Abstract
Y-family DNA polymerases carry out translesion synthesis past damaged DNA. DNA polymerases (pol) η and ι are usually uniformly distributed through the nucleus but accumulate in replication foci during S phase. DNA-damaging treatments result in an increase in S phase cells containing polymerase foci. Using photobleaching techniques, we show that polη is highly mobile in human fibroblasts. Even when localized in replication foci, it is only transiently immobilized. Although ubiquitination of proliferating cell nuclear antigen (PCNA) is not required for the localization of polη in foci, it results in an increased residence time in foci. polι is even more mobile than polη, both when uniformly distributed and when localized in foci. Kinetic modeling suggests that both polη and polι diffuse through the cell but that they are transiently immobilized for ∼150 ms, with a larger proportion of polη than polι immobilized at any time. Treatment of cells with DRAQ5, which results in temporary opening of the chromatin structure, causes a dramatic immobilization of polη but not polι. Our data are consistent with a model in which the polymerases are transiently probing the DNA/chromatin. When DNA is exposed at replication forks, the polymerase residence times increase, and this is further facilitated by the ubiquitination of PCNA.
INTRODUCTION
Most types of damage in cellular DNA block the progress of the replication fork because the highly stringent replicative DNA polymerases (pols) δ and ε are unable to accommodate the damaged bases in their active sites. An important mechanism for bypassing these replication blocks is by translesion synthesis (TLS), in which a low-stringency specialized polymerase is able to substitute for the blocked replicative polymerase (Friedberg et al., 2005). Most of these specialized TLS polymerases belong to the Y-family, whose members have a much more open structure than the B-family replicative polymerases (Yang and Woodgate, 2007). This enables them to accommodate damaged bases in their active sites, each Y-family polymerase having a different specificity for different types of altered bases. For example, polη can accommodate both bases of a cyclobutane pyrimidine dimer (CPD) in its active site and is able to replicate past a CPD with similar efficiency to an undamaged base (McCulloch et al., 2004). Moreover, in most cases it inserts the “correct” bases opposite the CPD (Masutani et al., 2000). Mutations in the POLH gene result in the variant form of xeroderma pigmentosum (XP-V) (Masutani et al., 1999; Johnson et al., 1999a). The high incidence of sunlight-induced skin cancer associated with this disorder probably results from a less efficient polymerase substituting for polη in its absence. When this substituting polymerase carries out TLS past UV photoproducts, it is presumed to be more error-prone than polη, resulting in a higher UV-induced mutation frequency, as seen in XP-V cells (Maher et al., 1976).
Polη and its paralogue polι are uniformly distributed throughout the cell nucleus in G2-M-G1 phases of the cell cycle. During S phase, both pols are localized in microscopically visible bright foci, representing replication factories (Kannouche et al., 2001, 2003). Treatments like UV and the inhibitor hydroxyurea (HU) result in an accumulation of cells in which polη and ι are localized in foci (Kannouche et al., 2001, 2003). These treatments reduce or block the progression of replication forks, slow down the passage through S phase and result in an increase in the proportion of S phase nuclei in the cell population. This accounts at least partially for the increased number of cells with polymerase foci.
The actual engagement of pol η and ι at the sites of stalled replication forks is mediated by the homotrimeric sliding clamp accessory protein PCNA. When the replication fork stalls, exposed single-stranded regions of DNA at the stalled forks activate the E3 ubiquitin ligase Rad18. Together with its E2 partner Rad6, Rad18 mono-ubiquitinates PCNA at the stalled fork on lysine-164 (Hoege et al., 2002; Kannouche et al., 2004; Watanabe et al., 2004). As well as having “PIP box” PCNA-binding motifs (Kannouche et al., 2001; Vidal et al., 2004), polη and ι both have ubiquitin-binding motifs in the C-terminal parts of the proteins (Bienko et al., 2005). Thus, when PCNA is ubiquitinated, its affinity for these polymerases is increased by virtue of these motifs, and this facilitates their binding to the stalled forks. This mechanism, deduced from in vivo studies, has recently been demonstrated for polη in a reconstituted in vitro system (Zhuang et al., 2008).
