Abstract
How scaffold proteins integrate signaling pathways with cytoskeletal components to drive axon outgrowth is not well understood. We report here that the multidomain scaffold protein Plenty of SH3s (POSH) regulates axon outgrowth. Reduction of POSH function by RNA interference (RNAi) enhances axon outgrowth in differentiating mouse primary cortical neurons and in neurons derived from mouse P19 cells, suggesting POSH negatively regulates axon outgrowth. Complementation analysis reveals a requirement for the third Src homology (SH) 3 domain of POSH, and we find that the actomyosin regulatory protein Shroom3 interacts with this domain of POSH. Inhibition of Shroom3 expression by RNAi leads to increased process lengths, as observed for POSH RNAi, suggesting that POSH and Shroom function together to inhibit process outgrowth. Complementation analysis and interference of protein function by dominant-negative approaches suggest that Shroom3 recruits Rho kinase to inhibit process outgrowth. Furthermore, inhibition of myosin II function reverses the POSH or Shroom3 RNAi phenotype, indicating a role for myosin II regulation as a target of the POSH–Shroom complex. Collectively, these results suggest that the molecular scaffold protein POSH assembles an inhibitory complex that links to the actin–myosin network to regulate neuronal process outgrowth.
INTRODUCTION
Neurite outgrowth is a dynamic and complex process. Axons undergo multiple cycles of extension and retraction, attachment and release, and attraction and repulsion. Guidance cues, intracellular signaling pathways, and cytoskeletal components are critical players in each of these events. Yet, how signaling pathways, cytoskeletal components, and extracellular cues are coordinately regulated during process outgrowth is still incompletely understood.
Plenty of SH3s (POSH) is a multidomain scaffold protein comprised of four Src homology (SH) 3 domains, a Rac binding domain, and a really interesting new gene (RING) domain. POSH assembles an active c-Jun NH2-terminal kinase (JNK) mitogen-activated protein kinase (MAPK) module composed of mixed lineage kinase (MLK) 1-3/dual leucine zipper kinase (DLK), MKK4/7, and JNK1/2 (Tapon et al., 1998; Xu et al., 2003). POSH interacts with activated Rac, which activates a subset of MLK family members, notably the Cdc42/Rac-interactive binding domain containing MLK1-3 (Tapon et al., 1998; Gallo and Johnson, 2002). Akt negatively regulates POSH-associated JNK signaling and Rac binding (Figueroa et al., 2003; Lyons et al., 2007). POSH functions as a proapoptotic protein in fibroblasts, mature neurons, and Xenopus embryos by activating JNK signaling (Tapon et al., 1998; Xu et al., 2003, 2005; Kim et al., 2005; Zhang et al., 2006). In addition to regulating apoptosis, roles for POSH in regulating calcium homeostasis, membrane trafficking, and human immunodeficiency virus production have been reported previously (Alroy et al., 2005; Kim et al., 2006; Tuvia et al., 2007). Furthermore, POSH was identified in a genetic modifier screen in Drosophila for mutants with morphological defects in dorsal appendages, suggesting a role for POSH in regulating cell shape change (Schnorr et al., 2001).
Shroom3 is an F-actin–binding protein that regulates apical constriction through a myosin II-dependent pathway (Hildebrand and Soriano, 1999; Haigo et al., 2003; Hildebrand, 2005). In Shroom3-deficient mice, the neural folds fail to converge at the dorsal midline and “mushroom” outward, resulting in exencephaly, acrania, facial clefting, and spinal bifida (Hildebrand and Soriano, 1999). Shroom family members are characterized by the presence of a conserved ASD2 (Apx/Shroom) domain (Staub et al., 1992; Hildebrand and Soriano, 1999; Fairbank et al., 2006; Etournay et al., 2007; Yoder and Hildebrand, 2007). In Shroom3, the ASD2 domain elicits apical constriction through the actomyosin network (Haigo et al., 2003; Hildebrand, 2005). Shroom3 regulates myosin II by recruiting Rho kinase (ROCK) through the ASD2 domain (Nishimura and Takeichi, 2008). A second conserved domain, the ASD1 domain, targets Shroom family members 1–3 to actin (Yoder and Hildebrand, 2007). Shroom3 is widely expressed during embryonic development. In addition to expression in the neural tube, Shroom3 is expressed in the forebrain, including the cortex, somites, heart, gut, and skeletal muscle (Hildebrand and Soriano, 1999). Thus, Shroom3 may have roles in other cellular events, in addition to apical constriction during neural tube closure.
In this report, we demonstrate that RNA interference (RNAi)-mediated inhibition of POSH in two neuronal model systems, differentiating P19 neurons and mouse primary cortical neurons, specifically enhances axon outgrowth, thus implicating POSH in negative regulation of axon outgrowth. Complementation analysis demonstrates a requirement for the third SH3 domain of POSH in process outgrowth inhibition, and we identify Shroom3 as a binding partner for the third SH3 domain of POSH. Endogenous Shroom3 and POSH coassociate and RNAi-mediated reduction of Shroom3, like POSH, enhances axon outgrowth in differentiating primary cortical neurons, suggesting that POSH and Shroom3 work in concert to negatively regulate process outgrowth. Shroom3 is an F-actin–binding protein that couples to a myosin II-dependent pathway (Haigo et al., 2003; Hildebrand, 2005). For Shroom3 to inhibit process outgrowth, both the actin binding and ASD2 domains are required, and the ASD2 domain couples to ROCK to inhibit process outgrowth. Finally, inhibition of myosin II suppresses the POSH or Shroom RNAi phenotype, suggesting that up-regulation of myosin II in POSH or Shroom RNAi neurons is the driving force behind the enhanced process outgrowth. Together, these results support the model that the multidomain scaffold protein POSH assembles an inhibitory complex, which includes the actomyosin regulatory protein Shroom3 and its partner ROCK, to regulate process outgrowth.
MATERIALS AND METHODS
Antibodies
Primary antibodies were as follows: neuronal β-III-tubulin (Covance Research Products, Princeton, NJ), microtubule-associated protein (MAP) 2 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), green fluorescent protein (GFP) (Invitrogen, Carlsbad, CA), POSH (M-290; Santa Cruz Biotechnology, Santa Cruz, CA), Shroom3 (T-17; Santa Cruz Biotechnology), nonmuscle myosin IIA heavy chain (Covance Research Products), nonmuscle myosin IIB heavy chain (Covance Research Products), and actin (Sigma-Aldrich, St. Louis, MO). Secondary antibodies included Alexa 488- and 594-conjugated antibodies (Invitrogen).
Yeast Two-Hybrid Library Screen
A yeast two-hybrid screen of a 9.5/10.5-d mouse embryo cDNA library was performed, as described previously, to identify proteins that interact with the third and fourth SH3 domains of POSH (amino acids 447-892) (Vojtek et al., 1993; Vojtek and Hollenberg, 1995).
RNAi
pCS2-SIBR and pUI4/5 are short interferring RNA (siRNA) expression vectors based on the miR-155 microRNA precursor RNA (Chung et al., 2006). The 22-nucleotide miR-155 sequence is replaced by an siRNA that is perfectly complementary to the target mRNA of interest. The siRNA precursor (the SIBR cassette) is expressed under the control of the RNA polymerase II simian cytomegalovirus (CMV) promoter in pCS2 or under control of the UbiquitinC promoter in pUI4/5. siRNAs and a fluorescent or selectable marker (GFP or puromycin) are expressed from a single vector based transcript in the UI4 vector. The UI5 vector is a derivative of UI4 in which an internal ribosome entry site (IRES) element has been positioned upstream of the coding region for GFP and downstream of a polylinker. The UI5 complementation vectors (see Figure 4) are created by inserting cDNAs encoding full-length POSH or POSH mutants deleted for various domains into the polylinker of UI5. Thus, the UI5 vectors express siRNAs, GFP, and POSH, wild-type or deletion mutants, from a single transcription unit. POSH, Shroom3 and myosin IIA siRNAs target sequences located within 3′-untranslated regions (UTRs). The sequence of the siRNAs are as follows: POSH1, 5′-UUACACAUCAGAACCCAGAGAG-3′; POSH2, 5′-UUGAAGAUCUGGAAGUCCACAG-3′; POSH6, 5′-UAAGCUUUCUCCGUGUCCCGUC-3′; Shrm3-1, 5′-UUUUGCCCAGGGCUUACAGGAG-3′; Shrm3-3, 5′-UUUGUAGUGAAAUCGCUUUCAG-3′; luciferase (Luc; control), 5′-UUUAUGAGGAUCUCUCUGAUUU-3′; myosin IIA-1 (Myh9), 5′-AUUAAAUAUAUUUGGUCCCCUA-3′; Robo-1, 5′-UAUUUUGCUAUCAAUUAGCGUG-3′; and EphrinB2-1, 5′-UAACACCCGAAUCCAUAGACGG-3′. POSH1+2 expresses both POSH1 and POSH2 siRNAs from two SIBR cassettes in tandem. The POSH6 and Luc SIBR cassettes have been published previously as POSH-2852 and Luc-1601 (Chung et al., 2006).
Figure 4.
