, proper chromosome oscillation/alignment, and kinetochore–microtubule attachments (West et al., 2001, 2002; Garcia et al., 2002a,b; Sanchez-Perez et al., 2005; Griffiths et al., 2008). Recently, it has become clear that these features are conserved in human kinesin-8 Kif18A (Zhu et al., 2005; Mayr et al., 2007; Stumpff et al., 2008). Previous work shows that Klp5 and Klp6 form a complex; colocalize to interphase microtubules, mitotic kinetochores, and nuclear spindles; and play noncomplementary roles in structural integrity of microtubules during both interphase and mitosis (West et al., 2001, 2002; Garcia et al., 2002a,b). In this study we unveil the physiological significance of mutual requirements of Klp5 and Klp6 using various types of mutant constructs. Our analysis shows that the existence of the N-terminal Klp5 and Klp6 kinesin domains in a trans configuration is necessary for at least three reasons. One is to prevent premature nuclear export of Klp5/6 molecules during mitosis. The second is for concerted control of microtubule dynamics, in particular frequency of catastrophe/rescue and dynamicity. The third is for spatiotemporal regulation of microtubule morphogenesis, especially at the interphase-to-mitosis transition.
Nuclear Entry and Retention of Klp5 and Klp6 during Mitosis
Our analysis indicates that Klp5 and Klp6 undergo constant shuttling between the cytoplasm and the nucleus. During interphase, their nuclear export exceeds import, whereas during mitosis this is reversed, leading to nuclear retention. Critically, this nuclear retention requires both Klp5 and Klp6. How is this regulation achieved? Although we show that Klp5 is not anchored in the cytoplasm during interphase via binding to cytoplasmic microtubules, Klp5 and Klp6, perhaps as a heterocomplex, might bind nuclear spindle microtubules or other nuclear substructures more tightly during mitosis. In line with this notion, we showed previously that nuclear localization of Klp5 is substantially diminished in alp14 mutants that are defective in mitotic spindle formation (Garcia et al., 2001, 2002a). Another possibility, not mutually exclusive, is that the association between Klp5 and Klp6 masks individual NESs, as described for oligomerized p53 or a heterocomplex of NF-AT and calcineurin (Stommel et al., 1999; Zhu and McKeon, 1999). Several attempts to identify the NES within Klp5 and Klp6 have not been successful. It is possible that Klp5 and Klp6 are exported to the cytoplasm by binding other factors that contain an NES. Determination of NESs within Klp5 and Klp6 or Klp5/6-interacting proteins would be the next step to understand the spatiotemporal control of Klp5/6 localization in detail.
Given that NLS activity of Klp5 is less efficient than that of Klp6 during interphase, another possibility, again not mutually exclusive, is that Klp5 nuclear import is coordinated with the cell cycle, i.e., its NLS is potentiated during mitosis. Consistently, the Klp5 NLS contains a consensus CDK site, SPKK (689–692), whereas the Klp6 NLS does not (Figure 2A). One feasible scenario is that CDK phosphorylation of this site augments NLS activity. If more Klp5 were imported into the nucleus during mitosis, this would result in enhanced formation of mitotic Klp5/6 complexes, which are retained in the nucleus.
Biochemical Activities of Klp5 and Klp6
Although the heterodimerization of kinesin-8 molecules has not been described in other organisms, the kinesin-2 molecules Kif3A and Kif3B are known to heterodimerize in human cells (Yamazaki et al., 1995; Zhang and Hancock, 2004). In this case, the two heads (kinesin domains) have distinct properties, a Kif3A/3A homodimer moves more slowly than the wild-type heterodimer, whereas a Kif3B/3B homodimer moves quickly but shows reduced processivity (Zhang and Hancock, 2004). It is therefore suggested that the Kif3B head accelerates detachment of the Kif3A head from the microtubule, thereby ensuring a balance between motility speed and processivity in the Kif3A/3B heterodimer. In addition, budding yeast Kar3 (kinesin-14) forms a heterodimer with a kinesin-like but nonmotor protein Vik1 (Allingham et al., 2007). Intriguingly Vik1 alone binds microtubules more tightly than Kar3 does and seems to play a cooperative role in microtubule decoration and motility of the Kar3/Vik1 heterodimer.
