Abstract
Neurons form elaborate subcellular structures such as dendrites, axons, cilia, and synapses to receive signals from their environment and to transmit them to the respective target cells. In the worm Caenorhabditis elegans, lack of the RFX transcription factor DAF-19 leads to the absence of cilia normally found on 60 sensory neurons. We now describe and functionally characterize three different isoforms of DAF-19. The short isoform DAF-19C is specifically expressed in ciliated sensory neurons and sufficient to rescue all cilia-related phenotypes of daf-19 mutants. In contrast, the long isoforms DAF-19A/B function in basically all nonciliated neurons. We discovered behavioral and cellular phenotypes in daf-19 mutants that depend on the isoforms daf-19a/b. These novel synaptic maintenance phenotypes are reminiscent of synaptic decline seen in many human neurodegenerative disorders. The C. elegans daf-19 mutant worms can thus serve as a molecular model for the mechanisms of functional neuronal decline.
INTRODUCTION
RFX proteins belong to the winged-helix family of transcription factors. They are defined by a 76-amino acid DNA-binding domain and are present in many eukaryotes. The genomes of Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Caenorhabditis elegans each harbor one RFX gene, Drosophila contains two, and five have been identified in mice and humans. Individual RFX proteins regulate related processes in several species, such as the cell cycle (Wu and McLeod, 1995; Huang et al., 1998; Otsuki et al., 2004), brain development and neuronal functions (Ma et al., 2006; Zhang et al., 2006), and ciliogenesis. Cilia develop as specialized subcellular structures with sensory or motile functions that project off many different cell types. Their structure and function have been investigated in mammals, Drosophila, and C. elegans. Initially, the characterization of the single C. elegans RFX transcription factor, DAF-19, had established for the first time a connection between RFX transcription factors and cilia development (Swoboda et al., 2000) and provided a basis for subsequent studies in other species. Drosophila dRFX is expressed in the peripheral nervous system, where it is essential for the proper function of ciliated type I sensory organs (Dubruille et al., 2002; Laurencon et al., 2007). Mammalian RFX3 is responsible for nodal cilia development, the specification of left-right asymmetry and the differentiation of ciliated ependymal cells in the brain (Bonnafe et al., 2004; Baas et al., 2006). Thus, the role of RFX transcriptions factors in ciliogenesis is conserved across species. In C. elegans, the gene daf-19 is expressed in ciliated sensory neurons mostly located in the head and tail of the worm (Swoboda et al., 2000). These neurons are the major source of input for environmental signals for the worm. daf-19 mutant worms are completely devoid of ciliated structures and are consequently unable to respond to environmental signals such as food, dauer pheromone, or nose touch (Perkins et al., 1986). Nevertheless, in the laboratory daf-19 mutants are viable and thus a suitable model to study ciliogenesis. We and others have identified a large number of direct DAF-19 target genes based on the presence of the x-box promoter sequence motif, the binding site for DAF-19. Their expression in ciliated sensory neurons was dependent on both daf-19 and the promoter x-box, and many of them are required for cilia structure and function (Blacque et al., 2005; Efimenko et al., 2005; Chen et al., 2006).
In the present study, we show that DAF-19 not only regulates the formation of cilia in sensory neurons but also is required for the maintenance of synaptic functions in the remainder of the nervous system. The hermaphrodite C. elegans nervous system consists of 302 neurons (60 of which are ciliated) that are connected via ∼7000 chemical synapses and 700 gap junctions (White et al., 1986). Chemical synapses are established either between neurons or between neurons and muscle cells, at the so-called neuromuscular junctions. Each synapse consists of three major areas: 1) the synaptic vesicle pool, made up of vesicles at various stages of the recycling process or ready for neurotransmitter release; 2) the presynaptic terminal, where synaptic vesicles fuse in a multistep process and release neurotransmitters into the synaptic cleft; and 3) the postsynaptic target area in the receiving neuron, the receptive field, in which neurotransmitter receptors cluster. The isolation of a large number of C. elegans synapse mutants has provided us with detailed knowledge about the function of the synapse, especially the life cycle of synaptic vesicles. Recent work addressed the hierarchical assembly of the presynaptic terminal, providing detailed insight into the interdependence of assembly steps at a molecular level (Dai et al., 2006; Patel et al., 2006). However, how the expression of individual synaptic components is regulated after their initial establishment and how their constant supplies are maintained, remains largely unknown.
Here, we present a detailed analysis of three different daf-19 transcripts. We show that the short isoform daf-19c is expressed in all ciliated sensory neurons and is sufficient to rescue ciliogenesis phenotypes of daf-19 mutants. The two long isoforms daf-19a/b are expressed in basically all nonciliated neurons. We describe novel behavioral and cellular phenotypes of daf-19. In particular, we demonstrate that DAF-19A/B are necessary to maintain expression levels of several synaptic proteins, which assigns DAF-19 a function in neurotransmission. Surprisingly, this reduced synaptic protein expression is rather mild at larval stages but declines progressively as adult daf-19 mutants age. Therefore, our study for the first time establishes a member of the RFX transcription factor family as a regulator of synaptic maintenance. Intriguingly, the synaptic defects in daf-19 mutants display strong parallels to the synaptic decline observed in human neurodegenerative disorders, suggesting that similar mechanisms may be affected.
MATERIALS AND METHODS
Strains and Culture Methods
Culture of C. elegans strains was carried out as described previously (Brenner, 1974). The strains and transgenes used in this work are summarized in Supplemental Table 4. All strains were grown at 20°C. At this temperature, daf-19 mutants display a highly penetrant Daf-c phenotype. However, ∼10% of the population does not activate the dauer formation program and can be used for experiments (Swoboda et al., 2000). Worms were picked singly at larval stage 4 (L4) before behavioral and paralysis assays that required a small number of worms (<50 animals/assay). Antibody stainings of mixed stage populations were performed on large batches of daf-19 worms. For all experiments that required large populations of staged worms (Western blot, quantitative polymerase chain reaction [PCR], and antibody stainings) or that involved the analysis of nonrescuing transgenes (transcriptional gfp fusions of x-box candidate genes, intron-gfp fusions, translational gfp fusions of synaptic genes), we used the daf-12 (sa204) background. The daf-12 mutation suppresses the Daf-c phenotype of daf-19 and prevents dauer formation.
Injection Constructs, Germ Line Transformation, and Green Fluorescent Protein (GFP) Expression Analyses
pGG20 and pGG21 contain the last 250 base pairs of daf-19 intron 3 and daf-19 intron 4 fused to gfp, respectively. The daf-19 rescue and deletion constructs pTJ803, pGG14, and pGG18 (see Figure 2) were derived from pTJ786 (daf-19 genomic plus 2.9-kb promoter). pGG67 is a genomic/cDNA fusion rescue construct specific for daf-19a (see Figure 4). Transcriptional gfp fusions of daf-19 were injected at 100 ng/μl and daf-19 rescue constructs were injected at 10 ng/μl. Synaptic markers and promoter gfp fusions were injected at 50–70 ng/μl. Adult hermaphrodites were transformed using standard techniques (Mello et al., 1991).
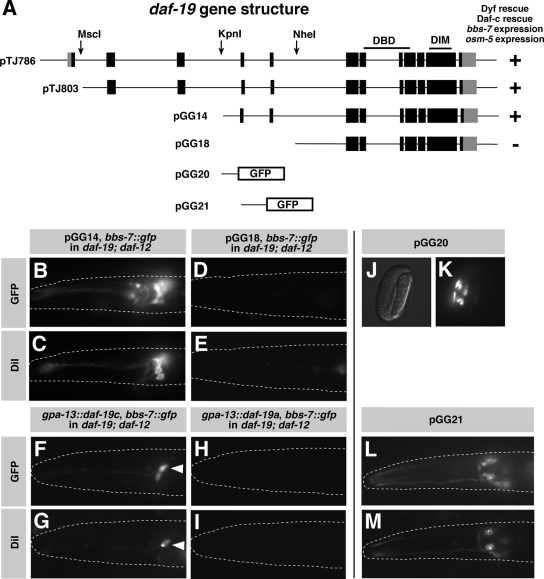
Figure 2.
daf-19c is transcribed from an internal promoter and regulates cilia formation. (A) Genomic organization of daf-19 and different deletion constructs. (B–E) daf-19 worms expressing pGG14 are rescued for the expression of the direct DAF-19 target cilia gene bbs-7::gfp and DiI fluorescent dye filling. daf-19 worms expressing pGG18 are not rescued. (F and G) daf-19 worms expressing daf-19c from the gpa-13 promoter are rescued for the expression of bbs-7::gfp and DiI fluorescent dye filling. The arrowhead depicts ASH; the other neuron in F is AWC. (H and I) Expression of daf-19a from the same promoter does not rescue. (J–M) Intron 3 (pGG20) and intron 4 (pGG21) contain regulatory elements driving gfp expression in ciliated sensory neurons in the embryo (J and K) and at all developmental stages (L), respectively. gfp expressing neurons were identified as ciliated sensory neurons by DiI fluorescent dye filling (M).