The microscopically visible replication foci presumably represent subnuclear structures at which replication-associated factors are concentrated. However, little is known about the nature of these structures or about the dynamics of the different factors that are localized in them. We have used high-resolution confocal microscopy and fluorescence recovery after photobleaching (FRAP) together with biochemical fractionation to give further insight into the relationship of polη and polι to the replication foci. Both polymerases were highly mobile within the nucleus, and interacted with immobile elements (most likely DNA) very transiently, with characteristic binding times of the order of 100–200 ms. Remarkably, we find that even when localized in foci, they remained highly mobile, with half-lives of <1 s. The foci thus represent dynamic “work stations” with polymerases entering and exiting continually, remaining in the foci for fractions of a second. We demonstrate that the two polymerases act independently, and we show that ubiquitination of PCNA facilitates but is not essential for accumulation of polη into the foci.
MATERIALS AND METHODS
Cell Lines and Culture Conditions
XP30RO SV40 transformed fibroblasts were transfected with enhanced green fluorescent protein (eGFP)-polη and eGFP-polη-pol-dead plasmids, and stable clones expressing the respective alleles of polη were isolated. All cell lines described in this article were grown in Eagle's minimal essential medium supplemented with 15% fetal calf serum. Cell lines were generated as described previously (Kannouche et al., 2001).
For global UV-irradiation, the cells were treated essentially as described previously (Kannouche et al., 2001) and irradiated, unless otherwise stated, with 15 Jm−2 UV-C before a further incubation for 7 h. For local UV-irradiation, cells were UV-irradiated with 120 Jm−2 through 5-μm pores of a polycarbonate filter. For HU treatment, the cells were incubated in 1 mM HU for 24 h. To inhibit the proteasome, the cells were preincubated for 1 h with 0.1 μM epoxomicin before UV-irradiation and incubated for a further 6 h in epoxomicin-containing medium after irradiation. DRAQ5 (Biostatus Limited, Leicestershire, United Kingdom) was used at the concentration of 10 μM and incubated with the cells for the duration of the experiment. Detectable DNA staining was visible already after a 3-min incubation.
Transfections and Plasmids
Plasmids were transfected into simian virus 40 (SV40)-transformed fibroblasts by using FuGENE 6 as described previously (Kannouche et al., 2001). eGFPpolη, eGFPpolι, eGFP-H2B, hRad18, and hRad18C28F were constructed in peGFP-C3 or pCDNA3.1 plasmids (Kannouche et al., 2004). eGFP-PCNA was subcloned in pCDNA3.1 by cutting a 1.6-kb fragment with XbaI and BamHI from pENeGFPPCNAL2 (a kind gift of Cristina Cardoso, Max Delbrück Center for Molecular Medicine, Berlin, Germany). To analyze the effect of Rad18 expression the cells were simultaneously cotransfected with monomeric red fluorescent protein (mRFP)-α-tubulin (a kind gift from Sally Wheatley) as a marker for transfected cells.
To generate the polη pol-dead mutant, amino acids D115 and E116 were mutated to alanine using the QuikChange kit (Stratagene, La Jolla, CA). The full coding region of polη was then sequenced to check for mutations.
For small interfering RNA (siRNA) knockdown, ONTARGET+ Smartpools (Dharmacon RNA Technologies, Lafayette, CO) containing four siRNAs against USP1 were used at 5 nM final concentration. The negative control (NTC) represents a pool of four siRNAs designed to have at least four mismatches for all the sequences present in the human genome. The cells were transfected in a 3-cm dish with siRNA by using Hiperfect (QIAGEN, Hilden, Germany) according to manufacturer's instructions using the fast forward procedure, and incubated for 48 h before analysis.
In Vivo Cell Imaging
Cells were plated at 5 × 105 cells/3-cm dish (MatTek, Ashland, MA) for at least 48 h before imaging. The cells were monitored under the microscope in a temperature-controlled chamber in 5% CO2 atmosphere.
All the FRAP analysis was performed on an LSM510 confocal microscope (Carl Zeiss, Jena, Germany) by using a 40× numerical aperture 1.3 differential interference contrast oil objective. Except otherwise stated, a region of 1.44 μm2 was monitored for 3 s (100 scans taken every 30 ms) before being bleached (1 iteration), and recovery of fluorescence was subsequently monitored for another 16.5 s (550 scans every 30 ms) using bidirectional scans. For the strip-FRAP, the monitored region was changed to a 2-μm strip positioned in the middle and spanning the whole nucleus. Using monodirectional scans, the cell was followed for 4 s before bleaching (200 scans every 20 ms, monodirectional) and 22 s after bleaching (1100 scans every 20 ms).
