Abstract
Proteins establish and maintain a distinct intracellular localization by means of targeting, retention, and retrieval signals, ensuring most proteins reside predominantly in one cellular location. The enzymes involved in the maturation of lamin A present a challenge to this paradigm. Lamin A is first synthesized as a 74-kDa precursor, prelamin A, with a C-terminal CaaX motif and undergoes a series of posttranslational modifications including CaaX processing (farnesylation, aaX cleavage and carboxylmethylation), followed by endoproteolytic cleavage by Zmpste24. Failure to cleave prelamin A results in progeria and related premature aging disorders. Evidence suggests prelamin A is imported directly into the nucleus where it is processed. Paradoxically, the processing enzymes have been shown to reside in the cytosol (farnesyltransferase), or are ER membrane proteins (Zmpste24, Rce1, and Icmt) with their active sites facing the cytosol. Here we have reexamined the cellular site of prelamin A processing, and show that the mammalian and yeast processing enzymes Zmpste24 and Icmt exhibit a dual localization to the inner nuclear membrane, as well as the ER membrane. Our findings reveal the nucleus to be a physiologically relevant location for CaaX processing, and provide insight into the biology of a protein at the center of devastating progeroid diseases.
INTRODUCTION
The nuclear envelope consists of an inner and outer membrane. The outer nuclear membrane (ONM) is continuous with the endoplasmic reticulum (ER) and connects with the inner nuclear membrane (INM) at the nuclear pore membrane (POM; Lusk et al., 2007). Integral membrane proteins that reside in the INM are initially inserted into the endoplasmic reticulum (ER) membrane and move via the POM to the INM where they maintain a concentrated localization by binding to chromatin or tethering to the nuclear lamina (Powell and Burke, 1990; Soullam and Worman, 1993; Holmer and Worman, 2001; Ohba et al., 2004; Lusk et al., 2007; Schirmer and Foisner, 2007). Until recently, conventional wisdom held that the localization of proteins to either the ER membrane or the INM is mutually exclusive. For example, proteins that are found within the INM, such as LAP1, are not generally found throughout the ER/ONM; likewise, ER proteins such as HMG-CoA reductase are strictly localized to the ER (Wright et al., 1988; Powell and Burke, 1990; Deng and Hochstrasser, 2006). It was recently shown that a notable exception to this paradigm is the yeast E3 ubiquitin ligase, Doa10p. Doa10p was long known to reside in the ER membrane (Swanson et al., 2001; Kreft et al., 2006). However, recent data indicate that Doa10p exhibits dual steady-state localizations, residing and functioning in both the ER membrane where it mediates ER-associated degradation of membrane and secretory proteins, and in the INM where it mediates degradation of the nuclear transcription factor MATα2 (Deng and Hochstrasser, 2006).
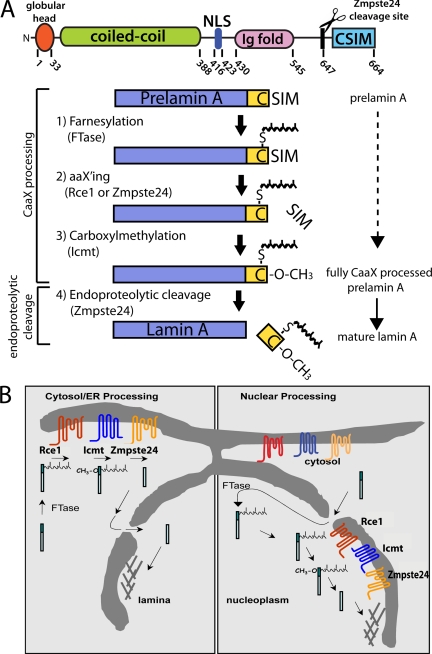
The present study documents another exception to the mutual exclusivity of ER membrane and INM localization with regard to the cellular CaaX-processing enzymes. CaaX processing constitutes a series of posttranslational modifications for a subset of cellular proteins (e.g., the oncoprotein Ras, the nuclear lamins, and the yeast mating pheromone a-factor) that contain a C-terminal CaaX motif (C, cysteine, a, aliphatic amino acid, X, any amino acid; Zhang and Casey, 1996; Young et al., 2005; Wright and Philips, 2006). CaaX processing affects the membrane binding affinity, localization, and functional activity of proteins (Gelb et al., 2006). This processing pathway is shown in the context of the mammalian nuclear scaffold protein lamin A in Figure 1A, steps 1–3. CaaX processing involves 1) farnesylation of the CaaX cysteine by farnesyltransferase (FTase), a soluble heterodimer consisting of α and β subunits, 2) cleavage of the aaX (by Rce1 or Zmpste24), and 3) carboxylmethylation by Icmt. The postprenylation CaaX-processing enzymes (Rce1, Zmpste24 and Icmt) are multispanning integral membrane proteins of the ER membrane oriented with their active site facing the cytoplasm. This ER localization has been well established in both yeast and mammalian cell systems (Dai et al., 1998; Romano et al., 1998; Schmidt et al., 1998; Corrigan et al., 2005; Wright and Philips, 2006).
Figure 1.
Schematics depicting the structure, steps and possible cellular sites of posttranslational processing of prelamin A. (A) Diagram showing the structural domains and corresponding amino acid numbers for human prelamin A (top). Processing steps and enzymes involved in the conversion of prelamin A to mature lamin A (bottom, see text for details). Steps 1–3 constitute CaaX-processing steps common to many proteins and are carried out by the enzymes indicated in parentheses, whereas step 4, mediated by Zmpste24, is an endoproteolytic cleavage event unique to prelamin A that cleaves the final 15 amino acids from prelamin A to yield mature lamin A. Note that the aaX'ing enzyme for prelamin A (either Rce1 or Zmpste24) has not yet been established. (B) Two opposing models for the intracellular location of prelamin A processing. Cytosol/ER processing (left) is supported by knowledge of CaaX enzyme localization to the cytosol (FTase) and ER membranes (Rce1, Icmt, Zmpste24) up to the time of this study. Nuclear processing (right) has been hypothesized based on lamin A localization and kinetic studies (Lehner et al., 1986; Goldman et al., 1992). Evidence for the presence of the processing enzymes on the INM is provided in the present study.
Of the two major types of nuclear lamin proteins (A- and B-type lamins), lamin A is distinctive in that after CaaX processing, the modified tail is cleaved off by Zmpste24 (Figure 1A, step 4; Bergo et al., 2002; Pendas et al., 2002). This final cleavage event is critically important for lamin A as several human disorders result from a lack of cleavage, including the premature aging disorder Hutchinson-Gilford Progeria Syndrome (HGPS) and the related progeroid disorders restrictive dermopathy (RD) and some forms of mandibuloacral dysplasia (MAD-B; Scaffidi et al., 2005; Capell and Collins, 2006; Ramirez et al., 2006; Young et al., 2006; Kudlow et al., 2007). It is notable that B-type lamins do not undergo endoproteolytic cleavage by Zmpste24 and retain their CaaX modifications, whereas lamin C (a splice variant of the lamin A/C gene) lacks a CaaX motif and is not modified at all.
Most proteins undergo farnesylation in the cytosol and postprenylation CaaX processing at the cytosolic face of the ER (Winter-Vann and Casey, 2005; Gelb et al., 2006). CaaX proteins that are processed by these enzymes have a variety of final intracellular localizations. For instance, Ras is located at the plasma membrane and endomembranes (Choy et al., 1999), Pex19 resides in the peroxisomal membrane (Gotte et al., 1998), and lamins and CENP-F are nuclear proteins (Liao et al., 1995; Ashar et al., 2000; Gruenbaum et al., 2005). Although it is possible that prelamin A and/or lamin B undergo processing in the cytosol and at the ER, followed by import into the nucleus via the nuclear localization sequence (NLS; Figure 1B, left), there is evidence to suggest that nuclear lamins may be an exception and that lamin A processing occurs within the nucleus (Figure 1B, right). For instance, kinetic studies suggest that nuclear import (observed to be ∼5 min for microinjected prelamin A protein) proceeds much more rapidly than the appearance of mature lamin A (t1/2 30–100 min; Lehner et al., 1986; Beck et al., 1990; Goldman et al., 1992). Furthermore, cleavage of prelamin A to mature lamin A has been observed to occur concomitantly with partitioning into a Triton X-100 (TX-100)–insoluble particulate subcellular fraction, thought to reflect incorporation into the nuclear lamina matrix (Lehner et al., 1986). These data strongly suggest nuclear processing of lamin A, and have contributed to a long-standing view in the nuclear field that posttranslational processing of lamin A occurs after nuclear import.