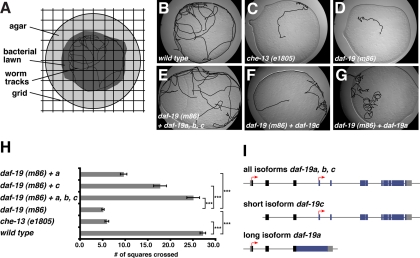
Figure 4.
DAF-19A and DAF-19C are required for complete rescue of the dwelling/roaming phenotype of daf-19 mutants. (A) Schematic visualizing the method for analyzing the dwelling/roaming assay. (B–G) Representative pictures of worm tracks on a bacterial lawn after 1 h (dwelling/roaming). Black lines visualize the worm tracks. (H) Quantification of the dwelling/roaming phenotype. Error bars show SEM values; ***p < 0.001, as detected by two-sample t test. (I) Genomic organization of the isoform-specific rescue constructs. Arrows mark the beginning of the three isoforms, respectively.
Behavioral Assays
Paralysis assays were performed on nematode growth medium agar plates containing 500 μM aldicarb or 100 μM levamisole. In addition, the resistance of daf-19 mutants to levamisole was confirmed at concentrations up to 1 mM (data not shown). At least 25–30 1-d-old adult worms were examined for each strain. Worms were classified as paralyzed when they did not move upon prodding with a pick three times in a row.
For dwelling/roaming assays, 1-d-old adult worms were transferred singly to fresh plates with a bacterial lawn of standardized size. After 1 h, worms were removed, each plate was put on a transparency with a grid (5 × 5 mm), and the number of squares that were filled with worm tracks was counted (Figure 4A). Each assay was repeated at least twice, with two independent lines for each transgene. More than 30 worms were examined in paralysis and dwelling/roaming assays.
DiI Staining, Microscopy, and Fluorescence Imaging
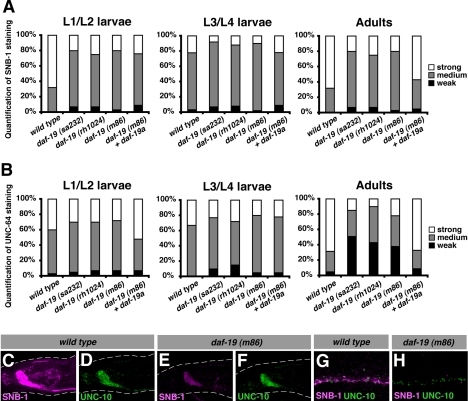
Fluorescent dye-filling assays with DiI were performed as described previously (Starich et al., 1995). For live imaging of GFP expression, worms were anesthetized in 0.1% sodium azide in M9 buffer and immobilized on a 2% agar pad. Differential interference contrast and fluorescence pictures were taken on an Axioplan 2 microscope (Carl Zeiss, Jena, Germany). We also used the microscope together with the OpenLab software (Improvision, Coventry, United Kingdom) for the analysis of expression levels of synaptic proteins (antibody stainings). Pictures of the comarker UNC-10 (unchanged between wild type and daf-19) and the synaptic protein under investigation were taken at fixed exposure times (optimized for the UNC-10 staining intensity). The intensity of the signal for the synaptic protein under these conditions was classified as “strong” when the picture was overexposed and as “weak” when the picture was underexposed (cf. Figures 6 and 7 and Supplemental Table 2).
Figure 6.
Mutations in daf-19 result in the down-regulation of the synaptic vesicle proteins SNB-1 and UNC-64 (cf. Table 1). (A and B) Quantification of SNB-1 (A) and UNC-64 (B) antibody stainings in mixed stage populations of wild type, daf-19, and rescued worms (see Supplemental Table 2). The unchanged UNC-10 staining was used as a reference. At least 40 animals were scored for each genotype and stage. (C–H) Confocal micrographs of adult worms stained with antibodies against SNB-1 (C, E, G, and H) and UNC-10 (D and F–H). C–F show the nerve ring in the head; G and H show a magnification of the ventral nerve cord. Genotypes are indicated above the panels. Representative pictures show SNB-1 staining classified as strong in wild type and weak in daf-19, both with regard to the unchanged UNC-10 signal.
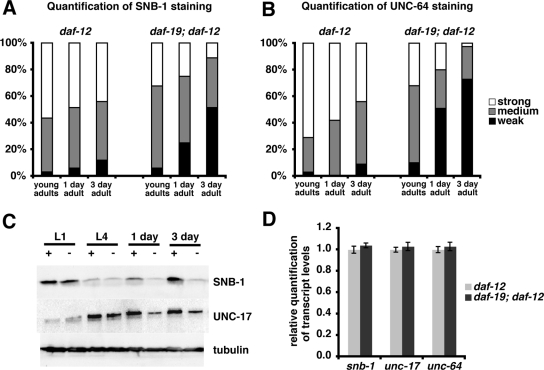
Figure 7.
Mutations in daf-19 result in the down-regulation of the synaptic vesicle proteins SNB-1, UNC-17, and UNC-64 but not of their transcripts. (A and B) Quantification of SNB-1 (A) and UNC-64 (B) antibody staining in staged adult daf-12 and daf-19; daf-12 worms. The unchanged UNC-10 staining was used as a reference. At least 40 animals were scored for each genotype and stage. (C) Western blots comparing SNB-1 and UNC-17 levels in protein extracts of staged daf-12 (+) and daf-19; daf-12 (−) worms. Within each developmental stage, proteins were isolated from an equal number of worms (note: this number varies for the different stages); 1 day and 3 day denote adults grown for 1 and 3 d after reaching L4, respectively. (D) Quantification of transcript levels of synaptic vesicle genes in daf-12 and daf-19; daf-12 adults by quantitative real-time PCR.
Northern Blot Analysis and RNase Protection Assay
Embryos were isolated from gravid wild-type adults grown on egg medium by hypochlorite treatment. Embryonic total RNA was extracted using TRIzol (Invitrogen, Paisley, United Kingdom). For Northern blot, radioactive probes were prepared using the Prime-It random labeling kit (Stratagene, La Jolla, CA) and purified over ProbeQant G50 columns (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). RNase protection assay: Radioactive probes were prepared according to the manufacturer's instructions (MAXIscript kit; Ambion, Austin, TX), and hybridization to 20 μg total RNA was carried out according to the instructions in the manual for the RPA III kit (Ambion).
Quantitative Real-Time PCR
We used TRIzol and the RNeasy kit (QIAGEN, Dorking, Surrey, United Kingdom) to extract total RNA from staged 2-d-old adult worms. All samples were checked for RNA integrity (Agilent 2100 Bioanalyzer; Agilent Technologies, Santa Clara, CA) and subjected to DNase digestion and single-strand cDNA synthesis (iScript; Bio-Rad, Hemel Hempstead, United Kingdom). Expression levels of selected genes were analyzed in an Applied Biosystems 7300 thermocycler (Applied Biosystems, Foster City, CA) by using actin as a reference gene. Each reaction was run in triplicates on two independent biological samples for each strain. All primers had a melting temperature of 58–60°C and produced a single amplicon. Data were analyzed using the Fast SDS software 1.3.1 (Applied Biosystems).