To avoid monitor bleaching, the laser was set to a power of 700 nW except during the bleaching iterations (140 μW). Raw fluorescence data were then background subtracted and normalized as described previously (Houtsmuller and Vermeulen, 2001). Briefly, the relative fluorescence was calculated as It/I0, where It represents the fluorescence intensity at time t, and I0 represents the average intensity of 20 points just before bleaching. Average measurements of at least 30 cells were used for each FRAP curve. The t0.5 was calculated by interpolation on the FRAP curves as the time required to reach half-fluorescence recovery (I0.5 = 0.5(Iend + Ibleach), where Iend is the average fluorescence of the last 20 points, and Ibleach is the fluorescence recorded immediately after the bleaching). The long-lasting immobile fraction is calculated as (1 − Iend)/(1 − Ibleach).
Half-Nucleus Bleaching Combined with Fluorescence Loss in Photobleaching (FLIP)-FRAP
For FLIP-FRAP, half of the nucleus was bleached for 2.4 s (4 iterations), after which the whole cell was imaged every 2 s for 50 s. To analyze the data, the FRAP (intensity of fluorescence in the whole of the bleached half-nucleus) was subtracted from the FLIP (intensity of fluorescence in the whole of the unbleached half-nucleus). The difference between FLIP and FRAP after bleaching was normalized to 1. The results are presented on a log scale, and the mobility of the protein is presented as the time necessary for the FRAP value to reach 90% of the prebleach value. Errors bars represent the SEs of the mean.
FRAP in Local Damage
The entire local damage was bleached in 0.7 s with two bleaching pulses, and the recovery of fluorescence monitored for by scanning the whole cell every second. The intensity of fluorescence in the local damage before bleaching was normalized to 1. Errors bars represent the SEs of the mean.
FRAP Data Modeling
For the model-based analysis of the FRAP data, raw FRAP curves were normalized to prebleach values and the best fitting curve (by ordinary least squares) was picked from a large set of computer simulated FRAP curves in which three parameters representing mobility properties were varied: diffusion rate (ranging from 0.04–25 μm2/s), immobile fraction (ranging from 0 to 90%), and time spent in immobile state (ranging from 0.1 to 300 s).
The Monte Carlo computer simulations used to generate FRAP curves for the fit were based on a model that simulates diffusion of molecules and binding to immobile elements in an ellipsoidal volume. The laser bleach pulse was simulated based on experimentally derived three-dimensional (3D) laser intensity profiles, which were used to determine the probability for each molecule to get bleached, considering their 3D position. The simulation of the FRAP curve was then run using discrete time steps corresponding to the experimental scan interval of 21 ms. Diffusion was simulated at each new time step t + Δt by deriving the new positions (xt+Δt, yt+Δt, zt+Δt) of all mobile molecules from their current positions (xt, yt, zt) by xt+Δt = xt + G(r1), yt+Δt = yt + G(r2), and zt+Δt = zt + G(r3), where ri is a random number (0 ≤ ri ≤ 1) chosen from a uniform distribution, and G(ri) is an inversed cumulative Gaussian distribution with μ = 0 and σ2 = 6DΔt, where D is the diffusion coefficient. Immobilization was derived from simple binding kinetics described by kon/koff = Fimm/(1 − Fimm), where Fimm is the relative number of immobile molecules. The probability for each particle to become immobilized is defined as Pimmobilize = kon = koff · Fimm/(1 − Fimm), where koff = 1/Timm, and Timm is the average time spent in the immobile state. The probability to be released is given by Pmobilize = koff = 1/Timm. In simulations of two immobile fractions with different kinetics, two immobilization/mobilization probabilities were evaluated at each unit time step. Simulations of the FRAP curve were performed at every unit time step by counting the number of unbleached molecules in the bleached region after simulations of diffusion and binding during that time step.