It is formally possible that prelamin A is farnesylated and CaaX processed at the ER membrane and then imported, as a membrane associated protein, around the nuclear pore and into the INM. In this case, final cleavage to yield mature lamin A may occur at the INM, consistent with the observations discussed above (Lehner et al., 1986). However, other published data suggests that the entire processing pathway for prelamin A can occur within the nucleus. Notably, nucleoplasmic accumulation of completely unprocessed prelamin A has been observed under a variety of conditions that block farnesylation, the first step of CaaX processing (Lutz et al., 1992; Sasseville and Raymond, 1995). On reversal of this block, lamin A was observed at the nuclear rim. Importantly, none of the above studies conclusively addressed whether, and where, the nuclear accumulated prelamin A is CaaX processed and cleaved. For example, prelamin A could be exported out of the nucleus, processed at the ER, and reimported by diffusion in the membrane around the nuclear pore. Alternatively, it remains possible that the prelamin A within the nucleus may simply incorporate into the lamina in its unprocessed form, because the previous studies suggested, but did not confirm, maturation into lamin A (Lutz et al., 1992; Sasseville and Raymond, 1995).
Here we have critically addressed the nuclear processing model and, as pointed to in previous studies, show that prelamin A can be entirely posttranslationally modified within the nuclear environment. Importantly, we demonstrate that this processing occurs without access to the cytosol or ER membranes. This finding has compelled us to reexamine the localization of the CaaX-processing enzymes and address the possibility of a dual ER/INM localization. We show that CaaX enzymes reside within the INM as well as the ER in both yeast and mammalian cells. Finally, we examine the physiological relevance of nuclear processing by comparing the maturation kinetics of lamin A constructs designed to be either nuclear-excluded (processed in cytosol/ER membrane) or nuclear-contained (processed in nucleoplasm/INM). We find that the kinetics of nuclear processing is similar to cytosol/ER kinetics. Taken together, our evidence supports the view that the nucleus is a physiological CaaX-processing compartment.
MATERIALS AND METHODS
Retroviral Plasmid Constructs
All inducible expression plasmids used in this study were constructed in a retroviral pMX vector containing a selectable hygromycin cassette and an inducible Tet-responsive promoter (TRE; Clontech, Mountain View, CA, pSM2277). Genes were amplified by PCR and standard subcloning techniques were used. All constructs were verified by DNA sequencing. Green fluorescent protein (GFP)-tagged prelamin A was constructed in pSM2277 by fusing GFP (pQBI25, Quantum, Durham, NC) to the 5′ end of human prelamin A open reading frame (ORF) to generate pSM2278. The C661S mutation in GFP-lamin A was introduced into pSM2278 using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions to create pSM2308. Lamin A tail fusion constructs, HAPK-LA388-664 (pSM2279) and HAPK-LA430-664 (pSM2280) were made by fusing hemagglutinin (HA)-tagged mouse pyruvate kinase, PCR amplified from NIH 3T3 cDNA, to the indicated amino acids of human prelamin A. To generate epitope-tagged Zmpste24 fusion constructs, a single HA epitope was inserted into mouse Zmpste24 at amino acid 469 (pSM2282), or directly at the C-terminus at amino acid 476 (pSM2281). Human Icmt was tagged at the C-terminus with a single HA epitope to create pSM2284. Human lamin B receptor was tagged at the C-terminus with RFP to create pSM2307. GFP+NLS was created by fusing amino acids 388–464 of human lamin A, containing the NLS, to the C-terminus of GFP to create pSM2286.
Retroviral Transduction and Creation of Stable, Inducible Cell Lines
All stable cell lines were created using retroviral transduction and selected and maintained as polyclonal lines. Retrovirus was generated by transient cotransfection of HEK293F cells with the packaging plasmid pCL-Eco and an expression construct as described in Naviaux et al. (1996). Cotransfections of HEK293F cells were performed using Fugene (Roche, Indianapolis, IN) according to the manufacturer's protocol. Plasmid DNA was prepared using a Qiagen Maxi Prep kit (Qiagen, Valencia, CA). Each plasmid (5 μg) was used to transfect 1 × 106 HEK293F cells plated in a 35-mm tissue culture dish. Media was changed 18 h after transfection; retrovirus was harvested 48 h after transfection. Retrovirus was used to infect target cells (NIH 3T3 or MEFs) at a 1:1 dilution in the presence of 8 μg/ml polybrene (Sigma, St. Louis, MO) to aid in virus-cell attachment. Media was changed 24 h after infection; selection for integration was applied 48 h after viral infection.
TetOff-inducible cell lines were created by infection of NIH 3T3, icmt−/− MEFs, or zmpste24−/− MEFs with retrovirus encoding the tetracycline-responsive element tTA (pRevTet-Off; Clontech) followed by selection for integration in 200 μg/ml geneticin (Invitrogen, Carlsbad, CA). Retrovirus encoding the expression construct was applied to a TetOff-inducible cell line to establish tetracycline-inducible gene expression. Cell lines were selected and maintained in DMEM supplemented with 10% fetal bovine serum, 400 μg/ml hygromycin (Invitrogen), 200 μg/ml geneticin (Invitrogen), and 10 μg/ml tetracycline (Sigma). Expression was induced by removal of tetracycline from the media for 24–48 h.
Cell Culture Conditions
Mouse NIH 3T3 fibroblasts, HeLa cells, zmpste24−/− MEFs, and icmt−/−MEFs (MEFs obtained from S. Young, UCLA) were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and 100 U/ml penicillin and streptomycin (Invitrogen). Farnesyltransferase inhibitor BMS-214662 (20 mM in DMSO; a gift from M. Gelb, University of Washington) was used in cell culture at a concentration of 1 μM. Cycloheximide (10 mg/ml stock solution dissolved in water; Sigma) was used at a final concentration of 20 μg/ml. We have found that this concentration is sufficient to completely inhibit protein translation in NIH 3T3 cells (J.B., C.H., S.M., our unpublished observation).
Heterokaryon Fusion Assay
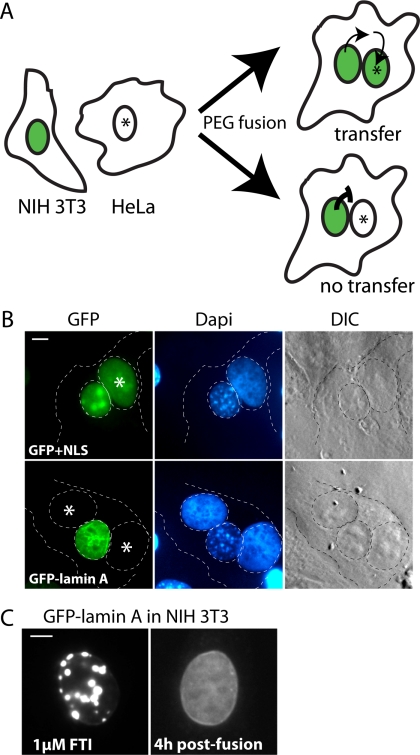
Heterokaryon fusion assays were performed as described previously with modifications (Pinol-Roma and Dreyfuss, 1992). To induce expression of either GFP-prelamin A or GFP+NLS, NIH 3T3 cell lines were incubated for 48 h in six-well plates in the presence of 1 μM farnesyltransferase inhibitor (FTI; BMS 214662) and the absence of tetracycline. HeLa cells, 1 × 106, were seeded on top of the NIH 3T3 cells and allowed to incubate for an additional 3 h. To initiate fusion between NIH 3T3 and HeLa cells, the cells were overlaid with prewarmed 50% PEG-8000 and incubated for 2 min at 37°C. Cells were washed four times with PBS, and incubated in DMEM containing 20 μg/ml cycloheximide and 10 μg/ml tetracycline for 4 h. Coverslips were rinsed with PBS, DAPI-stained for 5 min in 1 μg/ml Hoechst 33258, and mounted in Prolong Gold (Invitrogen) in preparation for standard fluorescence and light microscopy. Heterokaryons were identified by comparison of DAPI and phase images using a Zeiss Axioskop 2 (Zeiss, Thornwood, NY). Proximal nuclei with differential DAPI staining patterns characteristic of mouse cells (spotty) and human cells (diffuse) were identified, and confirmed to constitute heterokaryons arising from cell fusion by phase microscopy. Heterokaryons shown in Figure 3 are representative of those identified in three independent experiments.
Figure 3.