Antibody Production
cDNA fragments corresponding to DAF-19 amino acids 2–212 (for AbDAF19N) and 340–513 (for AbDAF19C), respectively, were expressed in BL21 (DE3) bacterial cells. Immunization of rabbits was carried out at Gramsch Laboratories (Schwabhausen, Germany). On Western blots, AbDAF19N detected a specific band of 120 kDa, by using wild-type protein extracts, corresponding to DAF-19A/B. This band was absent from protein extracts from daf-19 mutant worms (Supplemental Figure 1A). AbDAF19C was not suitable for Western blot analysis. On worm whole-mount stainings, both antibodies detected a signal in neuronal nuclei at all stages (Supplemental Figure 1, B and D). Aside from that, DAF-19 was also detectable in hypodermal cells at larval stages (data not shown).
Western Blot Analysis
Worms were staged by hypochlorite treatment of gravid adults. Western blots were incubated with AbDAF19N (1:250), anti-tubulin (YOL 1/34; 1:100), anti-UNC-17 (1:200), anti-SNB-1 (SN1; 1:200), horseradish peroxidase (HRP) anti-rat (1:10,000), and HRP anti-mouse (1:5000).
Antibody Staining
Staining with antibodies against UNC-29 and UNC-49 required permeabilization through freeze-fracture (Gally and Bessereau, 2003). For all other antibodies, whole-mount fixation and permeabilization were carried out as described previously (Finney et al., 1988). Worms were incubated with a 1:400 dilution of affinity-purified anti-DAF-19 antibodies. Other antibodies used were anti-OSM-5 (1:200), anti-SNB-1 (Ab1092; 1:2000), anti-SNB-1 (SN1; 1:200), anti-SNT-1 (R558; 1:100), anti-UNC-10 (RIM; 1:200), anti-UNC-13 (1:800), anti-UNC-17 (1:1000), anti-UNC-18 (G247; 1:100), anti-UNC-29 (1:200), anti-UNC-31 (1:200), anti-UNC-49 (1:800), anti-UNC-64 (Ab940; 1:5000), Alexa488 and Alexa546 (1:250: Jackson ImmunoResearch Laboratories, West Grove, PA), and Cy5 (1:1000; Rockland Immunochemicals, Gilbertsville, PA). The SN1 and RIM antibodies were obtained from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA). For all antibodies, we investigated the entire nervous system. However, for reasons of equal comparisons, we mainly focused on the head region/nerve ring. Confocal pictures of antibody stainings were taken on a TCS SP microscope (Leica, Wetzlar, Germany).
DNA Sequence Motif Searches
DNA sequences of C. elegans and Caenorhabditis briggsae synapse genes (3-kb promoter, the entire coding region and 1-kb downstream of the stop codon) were scanned for possible matches to an x-box consensus sequence RYYNYY(N)1-3RRNRRY with VectorNTI (Invitrogen). Candidate motifs were analyzed for 1) motifs that are conserved between both species and occur in several genes, or 2) motifs that occur in several C. elegans genes, in case the candidate motif lacked conservation in other nematodes species. Candidate motifs conserved between C. elegans and C. briggsae were not found.
To identify conserved motifs unrelated to the x-box, we searched 1.5 kb upstream of the ATG of the same genes. We scanned for motifs of 5–10, 8–14, and 10–16 nucleotides length using MEME (http://meme.sdsc.edu).
RESULTS
Evidence for a New daf-19 Transcript
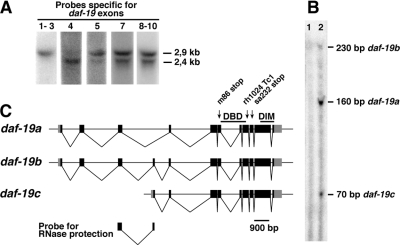
To study the subcellular localization and developmental dynamics of DAF-19, we generated antibodies against N- and C-terminal epitopes AbDAF19N and AbDAF19C, respectively. In wild-type worms, these two antibodies detected different DAF-19 expression patterns (see below), which suggested the existence of different DAF-19 isoforms. To determine the corresponding transcripts, we performed Northern blot experiments (Figure 1A). A probe specific for exons 1-3 hybridized to a single 2.9-kb band. This band corresponds to the two known transcripts, the long isoforms daf-19a/b, which differ only by the small, alternatively spliced exon 4 (Swoboda et al., 2000). In addition to the 2.9-kb band, probes specific for the remaining exons also detected a 2.4-kb transcript, which we termed short isoform daf-19c. To visualize all three isoforms daf-19a/b/c in one experiment, we conducted an RNase protection assay. A cDNA probe against exons 3–4 protected fragments of three different sizes, corresponding to the transcripts daf-19a (containing only exon 3), daf-19b (containing exons 3 and 4), and daf-19c (containing only exon 4) (Figure 1, B and C). Using 5′-rapid amplification of cDNA ends, we amplified a fragment that includes daf-19c exon 4 fused to the SL1 splice leader (data not shown). This is consistent with a trans-splice at the beginning of exon 4 followed by an ATG at position +9, in-frame with the remainder of the daf-19 transcript. In summary, these results show that in addition to the known long isoforms daf-19a/b, a third short isoform daf-19c exists that comprises SL1-spliced exons 4–12.
Figure 1.
Identification of a third, novel daf-19 transcript, daf-19c. (A) Northern blot analysis of total wild-type RNA. Probes against specific daf-19 exons (depicted above each lane) detect a novel daf-19 transcript of 2.4 kb. The two previously described transcripts daf-19a/b (Swoboda et al., 2000) differ only by ∼70 nt and run as one band of 2.9 kb. (B) RNase protection assay using an exons 3–4 probe visualizes all three daf-19 transcripts, which differ in the composition of exons 3 and 4 (lane 1, unspecific tRNA; lane 2, total RNA from wild-type worms). (C) Genomic organization of the three daf-19 transcripts. The arrows indicate the three daf-19 mutant alleles investigated. The RNA probe used for the RNase protection assay is indicated below. DBD, DNA-binding domain; DIM, dimerization domain.
DAF-19C Is Specifically Expressed in Ciliated Sensory Neurons and Regulates Ciliogenesis
A full-length genomic translational gfp fusion was shown to be sufficient to rescue the major cilia-related phenotypes of daf-19, dye-filling defective (Dyf) and dauer formation constitutive (Daf-c) (Swoboda et al., 2000). The identification of daf-19c raised the question about the functional significance of each transcript. To test for isoform-specific functions, we generated genomic deletion constructs and introduced them into a daf-19 mutant background (Figure 2A). Fragments of daf-19 lacking the promoter and the region up to intron 3 were still able to express DAF-19, as determined by staining with AbDAF19C (Supplemental Figure 2F). The expression was restricted to a small set of neurons in the head and the tail, a pattern reminiscent of ciliated sensory neurons. Consistent with the expression pattern, these constructs were sufficient to activate the expression of osm-5 and bbs-7, two well characterized, direct daf-19 target genes that are expressed in ciliated sensory neurons and function in cilia formation (Figure 2B and Supplemental Figure 2, A′ and A″) (Haycraft et al., 2001; Blacque et al., 2004). As expected, these daf-19 deletion constructs also rescued the Dyf and Daf-c phenotypes of daf-19 mutants (Figure 2, A and C; data not shown). By contrast, DNA constructs starting downstream of exon 4 failed to rescue (Figure 2, A, D, and E). We also expressed either daf-19a or daf-19c cDNAs from the gpa-13 promoter in a daf-19 mutant background. gpa-13 drives expression in five ciliated sensory neurons: ADF, ASH, AWC, PHA, and PHB (Jansen et al., 1999), out of which ASH (in the head) and PHA and PHB (in the tail) can be stained with the fluorescent dye DiI (Hedgecock et al., 1985). We found that gpa-13(p)::daf-19c, but not gpa-13(p)::daf-19a was sufficient to rescue cilia formation in sensory neurons as visualized by fluorescent dye DiI filling (Figure 2, F–I).