In all simulations, the size of the ellipsoid was based on the size of the nuclei, and the region used in the measurements determined the size of the simulated bleach region. The laser intensity profile using the simulation of the bleaching step was derived from confocal images stacks of chemically fixed nuclei containing green fluorescent protein (GFP) that were exposed to a stationary laser beam at various intensities and varying exposure times. The unit time step Δt corresponded to the experimental sample rate of 21 ms. The number of molecules in the simulations was 106, which was empirically determined by producing curves that closely approximate the data with comparable fluctuations.
Epifluorescence and Triton Extraction
Cells were seeded directly on a coverslip and irradiated the next day with 15 J/m2 before incubation for 7 h. Cells were then washed twice with phosphate-buffered saline (PBS) and fixed in 2% paraformaldehyde for 30 min before further washing in PBS and then mounted in VECTASHIELD (Vector Laboratories, Burlingame, CA) + 4,6-diamidino-2-phenylindole (DAPI). To extract the soluble proteins before fixation, the coverslips were washed in 0.2% Triton X as described previously (Kannouche and Lehmann, 2006).
Size Exclusion Chromatography
Cells were harvested from a 10-cm dish and lysed in 75 μl of buffer A20 (20 mM HEPES, pH 7.5, 20 mM NaCl, 1 mM MgCl2, 0.5% Triton X-100, and 1 μl/ml Benzonase [Sigma Chemical. Poole, Dorset, United Kingdom]). The extracts were incubated for 30 min on ice to allow DNA digestion by Benzonase. After incubation the extract was diluted in an equal volume of buffer A500 (same as buffer A20 but with 500 mM NaCl, 0.4 mM EDTA and 1 mM dithiothreitol). The extract was then spun down at 10,000 × g and filtered through a 0.2-μm pore VectaSpin Micro (Whatman, Maidstone, United Kingdom) before loading onto a 2.4-ml Superdex200 size exclusion column on a SMART system (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). By using standards of known Stokes radii run on the same column, the respective values for polη and polι were calculated by interpolation.
Glycerol Gradient
Cell extracts prepared as for the gel filtration were loaded on a 5-ml 15–35% glycerol gradient in buffer A260 (as described above but containing 260 mM NaCl). The gradient was centrifuged for 16 h in an AH650 swing-out rotor (Sorvall, Newton, CT) at 42,500 rpm, and finally 200 μl fractions were collected from the top. By using standards of known sedimentation coefficient alongside the extract, the respective values for polη and polι were calculated. The molecular weight (MW) was calculated as follows: MW = (6πηNaS)/(1 − υρ), where η is the viscosity of the medium, N is Avogadro's number, a is the Stokes radius, S is the sedimentation coefficient, υ is the partial specific volume of the protein, and ρ is the density of the medium.
Western Blot
Western blotting was performed on nitrocellulose and probed using the following antibodies: clones 7.1 and 13.1 (Roche Diagnostics, Mannheim, Germany) against GFP and PC10 (CRUK) against PCNA. Antibodies against full-length polη (Kannouche et al., 2001), against the C-terminal peptide of polι (a kind gift from Roger Woodgate, National Institutes of Health, Bethesda, MD; Kannouche et al., 2003), and against the N-terminal part of USP1 (a kind gift from Tony Huang, New York University, New York, NY; Huang et al., 2006) have been described previously.
RESULTS
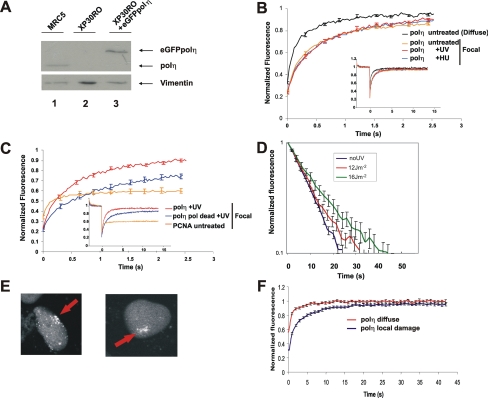
To measure the dynamics of polη in human cells, we used a cell line in which N-terminally tagged eGFP-polη was expressed in XP30RO cells (Kannouche et al., 2001). These cells contain a truncation mutation in the POLH gene close to the N terminus (Johnson et al., 1999a) and can be considered as polη null mutants. We previously showed that the eGFP-polη expressed in this cell line was able to correct the typical sensitivity of XP30RO to UV followed by treatment with caffeine (Kannouche et al., 2001). Using fluorescence-activated cell sorting, we selected a subpopulation of the cells in which the level of eGFP-polη was similar to that of endogenous polη in normal MRC5 cells. Figure 1A shows a Western blot of polη in this cell line, compared with that in the normal cell line MRC5. Polη expression levels remained stable over several weeks. There was no evidence, either by Western blotting or by the appearance of cytoplasmic autofluorescence, for any free eGFP protein (data not shown). We conclude that the quantitative fluorescence measurements described in the following sections were derived from cells that express full-length and biologically active GFP-tagged polη.