Heterokaryon fusion assay reveals prelamin A remains within the nucleus after FTI washout. (A) Schematic of the heterokaryon fusion assay used to examine the possibility of nuclear/cytosol shuttling of GFP-prelamin A after FTI washout. NIH 3T3 cells treated with FTI and expressing nuclear-accumulated GFP-prelamin A were fused to HeLa cells. After FTI washout, cells were incubated for 4 h and heterokaryons were examined to determine if nuclear transfer had occurred. NIH 3T3 cells expressing GFP fused to a NLS were used as a positive control for nuclear shuttling. (B) In heterokaryons observed 4 h after fusion, GFP+NLS efficiently transfers from a mouse nucleus to a HeLa nucleus (asterisk, top), whereas GFP-lamin A does not transfer to Hela nuclei (bottom). Results shown are representative of those obtained in three independent experiments. In each experiment, 4–6 heterokaryons (positively identified by phase contrast microscopy) were analyzed for each construct. (C) NIH 3T3 cells induced for expression of GFP-lamin A in the presence of 1 μM FTI before fusion with HeLa cells (left). At 4 h after fusion, the GFP-prelamin A chases from the foci to the nucleoplasm and the nuclear rim of the NIH 3T3 cells (right). Bar, 5 μm.
Digitonin Permeabilization and Immunofluorescence
Digitonin was used to selectively permeabilize the plasma membrane as described (Adam et al., 1990; Plutner et al., 1992) with some modifications. Cell lines expressing Zmpste24-HA, Icmt-HA, or lamin B receptor tagged to RFP (LBR-RFP) were grown on coverslips and washed two times with cold wash buffer (110 mM KOAc, 20 mM HEPES-KOH, pH 7.2, 2 mM MgOAc). Coverslips were incubated for 5 min in wash buffer containing 25 μg/ml digitonin. Cells were fixed by rinsing in wash buffer three more times, followed by incubation in 3.7% formaldehyde for 10 min. Coverslips were incubated for 30 min in block buffer (PBS containing 1% BSA) containing either 0.1% TX-100 or 0.1% TX-100 and 25 μg/ml digitonin. Primary antibodies were diluted in block buffer (1:1000 anti-HA 12CA5, Roche, Indianapolis, IN; 1:1000 anti-lamin A, Santa Cruz Biotechnology, Santa Cruz, CA, sc-20680; 1:500 anti-LBR, Epitomics, Burlingame, CA) and incubated with coverslips for 1 h. Coverslips were washed three times in block buffer, and secondary antibodies were applied for 1 h as follows: 1:500 CY3-conjugated goat anti-mouse (for HA) or anti-rabbit (for LBR; Jackson ImmunoResearch Laboratories, West Grove, PA) and 1:500 Alexa-Flour 350 goat anti-rabbit (for lamin A; Invitrogen). Coverslips were rinsed three times in block buffer and once in PBS, and mounted with Prolong Gold for viewing.
Yeast Theta Nuclei Assay
To determine if yeast Ste24p and Ste14p reside on the INM, theta nuclei were induced by overexpression of the nuclear pore complex protein Nup53p as described previously (Marelli et al., 2001; Deng and Hochstrasser, 2006). Wild-type yeast strain SM1058 (MATa, trp1, leu2, ura3, his4, can1) was cotransfected with a pSM2210 expressing Nup53p from the inducible CUP1 promoter (from R. Wozniak, University of Alberta), or an empty vector control, and a plasmid expressing GFP-Ste14p (pSM2138), or GFP-Ste24p (pSM2136). GFP-tagged Mga2p (from M. Hochstrasser, Yale University), expressed from the Gal-inducible promoter was transformed with either Nup53p or empty vector into a ufd1-1 strain to prevent proteosome dependent cleavage. Cells were grown overnight in selective media containing 2% glucose (or containing 2% galactose for expression of GFP-Mga2p) and 100 μM CuSO4 (for induction of Nup53p), and then diluted into early log phase and grown for an additional 4 h. Cells were viewed (>100) and scored for the frequency of theta nuclei from three independent experiments.
Western Blotting and Antibodies
Cells were directly lysed in SDS-PAGE sample buffer, heated to 65°C for 10 min, and briefly sonicated to disrupt chromatin. Standardly, ∼5 × 104 cell equivalents of lysate were loaded on 6% SDS-PAGE gels and subjected to electrophoresis. Western blots were blocked using TBST (100 mM Tris-HCL pH 8, 150 mM NaCl, 0.05% Tween-20) containing 5% Western Blocking Reagent (Roche). Primary antibodies used include 1:1000 polyclonal rabbit anti-lamin A (Santa Cruz, sc-20680), 1:4000 polyclonal goat anti-prelamin A (Santa Cruz, sc-6214). Secondary antibodies used were HRP-conjugated anti-rabbit (GE Healthcare, Waukesha, WI), and HRP-conjugated anti-goat (Santa Cruz, sc-2020) antibodies used at 1:5000. Secondary antibodies were detected using Lumi-light Western Blotting Substrate (Roche). Quantitation of Western blots was performed using ImageJ software (http://rsb.info.nih.gov/ij/).
RESULTS
GFP-Prelamin A Can Be Processed within the Nucleus
Mature lamin A is localized to the nucleus and is distributed between the nuclear rim and the nucleoplasm. The processing site for most CaaX proteins is the cytosolic face of the ER where the CaaX-processing enzymes are located (Figure 1B, left; Dai et al., 1998; Romano et al., 1998; Schmidt et al., 1998; Wright and Philips, 2006). Because the ER membrane localization of Zmpste24 and the CaaX-processing enzymes is incompatible with the previously postulated nuclear location for lamin A processing, we reexamined the cellular location of lamin A biogenesis in order to determine if the nucleus represents a CaaX-processing compartment (Figure 1B, right).
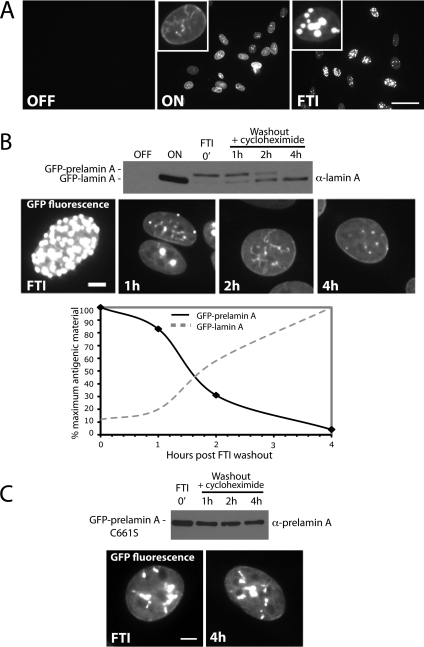
To investigate whether prelamin A can be processed entirely within the nuclear environment, we established a visual and biochemical assay to detect nuclear prelamin A processing. This assay utilizes a stable NIH 3T3 cell line engineered to inducibly express GFP-tagged prelamin A from the Tet Response Element (TET) when tetracycline is removed from the media (Clontech, TetOff). When uninduced, no signal is observed (Figure 2A, top left). When expression of GFP-prelamin A is induced for 48 h, GFP-lamin A is properly localized to the nucleoplasm and the nuclear rim (Figure 2A, top middle), and is properly processed (Figure 2B, top, ON). When induced in the presence of a farnesyltransferase inhibitor (1 μM FTI BMS-214662), the completely unprocessed form of GFP-prelamin A accumulates within the nucleus in large intranuclear foci (Figure 2A, top right). To assay for CaaX processing and subsequent Zmpste24-mediated endoproteolytic cleavage of this nuclear-accumulated and unprocessed pool of GFP-prelamin A, the FTI block was released by washout of the inhibitor. New protein synthesis was blocked with cycloheximide, and cell lysates were prepared and analyzed by Western blot at the indicated time points (Figure 2B). We found that proteolytic cleavage of prelamin A to the mature form under these conditions occurred with an estimated half-life of 100 min (Figure 2B, bottom), similar to what has been shown for prelamin A in mevinolin-treated cells (Beck et al., 1990). Persistent nuclear localization of GFP-lamin A was observed at all time points by microscopy (Figure 2B), strongly suggesting that the conversion to mature lamin A occurs entirely within the nucleus. Furthermore, nuclear maturation of prelamin A is dependent on CaaX processing, as GFP-prelamin A bearing a CaaX motif mutation (C661S) that prevents prenylation fails to be cleaved and fails to chase to the rim under similar conditions (Figure 2C).
Figure 2.