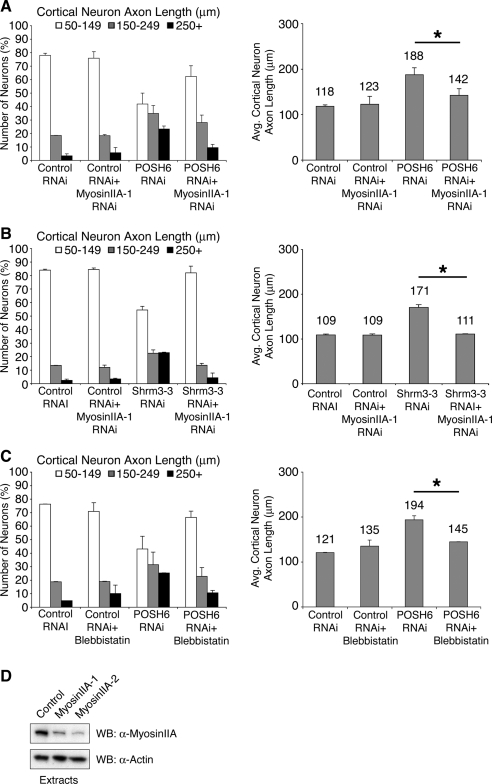
POSH RING and SH3-3 domains are required for complementation of the POSH RNAi process outgrowth phenotype. (A) Domain structure of POSH and POSH mutants. RING: Really interesting new gene, a conserved feature of a subfamily of E3 ubiquitin ligases. SH3, Src homology 3 protein–protein interaction domain. RBD, Rac binding domain. (B and C) Complementation analysis in P19 neurons indicates a requirement for the POSH RING domain (B) and the POSH SH3-3 domain (C). P19 cells were transfected with Ngn2 and an RNAi complementation vector, which simultaneously allows for RNAi, complementation analysis, and marker expression from a single transcript. POSH6 RNAi targets the 3′-UTR of endogenous POSH, allowing for complementation by expression of the POSH coding sequence, which lacks 3′-UTR sequences. POSH6 RNAi+POSH denotes a vector that expresses POSH6 siRNAs, full-length POSH, and GFP. POSH6 RNAi+POSHΔRING denotes a vector that expresses POSH6 siRNAs, a POSH mutant deleted for the RING domain, and GFP. Vectors expressing full-length or various deletion mutants of POSH (shown in A) were transfected into P19 cells along with Ngn2. Neurons were fixed and stained for GFP, which marks the transfected neurons 70–75 h after transfection. Process length was measured on 100 cells per condition per experiment for two to three independent experiments. The distribution of process lengths (50–149, 150–249, and 250+ μm; left) as well as average process lengths (right) is shown. Pairwise comparisons that are statistically significant (Wilcoxon's rank sum test, left; Student's t test, right): control RNAi, POSH6 RNAi; control RNAi, POSH6RNAi+POSHΔRING; POSH6 RNAi, POSH6RNAi+POSH; POSH6 RNAi+POSH, POSH6RNAi+POSHΔ3/4; POSH6 RNAi, POSH6 RNAi+POSHΔ4.
For testing the ability of the siRNAs to target endogenous POSH, Shroom3, or myosin IIA, Western analysis was performed on extracts prepared from transiently transfected, puromycin-selected P19 cells, as described previously for short hairpin siRNAs (Vojtek et al., 2003; Yu et al., 2003). The POSH mutants used in the complementation analysis are as follows: POSHΔRing, amino acids 53-892; POSHΔ4, amino acids 1-519; and POSHΔ3/4, amino acids 1-382.
Neuronal Differentiation Assays
P19 cells, grown in minimal essential medium-α (Invitrogen) supplemented with 7.5% calf serum (HyClone Laboratories, Logan, UT), 2.5% fetal bovine serum (FBS; HyClone Laboratories), and penicillin-streptomycin (Invitrogen), were plated the day before transfection to a density of 1.1 × 105 cells/35-mm dish. Cells were transfected with an expression vector for the neural basic helix-loop-helix protein (bHLH) Neurogenin1 (Ngn1) or Neurogenin2 (Ngn2), pCS2-Ngn1 or pUS-Ngn2 (1.5 μg), pCS2 SIBR control or pCS2 SIBR POSH siRNA expression vector (1.5 μg), and either pCS2-GFP or pUS2-GFP, a derivative of pCS2 that expresses GFP under the control of the human UbiquitinC promoter instead of simian CMV (0.75 μg) by using FuGENE (Roche Diagnostics, Indianapolis, IN) (Figure 1). In Figure 4, P19 cells were plated to a density of 0.9 × 104 cells/well of a 12-well dish and transfected the next day with 2 μg of total DNA (0.75 μg of Ngn2, 1.25 μg of pUI4/UI5 RNAi expression vector). In Figure 8, pUS2-R1C1, which expresses the Shroom3 binding domain of human Rock1, amino acids (aa) 698-957, was transfected at 850 ng/12-well. Four to 6 h after transfection, cells were resplit 1:5 or 1:6 to laminin-coated dishes. Eighteen to 20 h later, the cells were transferred to Opti-MEM (Invitrogen) supplemented with 1% FBS and penicillin-streptomycin. 3–4 d after transfection, cells were fixed in 3.7% formaldehyde-phosphate-buffered saline (PBS) and stained for GFP or neuronal markers of differentiation (Farah et al., 2000; Vojtek et al., 2003). Similar results were obtained when outgrowth assays were performed on poly-l-lysine or laminin-coated dishes.
Figure 1.
RNAi-mediated reduction of POSH expression leads to enhanced process outgrowth. (A) Inhibition of endogenous POSH by POSH siRNAs. Extracts were prepared from puromycin-selected P19 cells transiently transfected with POSH or control SIBR siRNA expression vectors and a GFP/puromycin expression vector. POSH1, POSH2, and POSH6 RNAi vectors express a single siRNA directed against a unique sequence in POSH. POSH1+2 expresses both POSH1 and POSH2 siRNAs from a single transcript. The RNAi control vector expresses an siRNA that targets luciferase; targeting luciferase has no functional consequences in P19 cells and this serves as a control for off-target effects. Endogenous POSH was detected in extracts by western blotting with an anti-POSH antibody (top). Western blotting for actin, loading control (bottom). (B–F) POSH RNAi enhances process outgrowth in P19 neurons. P19 cells were transiently transfected with expression vectors for the neural bHLH proteins to drive the differentiation program, GFP to mark the transfected cells, and RNAi vectors. Transfected cells were plated to laminin-coated dishes; neurons were fixed 72 h after transfection and stained for neural markers of differentiation. Photographs of neurons were captured with a digital camera and the length of the longest process per cell measured. One hundred cells were analyzed for each RNAi construct per condition per experiment in three independent experiments. (B) Representative images depicting the change in process length between control and POSH RNAi neurons. P19 neurons were transfected with Ngn2, GFP, and control or POSH6 RNAi vectors, fixed, and then stained for neuronal β-tubulin. (C) Average process length (±SD) of POSH RNAi neurons is enhanced relative to control (*p < 0.001, Student's t test). (D) Neurons were also classified on the basis of axon length: 50–149, 150–249, and 250 μm and greater (+), ±SD. The majority of control neurons have short processes (50–149 μm), whereas the population of neurons with long axons (250+ μm) was increased with POSH RNAi (p < 0.0002, Wilcoxon's rank sum test). (E) Representative images of P19 neurons transfected with Ngn1, GFP, and control or POSH1+2 RNAi vectors, fixed, and stained for MAP2. Thus, two different RNAi vectors expressing siRNAs targeting different POSH sequences (POSH6 or POSH1+2) give similar phenotypes, and similar phenotypes are observed with different bHLH proteins (Ngn2 or Ngn1) driving the differentiation program in P19 cells. (F) Representative images of P19 cells transfected with Ngn2, GFP, and control synthetic siRNA duplexes or POSH6 synthetic siRNA duplexes. Similar POSH RNAi phenotypes are induced by synthetic siRNAs (F) or siRNAs expressed in vivo (vector expressed) (B–E).
Figure 8.
Interference of ROCK interaction with Shroom3 leads to enhanced process outgrowth. P19 cells were transfected with a control vector, a POSH6 RNAi vector, or a vector expressing the Shroom3 binding region of hROCK1, denoted R1-C1. 54 h after transfection, differentiated P19 cells were treated with and without 10 μM of the ROCK inhibitor Y-27632 for 19 h. Cells were fixed, stained, and process length was measured. Ectopic expression of R1-C1 leads to enhanced process outgrowth compared with control neurons, similar to enhanced process outgrowth induced by RNAi-mediated reduction of POSH function. Ectopic expression of R1-C1 increases the population of neurons with long axons (250 μm) and decreases the population of neurons with short axons (50–149 μm), and average process length is increased as well (B). Blocking ROCK function in control neurons with Y-27632 also enhances process outgrowth, increasing the population of neurons with long axons and average process length. R1-C1–expressing or POSH RNAi neurons exhibit enhanced process outgrowth upon addition of Y-27632, but the response to the ROCK inhibitor is attenuated. This suggests that ROCK regulates process length in both a POSH–Shroom-dependent and -independent manner. (A) Distribution of process length. Wilcoxon's rank sum test: control ± Y-27632, control versus R1-C1, control versus POSH RNAi, p < 0.0002; R1-C1 ± Y-27632, POSH RNAi ± Y-27632, p < 0.003. (B) Average process length. * and control versus R1-C1, p < 0.0001, Student's t test.