Unlike Klp5, Vik1 lacks an ATP binding site, but the differing microtubule binding properties of Vik1 and Kar3 seem to be parallel with those of Klp5 and Klp6 reported in this study. We reproducibly observe more intense residual colocalization of Klp5 to microtubules upon LMB treatment than that of Klp6 (Figures 1, 2, and 4). This suggests that a major role of Klp5 lies in its ability to bind microtubules, whereas Klp6 may play other roles. The existence of Klp5 kinesin domains in a trans configuration (Klp5N+Klp5N) is deleterious to the cell, because cytoplasmic microtubules fail to disassemble due to remarkable reductions in microtubule dynamisms. Perhaps the Klp5 kinesin domains, especially in trans, bind/paint microtubules too tightly, stabilizing the straight, stable conformation of the tubulin protofilaments so that dissociation of tubulin subunits only occurs very slowly. Given that other kinesin-8 members have both microtubule depolymerizing and motor activities (Gupta et al., 2006; Varga et al., 2006; Mayr et al., 2007), one intriguing model is that Klp6 confers depolymerizing and/or motor activity and yet allows detachment of Klp5/6 molecules from the microtubule. Obviously, biochemical work, including in vitro microtubule binding assay and measurement of enzymatic activities, will be of critical importance for the future study of Klp5 and Klp6 kinesin-8.
Comparison of Fission Yeast Klp5/6 with Kinesin 8 Molecules from Other Organisms
Careful measurements of several parameters representing microtubule dynamics show that Klp5 and Klp6 play a key role in promoting catastrophe/rescue frequency and microtubule dynamicity. We do see reductions of these parameters in both single and double mutants; and importantly, the values are mostly similar, if not identical, between Δklp5, Δklp6 and Δklp5 Δklp6 strains. This result substantiates, although does not prove, the notion that the formation of the Klp5–Klp6 heterocomplex is vital to Klp5/6 function in vivo (Garcia et al., 2002b). Furthermore, it might imply that Klp5 and Klp6 are involved in promoting microtubule dynamics in general, in addition to putative microtubule depolymerizing activities. Like Klp5 and Klp6, budding yeast kinesin 8 Kip3 reportedly promotes catastrophe/rescue frequency (Gupta et al., 2006). Interestingly, however, Kip3 suppresses microtubule dynamicity and shrinkage rate, which we do not see in fission yeast kinesin 8. Human Kif18A suppresses shrinkage rate during mitosis (Stumpff et al., 2008), although it is also reported human kinesin 8 promotes this parameter instead (Mayr et al., 2007). Drosophila Klp67A inhibits spindle flux at the minus end (Buster et al., 2007). It seems that consensus properties of kinesin 8 molecules are that these molecules play no major roles in microtubule growth rate but promote frequencies of microtubule catastrophe and rescue. Whether these apparent divergent features of kinesin 8 could be explained by its intrinsic enzymatic activities, plus-end–directed motor and depolymerizing functions, or they might reflect species-specific biophysical properties of tubulin/microtubules per se is open for the future study.
Toward Understanding the Spatiotemporal Regulation of Microtubule Dynamics and Morphogenesis
We recently showed that fission yeast Alp7/TACC in complex with Alp14/TOG is a critical target of the Ran-GTPase machinery in bipolar spindle formation (Sato et al., 2004; Sato and Toda, 2007). But there is ample evidence suggesting that the Alp7–Alp14 complex is not the sole factor for Ran/importin-dependent spindle formation (Clarke and Sazer, 2007; Sato and Toda, 2007). Classical, canonical NLSs were identified in the C termini of Klp5 and Klp6, so both molecules are probably under the direct control of Ran and importins. In addition, other kinesins such as kinesin-5 (e.g., Eg5), kinesin-10 (e.g., kid), and kinesin-14 (e.g., XCTK2) are effectors for Ran-dependent spindle formation (Zheng, 2004), although it is not known whether kinesin-8 members in higher eukaryotes are regulated by the Ran system.