Prelamin A can be CaaX processed and endoproteolytically cleaved after preaccumulation within the nucleus. (A) An NIH 3T3 stable cell line was created to inducibly express GFP-prelamin A using the Clontech TetOff system. When expression is off, no signal is apparent. On induction (on), a nucleoplasmic and rim localization typical for lamin A is observed. When induced in the presence of farnesyltransferase inhibition (FTI), GFP-prelamin A accumulates within the nucleus in bright foci, as has also been documented by us and others (Capell et al., 2005; Glynn and Glover, 2005; Mallampalli et al., 2005; Toth et al., 2005; Yang et al., 2006). Bar, 50 μm. (B) Cycloheximide-chase analysis of preaccumulated nuclear prelamin A upon release from FTI block. Conversion of the GFP-prelamin A to GFP-lamin A corresponds with the disappearance of the prominent foci and appearance of GFP-lamin A at the nuclear rim. Bar, 5 μm. This experiment was performed three times with similar results, and a representative immunoblot is shown here. Quantitation of the above immunoblot reveals the half-life of maturation (disappearance of prelamin A and appearance of mature lamin A) to be approximately ∼100 min. (C) Maturation of nuclear-accumulated GFP-prelamin A is farnesylation dependent, as it does not occur for lamin A C661S. A C661S mutant, in which the CaaX motif cysteine has been mutated to serine, is not cleaved after nuclear preaccumulation and FTI washout (detected by anti-prelamin A antibody). Bar, 5 μm.
Interestingly, the distinctive intranuclear foci that accumulate under conditions of FTI treatment disappear upon inhibitor washout and GFP-lamin A chases to the nuclear rim and nucleoplasm over time, as seen in Figure 2B. Such large intranuclear foci have previously been observed under conditions of FTI treatment in several cell types, yet the nature and potential function of these prelamin A–containing foci remains unknown (Capell et al., 2005; Glynn and Glover, 2005; Mallampalli et al., 2005; Toth et al., 2005; Yang et al., 2005). Our data suggest that these foci represent an intermediate in biogenesis rather than dead-end aggregates, because lamin A chases from the foci to the nuclear rim over time in the absence of new protein synthesis (Figure 2B), but fails to chase to the rim when CaaX processing is blocked (Figure 2C).
Preaccumulated Nuclear-localized GFP-prelamin A Does Not Access the Cytosol after FTI Washout
We wanted to rule out the possibility that intranuclear accumulated GFP-prelamin A could access the cytosol, by diffusion or export, once the FTI had been removed. If GFP-prelamin A were able to undergo nuclear-cytoplasmic shuttling, then access to the cytosolic farnesyltransferase and the ER CaaX-processing enzymes could account for the processing we observed. To examine whether or not GFP-prelamin A undergoes nuclear export, processing, and reimport, we utilized a well-characterized heterokaryon fusion assay (Pinol-Roma and Dreyfuss, 1992; Gaubatz et al., 2001). This assay reveals nuclear-cytosolic shuttling by testing for transfer of a protein between the nuclei of two fused cells (one mouse, one human). Heterokaryons are formed using polyethylene glycol (PEG), and can be identified by the differential DAPI staining pattern of human and mouse cells (diffuse or spotty, respectively). Nuclear transfer is detected in this assay by observing transfer of a fluorescently tagged nuclear protein expressed in the mouse cell to the nucleus of the human cell, which does not express the protein (Figure 3A, top). When nuclear-cytoplasmic shuttling does not occur, fluorescence is restricted to the mouse cell nucleus (Figure 3A, bottom).
We fused the mouse NIH 3T3 cells expressing either 1) a positive control for nuclear transfer, GFP+NLS or 2) GFP-tagged human prelamin A (preaccumulated with FTI treatment) to human HeLa cells. After a brief incubation in PEG to allow fusion, the cells were rinsed to wash out the FTI, incubated in media containing cycloheximide, and heterokaryons were examined 4 h after washout. The control GFP+NLS efficiently transferred from the NIH 3T3 nuclei to the HeLa nuclei (Figure 3B, top) as expected, because it is small enough to diffuse in and out of nuclear pores. In contrast, no transfer of GFP-lamin A occurred in any of the heterokaryons observed from three independent experiments (Figure 3B, bottom). During this experiment, GFP-prelamin A chased from the foci to the nuclear rim of the NIH 3T3 cells as previously observed after FTI washout (Figure 3C), indicating that the removal of the drug was adequate to reverse the processing block. This result suggests that GFP-prelamin A does not access the cytosol through nuclear-cytosolic shuttling during the FTI washout period and provides convincing evidence that GFP-prelamin A can be processed entirely within the nucleus. Our results imply that Zmpste24 and the CaaX-processing enzymes must also be located in the nucleus, in addition to their established ER/cytosol localization. We further address this novel nuclear localization of the enzymes in detail below.
The C-Terminus of Zmpste24, Which Contains a Dilysine-like Motif, Is Critical for Establishing Its ER Localization
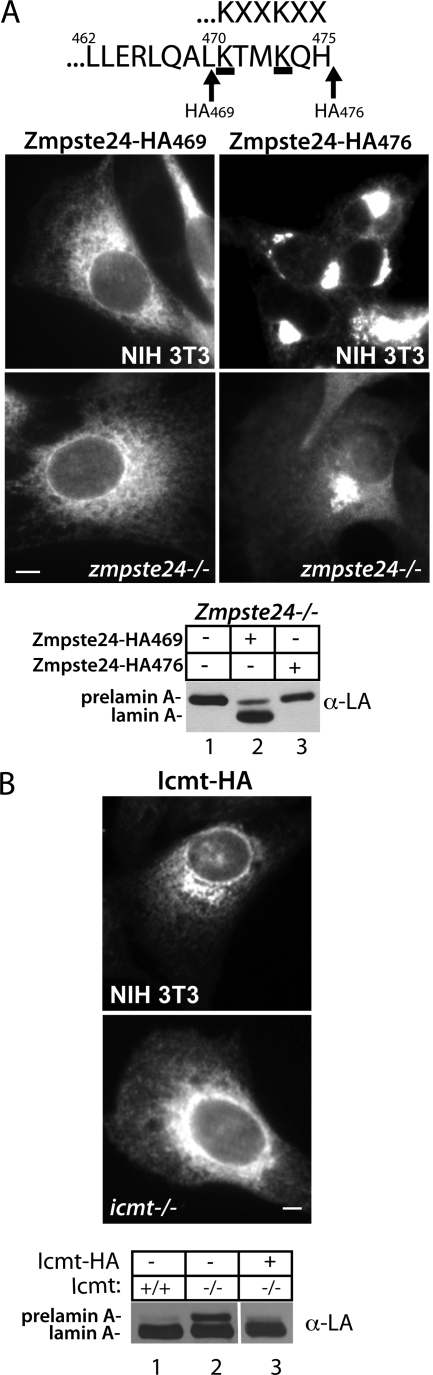
To explore the possibility that the prelamin A endoprotease Zmpste24 may be found within the INM in addition to its established ER membrane localization, we generated and characterized stable cell lines expressing an HA-tagged version of Zmpste24. Interestingly, we found that placing the HA tag directly at the C-terminus (Zmpste24-HA476) disrupted the normal ER localization of Zmpste24, and resulted in a perinuclear localization pattern resembling Golgi (Figure 4A, right) in NIH 3T3 or zmpste24−/− MEFs. A potential explanation for this result is the presence of a noncanonical dilysine ER retrieval motif at the C-terminus, which may function to maintain the steady-state localization of Zmpste24 to the ER. Well-characterized ER retrieval dilysine motifs for ER membrane proteins include KKXX and KXKXX (Jackson et al., 1990, 1993). In Zmpste24, the motif is KXXKXX (Figure 4A, top). When we inserted the HA tag subterminal to the ER retrieval motif (Zmpste24-HA469), the ER localization of Zmpste24 was apparent (Figure 4A, left), as has been previously shown for untagged Zmpste24 (Corrigan et al., 2005). Although this observation provides compelling suggestive evidence for the presence of a functional dilysine motif in Zmpste24, confirmation requires a mutational analysis of this region.
Figure 4.