Cortical primary progenitors were isolated from the dorsal telencephalon of mouse embryos at embryonic day (E) 14.5. Freshly dissociated cells were transfected using an AAD-1001 Nucleofector (Amaxa Biosystems, Gaithersburg, MD). In Figure 2, cells were transfected with UI4-luciferase control or UI4-POSH6 (6 μg). In Figure 6, cells were cotransfected with pCS2 SIBR POSH1+2, pCS2 SIBR Shroom 3-1, or pCS2 SIBR-luciferase (control luciferase siRNA) expression vector (4 μg) and a GFP expression vector (2.2–2.5 μg). In Figure 7A, dissociated cells were cotransfected with pUbc-Shroom3 ABD (aa 754-952) or pUS2 control expression vectors (5 μg) and pUS2+GFP expression vector (2 μg). In Figure 9, cells were cotransfected with UI4-POSH6 or UI4-luciferase RNAi expression vectors (3 μg) and either UI4-luciferase or UI4-myosin IIA RNAi expression vectors (3 μg). To enhance cell survival, untransfected cortical progenitors (0.4–0.7 × 105 cells per well) were plated to poly-l-lysine–laminin-coated 12-well dishes (Corning Life Sciences, Acton, MA), and then electroporated cells (0.6–1 × 107 cells total/electroporation; 5–9 × 105 cells/well) were layered over the untransfected cortical progenitors and allowed to differentiate in monolayer culture in serum-free media (L15 medium [Invitrogen] supplemented with 26 mM NaHCO3 [Sigma-Aldrich], 1% N2 [Invitrogen], 2% B27 [Invitrogen], 30 mM glucose [Sigma-Aldrich], and 1% penicillin/streptomycin). Then, 10 ng/ml basic fibroblast growth factor (R & D Systems, Minneapolis, MN) was added to the media for 2–3 h or overnight and then withdrawn to foster differentiation. Neuronal process outgrowth was monitored over time by visualization of GFP fluorescence. Twenty-four hours after nucleofection (day 2), the majority of GFP-positive, transfected cells had not yet extended processes. By day 3, the GFP-positive cells had extended processes, and process outgrowth continued into day 4. Cells were fixed 3 and 4 d after transfection and stained with anti-GFP antibody (Invitrogen).
Figure 2.
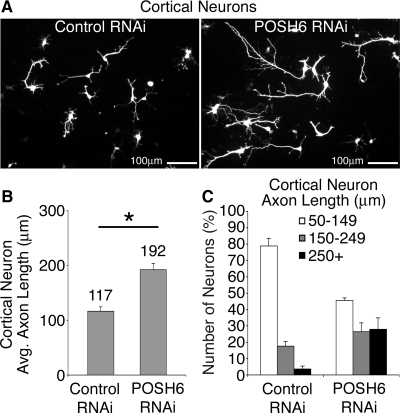
POSH regulates process outgrowth in primary cortical neurons. UI4-control or UI4-POSH RNAi expression vectors were introduced into cortical primary neurons prepared from E14.5 mouse embryos by nucleofection. In the UI4 vectors, a single vector-derived transcript mediates both RNAi and GFP marker expression. Nucleofected cells were plated to laminin–poly-l-lysine–coated dishes, and neurons were fixed and stained for GFP to enhance signal strength 72 h after nucleofection. Photographs of neurons were captured with a digital camera and the length of the longest process per cell measured. (A) Representative images depicting the change in axon length between control and POSH RNAi neurons 72 h after nucleofection. (B) Average process length of POSH RNAi primary cortical neurons is enhanced relative to control (*p < 0.043, Student's t test). (C) Neurons were also classified on the basis of axon length: the majority of control neurons have short processes (50–149 μm), whereas the population of neurons with long axons (250+ μm) was increased with POSH RNAi (p < 0.0002, Wilcoxon's rank sum test).
Figure 6.
Shroom3 regulates axon outgrowth in cortical primary neurons. (A) RNAi-mediated reduction of Shroom3. Extracts were prepared from puromycin selected P19 cells transiently transfected with Shroom3 or control RNAi expression vectors and a GFP/puromycin expression vector. Shrm3-3 targets a sequence in the 3′-UTR region of Shroom3, and is effective in reducing endogenous Shroom3 expression. Endogenous Shroom was detected in extracts by Western blotting with an anti-Shroom3 antibody (top). Western blotting for actin, loading control (bottom). (B and C) RNAi-mediated reduction of Shroom3 enhances axon outgrowth of primary cortical neurons. Control, POSH1+2, or Shrm3-3 RNAi expression vectors were introduced into cortical primary neurons prepared from E14.5 mouse embryos by nucleofection; a GFP expression vector was included to identify the transfected cells. Process length was measured on fixed, stained GFP-labeled neurons 3 and 4 d after nucleofection. (B) Shroom3 RNAi increases the percentage of neurons with long processes relative to control (p < 0.0002, two-tailed Wilcoxon's rank sum test). (C) Shroom3 RNAi also increases the average process length relative to control neurons (*p < 0.0001, Student's t test).
Figure 7.
Functional domains of Shroom3: requirement for ASD1/actin binding and ASD2/myosin regulatory domains. (A) Expression of dominant-negative Shroom3 ASD1/actin binding domain enhances axon outgrowth in primary cortical neurons. Primary cortical neurons were nucleofected with expression vectors for the ASD1/actin binding domain of Shroom3 or a vector control. The ASD1/ABD domain of Shroom3 functions as a dominant negative to block binding of endogenous Shroom3 to actin. GFP-positive neurons were fixed and stained 72 h after nucleofection, and process length was measured. The population of neurons with long axons (250 μm and greater) (p < 0.0004, Wilcoxon's rank sum test) as well as average process length (*p < 0.0001, Student's t test) was increased relative to control neurons upon expression of the ASD1/ABD domain, suggesting that the interaction of Shroom3 with actin is required for Shroom3 to function in process outgrowth inhibition. (B) Shroom3 RNAi complementation analysis: requirement for ASD2 domain. Primary cortical cells were nucleofected with expression vectors for wild-type or mutant Shroom3, GFP, and a Shroom3 RNAi vector. The Shroom3 RNAi vector expresses Shrm3-3, an siRNA that targets the 3′-UTR of endogenous Shroom3. The expression vectors for wild-type or mutant Shroom3 include only coding regions and, therefore, are not targeted by the RNAi construct. GFP-labeled neurons were fixed and stained 3 d after nucleofection, and process length was measured. Expression of wild-type Shroom3S (Shrm), but not Shroom3S deleted for the ASD2 domain (ShrmΔASD2), complements the Shroom3 RNAi-enhanced process outgrowth phenotype in primary cortical neurons, indicating a functional requirement for the Shroom3 ASD2 domain during process outgrowth inhibition. Left, p < 0.003, Wilcoxon's rank sum test, Shrm3 RNAi versus Shrm3 RNAi+Shrm. Right, *p < 0.001, Student's t test.
Figure 9.
Inhibition of myosin II decreases process length in POSH and Shroom3 RNAi neurons. (A and B) RNAi-mediated reduction in myosin IIA function reverses the POSH (A) or Shroom3 (B) RNAi-mediated enhanced process outgrowth phenotype in primary cortical neurons. Primary cortical progenitors were nucleofected with control, POSH, or Shroom3 RNAi vectors, with and without an RNAi vector that reduces myosin IIA function. Cells were cultured for 72 h on poly-l-lysine-laminin–coated coverslips, fixed, stained, imaged, and process length of GFP-labeled neurons was determined. Reduction of myosin IIA function shifts the distribution of axon lengths to control levels and decreases average process length to the levels of the control neurons for both POSH RNAi (A) and Shroom3 RNAi neurons (B). (C) Treatment of primary cortical neurons with the myosin II inhibitor blebbistatin reverses the POSH RNAi phenotype. Embryonic primary cortical progenitors were nucleofected with control or POSH RNAi vectors and cultured for 72 h on poly-l-lysine-laminin. At 72 h, the cultures were treated with DMSO (vehicle control) or blebbistatin, a pharmacological inhibitor of myosin II function (50 μM) for 2 h. Process length was determined on GFP-labeled fixed, stained neurons. Blebbistatin treatment alters the process length distribution of POSH RNAi neurons, shifting process distribution to levels of control neurons (C, left). Blebbistatin treatment also decreases the average process length of POSH RNAi neurons, to control levels (C, right). (A–C) Pairwise comparisons [POSH6 RNAi, POSH6 RNAi+myosin IIA RNAi; Shrm3-3 RNAi, Shrm3-3 RNAi+myosin IIA RNAi, POSH6 RNAi, POSH6 RNAi+blebbistatin] are statistically significant: Wilcoxon's rank sum test (left, A–C; p < 0.0001) and Student's t test (right, A–C; *p < 0.0001). (D) RNAi-mediated reduction of myosin IIA. Extracts were prepared from puromycin selected P19 cells transiently transfected with myosin IIA or control RNAi expression vectors and a GFP/puromycin expression vector. Myosin IIA-1/2 are two independent RNAi constructs targeting different sequences. Endogenous myosin IIA was detected in extracts by Western blotting with an anti-myosin IIA–specific antibody (top). Western blotting for actin, loading control (bottom).
Measurement of Process Length
To measure process length, photographs of fixed, stained neurons were captured with a digital camera, and the length of the longest process per cell was measured using the perimeter/length function in NIH Image 1.33 (Figure 1), the length function in NIH ImageJ 1.63 (Figure 6), or the measurement (polyline) function in MicroSuite Special Edition imaging software version 5.0 (Olympus, Tokyo, Japan). The longest process per cell is positive for Tau, an axonal marker. As needed, several photographs were joined in Adobe Photoshop (Adobe Systems, Mountain View, CA) to measure the length of the complete process. Processes 50 μm or greater were measured; 50 μm is ∼3 times the length of the cell body. Process length is determined in two or more independent experiments, 100 neurons per condition per experiment. Similar results for process length measurements are obtained if neurons are fixed and stained for GFP or for neuronal markers of differentiation (β-III-tubulin).