That a 5′-deleted genomic daf-19 fragment was able to express daf-19c and rescue ciliogenesis suggests that an internal promoter drives its expression. We generated gfp fusions to intronic sequences flanking exon 4 to investigate their expression patterns. A 250-base pair fragment upstream of exon 4 (Figure 2, J and K) and intron 4 (Figure 2, L and M) were sufficient to drive gfp expression in ciliated sensory neurons from the mid-embryonic stage to hatching and from the mid-embryo to adult stage, respectively. Intron 5 was unable to drive gfp expression (data not shown). We conclude that introns 3 and 4 contain promoter elements that are sufficient to initiate and maintain the expression of daf-19c. In summary, the novel isoform DAF-19C is specifically expressed in ciliated sensory neurons from its own promoter within the gene. daf-19c, in contrast to daf-19a, is sufficient to rescue the major, cilia-related phenotypes of daf-19 mutants.
DAF-19A/B Are Expressed in Nonciliated Neurons
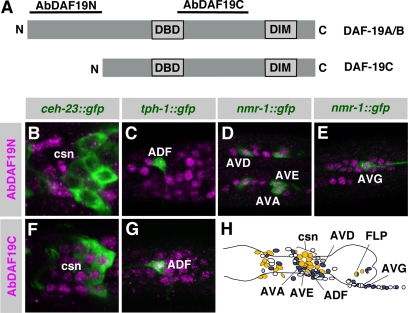
To elucidate the functions of DAF-19A/B, we analyzed their expression patterns in detail, by using antibodies against the N- and C-terminal regions of DAF-19, AbDAF19N, and AbDAF19C, respectively. Although the N-terminal antibody AbDAF19N recognizes epitopes unique to the long isoforms DAF-19A/B, the C-terminal antibody AbDAF19C recognizes the same epitopes common to isoforms DAF-19A/B/C (Figure 3A). To prove that AbDAF19N specifically detects DAF-19A/B and not C, we compared both antibodies on transgenic rescue lines expressing only DAF-19A or DAF-19C, respectively. As expected, we could detect DAF-19A with the N- and C-terminal antibodies, but DAF-19C only with the C-terminal antibody (Supplemental Figure 2, A–F). Stainings of wild-type worms with both antibodies detected DAF-19 in the majority of neuronal nuclei in the head and tail ganglia and in the ventral nerve cord. This signal was absent in all daf-19 mutant alleles tested (m86, m334, m407, rh1024, sa190, and sa232 affect all three isoforms equally; Swoboda et al., 2000), proving the specificity of both antibodies (Supplemental Figure 1, B–E). Although the AbDAF19N and AbDAF19C staining patterns overlapped in large parts, they were not identical. Posterior to the nerve ring, where the cell bodies of the amphid ciliated sensory neurons are located, we observed a group of cells, which stained only with AbDAF19C, but not with AbDAF19N (Supplemental Figure 1, B and D).
Figure 3.
An antibody specific for DAF-19A/B detects DAF-19 in all nonciliated neurons. (A) DAF-19 epitopes recognized by two different antibodies. Antibody AbDAF19N is specific for the long isoforms DAF-19A/B, whereas antibody AbDAF19C recognizes all three isoforms DAF-19A/B/C. (B–G) gfp reporter lines stained with antibodies against DAF-19. AbDAF19N detects DAF-19A/B only in nonciliated neurons (D and E) and not in ciliated sensory neurons (csn) (B and C). Ciliated sensory neurons express only DAF-19C. Because ciliated sensory neurons do not express DAF-19A/B, DAF-19C can be visualized by AbDAF19C (F and G). ceh-23::gfp marks ciliated sensory neurons, tph-1::gfp marks the serotonergic neuron ADF (ciliated), and nmr-1::gfp marks interneurons (nonciliated); see Supplemental Table 1 for details. (H) Schematic summary of all neurons investigated in the head region (blue, AbDAF19N; yellow, AbDAF19C; white, not determined neurons; and gray, no DAF-19 expression detected).
Our analysis revealed that DAF-19A/B are expressed in a larger number of neurons than DAF-19C, which is restricted to ciliated sensory neurons. From a rough cell count of all neurons that stained with AbDAF19N in the adult hermaphrodite, we estimate that DAF-19A/B are expressed in ∼200–240 neurons (data not shown). To understand where DAF-19A/B may exert their functions, we determined their expression patterns in detail. We stained gfp reporter lines, which mark subgroups of neurons, with anti-GFP and with AbDAF19N antibodies to determine whether they label the same neurons. Using nine different markers, we tested nearly half of all 302 neurons in the adult hermaphrodite, corresponding to ∼60 different classes of neurons (Figure 3H and Supplemental Table 1). We found that DAF-19A/B were expressed only in nonciliated neurons and not in ciliated sensory neurons (Figure 3). For example, ceh-23::gfp is expressed in many ciliated sensory neurons and the nonciliated neurons AIY and CAN. DAF-19A/B were detected in AIY and CAN but not in ciliated sensory neurons (Figure 3B; data not shown). Similarly, the nonciliated neurons marked with nmr-1::gfp stained with AbDAF-19N and therefore express DAF-19A/B (Figure 3, D and E). Thus, DAF-19C is specific for ciliated sensory neurons, and DAF-19A/B are specific for nonciliated neurons. In total, we found in 86 of 92 tested nonciliated neurons expression of DAF-19A/B, representing many different neuronal classes. In summary, DAF-19A/B are expressed in 200–240 nonciliated neurons and DAF-19C is expressed in 60 ciliated sensory neurons, which adds up to a basically pan-neuronal expression pattern of DAF-19 in the C. elegans hermaphrodite.
Dwelling/Roaming Behavior Depends on Multiple daf-19 Isoforms
Mutations in genes with broad neuronal expression often lead to the impaired movement of worms (UNCoordinated phenotype). daf-19 mutants move in a wild-type like manner and show no obvious Unc phenotype. We also tested daf-19 mutants in body bend assays to determine their movement speed, and we found that they can move as fast as wild type (data not shown). More specific aspects of C. elegans behavior (mating, feeding, egg laying, or patterns of movement) are usually dependent on or influenced by sensory abilities of the worm and thus depend on daf-19c. We did not identify a specific behavior that exclusively required nonciliated neurons or DAF-19A/B (data not shown). However, when performing body bend assays, we observed in daf-19 mutants severe defects in their dwelling/roaming behavior, which was dependent on all three DAF-19 isoforms. When put on a fresh plate seeded with bacteria, a single wild-type worm covers the entire bacterial lawn with tracks within a short time (dwelling/roaming) (Figure 4, A and B). In contrast, daf-19 mutants (we tested m86, rh1024, and sa232) move only for a short time and then start feeding locally (Figure 4D; data not shown). A similar behavior is observed in many cilia mutants, in which genes that are expressed exclusively in ciliated sensory neurons are mutated (e.g., che-13; Haycraft et al., 2003) and che-11 (Bell et al., 2006; Figure 4C; data not shown). To test the functions of the different DAF-19 isoforms in the dwelling/roaming behavior, we generated isoform-specific rescue constructs (Figure 4I). The dwelling/roaming phenotype of daf-19 mutants could partially be rescued by daf-19a or daf-19c (Figure 4, F and G). Complete rescue occurs only when all isoforms were present via a full-length genomic daf-19 construct (Figure 4, E and H). From these behavioral experiments, we conclude that the dwelling/roaming phenotype of daf-19 mutants is not merely caused by the lack of cilia, because the function of both the long and the short daf-19 isoforms are required.