Intriguingly, Alp7 and Alp14 shuttle between the cytoplasm and the nucleus in an NLS-dependent and LMB-sensitive manner (Sato and Toda, 2007), reminiscent of Klp5/6 localization patterns. Furthermore, Alp7/14 and Klp5/6 regulate microtubule dynamics and morphologies during both interphase and mitosis in an antagonistic but coordinated manner (Garcia et al., 2001, 2002a,b). Whereas Alp7/Alp14 acts mainly as a microtubule stabilizing/assembly factor, Klp5/6 plays a destabilizing role. We therefore envisage that common regulatory mechanisms, involving Ran-importin/exportin, exist to ensure proper spatiotemporal control of microtubule morphogenesis and possibly other processes. Elucidation of such mechanisms would lead to a better understanding of microtubule-dependent cell morphogenesis, mitotic spindle assembly, and coordinated chromosome segregation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eric Chang for EGFP-klp5 and EGFP-klp6 plasmids, Yuji Chikashige for pCST138/pYC19 (Kobe Advanced ICT Research Center, Japan), Minoru Yoshida for LMB, and Roger Y. Tsien (UCSD) for pRSETB-mCh. We thank Rafael Edgardo Carazo Salas (Cancer Research UK, London, United Kingdom) for technical support of the Deltavision microscope and Rob Cross and Muriel Erent (Marie Curie Research Institute, Surrey, United Kingdom) for discussion. Cancer Research UK (to T. T. and A. U.) and Medical Research Council (to A. U.) supported this work.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0224) on September 17, 2008.
REFERENCES
- Allingham J. S., Sproul L. R., Rayment I., Gilbert S. P. Vik1 modulates microtubule-Kar3 interactions through a motor domain that lacks an active site. Cell. 2007;128:1161–1172. doi: 10.1016/j.cell.2006.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K., Kume K., Kanai M., Goshima T., Miyahara K., Dhut S., Tee W. W., Hirata D., Toda T. The V260I mutation in fission yeast α-tubulin Atb2 affects microtubule dynamics and EB1-Mal3 localization and activates the Bub1 branch of the spindle checkpoint. Mol. Biol. Cell. 2006;17:1422–1436. doi: 10.1091/mbc.E05-08-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asbury C. L. Kinesin: world's tiniest biped. Curr. Opin. Cell Biol. 2005;17:89–97. doi: 10.1016/j.ceb.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Bähler J., Wu J., Longtine M. S., Shah N. G., McKenzie A., III, Steever A. B., Wach A., Philippsen P., Pringle J. R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Busch K. E., Brunner D. The microtubule plus end-tracking proteins mal3p and tip1p cooperate for cell-end targeting of interphase microtubules. Curr. Biol. 2004;14:548–559. doi: 10.1016/j.cub.2004.03.029. [DOI] [PubMed] [Google Scholar]
- Buster D. W., Zhang D., Sharp D. J. Poleward tubulin flux in spindles: regulation and function in mitotic cells. Mol. Biol. Cell. 2007;18:3094–3104. doi: 10.1091/mbc.E06-11-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter N. J., Cross R. A. Mechanics of the kinesin step. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- Clarke P. R., Sazer S. Mitosis: Ran scales the Alps of spindle formation. Curr. Biol. 2007;17:R643–R645. doi: 10.1016/j.cub.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Desai A., Mitchison T. J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Laskey R. A. Nuclear targeting sequences—a consensus? Trends Biochem. Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Dong C., Li Z., Alvarez R., Jr., Feng X.-H., Goldschmidt-Clermont P. J. Microtubule binding to smads may regulate TGFβ activity. Mol. Cell. 2000;5:27–34. doi: 10.1016/s1097-2765(00)80400-1. [DOI] [PubMed] [Google Scholar]
- Drummond D. R., Cross R. A. Dynamics of interphase microtubules in Schizosaccharomyces pombe. Curr. Biol. 2000;10:766–775. doi: 10.1016/s0960-9822(00)00570-4. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno M., Yoshida M., Mattaj I. W. CRM1 Is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Asano S., Nakamura T., Adachi M., Yoshida M., Yanagida M., Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Garcia M. A., Koonrugsa N., Toda T. Spindle-kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J. 2002a;21:6015–6024. doi: 10.1093/emboj/cdf611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M. A., Koonrugsa N., Toda T. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr. Biol. 2002b;12:610–621. doi: 10.1016/s0960-9822(02)00761-3. [DOI] [PubMed] [Google Scholar]