Caspase Inhibitor Assay
P19 cells were split to a density of 0.8 × 105/12 well plate and transfected 24 h later using TransIT (Mirus Bio, Madison, WI) with US2+mNgn2 (0.75 μg) and pUI4-GFP-Luc1601 (1.25 μg). Transfected cells were replated to laminin coated 12-well plates ∼5 h after transfection. Twenty-four hours after transfection the media were changed to differentiating media (Opti-MEM + 1% FBS) supplemented with the caspase inhibitor z-VAD-FMK (Promega, Madison, WI) (20 μM) or dimethyl sulfoxide (DMSO) (vehicle control). The cells were further incubated with the inhibitor for 48 h, fixed in 3.7% formaldehyde ∼72 h after transfection, and stained for neuronal β-III-tubulin (TuJ1 antibody; Covance Research Products) and GFP (Invitrogen). Process length was determined using the polyline function from MicroSuite Special Edition imaging software, version 5.0 (Olympus).
Detection of Active Caspases
P19 cells were split to a density of 1.5 × 105/12 well plate and transfected 5 h later using TransIT (Mirus) with US2+mNgn2 (0.75 μg) and pUI4-GFP-Luc1601 (1.25 μg) or pUI4-GFP-POSH6 (1.25 μg). Transfected P19s were replated to laminin-coated dishes 16 h after transfection then fixed in 3.7% formaldehyde ∼66 h after transfection, and stained with the rabbit cleaved-caspase-3 (Asp175) antibody (CST) overnight at 4°C. The percentage of cells undergoing apoptosis was determined by counting the cleaved caspase-3–positive/GFP-positive cells divided by the total number of GFP-transfected cells, using the point function from Olympus MicroSuite Special Edition imaging software, version 5.0.
Inhibitor Studies
Y-27632 (10 μM; Calbiochem, San Diego, CA), a ROCK1/2 inhibitor, was added 54 h after transfection and plating of P19s to laminin-coated dishes; P19 neurons were fixed and stained 73 h after transfection. Primary cortical neurons were incubated with blebbistatin, a pharmacological inhibitor of myosin II function (50 μM, racemic mixture of active and inactive enantiomers; Calbiochem) for 2 h before harvest ∼72 h after nucleofection.
Coassociation Assays
In Figure 5B, glutathione transferase (GST)-POSH SH3-3 and His-Shroom3 POSH binding domain (PBD; aa 300-422) fusion proteins were expressed from the vectors pGEX6P-1 (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) and pET-15b (Novagen, San Diego, CA), respectively, in Escherichia coli BL21. GST-POSH SH3-3 was purified with glutathione-Sepharose beads (GE Healthcare) and His-Shroom3 PBD with nickel-nitrilotriacetic acid His bind resin (Novagen). GST-POSH SH3-3 (1 μg) and His-Shrm PBD (1 μg) were incubated in Triton immunoprecipitation buffer (10 mM HEPES, pH 7.4, 100 mM NaCl, 1% Triton X-100, 2 mM EDTA, 0.1% β-mercaptoethanol, 1% aprotinin, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μM pepstatin, and 2 μM leupeptin). His-Shroom3 PBD was immunoprecipitated after an overnight incubation at 4°C with a rabbit anti-His antibody (CST), followed by addition of protein A/G-Sepharose (1:1; Sigma-Aldrich/GE Healthcare). Proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and detected by Western analysis with a mouse anti-GST antibody (Zymed Laboratories, South San Francisco, CA) and anti-His antibody. GST-POSH SH3-3 (1/10) and His-Shroom3 PBD (1/50) input proteins were included as loading controls.
Figure 5.
POSH and Shroom3 coassociate. (A) A yeast two-hybrid screen identified Shroom3 as an interacting partner for POSH. Alignment showing the domain structure of Shroom3L (aa 1-1986) and Shroom3S (aa 177-1986), the location of the Shroom3 two-hybrid isolates, and the number of times each class of isolate was recovered from the screen. PDZ, PSD-95/Dgl/ZO-1 (aa 1-106). ASD1/2, Apx/Shroom domain 1/2 (aa 927-1028 and aa 1659–1983, respectively). ABD, F-actin binding domain (aa 754-952). EVH1 BS, Enabled/vasodilator-stimulated phosphoprotein homology 1 domain binding site (aa 1533-1539). PDZ BS, PDZ domain binding site (aa 1983-1986). (B) Direct interaction between POSH and Shroom3. GST-POSH SH3-3 or GST and His epitope-tagged Shroom3 POSH binding domain (PBD) fusion proteins were purified from bacteria, allowed to bind in vitro, then His-Shroom3 was immunoprecipitated (IP). His-Shroom3 and coassociated GST proteins were detected by Western blot analysis (WB) with antibodies directed against the epitope tags after SDS-PAGE. (C) Endogenous POSH and Shroom3 coassociate. Shroom3 was immunoprecipitated from E16.5 mouse embryonic brain extracts (left) or IMR32 human neuroblastoma cells (right). An irrelevant goat polyclonal antibody was used as a specificity control (Ctrl). After SDS-PAGE, Western blot analysis was used to detect Shroom3 and coassociated POSH.
In Figure 5C, E16.5 mouse embryo brains were homogenized in PBST (+) buffer (PBS, pH 7.4, 2 mM EDTA, 0.2 mM PMSF, 0.1% β-mercaptoethanol, 1% aprotinin, 1% Triton X-100, 2 μM leupeptin, and 1 μM pepstatin). Homogenates were sonicated, passed through cheesecloth, and centrifuged twice at 14,000 rpm at 4°C for 30 min. IMR-32 cell extracts were prepared in modified radioimmunoprecipitation assay buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 2 mM EDTA, 1% aprotinin, 1 mM PMSF, 1 mM sodium orthovanadate, 2 μM leupeptin, and 1 μM pepstatin) and subjected to sonication. After sonication, Triton X-100 was added to a final concentration of 1%; insoluble material was removed by centrifugation at 14,000 rpm at 4°C. Clarified extracts were precleared with Sepharose beads for 30 min at 4°C. Brain homogenates and IMR-32 extracts were incubated overnight at 4°C with 2 μg of goat polyclonal Shroom3 antibody (T-17; Santa Cruz Biotechnology) prebound to protein A/G-Sepharose (Sigma-Aldrich/GE Healthcare). For the negative control immunoprecipitations, brain homogenates and IMR-32 extracts were incubated with a goat polyclonal antibody (E-20; Santa Cruz Biotechnology), prebound to protein A/G-Sepharose. Proteins were resolved by SDS-PAGE and detected by Western analysis.
RESULTS
POSH mRNA is present throughout the developing CNS of E14.5 mouse embryos (data not shown). The expression of POSH in the developing CNS suggests that POSH may play a role in neuronal differentiation. To investigate the role of endogenous POSH during neuronal differentiation, we used RNAi to reduce POSH function. siRNAs targeting murine POSH were expressed in cells by using the CS2+SIBR RNAi expression vector. The Short Interfering BIC-derived RNA (SIBR) vectors generate siRNAs from an RNA polymerase II-transcribed synthetic microRNA precursor based on the miR-155 microRNA precursor present in the BIC noncoding RNA (Chung et al., 2006). Three siRNAs, POSH1, POSH2, or POSH6, each targeting a different sequence in the POSH 3′-UTR, were expressed in the CS2+SIBR vector. The POSH1 and POSH2 siRNAs also were expressed in a single transcription unit in the same vector (POSH1+2). The control RNAi vector expresses an siRNA targeting luciferase. To test the effectiveness of the siRNAs at reducing the endogenous levels of POSH, we performed Western blot analysis on extracts from mouse P19 cells transiently transfected with control, CS2+SIBR POSH1, CS2+SIBR POSH2, CS2+SIBR POSH1+2, or CS2+SIBR POSH6 RNAi expression vectors and biCS5puro/GFP (puromycin/green fluorescent protein). The inclusion of the biCS5puro/GFP vector permitted the selection of the transiently transfected population by using puromycin (Vojtek et al., 2003; Yu et al., 2003). siRNAs against POSH were effective at reducing the endogenous level of POSH protein (Figure 1A).