daf-19 Mutants Are Resistant to Aldicarb and Levamisole
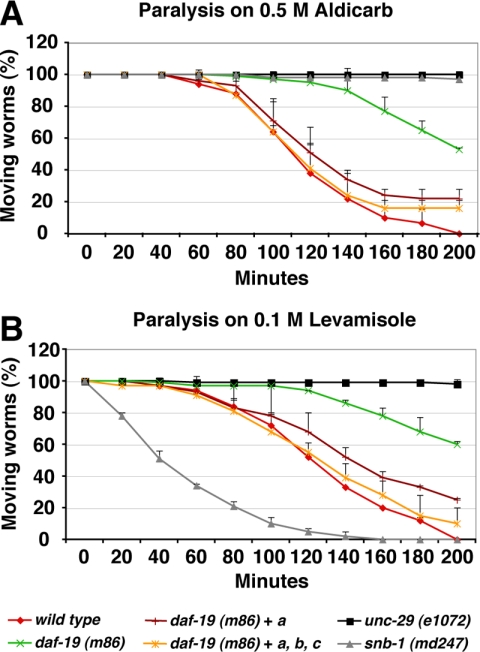
DAF-19C regulates cilia formation in ciliated sensory neurons. Do DAF-19A/B regulate an analogous, common function in nonciliated neurons? Neurons must establish synaptic connections to multiple partners to guarantee the correct wiring and function of the neuronal network. To test for connectivity, we visualized the nervous system with the pan-neuronal marker unc-104::gfp and other markers. daf-19 mutants develop a grossly normal neuronal network that includes all the required neurons and processes (data not shown). To examine the efficiency of synaptic transmission, we exposed wild type and daf-19 mutants to the pharmacological substances aldicarb and levamisole. Aldicarb, an acetylcholine esterase inhibitor, leads to the accumulation of acetylcholine in the synaptic cleft and the paralysis of wild-type animals. daf-19 mutants (m86, rh1024, and sa232) showed moderate, but statistically significant, resistance to aldicarb compared with wild-type worms (Figure 5A; data not shown). This resistance could be the result of a presynaptic defect (synthesis or release of acetylcholine [ACh]), or a postsynaptic defect (response to ACh). To determine whether daf-19 mutants had any postsynaptic deficiencies, we tested daf-19 mutants on levamisole, an acetylcholine receptor agonist, which activates cholinergic receptors independent of presynaptic input. Strikingly, daf-19 mutants (m86, rh1024, and sa232) were also resistant to levamisole compared with wild type (Figure 5B; data not shown). To exclude that these phenotypes are caused by the lack of cilia, we performed paralysis assays on the cilia mutants che-11, che-13, and osm-5. None of them showed the same phenotype as daf-19 mutants; rather, they behaved similar to wild type (Supplemental Figure 3A; data not shown). Furthermore, the resistance of daf-19 to both aldicarb and levamisole was rescued by a genomic daf-19 fragment or by daf-19a alone (Figure 5). This directly demonstrates that the long isoform DAF-19A is required to regulate synaptic transmission. Finally, because levamisole is thought to mainly act on postsynaptic acetylcholine receptors at neuromuscular junctions, we tested the function of DAF-19A in body wall muscles. Ectopic expression of daf-19a in muscle tissue did not alter the resistance of daf-19 mutants to levamisole (Supplemental Figure 3B). Together, our experiments uncover a hitherto undescribed neuronal function of daf-19a in synaptic signal transmission.
Figure 5.
Paralysis assays on aldicarb and levamisole. daf-19 mutants show resistance to aldicarb (A) and levamisole (B) in paralysis assays. daf-19a rescues to a similar extent as a genomic full-length fragment. Error bars show SEM values.
Diminished Expression of Synaptic Vesicle Proteins in daf-19 Mutants
To elucidate the reason for the reduced synaptic transmission efficiency in daf-19 mutants, we investigated the expression and localization of several types of neuronal proteins that may explain the aldicarb and levamisole phenotypes of daf-19 mutants (Table 1). The expression of general neuronal proteins (JNK-1 and UNC-104) and pre- and postsynaptic proteins (SYD-1, UNC-10, UNC-13, UNC-18, UNC-31, GLR-1, UNC-29, UNC-43, and UNC-49) did not differ between wild type and daf-19 mutants. These results suggest that the overall abundance of synapses and synaptic proteins is not affected in daf-19 mutants.
Table 1.
Comparison of general neuronal and synaptic markers for their expression in wild-type and daf-19 mutant adult worms
| Protein | Molecular function or similarity | Detection method | daf-19 (m86) vs. wild type |
|---|---|---|---|
| General neuronal proteins | |||
| JNK-1 | Serine/threonine kinase | gfp reporter | No change |
| UNC-104 | Kinesin-like protein | gfp reporter | No change |
| Pre- and postsynaptic proteins | |||
| SYD-1 | PDZ and rhoGAP domain protein | gfp reporter | No change |
| UNC-10 | Rim1 homologue | Antibody | No change |
| UNC-13 | Neurotransmitter release regulator | Antibody | No change |
| UNC-18 | Sec1 homologue | Antibody | No change |
| UNC-31 | PH-domain protein | Antibody | No change |
| GLR-1 | Glutamate receptor | gfp reporter | No change |
| UNC-29 | Acetylcholine receptor | Antibody | No change |
| UNC-43 | CaM kinase II | gfp reporter | No change |
| UNC-49 | GABA receptor | Antibody | No change |
| UNC-64 | Syntaxin | Antibody | Reduced |
| Synaptic vesicle proteins | |||
| IDA-1 | Tyr phosphatase-like receptor | gfp reporter | Reduced |
| SNB-1 | Synaptobrevin, v-SNARE | Antibody, gfp reporter | Reduced |
| SNG-1 | Synaptogyrin | gfp reporter | No change |
| SNT-1 | Synaptotagmin | Antibody | Reduced |
| UNC-17 | Vesicular acetylcholine transporter | Antibody | Reduced |
All GFP reporters are translational fusions to gfp.
However, we also found proteins whose abundance was reduced in daf-19 mutants. Of all pre- and postsynaptic proteins tested only one component of the presynaptic terminal, UNC-64/syntaxin, was reduced in daf-19 mutants compared with wild type (Figure 6B and Supplemental Table 2). UNC-64 is a plasma membrane receptor for intracellular vesicles and part of the core synaptic vesicle fusion machinery, involved in the release of neurotransmitters. Among the synaptic vesicle markers investigated, all with the exception of SNG-1/synaptogyrin were reduced in daf-19 mutants: IDA-1 (tyrosine phosphatase-like receptor that interacts with UNC-31 and UNC-64), UNC-17 (acetylcholine transporter), SNB-1 (synaptobrevin, v-SNARE/vesicular soluble N-ethylmaleimide-sensitive factor attachment protein receptor), and SNT-1 (calcium-dependent phospholipid-binding protein) (Figure 6, A, C–H; Table 1; and Supplemental Table 2). To ensure that these observations were not a result of staining artifacts, we analyzed in each individual animal in parallel the expression of UNC-10, which remained unchanged between wild type and daf-19. Thus, our analysis discovered a so far undescribed daf-19 phenotype, the reduced expression of selective synaptic components. Interestingly, the analysis of mixed stage populations revealed that this reduction was prominent, particularly at adult stages. To analyze this observation in detail, we performed the same analysis on staged worms at different times during adulthood. Intriguingly, the difference of SNB-1 and UNC-64 levels between wild type and daf-19 became stronger as they progressed through adulthood (Figure 7, A and B). Corroborating evidence for the gradual reduction of synaptic proteins in the absence of DAF-19 was obtained by Western blot analysis. Protein extract from daf-19 (−) worms contained less SNB-1 and UNC-17 compared with daf-19 (+) worms and this difference increased dramatically with the age of the worms (Figure 7C). We were interested in whether this stage-related observation correlated with behavioral phenotypes, and we compared L4 larvae and adults in dwelling/roaming and aldicarb assays. In both experiments, daf-19 mutant adult worms showed stronger phenotypes than L4 larvae (data not shown). Reduced neuronal expression of SNB-1 and UNC-64 could be rescued by a full-length genomic daf-19 rescue fragment that expresses all three isoforms as well as by isoform-specific rescue for daf-19a alone (Figure 6, A and B; data not shown). These results support those obtained in the paralysis assays (Figure 5), in which DAF-19A could rescue the resistance of daf-19 mutants to aldicarb and levamisole. Furthermore, the rescue by DAF-19A alone indicates that the reduction of SNB-1 and UNC-64 in nonciliated neurons is not merely a result of the lack of cilia and consequently the lack of environmental stimuli. To support this notion, we investigated SNB-1 and UNC-64 expression in the cilia mutants che-11 and che-13 and we found that protein levels were similar to wild type (Supplemental Table 2). Therefore, we conclude that the reduced SNB-1 and UNC-64 levels seen in daf-19 mutant adults are not caused by the lack of cilia or lack of sensory input but are a consequence of the absence of DAF-19A/B.