HA-tagged Zmpste24 and Icmt are functional and localization of Zmpste24 to the ER membrane is dependent on its C-terminus. (A) Localization of tagged versions of Zmpste24 in stable NIH 3T3 cells or zmpste24−/− MEFs by indirect IF with α-HA antibodies. Placement of a HA tag subterminal to a potential dilysine ER retrieval motif (KXXKXXCOOH; Zmpste24-HA469) does not disrupt the normal ER localization of Zmpste24 (left) and restores the cell's ability to cleave prelamin A in zmpste24−/− MEFs (lane 2). In contrast, placement of a HA epitope tag at the C-terminus of Zmpste24 (Zmpste24-HA476) disrupts ER retrieval, resulting in mislocalization to the Golgi (right). Golgi-localized Zmpste24-HA476 fails to restore the ability to cleave prelamin A to zmpste24−/− MEFs (lane 3). (B) Indirect IF and functional analysis of HA-tagged Icmt stably expressed in NIH 3T3 and icmt−/− MEFs. HA-tagged Icmt fully restores the ability of prelamin A to be efficiently processed to mature lamin A in icmt−/− MEFs (compare lane 2 and lane 3). Bar, 5 μm.
To ensure that epitope-tagged Zmpste24 is functional, we expressed Zmpste24-HA469 and Zmpste24-HA476 in MEFs deficient in Zmpste24 activity (zmpste24−/− MEFs), and assayed for the ability to restore prelamin A endoproteolytic cleavage (Figure 4A, bottom). We found that the ability to cleave prelamin A was restored only with the ER-localized Zmpste24-HA469, but not with the Golgi-localized Zmpste24-HA476, thus indicating that the HA tag placed subterminal to the ER retrieval motif did not disrupt function. It is possible that Golgi-localized Zmpste24-HA476 may not be functional either because of its mislocalization away from the prelamin A substrate or its improper folding and activity at the Golgi.
Zmpste24 and Icmt Are Localized to the INM and Nucleoplasm
We noted that Zmpste24-HA469 appeared to exhibit a pronounced nuclear rim-staining pattern by indirect IF, in addition to the extensive reticular ER pattern (Figure 4A). This rim staining could be attributed to either ONM or INM localization (or both), because under normal conditions the ONM cannot be distinguished from the INM at the light microscope level. In Zmpste24-HA469 the HA tag resides in the cytosol. However, if a portion of Zmpste24 were localized to the INM, the HA tag would also reside within the nucleoplasm.
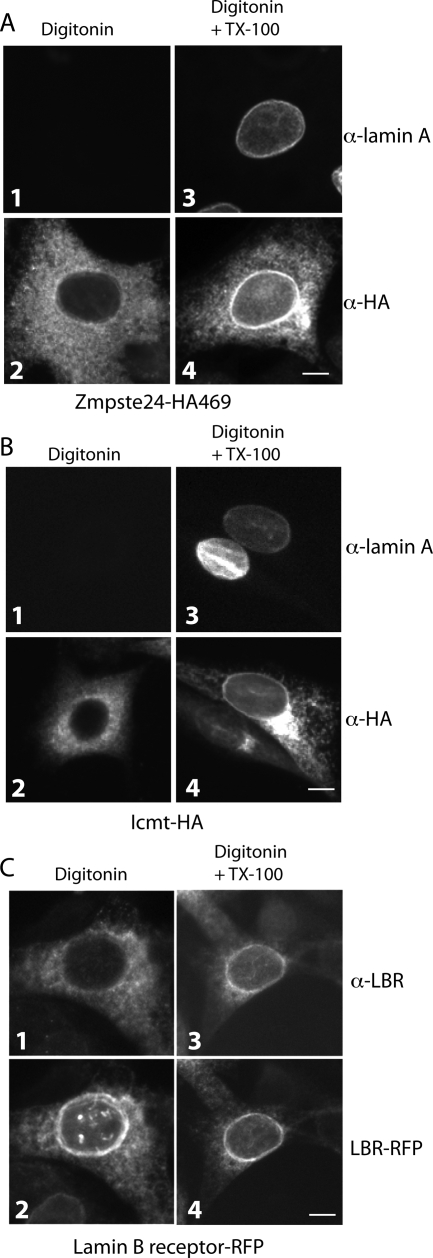
To determine if Zmpste24-HA469 localizes to the INM in addition to the ER membrane, we used standard indirect IF and compared the HA immunostaining pattern in digitonin and TX-100–permeabilized cells. Digitonin permeabilizes the plasma membrane, but not the nuclear envelope (Adam et al., 1990; Hieda et al., 2008). This is in contrast to TX-100, which permeabilizes all cellular membranes. Thus, lamin A staining is absent in digitonin-permeabilized cells (Figure 5A, panel 1), but is present when TX-100 is included (Figure 5A, panel 3). For Zmpste24-HA469, ER staining is apparent with both digitonin and digitonin/TX-100 permeabilization (Figure 5A, panels 2 and 4). Thus, the rim staining is indicative of an INM localization of Zmpste24-HA469.
Figure 5.
Zmpste24 and Icmt exhibit a novel dual localization to the INM, in addition to the ER. (A) Stable zmpste24−/− MEFs expressing Zmpste24-HA469 were permeabilized with digitonin and stained with anti-lamin A (panel 1) or anti-HA (panel 2). Digitonin-permeabilized cells processed in parallel, but in the presence of TX-100, allow antibody access to the INM, as revealed by anti-lamin A staining (panel 3) and the prominent nuclear rim staining displayed by Zmpste24 (panel 4). (B) Stable icmt−/− MEFs expressing Icmt-HA were processed for indirect IF as in A. Prominent nuclear rim staining for lamin A (panel 3) and Icmt (panel 4) is absent in cells permeabilized with digitonin only and is apparent in the presence of TX-100, indicating an INM localization as well as an ER localization (panel 2) for Icmt. (C) NIH 3T3 cells stably expressing RFP-tagged lamin B receptor (LBR-RFP) were permeabilized with digitonin as in A, except anti-LBR antibodies were used in the presence and absence of TX-100. Note the rim-localized portion of LBR-RFP (panels 2 and 4), a well-characterized INM localized protein, is only accessible to anti-LBR antibodies in the presence of TX-100 (panel 3). In all experiments, images are representative of >70% of cells observed. Bar, 5 μm.
We also sought to determine if Icmt, like Zmpste24, resides on the INM in addition to the ER. We generated stable cell lines expressing C-terminally HA-tagged Icmt in both icmt−/− MEFs as well as NIH 3T3 fibroblasts (Figure 4B). As expected, Icmt-HA localized predominantly to the ER in both cell lines. We also observed that a portion of Icmt-HA localized to the Golgi in both lines, as has been seen before in transient transfections (Dai et al., 1998) and which we attribute to overexpression. As noted before, icmt−/− MEFs show a partial block in prelamin A processing (Young et al., 2005), and exhibit a small accumulation of prelamin A at steady state (Figure 4B, lane 2). Expression of Icmt-HA in icmt−/− MEFs completely restored the processing of prelamin A to mature, indicating that Icmt-HA is functional (Figure 4B, compare lane 3 with lane 2). Selective permeabilization experiments using digitonin revealed a prominent nuclear rim staining that was absent in cells permeabilized solely with digitonin, but was readily apparent when TX-100 was included (Figure 5B, panels 2 and 4). Our results suggest that Icmt, like Zmpste24, is dually localized and resides in the INM as well as in the ER.
As an additional control, we also analyzed a cell line expressing the exclusively localized INM protein, lamin B receptor (LBR), tagged with red fluorescent protein (RFP) at its C-terminus. When viewed by direct fluorescence microscopy, LBR-RFP localizes at the nuclear rim, with some ER signal due to overexpression (Figure 5C, panels 2 and 4). However, antibodies to a nucleoplasmic region of LBR do not decorate the population of LBR-RFP present in the nuclear rim when permeabilized with digitonin (Figure 5C, panel 1). In contrast, anti-LBR antibodies clearly also stain the nuclear rim when TX-100 is included (Figure 5C, panel 3).