To investigate the role of endogenous POSH during neuronal differentiation, we introduced an expression vector for the neural basic helix-loop-helix protein (bHLH) Neurogenin 2 (Ngn2) into uncommitted P19 cells together with CS2+SIBR vectors that express POSH siRNAs or a control expression vector. In addition, the cells were cotransfected with a vector that expresses GFP, allowing the transfected cell population to be readily identified. P19 cells are uncommitted mouse embryonal carcinoma cells that undergo neuronal differentiation after treatment with retinoic acid or after introduction of a neural bHLH protein (McBurney, 1993; Farah et al., 2000). Ngn1- (Figure 1, B–D and F) or Ngn2 (Figure 1E)-transfected P19 cells adopted a neuronal morphology and expressed neuronal markers of differentiation, including neuronal β-tubulin (Figure 1, B–D and F) and MAP2 (Figure 1E). Neurons transfected with the POSH6 RNAi expression vector exhibited a 2.5-fold increase in average process length compared with control cells (Figure 1, B and C). In addition, POSH6 RNAi resulted in an increase in the number of neurons with long processes and a corresponding decrease in the number of neurons with short processes: 73% of POSH6 RNAi neurons had process lengths of 250 μm or greater compared with 7% in the control, whereas 5% of POSH6 RNAi neurons had process lengths of 50–149 μm compared with 62% in the control (Figure 1D). Similar results were obtained with additional siRNAs directed against different sequences in the POSH mRNA: POSH1+2 (Figure 1E) and POSH7 (data not shown). In Figure 1E, neurons were generated with a second bHLH protein, Ngn1. POSH1+2 RNAi had similar effects on process outgrowth in P19 neurons generated with Ngn1, as observed for Ngn2. In addition to using DNA vectors, we also tested synthetic siRNA duplexes directed against POSH. Neurons derived from cells cotransfected with Ngn2 and POSH6 synthetic 22-nt siRNA duplexes exhibited increased axon lengths compared with neurons derived from P19 cells transfected with Ngn2 and Luc control synthetic 22-nt siRNA duplexes (Figure 1F). Reduction of POSH function specifically enhances process length; the number of processes per cell is not altered in POSH RNAi neurons (average number of processes per cell: 1.41 ± 0.56 P19 control neurons; 1.42 ± 0.54 POSH6 RNAi neurons). Collectively, these results, as well as POSH RNAi complementation studies described below, indicate that increased process length in neurons transfected with POSH siRNAs specifically reflects inhibition of endogenous POSH.
To determine the role of POSH in process outgrowth in primary neurons, siRNA expression vectors targeting POSH were introduced into differentiating cortical neurons isolated from E14.5 mouse embryos. UI4 GFP SIBR Luc RNAi control and UI4 GFP SIBR POSH6 RNAi expression vectors were introduced by nucleofection into the primary cortical neurons in vitro. The UI4 vectors express a single vector derived transcript for both RNAi and GFP expression under control of the UbiquitinC RNA polymerase II promoter (Chung et al., 2006). Four days after nucleofection and dissociation into monolayer culture, process outgrowth was analyzed in GFP-labeled cortical neurons. As observed for P19 neurons, POSH RNAi decreased the number of primary cortical neurons with short axons and increased the number of neurons with long axons: 27% POSH RNAi neurons had axon lengths >250 μm in contrast to 4% Luc RNAi control neurons (Figure 2, A and B). Dendrite length was similar in control and POSH RNAi neurons (24.6 μm in control; 23.2 μm in POSH RNAi).
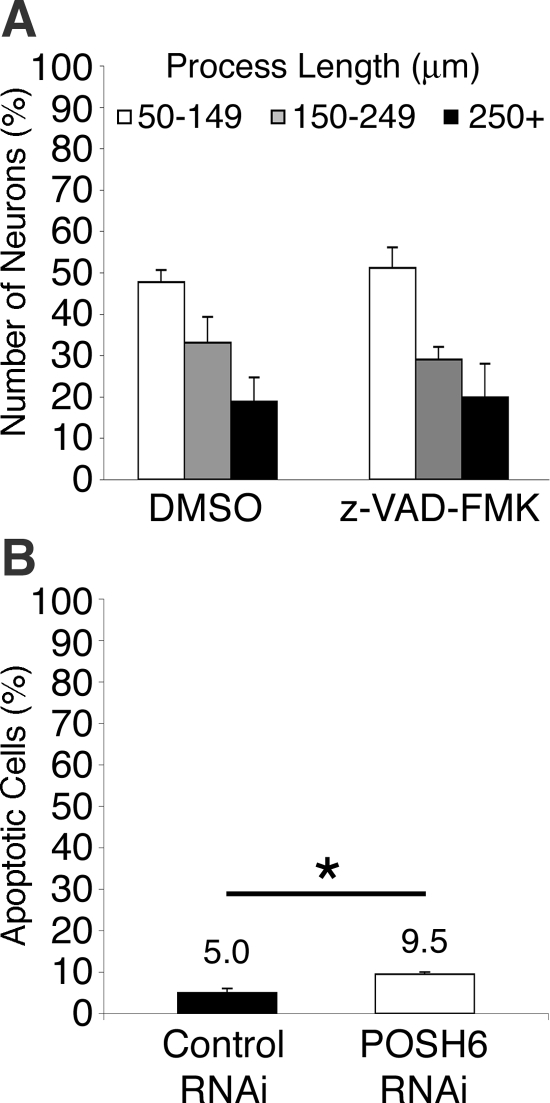
Earlier studies report a proapoptotic role for POSH (Tapon et al., 1998; Xu et al., 2003, 2005; Kim et al., 2005; Zhang et al., 2006). If POSH or other proapoptotic proteins limit process outgrowth by an apoptotic mechanism, then blocking the function of the apoptotic proteins with z-VAD-FMK, an inhibitor of apoptosis-inducing caspases, should lead to enhanced process outgrowth. However, the addition of z-VAD-FMK does not increase process length in P19 neurons, indicating that programmed cell death is not limiting process outgrowth (Figure 3A). In addition, the number of apoptotic cells was determined in control RNAi and POSH RNAi P19 cells, using indirect immunofluorescence to detect anti-caspase-3–positive cells. POSH RNAi resulted in a twofold increase in apoptosis, suggesting that POSH RNAi cells are modestly prone to enhanced apoptosis rather than cell survival (Figure 3B). These observations suggest that POSH RNAi does not indirectly increase process length by enhancing cell survival.
Figure 3.
Apoptotic mechanisms do not negatively regulate process length in P19 neurons. (A) Inhibition of cell death with z-VAD-FMK has little effect on process length of P19 control neurons. P19 cells were transfected with Ngn2 and control GFP expression vectors. Transfected cells were plated to laminin-coated dishes. The caspase inhibitor z-VAD-FMK (20 μM) or DMSO (vehicle control) was added 24 h after transfection. Neurons were fixed and stained for GFP 72 h after transfection and process length determined. (B) The number of apoptotic cells is increased in POSH RNAi neurons. A decrease in apoptotic cell number would be expected if POSH RNAi enhanced process length indirectly by promoting survival. P19 cells transfected with Ngn2 and control or POSH RNAi expression vectors were plated to laminin coated dishes, and 72 h after transfection, they were fixed and stained for active caspase-3 and GFP. The percentage of apoptotic cells is the number of transfected active caspase-3–positive cells divided by the number of transfected cells × 100. (*p < 0.015, Student's t test).
POSH is a multidomain protein, with an N-terminal RING domain, four Src homology 3 domains (SH3-1 to SH3-4, named in order of location in the protein, N to C), and a Rac binding domain [RBD]; Figure 4A). Complementation analysis in POSH RNAi neurons was performed to identify domains that mediate the function of POSH in process outgrowth. To facilitate complementation analysis, a vector that simultaneously allows for RNAi, complementation analysis, and marker expression from a single vector transcript was designed. The POSH6 siRNA targets 3′-UTR sequences, enabling complementation by expression of the POSH coding region. In addition, in the complementation vector, an IRES driven GFP coding sequence is inserted downstream of the POSH coding sequence, allowing for expression of both POSH and GFP proteins. Vectors expressing full-length POSH or POSH mutants were cotransfected into P19 cells with Ngn2 to drive differentiation. Process outgrowth was scored 70–75 h after transfection. Wild-type POSH suppresses the POSH RNAi phenotype, shifting the process length distribution to control levels (Figure 4B). POSH mutants lacking the RING domain fail to complement, suggesting that the RING domain is required for POSH function (Figure 4B). POSH mutants lacking SH3-4 complement the POSH RNAi phenotype, indicating that the fourth SH3 domain is not required for POSH to regulate process outgrowth (Figure 4C). In contrast, POSH mutants lacking both SH3-3 and SH3-4 fail to complement the POSH RNAi phenotype, suggesting that the third SH3 domain is required for POSH function (Figure 4C).
To gain insight into the molecular mechanism of POSH function, we identified POSH interacting proteins, by using a yeast two-hybrid approach (Vojtek et al., 1993; Vojtek and Hollenberg, 1995). Multiple overlapping clones of the F-actin–binding protein Shroom3 were identified. We identified five isolates that encode amino acids 300-422 of Shroom3 and 2 isolates that encode amino acids 295–444 of Shroom3 (Figure 5A). The overlap among the two-hybrid isolates identifies amino acids 300-422 of Shroom3 as being the minimal POSH binding domain (PBD). This domain is evolutionarily conserved and is present in both Shroom3 isoforms, Shroom3L (L, long) and Shroom3S (S, short). Shroom3L contains an amino-terminal postsynaptic density 95/disc-large/zona occludens (PDZ) domain that Shroom3S lacks. The region of Shroom3 identified in the screen has not previously been assigned a function; this region is located upstream of the F-actin binding domain in Shroom3L and Shroom3S and downstream of the PDZ domain in Shroom3L.
To confirm the yeast two-hybrid results, we tested the coassociation of bacterially expressed and purified POSH and Shroom3 interaction domains. Histidine (His) epitope-tagged Shroom3 PBD interacts with GST-POSH SH3-3 (amino acids 447-550) (Figure 5B), indicating that the two proteins directly interact in vitro. To determine whether endogenous POSH and Shroom3 coassociate, Shroom3 was immunoprecipitated from extracts prepared from embryonic day 16.5 mouse brains or from IMR-32 human neuroblastoma cells. POSH was detected in the Shroom3 immunoprecipitates by Western analysis after SDS-PAGE, demonstrating that endogenous Shroom3 and POSH coassociate (Figure 5C).