In summary, these experiments show that daf-19 mutants have reduced levels of several synaptic proteins (e.g., SNB-1 and UNC-64). snb-1 and unc-64 mutants are resistant to aldicarb and levamisole, suggesting that their gradual loss in daf-19 mutants directly causes changes in synaptic transmission. Interestingly, the reduced expression of synaptic proteins in daf-19 mutants affects mostly components of the synaptic vesicle pool and is increasingly evident at adult stages, whereas larval stages are not or only mildly affected. Thus, the synaptic defects seen in daf-19 mutants are likely not caused by early developmental deficiencies. We speculate that they are the consequence of a problem arising during the maintenance of synaptic protein expression in the aging adult.
DAF-19A/B Regulate Synaptic Protein Expression Indirectly
DAF-19C regulates target cilia genes directly through a conserved promoter motif, the x-box. Are synaptic genes regulated by DAF-19A/B in a similar manner? Because all DAF-19 isoforms contain the same DNA binding domain (Figures 1C and 3A), we reasoned that they could bind to overall very similar DNA sequence motifs and that direct target genes for DAF-19A/B are included in published lists of predicted x-box genes (Blacque et al., 2005; Efimenko et al., 2005; Chen et al., 2006). We filtered those lists for all genes with functions at synapses or in vesicle formation/transport (Supplemental Table 3). ida-1, snb-1, snt-1, unc-17, and unc-64 were not among them. In addition, we searched those five genes for degenerated, x-box-like or other conserved sequence motifs. None of these searches revealed any common motifs (data not shown), suggesting that they do not harbor a binding site for DAF-19A/B. To search for other possible direct DAF-19A/B targets, we checked the expression of multiple candidates from the above-mentioned lists for their dependence on the transcription factor. None of them was affected in daf-19 mutants (Supplemental Table 3). To finally test whether snb-1, unc-17, and unc-64 are directly or indirectly regulated at the transcript level, we compared their expression levels by quantitative real-time PCR. We did not detect any difference between wild type and daf-19 mutants in transcript levels of these three genes (Figure 7D). Thus, we conclude that DAF-19A/B do not regulate synaptic genes at the transcriptional level. We speculate that DAF-19A/B maintain synaptic protein expression in nonciliated neurons via an indirect mechanism, yet to be discovered (Figure 8).
Figure 8.
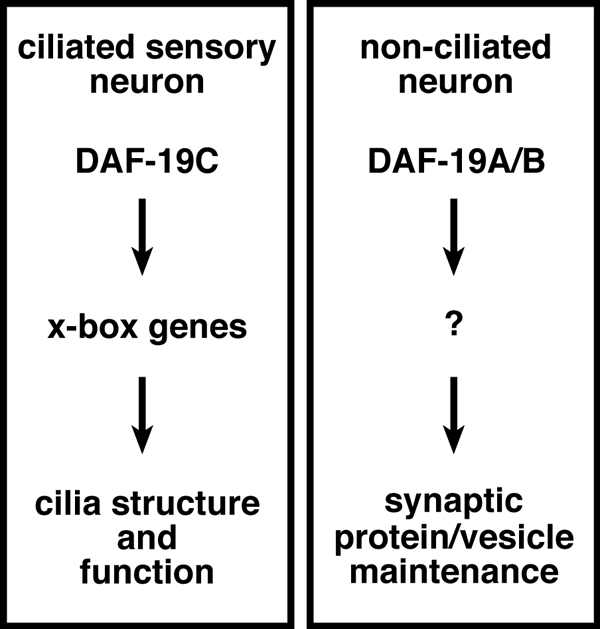
DAF-19A/B and DAF-19C execute distinct functions in synapses and cilia, respectively.
DISCUSSION
Different DAF-19 Isoforms Have Distinct Functions in Subsets of Neurons
C. elegans DAF-19 was shown to regulate the expression of genes required for the structure and function of cilia (Swoboda et al., 2000). Here, we identified a novel short transcript daf-19c that lacks exons 1-3. This short isoform DAF-19C is specifically expressed in ciliated sensory neurons from an internal promoter, and it is sufficient to rescue all cilia-related phenotypes of daf-19 mutants (Dyf, Daf-c, expression of cilia-specific, direct target genes). In contrast, the long isoforms DAF-19A/B are expressed from a different promoter in almost all nonciliated neurons, resulting in a basically pan-neuronal expression pattern of DAF-19. This expression of multiple isoforms via the so-called two-promoter system is common to many genes in C. elegans and crucial for the execution of their isoform-specific functions (Choi and Newman, 2006). We discovered that daf-19 mutants are resistant to the pharmacological substances aldicarb and levamisole, both of which modulate cholinergic synaptic transmission and lead to paralysis. The reason for this resistance was found in strongly reduced levels of synaptic vesicle proteins that were observed in adult but not juvenile animals. In addition, the lack of DAF-19 results in impaired dwelling/roaming behavior of the worm. These phenotypes can be rescued by the long isoform DAF-19A and therefore implicate a novel role of DAF-19 in the maintenance of synaptic neurotransmission.
How Do the Different DAF-19 Isoforms Activate Different Groups of Target Genes?
A large number of direct target genes has been identified for the cilia-specific short isoform DAF-19C. All those genes have in common that they 1) are expressed and function in ciliated sensory neurons and 2) contain an x-box promoter motif. Direct target genes of DAF-19A/B in nonciliated neurons currently remain unidentified. Furthermore, DAF-19A is not sufficient to replace DAF-19C in ciliated sensory neurons, indicating that these isoforms activate different target genes. Therefore, what determines the respective functions of the different isoforms?
First, the x-box DNA sequence motifs bound by DAF-19A/B could vary slightly but significantly from the motifs bound by DAF-19C. In C. elegans, the consensus in cilia-specific x-box genes contains a defined spacer of two central nucleotides (Efimenko et al., 2005), whereas the consensus sequence for hRFX has a variable spacer of zero to three nucleotides (Emery et al., 1996; Gajiwala et al., 2000). It is possible that the larger DAF-19A/B also could bind a consensus sequence with no or three spacer nucleotides, like hRFX proteins do. Alternatively, DAF-19A/B could act on x-box motifs in positions different from hitherto proven x-box motifs (i.e., >250 base pairs upstream of the ATG or within introns).
In another scenario, DAF-19–interacting proteins could decide which genes can be transcribed. DAF-19A/B contain an N-terminal part encoded by exons 1-3 lacking in DAF-19C. This N-terminal extension might serve as a site for protein interactions through which isoform-specific binding partners regulate the affinity to synaptic x-box genes instead of cilia x-box genes. Interestingly, RFX genes in all eukaryotes encode proteins of a size similar to the long isoforms DAF-19A/B, having a long N-terminal part upstream of the DNA binding domain. In addition, for some RFX genes, such as daf-19, alternative splicing of different isoforms has been demonstrated (e.g., Zhang et al., 2006). However, the protein part encoded by daf-19 exons 1-3 is not highly conserved at the amino acid level across species. Conservation between RFX proteins of different organisms could thus exist at a structural level. We assume that the N-terminal part of the protein, despite the lack of any assigned conserved domains, is important for the specific function of DAF-19A/B and other RFX proteins. It will thus be essential to characterize the function of the protein domains encoded by exons 1-3.
DAF-19A/B Are Required for Pre- and Postsynaptic Functions in Neurons
We discovered novel daf-19 mutant phenotypes that are caused by the lack of DAF-19A/B and suggest pre- and postsynaptic maintenance defects in neurotransmission. In agreement with these defects, we found that the abundance of several synaptic proteins, especially SNB-1, UNC-17, and UNC-64, was gradually reduced during adulthood. Three characteristics set the synaptic defects of daf-19 apart from all other synapse mutants identified so far: 1) Intriguingly, the decline of synaptic protein levels was most prominently seen in adult worms, whereas larval stages were hardly affected. 2) In neurons both pre- and postsynaptic functions are affected. 3) Because DAF-19A/B are expressed in neurons but not in muscles, it is likely that muscular postsynaptic terminals are intact. The absence of DAF-19 in muscular tissue indicates that the protein does not have a function in muscle cells. This explains why ectopic expression of daf-19 in body wall muscles does not rescue the levamisole-induced paralysis phenotype of daf-19 mutants. The facts listed above also help explain why daf-19 mutants do not have a severe Unc phenotype and are only moderately resistant to paralyzing substances such as aldicarb and levamisole as opposed to the complete resistance seen, for example, in the Unc mutants unc-29, unc-64, or snb-1 (Nonet et al., 1998; Saifee et al., 1998).