Figure 1.
Dynamics of eGFP-polη in living cells. (A) Western blot of the XP30RO-eGFP-polη cell line used in this study (lane 3), compared with MRC5 (lane 1) and XP30RO (lane 2). (B) Comparison of FRAP curves (relative fluorescence recovery plotted against time) of eGFP-polη uniformly distributed in untreated XP30RO-eGFP-polη cells and in foci in S phase cells from untreated, UV-treated, and HU-treated cells. (C) Fluorescence recovery of “pol-dead” mutant (blue) and wild-type polη (red) in foci (7 h after 15 Jm−2 UV-C). Also shown is the FRAP curve for eGFP-PCNA in foci (orange), showing large immobile fraction. (D) FLIP-FRAP analysis of eGFP-polη. Cells were not irradiated (no UV, mean of 63 cells) or globally irradiated with 12 (mean of 50 cells) and 16 Jm−2 (mean of 27 cells). Five hours later, half-nucleus bleaching associated with FLIP-FRAP analysis was performed. The data were normalized as described in Materials and Methods. The error bars represent the SE of the mean. (E) eGFP-polη accumulated at site of local irradiation. (F) Five hours after local irradiation, the area of local damage was entirely bleached, the recovery of fluorescence was measured in the bleached area and normalized to the level of fluorescence in the whole nucleus. Control cells represent cells in which no local damage was inflicted, but in which a square of the same size as irradiated cells was bleached.
In previous work, we showed that eGFP-polη transfected into human fibroblasts was uniformly distributed throughout the nucleus outside S phase but that it accumulated in bright foci representing replication factories in S phase cells. In cells treated with 15 Jm−2 UV-irradiation and incubated for 7 h or with 1 mM HU for 24 h, the number of cells in which polη was located in foci increased substantially, partly or wholly because of the accumulation of S phase cells after these treatments (Kannouche et al., 2001). Supplemental Figure S1 shows stills from a confocal time-lapse series in which the stable cell line was UV-irradiated through a micropore filter to generate localized damage within the nucleus (Volker et al., 2001). Using this procedure, proteins involved in processing of DNA damage accumulate at the sites of the localized irradiation. We found that, throughout S phase, eGFP-polη accumulated at the sites of local damage. Within the damaged area polη accumulated in a focal pattern because of the stalling of replication forks (Kannouche et al., 2001). In contrast, in G2 the eGFP-polη neither accumulated at the local damage nor was it in bright foci, but it became uniformly distributed through the nucleus. These data confirm the S phase-specific function of polη.
We have used FRAP to measure the mobility of polη under different conditions. We photobleached a small square of the nucleus and measured the rate of recovery of fluorescence within the square. Polη that was uniformly distributed in the nucleus (i.e., in G1 or G2 cells) relocalized into the bleached area extremely rapidly with a t0.5 of 0.15 s (Figure 1B, polη untreated-diffuse), indicating that it is highly mobile within the nucleus.
We next photobleached the eGFP-polη within a focus in an S phase nucleus by aligning the square over a visible focus in S phase cells (Box-FRAP). We used a square as small as possible so that the focus filled almost the whole area of the square. In this situation, the recovery rate was reduced about two-fold (Figure 1B). The t0.5 was still very short, 0.33 s. The mobility of polη in foci generated in cells irradiated with UV-irradiation or following HU treatment was indistinguishable from that in an unperturbed S phase (Figure 1B). Thus, surprisingly, even when associated with microscopically visible structures, the majority of the polη molecules within the focus remained highly mobile. Examination of the curves in Figure 1B at later times (up to 15 s; see inset) suggests that at most only 7% of the molecules were immobilized for a long period (see Materials and Methods for definitions of t0.5 and immobile fraction). In contrast to the highly dynamic association of eGFP-polη, we observed a relatively large (∼60%) fraction of eGFP-PCNA (Figure 1C), in which proteins were significantly immobilized for long periods (see Materials and Methods for calculation of long-lasting immobile fractions), in line with previous studies (Sporbert et al., 2002; Essers et al., 2005).This demonstrates that our system was capable of detecting immobilized proteins.