Nuclear and Cytosolic/ER Processing Kinetics Are Similar
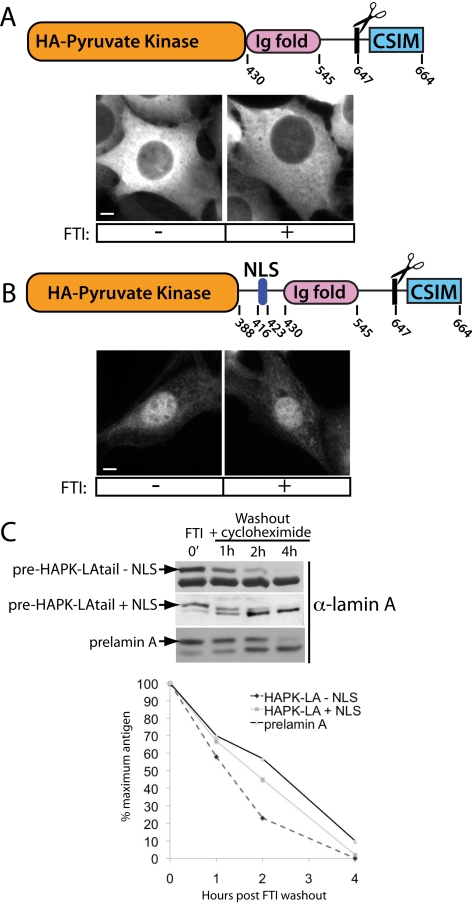
To determine if the nucleus represents a physiologically relevant CaaX-processing compartment we compared the kinetics of lamin A biogenesis inside and outside of the nucleus. To do so, we generated isogenic truncated versions of lamin A based on previously published constructs (Hennekes and Nigg, 1994). One version is unable to enter the nucleus (Figure 6A, top), whereas the other is nuclear (Figure 6B, top). In the former construct, the lamin A tail (aa 430–664) was fused to HA-tagged pyruvate kinase, resulting in a protein too large to diffuse into the nucleus through the nuclear pores (HAPK-LAtail − NLS). The construct lacks the coiled-coil region required for filament formation, preventing polymerization with endogenous lamin A that could result in piggybacking into the nucleus. In the latter case, the NLS of lamin A was added to promote nuclear import (HAPK-LAtail + NLS). On induction, HAPK-LAtail − NLS is excluded from the nucleus as expected (Figure 6A, bottom), whereas the HAPK-LAtail + NLS is nuclear (Figure 6B, bottom).
Figure 6.
Kinetics of complete postttranslational processing of prelamin A to mature is similar inside and outside of the nucleus. (A) Schematic of the fusion protein used to test the maturation kinetics of a cytosolic lamin A fusion (top). The region encoding the N-terminal coiled-coil domain and NLS of lamin A was replaced with HA-tagged pyruvate kinase to prevent directed import and diffusional access to the nucleus. Deletion of the coiled-coil domain is necessary to prevent dimerization with endogenous lamin A and “piggybacking” into the nucleus. Cytosolic localization is maintained when expressed both with and without FTI treatment (bottom). (B) Schematic of the equivalent nuclear-localized fusion used to assay nuclear maturation kinetics. This construct was designed to fuse amino acids 388–664 to pyruvate kinase, and includes the region containing the NLS. This fusion protein localizes to the nucleoplasm in both FTI-treated and untreated cells (bottom). Note the absence of the coiled-coil region of lamin A in this construct likely accounts for the diffuse nucleoplasmic staining and absence of foci in the presence of FTI. Bar, 5 μm. (C) After FTI washout, the maturation kinetics of both nuclear-excluded and nuclear-contained lamin A fusion proteins are similar to the kinetics of endogenous prelamin A maturation. This experiment was performed three times, and a representative immunoblot is shown here. The immunoblots shown here were quantitated, and the amount of prelamin A remaining over time is indicated in the graph.
We assayed the kinetics of processing of these constructs by performing a cycloheximide chase after FTI washout. We found that the disappearance of the prelamin form of the nuclear and cytosolic fusions occurred with kinetics that are similar to each other and to endogenous lamin A (Figure 6C). Interestingly, the cytosolic fusion protein (HAPK-LAtail − NLS) does not completely accumulate in the precursor form in the presence of FTI, compared with the nuclear fusion protein and endogenous lamin A (Figure 6C, 0-min time point). This could reflect an incomplete block of FTase activity in the cytosol by FTI. Our finding that the kinetics of processing are similar for a lamin A fusion protein artificially placed either inside of, or outside of, the nuclear environment, and that both are similar to endogenous lamin A, demonstrates the physiological relevance of nuclear processing.
The Yeast CaaX-processing Enzymes Ste14p and Ste24p Also Reside on the INM
It was recently shown that the yeast integral membrane E3 ubiquitin ligase, Doa10p, localizes to the INM, in addition to its well-established ER membrane localization (Deng and Hochstrasser, 2006). This result is the first example in yeast of a functional ER-localized protein also residing and functioning within the INM, with no obvious targeting mechanism (i.e., no known NLS) and with no specific concentration within the INM.
Because our results indicate that the CaaX-processing enzymes are localized to both the ER and INM in mammalian cells, we wanted to reexamine the established ER localization of the yeast Icmt and Zmpste24 homologues (Ste14p and Ste24p, respectively). Specifically, we sought to determine whether they exhibit a similar dual localization pattern as their mammalian counterparts. Although yeast are not predicted to have nuclear lamins, there are several CaaX proteins in yeast that contain a predicted NLS.
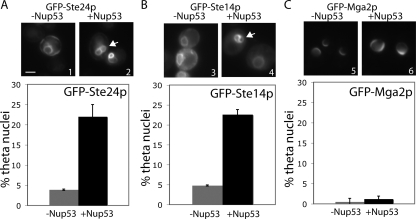
To examine the potential localization of the CaaX enzymes to the INM in yeast we used the “theta nuclei” assay (Deng and Hochstrasser, 2006). This assay takes advantage of the observation that overexpression of nuclear pore proteins, such as Nup53p, causes massive proliferation of the INM (Marelli et al., 2001). The INM transects the two sides of the nucleus in some cells under these circumstances. Proteins that localize on the INM under these conditions would be seen to form the Greek letter theta, θ, thus giving this assay its name. When we looked at the localization of GFP-tagged Ste24p and Ste14p under conditions of Nup53 overexpression, we found that a significant number of cells exhibited theta nuclei, indicating an INM localization (Figure 7, A and B). The negative control, the ER-bound membrane protein Mga2p, tested negative in this assay (Figure 7C). We also examined the integral ER membrane protein, Hrd1p, in this assay and found that, as has been previously published, it also tests negative (J.B., S.M., our unpublished observation; Deng and Hochstrasser, 2006). This result provides evidence for the first time that the yeast CaaX-processing enzymes also reside within the nuclear environment in the INM, in addition to their well-established ER localization. Importantly, we have confirmed that the dual localization of the CaaX-processing enzymes is conserved from yeast to mammals. This observation is particularly intriguing in light of the absence of nuclear lamin homologues in yeast, but suggests that yeast CaaX proteins with an NLS could be processed in the nucleus.
Figure 7.
Yeast Ste24p and Ste14p show an INM localization pattern using a theta (θ) nuclei assay. Overexpression of the nuclear pore complex protein Nup53 causes proliferation of the INM, such that the INM develops a figure eight (i.e., theta-like) structure (arrows). Theta nuclei are observed with a GFP-tagged protein that resides solely in the INM or in both the INM and ER, but not observed with a strictly ER membrane protein such as Mga2p. Yeast cells constitutively expressing the indicated GFP-tagged constructs were induced (panels 2, 4, and 6) or not (panels 1, 3, and 5) for high level Nup53p expression. >100 nuclei were counted in three separate experiments. Bar, 2 μm.
DISCUSSION
This study resolves a paradoxical issue regarding the intracellular location of prelamin A maturation. Early evidence had suggested that prelamin A is processed within the nucleus (Lehner et al., 1986; Beck et al., 1990; Goldman et al., 1992; Lutz et al., 1992; Sasseville and Raymond, 1995). However, the postprenylation CaaX-processing enzymes Icmt, Zmpste24, and possibly Rce1 are known to be integral membrane ER-resident proteins with their active sites facing the cytosol, suggesting that prelamin A could be processed before nuclear import (Romano et al., 1998; Schmidt et al., 1998; Corrigan et al., 2005). Here we show that the CaaX-processing enzymes are dually localized to the INM, in addition to their previously characterized ER membrane localization. Our data reconciles the paradox surrounding nuclear lamin A processing, and indicates that the nucleus is a CaaX-processing compartment, in addition to the ER membrane where Ras, Rho, a-factor, and other CaaX proteins are known to be modified (Gelb et al., 2006; Wright and Philips, 2006).
Our findings provide a mechanistic and logistical basis for earlier evidence suggesting that the nuclear import of prelamin A via its strong NLS precedes its processing (Lehner et al., 1986; Lutz et al., 1992; Sasseville and Raymond, 1995). Confirmation of nuclear processing of lamin A also significantly impacts speculation of the as-yet-undetermined fate of the lamin A tail that is released by Zmpste24-mediated endoproteolytic cleavage (Figure 1A, step 4). Whether the tail serves a cellular function, or is degraded, the production of the free prenylated, carboxylmethylated 15-mer tail within the nucleus would now need to be considered.