Although our paralysis experiments using levamisole revealed deficiencies at postsynaptic terminals in daf-19 mutants, we currently do not know their cause. All postsynaptic proteins checked were unchanged in daf-19 mutants. It is unlikely that the presynaptic effects found induce an indirect postsynaptic defect (resistance to levamisole) through a feedback mechanism. In that case, daf-19 mutants should on levamisole phenocopy other presynaptic mutants, such as snb-1. We therefore hypothesize that in addition to the presynaptic proteins we describe, so far unidentified postsynaptic molecules are also affected by the lack of DAF-19.
Maintaining Synaptic Protein Expression: A Novel Role for DAF-19A/B
Several screens have been performed that used SNB-1::GFP as synaptic vesicle marker (Zhen and Jin, 1999; Schaefer et al., 2000; Zhen et al., 2000; Crump et al., 2001; Shen and Bargmann, 2003). Others investigated genes with predicted roles in synaptic functions (Sieburth et al., 2005), synaptic vesicle recycling and transport (Koushika et al., 2004; Dittman and Kaplan, 2006). These screens uncovered genes required for the localization of SNB-1::GFP at the synapse but not for the maintenance of SNB-1 function. Therefore, daf-19 is the first C. elegans mutant that shows a strong reduction of several synaptic proteins, especially during the later phases of adulthood. This suggests that DAF-19A/B are required for the maintenance of synaptic components rather than for their expression during development.
We identified several synaptic vesicle proteins that are reduced upon loss of daf-19. Two possible scenarios could explain these findings: 1) DAF-19A/B have an influence on synaptic vesicle biogenesis/recycling, or 2) DAF-19A/B regulate a neuronal gene or process that is required for synaptic vesicle protein expression or maintenance. If a general reduction of synaptic vesicles was taking place, one would expect all vesicle proteins to be reduced to similar extents. Although we formally cannot rule out this possibility, the various degrees of reduction between different vesicle proteins (strong reduction of SNB-1 and UNC-64, mild reduction of UNC-17, and no reduction of SNG-1) argue against a general vesicle problem and indicate that these proteins are regulated differentially. Work from mammalian systems supports the notion of individual regulation of synapse components (Shimohama et al., 1998). Furthermore, the increase of synaptic proteins during neuronal development is not due to the increase of the transcriptional rate, but it is regulated at the level of protein stability (Daly and Ziff, 1997). Because most synaptic proteins are highly conserved, it is very likely that also in C. elegans the expression, maintenance, or both of synaptic proteins is individually regulated. We hypothesize that if DAF-19A/B regulate synaptic protein expression, they execute this function indirectly at a posttranscriptional level, because transcript abundance of the corresponding genes in daf-19 mutants were similar to wild type (Figure 8).
Cilia development is an essential process regulated by RFX transcription factors across species. Is it similar with regard to the functional maintenance of synapses? Although brain defects have been reported for Rfx3- and Rfx4_v3-deficient mice (Baas et al., 2006; Zhang et al., 2006), embryonic lethality precluded the analysis of late brain defects. Our analysis of daf-19 mutants suggests that the specific investigation of synapse-related functions of RFX transcription factors in other organisms is relevant to synaptic maintenance.
The C. elegans daf-19 Mutant: A New Disease Model for Functional Synaptic Decline?
Deregulation of synaptic proteins has been described for several neurological diseases, such as Huntington's disease (Morton et al., 2001) or Alzheimer's disease (Sze et al., 2000; Reddy et al., 2005). Research concerning neurodegeneration nowadays increasingly focuses on the loss of synaptic proteins, which is thought to trigger synaptic loss (Selkoe, 2002). The phenotypes seen in daf-19 mutants show parallels to the loss of synaptic proteins described for neurodegenerative diseases. RFX transcription factors as well as the majority of synaptic proteins in C. elegans are highly conserved, which suggests that synaptic protein stability in different organisms may be similarly regulated. Therefore, C. elegans and the daf-19 mutant in particular may in the future prove to be a useful model system to experimentally dissect the mechanisms that maintain synaptic function.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jean-Louis Bessereau, Mike Nonet, Ken Miller, James Rand, Courtney Haycraft, Christos Samakovlis, Piali Sengupta, Massimo Hilliard, John Hutton, Naoki Hisamoto, Jonathan Scholey, Chris Rongo, Adam Antebi, Yishi Jin, and the Caenorhabditis Genetics Center for providing worm strains and reagents; Maria Trieb for technical help; and Kirsten Senti, Mike Ailion, and members of the Bessereau, Barr, and Jorgensen laboratories for valuable comments. Work in the laboratory of P. S. is supported by grants from the Swedish Research Council and from the Swedish Foundation for Strategic Research. G. S. was supported by DOC, the doctoral scholarship program of the Austrian Academy of Sciences.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-04-0416) on October 8, 2008.
REFERENCES
- Baas D., Meiniel A., Benadiba C., Bonnafe E., Meiniel O., Reith W., Durand B. A deficiency in RFX3 causes hydrocephalus associated with abnormal differentiation of ependymal cells. Eur J. Neurosci. 2006;24:1020–1030. doi: 10.1111/j.1460-9568.2006.05002.x. [DOI] [PubMed] [Google Scholar]
- Bell L. R., Stone S., Yochem J., Shaw J. E., Herman R. K. The molecular identities of the Caenorhabditis elegans intraflagellar transport genes dyf-6, daf-10 and osm-1. Genetics. 2006;173:1275–1286. doi: 10.1534/genetics.106.056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O. E., et al. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 2004;18:1630–1642. doi: 10.1101/gad.1194004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O. E., et al. Functional genomics of the cilium, a sensory organelle. Curr. Biol. 2005;15:935–941. doi: 10.1016/j.cub.2005.04.059. [DOI] [PubMed] [Google Scholar]
- Bonnafe E., et al. The transcription factor RFX3 directs nodal cilium development and left-right asymmetry specification. Mol. Cell. Biol. 2004;24:4417–4427. doi: 10.1128/MCB.24.10.4417-4427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., et al. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., et al. Identification of ciliary and ciliopathy genes in Caenorhabditis elegans through comparative genomics. Genome Biol. 2006;7:R126. doi: 10.1186/gb-2006-7-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Newman A. P. A two-promoter system of gene expression in C. elegans. Dev. Biol. 2006;296:537–544. doi: 10.1016/j.ydbio.2006.04.470. [DOI] [PubMed] [Google Scholar]
- Crump J. G., Zhen M., Jin Y., Bargmann C. I. The SAD-1 kinase regulates presynaptic vesicle clustering and axon termination. Neuron. 2001;29:115–129. doi: 10.1016/s0896-6273(01)00184-2. [DOI] [PubMed] [Google Scholar]
- Dai Y., Taru H., Deken S. L., Grill B., Ackley B., Nonet M. L., Jin Y. SYD-2 Liprin-alpha organizes presynaptic active zone formation through ELKS. Nat. Neurosci. 2006;9:1479–1487. doi: 10.1038/nn1808. [DOI] [PubMed] [Google Scholar]
- Daly C., Ziff E. B. Post-transcriptional regulation of synaptic vesicle protein expression and the developmental control of synaptic vesicle formation. J. Neurosci. 1997;17:2365–2375. doi: 10.1523/JNEUROSCI.17-07-02365.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittman J. S., Kaplan J. M. Factors regulating the abundance and localization of synaptobrevin in the plasma membrane. Proc. Natl. Acad. Sci. USA. 2006;103:11399–11404. doi: 10.1073/pnas.0600784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubruille R., Laurencon A., Vandaele C., Shishido E., Coulon-Bublex M., Swoboda P., Couble P., Kernan M., Durand B. Drosophila regulatory factor X is necessary for ciliated sensory neuron differentiation. Development. 2002;129:5487–5498. doi: 10.1242/dev.00148. [DOI] [PubMed] [Google Scholar]