Because POSH and Shroom3 coassociate, we reasoned that Shroom3 might mediate, at least in part, the actions of POSH on process outgrowth. To determine whether Shroom3 regulates process outgrowth in a manner similar to POSH, siRNA expression vectors targeting Shroom3 (Figure 6A) were introduced into cortical neurons isolated from E14.5 mouse embryos. CS2+SIBR control, CS2+SIBR POSH1+2, and CS2+SIBR Shroom3 expression vectors were introduced by nucleofection into the primary cortical neurons in vitro, together with a GFP expression vector to label the transfected cells. CS2+SIBR Shroom3-3 expresses an siRNA targeting the Shroom3 3′-UTR. Three and 4 d after nucleofection and dissociation into monolayer culture, process outgrowth was analyzed in GFP-labeled neurons. Shroom3 RNAi increased the number of neurons with long axons and correspondingly decreased the number of neurons with short axons (Figure 6B). By day 4, 82% of Shroom3 RNAi neurons had axon lengths of 250 μm or greater compared with 38% of control RNAi neurons. One percent of Shroom3 RNAi neurons had axon lengths of 50–149 μm compared with 18% of control RNAi neurons. Similar effects on process length were observed with a second Shroom3 siRNA expression vector (data not shown). Like Shroom RNAi, on day 4, 4% of POSH RNAi neurons had short axons, 50–149 μm in length, compared with 18% of control RNAi neurons, whereas 76% of the POSH RNAi neurons had axon lengths of 250 μm or greater compared with 38% in the control (Figure 6B). Process length changes were observed on both day 3 and day 4, with day 4 exhibiting a further increase in average process length, as expected if process outgrowth is the measured parameter in this assay (Figure 6C). The observation that RNAi-mediated inhibition of either POSH or Shroom3 leads to a dramatic increase in axon length, together with the observation that POSH and Shroom3 coimmunoprecipitate from embryonic brain extracts, supports a key role for the POSH–Shroom3 complex in regulating axonal morphogenesis.
Shroom3 has been reported previously to regulate apical constriction. Regulation of this cell shape change requires the ASD1/actin binding domain and the ASD2/myosin regulatory domain. To determine whether apical constriction and inhibition of process outgrowth have similar or distinct requirements for the Shroom3 functional domains, we assessed the requirement for each of the Shroom3 domains during process outgrowth inhibition. To test for a requirement for the Shroom3 actin binding domain, we ectopically expressed this domain (Shrm ABD 754-952) in primary cortical neurons. In Xenopus, ectopic expression of the Shroom3 actin binding domain acts in a dominant negative manner and inhibits Shroom3 induced actin accumulation, hingepoint formation, and neural tube closure in Xenopus embryos (Haigo et al., 2003). Ectopic expression of the Shroom3 F-actin binding domain decreased the number of neurons with short axons and increased the number of neurons with long axons (Figure 7A), suggesting that Shroom3's ability to negatively regulate axon outgrowth is coupled to its ability to bind F-actin. To test for a requirement for the Shroom3 ASD2 domain, an RNAi complementation assay in primary cortical neurons was used. Shroom3-3, a specific Shroom3 RNAi construct, targets 3′-UTR sequences, enabling complementation by expression of the Shroom3 coding region. Expression of Shoom3S, the short version of Shroom3 that lacks the PDZ domain present in ShroomL, complements the Shroom RNAi phenotype, whereas expression of Shroom3ΔASD2, a Shroom3 protein, lacking the ASD2 domain, fails to complement (Figure 7B). These observations suggest that the PDZ domain is not required for Shroom3 function in process outgrowth inhibition and support a role for the Shroom3 ASD2 domain in process outgrowth inhibition.
Recent studies suggest that Shroom3 recruits ROCK to regulate apical constriction. ROCK binds the Shroom3 ASD2 domain through amino acids 698-957, denoted RI-CI (Nishimura and Takeichi, 2008). Ectopic expression of RI-CI acts in a dominant negative manner to block apical constriction, by blocking the ability of endogenous ROCK to bind Shroom3 (Nishimura and Takeichi, 2008). To determine whether ROCK functions with Shroom3 to inhibit process outgrowth, P19s were transfected with RI-CI, and its effects on process length were assessed. Process length is enhanced in RI-CI expressing neurons relative to control, suggesting that Shroom3 acts through ROCK to inhibit process outgrowth (Figure 8). Inhibition of ROCK with the pharmacological inhibitor Y-27632 enhances neuronal process outgrowth in control neurons (2.2-fold), consistent with previous studies that demonstrate a role for ROCK as an inhibitor of axon outgrowth and regeneration after injury. Addition of the ROCK inhibitor enhances process outgrowth in RI-CI–expressing neurons and in POSH RNAi neurons, 1.37-fold and 1.45-fold, respectively, but the enhancement of process outgrowth is attenuated relative to control neurons (Figure 8). The observation that the enhancement of outgrowth in RI-CI–expressing neurons and POSH RNAi neurons is attenuated relative to control suggests that ROCK regulates process length in both a POSH–Shroom-dependent and -independent manner.
Previous studies suggest that Shroom, by localizing ROCK, regulates myosin II distribution and/or activity to promote apical constriction (Hildebrand, 2005; Nishimura and Takeichi, 2008). Myosin II has been implicated as a key player in process outgrowth through its ability to regulate process extension, retraction, adhesion, and detachment (Wylie and Chantler, 2001; Brown and Bridgman, 2003; Chantler and Wylie, 2003; Medeiros et al., 2006; Even-Ram et al., 2007; Ketschek et al., 2007). Therefore, RNAi was used to determine whether a reduction of myosin II function would alter POSH RNAi or Shroom RNAi process length relative to control cortical neurons. Reduction of myosin IIA by RNAi altered process length in POSH RNAi neurons: 23% POSH RNAi neurons had processes 250 μm or greater in length, whereas 10% neurons deficient in both POSH and myosin IIA function had processes 250 μm or greater (p < 0.0012; Figure 9A). The ability of myosin IIA inhibition to suppress or reverse the POSH RNAi phenotype is independent of plating conditions, because similar results were observed when neurons were grown on poly-l-lysine (data not shown) or poly-l-lysine and laminin. Myosin IIA RNAi also altered process length in Shroom RNAi neurons: 23% Shroom RNAi neurons had processes 250 μm or greater in length, whereas 4% neurons deficient in both Shroom and myosin IIA function had processes 250 μm or greater (p < 0.0012; Figure 9B). Reduction of myosin IIA by RNAi had no effect on process length of Luc RNAi control neurons (Figure 9, A and B). Reduction of myosin IIB function by RNAi reduced cell viability in both control and POSH RNAi neurons, preventing an assessment of the role of myosin IIB as an effector of POSH function. To confirm the involvement of myosin II activity, primary cortical neurons were treated for 2 h with blebbistatin, a pharmacological inhibitor of myosin II (Kovacs et al., 2004), and process length was measured at the end of the 2-h window. Blebbistatin-treated POSH RNAi neurons exhibited a decrease in process length relative to DMSO (vehicle control)-treated POSH RNAi neurons: 25% POSH RNAi DMSO-treated cortical neurons have processes 250 μm or greater in length, whereas 11% POSH RNAi blebbistatin-treated neurons deficient in myosin II activity had processes 250 μm or greater (p < 0.003; Figure 9C). Average process length of POSH RNAi neurons decreased by ∼50 μm in 2 h; because POSH RNAi neurons extend at an estimated rate of 5 μm/h, blebbistatin promoted retraction or decreased adherence is likely to contribute to this substantial decrease in process length, in addition to inhibition of process outgrowth.
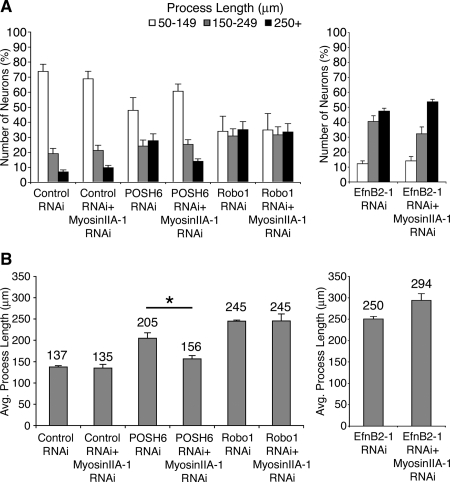
Robo and EphrinB2 are components of inhibitory pathways that negatively regulate process outgrowth (Tessier-Lavigne and Goodman, 1996; Huber et al., 2003). RNAi-mediated decrease in Robo or EphrinB2 function led to enhanced process outgrowth in P19 neurons (Figure 10), consistent with their roles as negative regulators of process outgrowth. We assessed whether myosin IIA RNAi could reverse enhanced process outgrowth phenotypes resulting from RNAi-mediated inhibition of either Robo or EphrinB2 function. In contrast to POSH and Shroom, myosin IIA RNAi did not reverse the Robo RNAi or EphrinB2 RNAi phenotypes. These observations, together with the observation that myosin IIA RNAi does not inhibit process outgrowth of cells transfected with control vectors, indicate that myosin IIA RNAi does not simply function as a general suppressor of process outgrowth, but instead it specifically reverses the effects of POSH and Shroom3 RNAi. Collectively, these results suggest that up-regulation of myosin IIA function may be the driving force behind the increase in length in neurons deficient in POSH or Shroom function.
Figure 10.