The dual localization of Zmpste24 and Icmt that we document here is a somewhat surprising finding, as most integral membrane proteins are thought to localize either to the ER or the INM, but not to both. However, the integral membrane yeast protein Doa10p provides a precedence for dual localization and has been shown to function in both the ER and INM (Deng and Hochstrasser, 2006). For integral membrane proteins strictly localized to the INM such as mammalian LEM domain proteins (Lap isoforms, Emerin, Man1) and lamin B receptor, as well as yeast Ire1p, Heh1p, and Heh2p, the requirements for INM targeting seem to be very similar to those for soluble nuclear proteins (Worman and Courvalin, 2000; King et al., 2006; Lusk et al., 2007). Their nuclear import is directed by a NLS and is also dependent on the function of nuclear pore complex–associated proteins (Nups). Ire1p, a regulator of the unfolded protein response and previously thought be an ER membrane protein, has been shown to contain a NLS and preferentially localize to the INM (Goffin et al., 2006). In contrast, like Doa10p, Zmpste24 and Icmt contain no obvious NLS as predicted by the PSORT II algorithm and do not concentrate in the INM, yet they also reside and are active within the nucleus, as shown here. The mechanisms by which these proteins reach the INM can now be experimentally addressed.
In this study, we have not directly addressed the localization of the soluble α and β subunits of FTase. FTase is thought to be largely cytosolic, although both subunits are small enough to diffuse through nuclear pores (Reiss et al., 1990). Purification methods for nuclei are ideal for retaining any nuclear proteins that are physically connected to nuclear structures such as the lamina or chromatin, but diffusible nuclear proteins can be lost, thus precluding the possibility of obtaining direct evidence for a nuclear pool of FTase. It should be noted that indirect IF using FTase antibodies has been reported to show a predominantly nuclear localization (Sinensky et al., 1994). However, our analysis using FTase antibodies does not reveal an unequivocal, concentrated nuclear signal, making the previously published indirect IF data difficult to interpret (J.B. and S.M., unpublished observation). Our observation that a prelamin A mutant that cannot be prenylated (C661S) is not cleaved after nuclear accumulation (Figure 2C) strongly argues that FTase activity is required for lamin A processing occurring within the nucleus.
Interestingly, a comprehensive study by Schirmer and colleagues to identify nuclear envelope proteins did not uncover Zmpste24 and Icmt (Schirmer et al., 2003; Schirmer and Gerace, 2005). This study used a subtractive approach to identify proteins that were enriched within nuclear envelopes. Proteins that were also found within the ER/microsome fraction, which is not completely separable from nuclei, were excluded. These criteria of specifically seeking proteins enriched in the INM would have eliminated the dually localized Zmpste24 and Icmt, and explains why they were not previously identified as resident INM proteins.
Here we have also extended our finding of the dual localization of Zmpste24 and Icmt in mammalian cells to that of the yeast homologues, Ste24p and Ste14p. We find using the theta nuclei assay (Deng and Hochstrasser, 2006), that the yeast homologues are also localized to the inner nuclear membrane. Future work will focus on determining the requirements for INM localization of these CaaX-processing enzymes. Importantly, the role of Nup proteins or the presence of large cytosolic domains to influence the ability of Zmpste24 or Icmt to access the INM can be addressed by taking advantage of the genetic tractability of yeast. The finding that dual localization of these proteins is conserved from yeast to mammalian cells will allow the use of yeast as a model system in these endeavors.
ACKNOWLEDGMENTS
We thank Stephen Young for zmpste24−/− and icmt−/− MEFs, Michael Gelb for the FTI BMS-214662, Thomas Glover (University of Michigan) for retroviral reagents, and members of the Michaelis lab and Michael Matunis for helpful discussions. We thank Peter Espenshade, Gregory Huyer, and Meredith Metzger for a critical reading of the manuscript. Funding was provided by a grant from the Progeria Foundation (J.B. and S.M.) and National Institutes of Health Grant GM-41223.
Abbreviations used:
- ER
endoplasmic reticulum
- FTase
farnesyltransferase
- FTI
farnesyltransferase inhibitor
- HA
hemagglutinin
- HGPS
Hutchinson-Gilford Progeria Syndrome
- Icmt
isoprenylcysteine carboxylmethyltransferase
- INM
inner nuclear membrane
- MEF
mouse embryonic fibroblast
- NLS
nuclear localization sequence
- ONM
outer nuclear membrane
- Zmpste24
zinc metalloprotease STE24 homolog.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-07-0704) on October 15, 2008.
REFERENCES
- Adam S. A., Marr R. S., Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashar H. R., James L., Gray K., Carr D., Black S., Armstrong L., Bishop W. R., Kirschmeier P. Farnesyl transferase inhibitors block the farnesylation of CENP-E and CENP-F and alter the association of CENP-E with the microtubules. J. Biol. Chem. 2000;275:30451–30457. doi: 10.1074/jbc.M003469200. [DOI] [PubMed] [Google Scholar]
- Beck L. A., Hosick T. J., Sinensky M. Isoprenylation is required for the processing of the lamin A precursor. J. Cell Biol. 1990;110:1489–1499. doi: 10.1083/jcb.110.5.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergo M. O., et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc. Natl. Acad. Sci. USA. 2002;99:13049–13054. doi: 10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell B. C., Collins F. S. Human laminopathies: nuclei gone genetically awry. Nat. Rev. Genet. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- Capell B. C., Erdos M. R., Madigan J. P., Fiordalisi J. J., Varga R., Conneely K. N., Gordon L. B., Der C. J., Cox A. D., Collins F. S. Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 2005;102:12879–12884. doi: 10.1073/pnas.0506001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy E., Chiu V. K., Silletti J., Feoktistov M., Morimoto T., Michaelson D., Ivanov I. E., Philips M. R. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- Corrigan D. P., Kuszczak D., Rusinol A. E., Thewke D. P., Hrycyna C. A., Michaelis S., Sinensky M. S. Prelamin A endoproteolytic processing in vitro by recombinant Zmpste24. Biochem. J. 2005;387:129–138. doi: 10.1042/BJ20041359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q., Choy E., Chiu V., Romano J., Slivka S. R., Steitz S. A., Michaelis S., Philips M. R. Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J. Biol. Chem. 1998;273:15030–15034. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- Deng M., Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 2006;443:827–831. doi: 10.1038/nature05170. [DOI] [PubMed] [Google Scholar]
- Gaubatz S., Lees J. A., Lindeman G. J., Livingston D. M. E2F4 is exported from the nucleus in a CRM1-dependent manner. Mol. Cell. Biol. 2001;21:1384–1392. doi: 10.1128/MCB.21.4.1384-1392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb M. H., Brunsveld L., Hrycyna C. A., Michaelis S., Tamanoi F., Van Voorhis W. C., Waldmann H. Therapeutic intervention based on protein prenylation and associated modifications. Nat. Chem. Biol. 2006;2:518–528. doi: 10.1038/nchembio818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn M. W., Glover T. W. Incomplete processing of mutant lamin A in Hutchinson-Gilford progeria leads to nuclear abnormalities, which are reversed by farnesyltransferase inhibition. Hum. Mol. Genet. 2005;14:2959–2969. doi: 10.1093/hmg/ddi326. [DOI] [PubMed] [Google Scholar]
- Goffin L., et al. The unfolded protein response transducer Ire1p contains a nuclear localization sequence recognized by multiple beta importins. Mol. Biol. Cell. 2006;17:5309–5323. doi: 10.1091/mbc.E06-04-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman A. E., Moir R. D., Montag-Lowy M., Stewart M., Goldman R. D. Pathway of incorporation of microinjected lamin A into the nuclear envelope. J. Cell Biol. 1992;119:725–735. doi: 10.1083/jcb.119.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotte K., Girzalsky W., Linkert M., Baumgart E., Kammerer S., Kunau W. H., Erdmann R. Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol. Cell. Biol. 1998;18:616–628. doi: 10.1128/mcb.18.1.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Margalit A., Goldman R. D., Shumaker D. K., Wilson K. L. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- Hennekes H., Nigg E. A. The role of isoprenylation in membrane attachment of nuclear lamins. A single point mutation prevents proteolytic cleavage of the lamin A precursor and confers membrane binding properties. J. Cell Sci. 1994;107(Pt 4):1019–1029. doi: 10.1242/jcs.107.4.1019. [DOI] [PubMed] [Google Scholar]