To determine whether the catalytic activity of polη might affect its mobility, we generated an XP30RO cell line expressing eGFP-polη in which amino acids (aa) D115 and E116, shown to be vital for catalytic activity (Johnson et al., 1999b), were mutated to alanines. This mutation allows the incoming dNTP to bind but cannot support the formation of the phosphodiester bond (Li et al., 1998). The mobility of this “pol dead” polη mutant, when distributed uniformly in the nucleus, was identical to that of wild-type polη (data not shown), but interestingly, its mobility in foci was about twofold lower than that of wild-type polη, with a t0.5 of ∼0.67 s and a long-lasting immobile fraction of 15% (Figure 1C).
As an alternative methodology, we have also used FLIP-FRAP in which we bleached half the nucleus. We then measured both the rate of reduction in fluorescence intensity of the unbleached half (FLIP) and the rate of recovery in the bleached half of the nucleus (FRAP). With this technique, we are able to analyze the overall mobility in the whole of the nucleus, providing the collective mobility of polη in a large number of foci, in contrast to the mobility within a single focus in the experiments described above. As with bleaching of a small square, we observed rapid redistribution of polη. The difference between FLIP and FRAP immediately after bleaching was normalized to 1, and, in Figure 1D, at different times after bleaching, the normalized difference between the FLIP and FRAP is presented on a log scale. With nuclei in which polη was uniformly distributed, polη had returned to 90% of the prebleach distribution (i.e., normalized fluorescence = 0.1) in 25 s (Figure 1D). Using this FLIP-FRAP analysis, we have examined the effect of different doses of UV on the mobility of polη in nuclei containing focal polη. We compared polη mobility in these cells with its mobility when diffusely distributed in untreated cells. A UV dose response was observed, with increasing delay in polη redistribution due to transient immobilization to subnuclear structures (Figure 1D). Higher UV doses resulted in a more pronounced delay in redistribution, reaching a maximum after irradiation with 16 Jm−2, with a redistribution time of about 45 s, compared with ∼25 s in untreated cells not exhibiting foci. This approximate doubling of the redistribution time agrees well with the approximately two-fold decrease in mobility in the Box-FRAP data presented in Figure 1B. These data suggest that with increasing UV-doses, as expected, more substrate sites (i.e., stalled forks) were created that transiently bind a larger pool of the resident polη molecules but that the average binding time within a single focus is not affected by an increasing number replication blocks.
In a further variation, we UV-irradiated cells through a micropore filter to produce localized damage in the nucleus (Volker et al., 2001). Five hours after irradiation, we selected cells in which polη had accumulated in foci at the sites of local damage (examples shown in Figure 1E), and we bleached the entire site of local damage. As control, we bleached an identical area in a nucleus in which no local damage had been inflicted. Relocalization into the bleached damaged site was again approximately two-fold slower than into undamaged areas (Figure 1F). We conclude from these different photobleaching studies that polη is highly mobile within the nucleus and that its mobility is only slightly reduced within replication foci.