RNAi-mediated reduction of myosin IIA selectively attenuates POSH RNAi long process outgrowth phenotype and does not attenuate the long process outgrowth phenotype of RNAi-mediated decrease in the axon guidance cues Robo1 and EphrinB2 (EfnB2). P19 cells were transfected with expression vectors for Ngn2 and RNAi vectors targeting POSH, Robo, EphrinB2, with and without a second RNAi vector targeting myosin IIA. Cells were cultured on laminin for 72 h, fixed, stained, and process length of GFP-labeled transfected neurons was determined. Robo and Ephrin B2 RNAi neurons exhibit enhanced process length, consistent with their roles as regulators of process outgrowth. No significant changes in distribution of process lengths (A) or average process lengths (B) are evident when myosin IIA function is reduced in the Robo or EphrinB2 RNAi neurons, in contrast to POSH RNAi neurons (*p < 0.0001, Student's t test).
DISCUSSION
The POSH scaffold protein is expressed in the mouse CNS during embryonic development. Here, we demonstrate a role for POSH in regulating neuronal process outgrowth. P19 neurons with RNAi-mediated inhibition of POSH exhibit an increase in process length. Likewise, RNAi reduction of POSH function in embryonic primary cortical neurons leads to an increase in axon length. Collectively, these observations support a role for POSH in negatively regulating process outgrowth.
Although previous studies have reported that POSH promotes apoptosis in various cell types, including mature neurons (Tapon et al., 1998; Xu et al., 2003, 2005; Kim et al., 2005; Zhang et al., 2006), increased process length in the differentiating POSH RNAi neurons is not due to enhanced survival resulting from the loss of a proapoptotic protein. First, the percentage of apoptotic cells was modestly increased with POSH RNAi, not decreased as expected if POSH RNAi enhanced cell survival in differentiating neurons. Second, pharmacological inhibition of proapoptotic caspases did not increase process length under the experimental conditions used here. How POSH regulates these two distinct biological processes remains to be discovered, but one possibility is that the ability of POSH to function in one pathway or another is a consequence of different partners bound to POSH.
POSH is composed of multiple protein interaction domains. Complementation analysis supports a role for the third SH3 domain of POSH in regulating process length, and we show that the SH3-3 domain of POSH binds Shroom3. We also report a requirement for the POSH RING domain, a characteristic feature of a subfamily of E3 ubiquitin ligases. Membrane trafficking, apoptosis, calcium homeostasis, and process outgrowth inhibition each require the RING domain of POSH, suggesting a conserved function for this domain (Alroy et al., 2005; Tsuda et al., 2005; Kim et al., 2006; Tuvia et al., 2007). POSH interacts with JNK pathway signaling components (Tapon et al., 1998; Xu et al., 2003), the AKT serine threonine kinase (Figueroa et al., 2003; Lyons et al., 2007), and the small GTPase Rac (Tapon et al., 1998). All are known regulators of cytoskeletal dynamics; however, their contribution to POSH-mediated process outgrowth regulation remains to be determined.
We identified Shroom3, an F-actin binding protein that regulates apical constriction through a myosin II-dependent pathway (Hildebrand and Soriano, 1999; Haigo et al., 2003; Hildebrand, 2005), as a POSH-interacting partner by using a yeast two-hybrid screen. We screened a high complexity library (5–10 million clones) and recovered multiple overlapping Shroom3 isolates. In addition, when this same high-complexity library was screened for Shroom3-interacting partners, we recovered POSH (Figueroa and Vojtek, unpublished observations). POSH and Shroom3 are expressed in overlapping regions of the developing brain. Furthermore, we demonstrate that endogenous POSH and Shroom3 coassociate during embryonic neural development, suggesting that complex formation between these two proteins may have functional consequences. Finally, we show that primary cortical neurons deficient in Shroom3 function exhibit a phenotype strikingly similar to the POSH RNAi neurons, namely, exuberant axon outgrowth. Together, the biochemical and molecular genetic studies reported here suggest that POSH and Shroom3 work in concert to negatively regulate axon outgrowth.
Shroom3 requires the function of the ASD1 and ASD2 domains for process outgrowth inhibition. The ASD1 domain is thought to function as a localization domain, to link Shroom3 to the actin cytoskeleton (Haigo et al., 2003; Hildebrand, 2005; Dietz et al., 2006). The ASD2 domain regulates myosin II activity and/or localization, likely by recruiting ROCK (Hildebrand, 2005; Nishimura and Takeichi, 2008). Consistent with a proposed role for myosin function, we demonstrate that RNAi-mediated inhibition of myosin IIA or blebbistatin-mediated inhibition of myosin II activity reverses the POSH and Shroom3 RNAi phenotypes. This result suggests that increased myosin II activity may be the driving force behind the increase in length in the POSH or Shroom3 RNAi phenotypes. The POSH–Shroom3 complex is likely to act through ROCK, which binds the ASD2 domain of Shroom3. Consistent with this, we show that ectopic expression of the domain of ROCK that binds Shroom3, which functions as a dominant negative by blocking the association of endogenous ROCK with the Shroom3 ASD2 domain (Nishimura and Takeichi, 2008), increases process length. ROCK regulates myosin II activity by phosphorylating myosin light chain and myosin light chain phosphatase (Riento and Ridley, 2003). The POSH–Shroom3 complex, by regulating ROCK function, may regulate myosin II. Alternately, POSH may not directly regulate myosin IIA activity; rather, a reduction of myosin IIA function may compensate for a shift in balance among cytoskeletal forces engendered by a reduction in POSH or Shroom3 function. Such a shift in balance among cytoskeletal forces also could be mediated by ROCK, which regulates multiple effector pathways (Riento and Ridley, 2003; Gallo, 2006). In addition to regulating myosin activity, ROCK phosphorylates LIM kinase, which regulates actin filament assembly, and CRMP2, which regulates microtubule assembly, among other effector proteins (Riento and Ridley, 2003). Loss of ROCK function in one specific complex, perhaps reducing signals through a particular effector pathway, could shift the balance among cytoskeletal forces. Understanding in greater detail which substrates ROCK phosphorylates when associated with the POSH complex will likely provide additional insight into the mechanism by which POSH and Shroom3 inhibit process outgrowth and further define the role of myosin IIA in this process.
POSH inhibits process outgrowth through the actomyosin regulatory protein Shroom3. POSH also regulates membrane trafficking and calcium homeostasis (Alroy et al., 2005; Kim et al., 2006; Tuvia et al., 2007). The interplay between the actomyosin network, membrane trafficking, calcium signaling events, and inhibition of process outgrowth remain to be determined. POSH may regulate process outgrowth through its ability to regulate membrane trafficking and/or calcium homeostasis, in addition to regulating the actomyosin network. Alternately, POSH by regulating the actomyosin network may indirectly regulate membrane trafficking and/or calcium levels.
Previous studies demonstrate that Shroom3 regulates apical constriction and that two conserved domains, the ASD1/actin binding domain and the ASD2 domains, are each required for Shroom3 to promote apical constriction (Haigo et al., 2003; Hildebrand, 2005; Dietz et al., 2006; Nishimura and Takeichi, 2008). Process outgrowth inhibition also requires these two conserved domains of Shroom3. Thus, two distinct cell shape changes have similar requirements for Shroom3. Intriguingly, myosin II activity is required for Shroom3-promoted apical constriction, whereas repression of myosin II activity seems to be required for POSH–Shroom3-mediated inhibition of process outgrowth. As discussed above, repression of myosin II activity may be direct or indirect, with Rho kinase a likely key player. A further distinguishing characteristic of the two cell shape changes is a requirement for different Ras family members. In Xenopus, disruption of Rap1 signaling blocks Shroom3-mediated apical constriction (Haigo et al., 2003), whereas Rac is linked to POSH (Tapon et al., 1998; Figueroa et al., 2003; Xu et al., 2003). Together, these results highlight the role that scaffold protein dependent signaling events play in mediating specific biological outcomes.
Collectively, the results presented here suggest that the molecular scaffold protein POSH assembles an inhibitory complex that links, directly or indirectly, to the actin-myosin network to regulate neuronal process outgrowth. Also, these findings reveal a role for Shroom3 in axon outgrowth, in addition to its previously described role in apical constriction. As a negative regulator of process outgrowth, the POSH–Shroom3 complex may represent a useful target to overcome neurite outgrowth inhibition.
ACKNOWLEDGMENTS
We thank Samantha Tarras and Lawrence Tsoi for assistance with the yeast two-hybrid screen, Jeff Hildebrand for providing Shroom3 expression vectors, and Monica Deo for in situ hybridization analysis of POSH expression. This work was supported by National Institute of Mental Health grant MH-073085 (to A.B.V.), National Institutes of Health grant NS-38698 (to D.L.T.), and the American Cancer Society research scholar grant RSG-01-177-01-MGO (to A.B.V.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0231) on October 1, 2008.