- Garcia M. A., Vardy L., Koonrugsa N., Toda T. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 2001;20:3389–3401. doi: 10.1093/emboj/20.13.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M. K., Odde D. J., Bloom K. Kinesin-8 molecular motors: putting the brakes on chromosome oscillations. Trends Cell Biol. 2008;18:307–310. doi: 10.1016/j.tcb.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D., Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Griffiths K., Masuda H., Dhut S., Toda T. Fission yeast dam1–A8 mutant is resistant to and rescued by an anti-microtubule agent. Biochem. Biophys. Res. Commun. 2008;368:670–676. doi: 10.1016/j.bbrc.2008.01.156. [DOI] [PubMed] [Google Scholar]
- Gupta M. L., Jr., Carvalho P., Roof D. M., Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Haller K., Rambaldi I., Daniels E., Featherstone M. Subcellular localization of multiple PREP2 isoforms is regulated by actin, tubulin, and nuclear export. J. Biol. Chem. 2004;279:49384–49394. doi: 10.1074/jbc.M406046200. [DOI] [PubMed] [Google Scholar]
- Howard J., Hyman A. A. Microtubule polymerases and depolymerases. Curr. Opin. Cell Biol. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Kline-Smith S. L., Walczak C. E. Mitotic spindle assembly and chromosome segregation; refocusing on microtubule dynamics. Mol. Cell. 2004;15:317–327. doi: 10.1016/j.molcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Kutay U., Güttinger S. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 2005;15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Lawrence C. J., et al. A standardized kinesin nomenclature. J. Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chang E. C. Schizosaccharomyces pombe Ras1 effector, Scd1, interacts with Klp5 and Klp6 kinesins to mediate cytokinesis. Genetics. 2003;165:477–488. doi: 10.1093/genetics/165.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H., Miyamoto R., Haraguchi T., Hiraoka Y. The carboxy-terminus of Alp4 alters microtubule dynamics to induce oscillatory nuclear movement led by the spindle pole body in Schizosaccharomyces pombe. Genes Cells. 2006;11:337–352. doi: 10.1111/j.1365-2443.2006.00947.x. [DOI] [PubMed] [Google Scholar]
- Matsuyama A., et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Mayr M. I., Hummer S., Bormann J., Gruner T., Adio S., Woehlke G., Mayer T. U. The human kinesin Kif18A Is a motile microtubule depolymerase essential for chromosome congression. Curr. Biol. 2007;17:489–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Miki H., Okada Y., Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Mitchison T. J., Salmon E. D. Mitosis: a history of division. Nat. Cell Biol. 2001;3:E17–E21. doi: 10.1038/35050656. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analyses of fission yeast. Schizosaccharomyces pombe. Methods Enzymol. 1991;194:773–782. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B., Bachelerie F., Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Poon I. K., Jans D. A. Regulation of nuclear transport: central role in development and transformation? Traffic. 2005;6:173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S., Mishra M., Balasubramanian M. K. Schizosaccharomyces pombe homolog of Survivin, Bir1p, exhibits a novel dynamic behavior at the spindle mid-zone. Genes Cells. 2006;11:815–827. doi: 10.1111/j.1365-2443.2006.00980.x. [DOI] [PubMed] [Google Scholar]
- Sagolla M. J., Uzawa S., Cande W. Z. Individual microtubule dynamics contribute to the function of mitotic and cytoplasmic arrays in fission yeast. J. Cell Sci. 2003;116:4891–4903. doi: 10.1242/jcs.00796. [DOI] [PubMed] [Google Scholar]