- Efimenko E., Bubb K., Mak H. Y., Holzman T., Leroux M. R., Ruvkun G., Thomas J. H., Swoboda P. Analysis of xbx genes in C. elegans. Development. 2005;132:1923–1934. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- Emery P., Strubin M., Hofmann K., Bucher P., Mach B., Reith W. A consensus motif in the RFX DNA binding domain and binding domain mutants with altered specificity. Mol. Cell. Biol. 1996;16:4486–4494. doi: 10.1128/mcb.16.8.4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney M., Ruvkun G., Horvitz H. R. The C. elegans cell lineage and differentiation gene unc-86 encodes a protein with a homeodomain and extended similarity to transcription factors. Cell. 1988;55:757–769. doi: 10.1016/0092-8674(88)90132-8. [DOI] [PubMed] [Google Scholar]
- Gajiwala K. S., Chen H., Cornille F., Roques B. P., Reith W., Mach B., Burley S. K. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature. 2000;403:916–921. doi: 10.1038/35002634. [DOI] [PubMed] [Google Scholar]
- Gally C., Bessereau J. L. GABA is dispensable for the formation of junctional GABA receptor clusters in Caenorhabditis elegans. J. Neurosci. 2003;23:2591–2599. doi: 10.1523/JNEUROSCI.23-07-02591.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft C. J., Schafer J. C., Zhang Q., Taulman P. D., Yoder B. K. Identification of CHE-13, a novel intraflagellar transport protein required for cilia formation. Exp. Cell Res. 2003;284:251–263. doi: 10.1016/s0014-4827(02)00089-7. [DOI] [PubMed] [Google Scholar]
- Haycraft C. J., Swoboda P., Taulman P. D., Thomas J. H., Yoder B. K. The C. elegans homolog of the murine cystic kidney disease gene Tg737 functions in a ciliogenic pathway and is disrupted in osm-5 mutant worms. Development. 2001;128:1493–1505. doi: 10.1242/dev.128.9.1493. [DOI] [PubMed] [Google Scholar]
- Hedgecock E. M., Culotti J. G., Thomson J. N., Perkins L. A. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev. Biol. 1985;111:158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- Huang M., Zhou Z., Elledge S. J. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- Jansen G., Thijssen K. L., Werner P., van der Horst M., Hazendonk E., Plasterk R. H. The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat. Genet. 1999;21:414–419. doi: 10.1038/7753. [DOI] [PubMed] [Google Scholar]
- Koushika S. P., Schaefer A. M., Vincent R., Willis J. H., Bowerman B., Nonet M. L. Mutations in Caenorhabditis elegans cytoplasmic dynein components reveal specificity of neuronal retrograde cargo. J. Neurosci. 2004;24:3907–3916. doi: 10.1523/JNEUROSCI.5039-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurencon A., Dubruille R., Efimenko E., Grenier G., Bissett R., Cortier E., Rolland V., Swoboda P., Durand B. Identification of novel regulatory factor X (RFX) target genes by comparative genomics in Drosophila species. Genome Biol. 2007;8:R195. doi: 10.1186/gb-2007-8-9-r195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K., Zheng S., Zuo Z. The transcription factor regulatory factor X1 increases the expression of neuronal glutamate transporter type 3. J. Biol. Chem. 2006;281:21250–21255. doi: 10.1074/jbc.M600521200. [DOI] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton A. J., Faull R. L., Edwardson J. M. Abnormalities in the synaptic vesicle fusion machinery in Huntington's disease. Brain Res. Bull. 2001;56:111–117. doi: 10.1016/s0361-9230(01)00611-6. [DOI] [PubMed] [Google Scholar]
- Nonet M. L., Saifee O., Zhao H., Rand J. B., Wei L. Synaptic transmission deficits in Caenorhabditis elegans synaptobrevin mutants. J. Neurosci. 1998;18:70–80. doi: 10.1523/JNEUROSCI.18-01-00070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuki K., Hayashi Y., Kato M., Yoshida H., Yamaguchi M. Characterization of dRFX2, a novel RFX family protein in Drosophila. Nucleic Acids Res. 2004;32:5636–5648. doi: 10.1093/nar/gkh895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M. R., Lehrman E. K., Poon V. Y., Crump J. G., Zhen M., Bargmann C. I., Shen K. Hierarchical assembly of presynaptic components in defined C. elegans synapses. Nat. Neurosci. 2006;9:1488–1498. doi: 10.1038/nn1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L. A., Hedgecock E. M., Thomson J. N., Culotti J. G. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Reddy P. H., Mani G., Park B. S., Jacques J., Murdoch G., Whetsell W., Jr., Kaye J., Manczak M. Differential loss of synaptic proteins in Alzheimer's disease: implications for synaptic dysfunction. J. Alzheimers Dis. 2005;7:103–117. doi: 10.3233/jad-2005-7203. discussion 173–180. [DOI] [PubMed] [Google Scholar]
- Saifee O., Wei L., Nonet M. L. The Caenorhabditis elegans unc-64 locus encodes a syntaxin that interacts genetically with synaptobrevin. Mol. Biol. Cell. 1998;9:1235–1252. doi: 10.1091/mbc.9.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A. M., Hadwiger G. D., Nonet M. L. rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron. 2000;26:345–356. doi: 10.1016/s0896-6273(00)81168-x. [DOI] [PubMed] [Google Scholar]
- Selkoe D. J. Alzheimer's disease is a synaptic failure. Science. 2002;298:789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- Shen K., Bargmann C. I. The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell. 2003;112:619–630. doi: 10.1016/s0092-8674(03)00113-2. [DOI] [PubMed] [Google Scholar]
- Shimohama S., Fujimoto S., Sumida Y., Akagawa K., Shirao T., Matsuoka Y., Taniguchi T. Differential expression of rat brain synaptic proteins in development and aging. Biochem. Biophys. Res. Commun. 1998;251:394–398. doi: 10.1006/bbrc.1998.9480. [DOI] [PubMed] [Google Scholar]
- Sieburth D., et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- Starich T. A., Herman R. K., Kari C. K., Yeh W. H., Schackwitz W. S., Schuyler M. W., Collet J., Thomas J. H., Riddle D. L. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics. 1995;139:171–188. doi: 10.1093/genetics/139.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda P., Adler H. T., Thomas J. H. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell. 2000;5:411–421. doi: 10.1016/s1097-2765(00)80436-0. [DOI] [PubMed] [Google Scholar]
- Sze C. I., Bi H., Kleinschmidt-DeMasters B. K., Filley C. M., Martin L. J. Selective regional loss of exocytotic presynaptic vesicle proteins in Alzheimer's disease brains. J. Neurol. Sci. 2000;175:81–90. doi: 10.1016/s0022-510x(00)00285-9. [DOI] [PubMed] [Google Scholar]
- White J. G., Southgate E., Thomson J. N., Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wu S. Y., McLeod M. The sak1+ gene of Schizosaccharomyces pombe encodes an RFX family DNA-binding protein that positively regulates cyclic AMP-dependent protein kinase-mediated exit from the mitotic cell cycle. Mol. Cell. Biol. 1995;15:1479–1488. doi: 10.1128/mcb.15.3.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Stumpo D. J., Graves J. P., DeGraff L. M., Grissom S. F., Collins J. B., Li L., Zeldin D. C., Blackshear P. J. Identification of potential target genes for RFX4_v3, a transcription factor critical for brain development. J. Neurochem. 2006;98:860–875. doi: 10.1111/j.1471-4159.2006.03930.x. [DOI] [PubMed] [Google Scholar]
- Zhen M., Huang X., Bamber B., Jin Y. Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron. 2000;26:331–343. doi: 10.1016/s0896-6273(00)81167-8. [DOI] [PubMed] [Google Scholar]
- Zhen M., Jin Y. The liprin protein SYD-2 regulates the differentiation of presynaptic termini in C. elegans. Nature. 1999;401:371–375. doi: 10.1038/43886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.