Role of PCNA-Ubiquitination
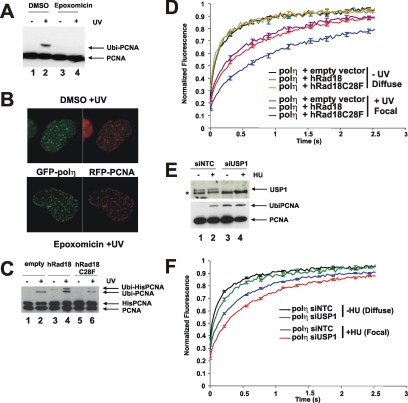
Polη has a PIP box binding motif for interaction with PCNA (Haracska et al., 2001; Kannouche et al., 2001), and it is likely that PCNA plays a role in assisting polη to find its substrate. After exposure of cells to UV-irradiation or other agents that block progression of the replication fork, PCNA becomes mono-ubiquitinated on lysine-164 at the sites of stalled forks, a reaction mediated by the Rad6–Rad18 ubiquitination system (Hoege et al., 2002; Kannouche et al., 2004; Watanabe et al., 2004). It is widely assumed, but without direct evidence, that ubiquitination of PCNA is required for localization of polη in replication foci. Dantuma et al. (2006) reported that treatment of cells with the general proteasome inhibitor MG132 induced a depletion of the free ubiquitin pool and a concomitant reduction of mono-ubiquitinated target proteins such as ubiquitinated histones. We observed similar effects on UV-irradiation–induced PCNA mono-ubiquitination when cells were treated with either MG132 (data not shown) or with another proteasome inhibitor epoxomicin (Figure 2A). Remarkably, polη accumulated in foci to a similar extent in UV-irradiated MRC5 cells treated with or without epoxomicin (Figure 2B), indicating that ubiquitination of PCNA is not essential for polη foci formation.
Figure 2.
Ubiquitination of PCNA and polη mobility. (A) MRC5 cells were UV-irradiated (15 Jm−2) and incubated for 6 h with epoxomicin. PCNA was analyzed by Western blotting. (B) MRC5 cells transfected with eGFP-polη and mRFP-PCNA were UV irradiated and incubated either in the presence or absence of epoxomicin. Six hours later, the cells were fixed and analyzed by autofluorescence. (C) MRC5 cells were transfected with empty vector, wild-type Rad18, or C28F mutant together with His-tagged PCNA, treated with or without UV, and then incubated for 6 h before analysis by Western blotting. (D) XP30RO-eGFP-polη cells were cotransfected with either wild-type or C28F mutant Rad18 together with mRFP-tubulin to identify the transfected cells. The following day, they were unirradiated or UV irradiated, and the mobility of eGFP-polη was measured using FRAP. (E) Western blot showing increased ubiquitination of PCNA in XP30RO-eGFP-polη cells in which USP1 was depleted by siRNA (lanes 3 and 4). Nontargeting control (siNTC, lanes 1 and 2). (F) Effect of siUSP1 on mobility of eGFP-polη in foci in HU-treated cells.
These findings do not however rule out the possibility that ubiquitination of PCNA affects the dynamics of polη in foci. Because inhibition of the proteasome is likely to have many pleiotropic effects, it would be difficult to interpret dynamic experiments making use of this inhibitor. An alternative way of preventing PCNA ubiquitination is by depletion of Rad18, by using siRNA (Kannouche et al., 2004). However, Rad18 interacts physically with polη and is required for the accumulation of polη in foci, independently from its role in PCNA ubiquitination (Watanabe et al., 2004); so, this approach also could not be used. Instead, we looked at the effect of overexpressing Rad18 in our eGFP-polη–expressing cells and measured the mobility of polη, both uniformly distributed and in foci. Overexpression of Rad18 has been reported to cause increased PCNA ubiquitination (Huang et al., 2006; Davies et al., 2008). To test whether this was also the case in our experimental system, we contransfected His-PCNA and Rad18. The use of His-PCNA was needed because the low transfection efficiency of our cell lines made it impossible to detect any changes in endogenous PCNA. In the overexpressing cells, there was an increase in the level of ubiquitination of His-tagged PCNA, especially after UV-irradiation (Figure 2C, compare lanes 4 and 2).
Overexpression of Rad18 (together with mRFP-α-tubulin, used as transfection marker) had no effect on the mobility of uniformly distributed polη (Figure 2D). In contrast, there was a decrease in the mobility of polη in foci (Figure 2D). To determine whether this effect of Rad18 was mediated by ubiquitination of PCNA or by binding to polη, we mutated the RING finger motif of Rad18 that is required for its ubiquitin ligase activity and the ubiquitination of PCNA but is not involved in direct interaction of Rad18 with polη (Watanabe et al., 2004). Using the Rad18-C28F mutation (Tateishi et al., 2000), in which the E3 ubiquitin ligase activity is inactivated, levels of ubiquitinated PCNA were the same as in mock-transfected cells (Figure 2C, lane 6), and the reduction in mobility of focal polη was abolished (Figure 2D).