- Sanchez-Perez I., Renwick S. J., Crawley K., Karig I., Buck V., Meadows J. C., Franco-Sanchez A., Fleig U., Toda T., Millar J. B. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J. 2005;24:2931–2943. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Dhut S., Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- Sato M., Toda T. Alp7/TACC is a crucial target in Ran-GTPase-dependent spindle formation in fission yeast. Nature. 2007;447:334–337. doi: 10.1038/nature05773. [DOI] [PubMed] [Google Scholar]
- Sato M., Vardy L., Garcia M. A., Koonrugsa N., Toda T. Interdependency of fission yeast Alp14/TOG and coiled coil protein Alp7 in microtubule localization and bipolar spindle formation. Mol. Biol. Cell. 2004;15:1609–1622. doi: 10.1091/mbc.E03-11-0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Stommel J. M., Marchenko N. D., Jimenez G. S., Moll U. M., Hope T. J., Wahl G. M. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J., von Dassow G., Wagenbach M., Asbury C., Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso R. J., Jordan M. A., Farrell K. W., Matsumoto B., Wilson L. Kinetics stabilization of microtubule dynamic instability in vitro by vinblastin. Biochemistry. 1993;32:1285–1293. doi: 10.1021/bi00056a013. [DOI] [PubMed] [Google Scholar]
- Vale R. D., Reese T. S., Sheetz M. P. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V., Helenius J., Tanaka K., Hyman A. A., Tanaka T. U., Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Veal E. A., Day A. M., Morgan B. A. Hydrogen peroxide sensing and signaling. Mol. Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- West R. R., Malmstrom T., McIntosh J. R. Kinesins klp5+ and klp6+ are required for normal chromosome movement in mitosis. J. Cell Sci. 2002;115:931–940. doi: 10.1242/jcs.115.5.931. [DOI] [PubMed] [Google Scholar]
- West R. R., McIntosh J. R. Novel interactions of fission yeast kinesin 8 revealed through in vivo expression of truncation alleles. Cell Motil. Cytoskeleton. 2008;65:626–640. doi: 10.1002/cm.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R. R., Malmstrom T., Troxell C. L., McIntosh J. R. Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol. Biol. Cell. 2001;12:3919–3932. doi: 10.1091/mbc.12.12.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L. Microtubule-depolymerizing kinesins. Curr. Opin. Cell Biol. 2005;17:82–88. doi: 10.1016/j.ceb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Yamashita A., Sato M., Fujita A., Yamamoto M., Toda T. The roles of fission yeast Ase1 in mitotic cell division, meiotic nuclear oscillation and cytokinesis checkpoint signaling. Mol. Biol. Cell. 2005;16:1378–1395. doi: 10.1091/mbc.E04-10-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H., Nakata T., Okada Y., Hirokawa N. KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J. Cell Biol. 1995;130:1387–1399. doi: 10.1083/jcb.130.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A., Selvin P. R. Kinesin: walking, crawling or sliding along? Trends Cell Biol. 2005;15:112–120. doi: 10.1016/j.tcb.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Yoneda Y. Nucleocytoplasmic protein traffic and its significance to cell function. Genes Cells. 2000;5:777–787. doi: 10.1046/j.1365-2443.2000.00366.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Hancock W. O. The two motor domains of KIF3A/B coordinate for processive motility and move at different speeds. Biophys. J. 2004;87:1795–1804. doi: 10.1529/biophysj.104.039842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y. G protein control of microtubule assembly. Annu. Rev. Cell Dev. Biol. 2004;20:867–894. doi: 10.1146/annurev.cellbio.20.012103.094648. [DOI] [PubMed] [Google Scholar]
- Zhu J., McKeon F. NF-AT activation requires suppression of Crm1-dependent export by calcineurin. Nature. 1999;398:256–260. doi: 10.1038/18473. [DOI] [PubMed] [Google Scholar]
- Zhu C., Zhao J., Bibikova M., Leverson J. D., Bossy-Wetzel E., Fan J. B., Abraham R. T., Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell. 2005;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S., Chang F. Effects of γ-tubulin complex proteins on microtubule nucleation and catastrophe in fission yeast. Mol. Biol. Cell. 2005;16:2719–2733. doi: 10.1091/mbc.E04-08-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.