REFERENCES
- Alroy I., et al. The trans-Golgi network-associated human ubiquitin-protein ligase POSH is essential for HIV type 1 production. Proc. Natl. Acad. Sci. USA. 2005;102:1478–1483. doi: 10.1073/pnas.0408717102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J., Bridgman P. C. Role of myosin II in axon outgrowth. J. Histochem. Cytochem. 2003;51:421–428. doi: 10.1177/002215540305100403. [DOI] [PubMed] [Google Scholar]
- Chantler P. D., Wylie S. R. Elucidation of the separate roles of myosins IIA and IIB during neurite outgrowth, adhesion and retraction. IEE Proc. Nanobiotechnol. 2003;150:111–125. doi: 10.1049/ip-nbt:20031076. [DOI] [PubMed] [Google Scholar]
- Chung K. H., Hart C. C., Al-Bassam S., Avery A., Taylor J., Patel P. D., Vojtek A. B., Turner D. L. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz M. L., Bernaciak T. M., Vendetti F., Kielec J. M., Hildebrand J. D. Differential actin-dependent localization modulates the evolutionarily conserved activity of Shroom family proteins. J. Biol. Chem. 2006;281:20542–20554. doi: 10.1074/jbc.M512463200. [DOI] [PubMed] [Google Scholar]
- Etournay R., Zwaenepoel I., Perfettini I., Legrain P., Petit C., El-Amraoui A. Shroom2, a myosin-VIIa- and actin-binding protein, directly interacts with ZO-1 at tight junctions. J. Cell Sci. 2007;120:2838–2850. doi: 10.1242/jcs.002568. [DOI] [PubMed] [Google Scholar]
- Even-Ram S., Doyle A. D., Conti M. A., Matsumoto K., Adelstein R. S., Yamada K. M. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat. Cell Biol. 2007;9:299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- Fairbank P. D., Lee C., Ellis A., Hildebrand J. D., Gross J. M., Wallingford J. B. Shroom2 (APXL) regulates melanosome biogenesis and localization in the retinal pigment epithelium. Development. 2006;133:4109–4118. doi: 10.1242/dev.02563. [DOI] [PubMed] [Google Scholar]
- Farah M. H., Olson J. M., Sucic H. B., Hume R. I., Tapscott S. J., Turner D. L. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Figueroa C., Tarras S., Taylor J., Vojtek A. B. Akt2 negatively regulates assembly of the POSH-MLK-JNK signaling complex. J. Biol. Chem. 2003;278:47922–47927. doi: 10.1074/jbc.M307357200. [DOI] [PubMed] [Google Scholar]
- Gallo G. RhoA-kinase coordinates F-actin organization and myosin II activity during semaphorin-3A-induced axon retraction. J. Cell Sci. 2006;119:3413–3423. doi: 10.1242/jcs.03084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo K. A., Johnson G. L. Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nat. Rev. Mol. Cell Biol. 2002;3:663–672. doi: 10.1038/nrm906. [DOI] [PubMed] [Google Scholar]
- Haigo S. L., Hildebrand J. D., Harland R. M., Wallingford J. B. Shroom induces apical constriction and is required for hingepoint formation during neural tube closure. Curr. Biol. 2003;13:2125–2137. doi: 10.1016/j.cub.2003.11.054. [DOI] [PubMed] [Google Scholar]
- Hildebrand J. D. Shroom regulates epithelial cell shape via the apical positioning of an actomyosin network. J. Cell Sci. 2005;118:5191–5203. doi: 10.1242/jcs.02626. [DOI] [PubMed] [Google Scholar]
- Hildebrand J. D., Soriano P. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell. 1999;99:485–497. doi: 10.1016/s0092-8674(00)81537-8. [DOI] [PubMed] [Google Scholar]
- Huber A. B., Kolodkin A. L., Ginty D. D., Cloutier J. F. Signaling at the growth cone: ligand-receptor complexes and the control of axon growth and guidance. Annu. Rev. Neurosci. 2003;26:509–563. doi: 10.1146/annurev.neuro.26.010302.081139. [DOI] [PubMed] [Google Scholar]
- Ketschek A. R., Jones S. L., Gallo G. Axon extension in the fast and slow lanes: substratum-dependent engagement of myosin II functions. Dev. Neurobiol. 2007;67:1305–1320. doi: 10.1002/dneu.20455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G. H., Park E., Han J. K. The assembly of POSH-JNK regulates Xenopus anterior neural development. Dev. Biol. 2005;286:256–269. doi: 10.1016/j.ydbio.2005.07.033. [DOI] [PubMed] [Google Scholar]
- Kim G. H., Park E., Kong Y. Y., Han J. K. Novel function of POSH, a JNK scaffold, as an E3 ubiquitin ligase for the Hrs stability on early endosomes. Cell Signal. 2006;18:553–563. doi: 10.1016/j.cellsig.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kovacs M., Toth J., Hetenyi C., Malnasi-Csizmadia A., Sellers J. R. Mechanism of blebbistatin inhibition of myosin II. J. Biol. Chem. 2004;279:35557–35563. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- Lyons T. R., Thorburn J., Ryan P. W., Thorburn A., Anderson S. M., Kassenbrock C. K. Regulation of the pro-apoptotic scaffolding protein POSH by Akt. J. Biol. Chem. 2007;282:21987–21997. doi: 10.1074/jbc.M704321200. [DOI] [PubMed] [Google Scholar]
- McBurney M. W. P19 embryonal carcinoma cells. Int. J. Dev. Biol. 1993;37:135–140. [PubMed] [Google Scholar]
- Medeiros N. A., Burnette D. T., Forscher P. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat. Cell Biol. 2006;8:215–226. doi: 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
- Nishimura T., Takeichi M. Shroom3-mediated recruitment of Rho kinases to the apical cell junctions regulates epithelial and neuroepithelial planar remodeling. Development. 2008;135:1493–1502. doi: 10.1242/dev.019646. [DOI] [PubMed] [Google Scholar]
- Riento K., Ridley A. J. Rocks: multifunctional kinases in cell behaviour. Nat. Rev. Mol. Cell Biol. 2003;4:446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- Schnorr J. D., Holdcraft R., Chevalier B., Berg C. A. Ras1 interacts with multiple new signaling and cytoskeletal loci in Drosophila eggshell patterning and morphogenesis. Genetics. 2001;159:609–622. doi: 10.1093/genetics/159.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub O., Verrey F., Kleyman T. R., Benos D. J., Rossier B. C., Kraehenbuhl J. P. Primary structure of an apical protein from Xenopus laevis that participates in amiloride-sensitive sodium channel activity. J. Cell Biol. 1992;119:1497–1506. doi: 10.1083/jcb.119.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N., Nagata K., Lamarche N., Hall A. A new rac target POSH is an SH3-containing scaffold protein involved in the JNK and NF-kappaB signalling pathways. EMBO J. 1998;17:1395–1404. doi: 10.1093/emboj/17.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier-Lavigne M., Goodman C. S. The molecular biology of axon guidance. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- Tsuda M., Langmann C., Harden N., Aigaki T. The RING-finger scaffold protein Plenty of SH3s targets TAK1 to control immunity signalling in Drosophila. EMBO Rep. 2005;6:1082–1087. doi: 10.1038/sj.embor.7400537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuvia S., Taglicht D., Erez O., Alroy I., Alchanati I., Bicoviski V., Dori-Bachash M., Ben-Avraham D., Reiss Y. The ubiquitin E3 ligase POSH regulates calcium homeostasis through spatial control of Herp. J. Cell Biol. 2007;177:51–61. doi: 10.1083/jcb.200611036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek A. B., Hollenberg S. M. Ras-Raf interaction: two-hybrid analysis. Methods Enzymol. 1995;255:331–342. doi: 10.1016/s0076-6879(95)55036-4. [DOI] [PubMed] [Google Scholar]
- Vojtek A. B., Hollenberg S. M., Cooper J. A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Vojtek A. B., Taylor J., DeRuiter S. L., Yu J. Y., Figueroa C., Kwok R. P., Turner D. L. Akt regulates basic helix-loop-helix transcription factor-coactivator complex formation and activity during neuronal differentiation. Mol. Cell Biol. 2003;23:4417–4427. doi: 10.1128/MCB.23.13.4417-4427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie S. R., Chantler P. D. Separate but linked functions of conventional myosins modulate adhesion and neurite outgrowth. Nat. Cell Biol. 2001;3:88–92. doi: 10.1038/35050613. [DOI] [PubMed] [Google Scholar]
- Xu Z., Kukekov N. V., Greene L. A. POSH acts as a scaffold for a multiprotein complex that mediates JNK activation in apoptosis. EMBO J. 2003;22:252–261. doi: 10.1093/emboj/cdg021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Kukekov N. V., Greene L. A. Regulation of apoptotic c-Jun N-terminal kinase signaling by a stabilization-based feed-forward loop. Mol. Cell Biol. 2005;25:9949–9959. doi: 10.1128/MCB.25.22.9949-9959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder M., Hildebrand J. D. Shroom4 (Kiaa1202) is an actin-associated protein implicated in cytoskeletal organization. Cell Motil. Cytoskeleton. 2007;64:49–63. doi: 10.1002/cm.20167. [DOI] [PubMed] [Google Scholar]
- Yu J. Y., Taylor J., DeRuiter S. L., Vojtek A. B., Turner D. L. Simultaneous inhibition of GSK3alpha and GSK3beta using hairpin siRNA expression vectors. Mol. Ther. 2003;7:228–236. doi: 10.1016/s1525-0016(02)00037-0. [DOI] [PubMed] [Google Scholar]
- Zhang Q. G., Han D., Xu J., Lv Q., Wang R., Yin X. H., Xu T. L., Zhang G. Y. Ischemic preconditioning negatively regulates plenty of SH3s-mixed lineage kinase 3-Rac1 complex and c-Jun N-terminal kinase 3 signaling via activation of Akt. Neuroscience. 2006;143:431–444. doi: 10.1016/j.neuroscience.2006.07.049. [DOI] [PubMed] [Google Scholar]