- Hieda M., Isokane M., Koizumi M., Higashi C., Tachibana T., Shudou M., Taguchi T., Hieda Y., Higashiyama S. Membrane-anchored growth factor, HB-EGF, on the cell surface targeted to the inner nuclear membrane. J. Cell Biol. 2008;180:763–769. doi: 10.1083/jcb.200710022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmer L., Worman H. J. Inner nuclear membrane proteins: functions and targeting. Cell Mol. Life Sci. 2001;58:1741–1747. doi: 10.1007/PL00000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M. R., Nilsson T., Peterson P. A. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M. R., Nilsson T., Peterson P. A. Retrieval of transmembrane proteins to the endoplasmic reticulum. J. Cell Biol. 1993;121:317–333. doi: 10.1083/jcb.121.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M. C., Lusk C. P., Blobel G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- Kreft S. G., Wang L., Hochstrasser M. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI) J. Biol. Chem. 2006;281:4646–4653. doi: 10.1074/jbc.M512215200. [DOI] [PubMed] [Google Scholar]
- Kudlow B. A., Kennedy B. K., Monnat R. J., Jr. Werner and Hutchinson-Gilford progeria syndromes: mechanistic basis of human progeroid diseases. Nat. Rev. Mol. Cell Biol. 2007;8:394–404. doi: 10.1038/nrm2161. [DOI] [PubMed] [Google Scholar]
- Lehner C. F., Furstenberger G., Eppenberger H. M., Nigg E. A. Biogenesis of the nuclear lamina: in vivo synthesis and processing of nuclear protein precursors. Proc. Natl. Acad. Sci. USA. 1986;83:2096–2099. doi: 10.1073/pnas.83.7.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H., Winkfein R. J., Mack G., Rattner J. B., Yen T. J. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J. Cell Biol. 1995;130:507–518. doi: 10.1083/jcb.130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk C. P., Blobel G., King M. C. Highway to the inner nuclear membrane: rules for the road. Nat. Rev. Mol. Cell Biol. 2007;8:414–420. doi: 10.1038/nrm2165. [DOI] [PubMed] [Google Scholar]
- Lutz R. J., Trujillo M. A., Denham K. S., Wenger L., Sinensky M. Nucleoplasmic localization of prelamin A: implications for prenylation-dependent lamin A assembly into the nuclear lamina. Proc. Natl. Acad. Sci. USA. 1992;89:3000–3004. doi: 10.1073/pnas.89.7.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallampalli M. P., Huyer G., Bendale P., Gelb M. H., Michaelis S. Inhibiting farnesylation reverses the nuclear morphology defect in a HeLa cell model for Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 2005;102:14416–14421. doi: 10.1073/pnas.0503712102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli M., Lusk C. P., Chan H., Aitchison J. D., Wozniak R. W. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J. Cell Biol. 2001;153:709–724. doi: 10.1083/jcb.153.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naviaux R. K., Costanzi E., Haas M., Verma I. M. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T., Schirmer E. C., Nishimoto T., Gerace L. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J. Cell Biol. 2004;167:1051–1062. doi: 10.1083/jcb.200409149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendas A. M., et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat. Genet. 2002;31:94–99. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S., Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Plutner H., Davidson H. W., Saraste J., Balch W. E. Morphological analysis of protein transport from the ER to Golgi membranes in digitonin-permeabilized cells: role of the P58 containing compartment. J. Cell Biol. 1992;119:1097–1116. doi: 10.1083/jcb.119.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell L., Burke B. Internuclear exchange of an inner nuclear membrane protein (p55) in heterokaryons: in vivo evidence for the interaction of p55 with the nuclear lamina. J. Cell Biol. 1990;111:2225–2234. doi: 10.1083/jcb.111.6.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez C. L., Cadinanos J., Varela I., Freije J. M., Lopez-Otin C. Human progeroid syndromes, aging and cancer: new genetic and epigenetic insights into old questions. Cell Mol. Life Sci. 2006;64:155–170. doi: 10.1007/s00018-006-6349-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss Y., Goldstein J. L., Seabra M. C., Casey P. J., Brown M. S. Inhibition of purified p21ras farnesyl:protein transferase by Cys-AAX tetrapeptides. Cell. 1990;62:81–88. doi: 10.1016/0092-8674(90)90242-7. [DOI] [PubMed] [Google Scholar]
- Romano J. D., Schmidt W. K., Michaelis S., et al. The Saccharomyces cerevisiae prenylcysteine carboxyl methyltransferase Ste14p is in the endoplasmic reticulum membrane. Mol. Biol. Cell. 1998;9:2231–2247. doi: 10.1091/mbc.9.8.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasseville A. M., Raymond Y. Lamin A precursor is localized to intranuclear foci. J. Cell Sci. 1995;108(Pt 1):273–285. doi: 10.1242/jcs.108.1.273. [DOI] [PubMed] [Google Scholar]
- Scaffidi P., Gordon L., Misteli T. The cell nucleus and aging: tantalizing clues and hopeful promises. PLoS Biol. 2005;3:e395. doi: 10.1371/journal.pbio.0030395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer E. C., Florens L., Guan T., Yates J. R., 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- Schirmer E. C., Foisner R. Proteins that associate with lamins: many faces, many functions. Exp. Cell Res. 2007;313:2167–2179. doi: 10.1016/j.yexcr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Schirmer E. C., Gerace L. The nuclear membrane proteome: extending the envelope. Trends Biochem. Sci. 2005;30:551–558. doi: 10.1016/j.tibs.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Schmidt W. K., Tam A., Fujimura-Kamada K., Michaelis S. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc. Natl. Acad. Sci. USA. 1998;95:11175–11180. doi: 10.1073/pnas.95.19.11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinensky M., Fantle K., Trujillo M., McLain T., Kupfer A., Dalton M. The processing pathway of prelamin A. J. Cell Sci. 1994;107(Pt 1):61–67. doi: 10.1242/jcs.107.1.61. [DOI] [PubMed] [Google Scholar]
- Soullam B., Worman H. J. The amino-terminal domain of the lamin B receptor is a nuclear envelope targeting signal. J. Cell Biol. 1993;120:1093–1100. doi: 10.1083/jcb.120.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R., Locher M., Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth J. I., Yang S. H., Qiao X., Beigneux A. P., Gelb M. H., Moulson C. L., Miner J. H., Young S. G., Fong L. G., et al. Blocking protein farnesyltransferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc. Natl. Acad. Sci. USA. 2005;102:12873–12878. doi: 10.1073/pnas.0505767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter-Vann A. M., Casey P. J. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat. Rev. Cancer. 2005;5:405–412. doi: 10.1038/nrc1612. [DOI] [PubMed] [Google Scholar]
- Worman H. J., Courvalin J. C. The inner nuclear membrane. J. Membr. Biol. 2000;177:1–11. doi: 10.1007/s002320001096. [DOI] [PubMed] [Google Scholar]
- Wright L. P., Philips M. R. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J. Lipid Res. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- Wright R., Basson M., D'Ari L., Rine J. Increased amounts of HMG-CoA reductase induce “karmellae”: a proliferation of stacked membrane pairs surrounding the yeast nucleus. J. Cell Biol. 1988;107:101–114. doi: 10.1083/jcb.107.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. H., et al. Blocking protein farnesyltransferase improves nuclear blebbing in mouse fibroblasts with a targeted Hutchinson-Gilford progeria syndrome mutation. Proc. Natl. Acad. Sci. USA. 2005;102:10291–10296. doi: 10.1073/pnas.0504641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. H., Meta M., Qiao X., Frost D., Bauch J., Coffinier C., Majumdar S., Bergo M. O., Young S. G., Fong L. G. A farnesyltransferase inhibitor improves disease phenotypes in mice with a Hutchinson-Gilford progeria syndrome mutation. J. Clin. Invest. 2006;116:2115–2121. doi: 10.1172/JCI28968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S. G., Fong L. G., Michaelis S. Prelamin A, Zmpste24, misshapen cell nuclei, and progeria—new evidence suggesting that protein farnesylation could be important for disease pathogenesis. J. Lipid. Res. 2005;46:2531–2558. doi: 10.1194/jlr.R500011-JLR200. [DOI] [PubMed] [Google Scholar]
- Young S. G., Meta M., Yang S. H., Fong L. G. Prelamin A farnesylation and progeroid syndromes. J. Biol. Chem. 2006;281:39741–39745. doi: 10.1074/jbc.R600033200. [DOI] [PubMed] [Google Scholar]
- Zhang F. L., Casey P. J. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]