USP1 is a deubiquitinating enzyme (DUB), which removes the ubiquitin from ubiquitinated PCNA (Huang et al., 2006). Depletion of USP1 by using siRNA results in increased levels of ubiquitinated PCNA in undamaged cells (Huang et al., 2006; Figure 2E, bottom, lane 3). In these USP1-depleted cells, the mobility of uniformly distributed GFP-polη was slightly reduced; in foci in HU-treated cells, it was reduced to a similar level to that in the cells overexpressing Rad18 (Figure 2F). (Note that we could not use UV in these experiments as UV-irradiation results in disappearance of USP1 from the cell. This is not seen after HU treatment; Huang et al., 2006 and our unpublished data.) Together, these results suggest that although ubiquitination of PCNA is not required for accumulation of polη into replication factories, it results in an increased residence time of polη in the factories.
Mobility of polι
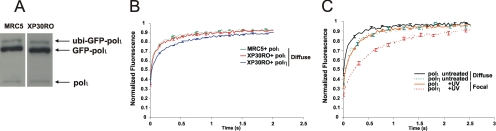
Polι is a paralogue of polη (Tissier et al., 2000), but its precise function remains to be established. In previous work, we showed that polη could physically interact with polι, although we could not demonstrate such an interaction in human cell lysates (Kannouche et al., 2003). Polι accumulates in replication foci in an identical manner to polη, and this accumulation is substantially dependent on the presence of polη, because it was greatly reduced in XP-V cells (Kannouche et al., 2003). To investigate the intracellular relationship between polη and ι further, we established stable MRC5 and XP-V XP30RO cell lines expressing eGFP-polι. The levels of polι expression are shown in Figure 3A and are approximately 4 times the endogenous level (see Supplemental Figure S3 for calculation). This is the minimum level of expression that enables us to visualize the eGFP-polι foci. However, by comparing cells expressing different levels of eGFP-polι, we ascertained that the mobility of polι was independent of its expression level. We compared the mobility of polι with that of polη. We found that polι was even more mobile than polη, with a t0.5 of only 90 ms when uniformly distributed, this mobility being similar in MRC5 and XP30RO cells and therefore independent of the presence of polη (Figure 3B). As with polη, the mobility of polι was somewhat decreased in replication foci (t0.5 of 200 ms), but it remained more mobile than polη (Figure 3C). These data do not support the idea that the two polymerases exist in the same complex within the cell (although they do not rule out the possibility that a small subfraction might be associated).
Figure 3.
Polι is more mobile than polη. (A) Western blot with anti-polι of lysates from stable cell lines expressing eGFP-polι. (Note that the slow mobility band is the previously reported ubiquitinated form of polι; Bienko et al., 2005.) (B) Comparison of mobilities of eGFP-polι in stable cell lines of MRC5 and XP30RO expressing GFP-polι. (C) Comparison of mobilities of polι distributed uniformly in unirradiated cells and in foci in irradiated cells. Data for polη from Figure 1B are also shown (as dotted curves) for comparison.
Because polη and ι have very similar molecular weights, if they exist in the cell as freely diffusible monomers, their redistribution kinetics should be very similar. There are two possible explanations for the different kinetics. The first possibility is that when uniformly distributed, both polymerases are components of protein complexes that are freely diffusible within the cell and the polη complex is larger than the polι complex. Alternatively, the polymerases spend a proportion of their time transiently immobilized. To distinguish between these alternatives, we have applied Monte Carlo simulations to the redistribution kinetics of uniformly distributed polη and ι. The best fits to the data are shown in Supplemental Figure S2 and Table 1, and they are derived from a model in which both polymerases diffuse through the cell but are transiently immobilized. As shown in Table 1, the diffusion coefficients of the two polymerases inside the cell are quite similar, but it is the proportion of transiently immobilized polη (48%) that is much greater than that of polι (17.5%) and accounts for the slower redistribution of polη than polι. The immobilization time is ∼150 ms for both.
Table 1.
Mobility parameters of polη and i
| Diffusion coefficient | Immobile fraction | Binding time | |
|---|---|---|---|
| Polη | 7.4 ± 1.6 | 0.48 ± 0.11 | 0.15 ± 0.07 |
| Polι | 8.9 ± 1.4 | 0.175 ± 0.11 | 0.14 ± 0.08 |
These parameters were based on modeling the raw FRAP data as described in Materials and Methods.
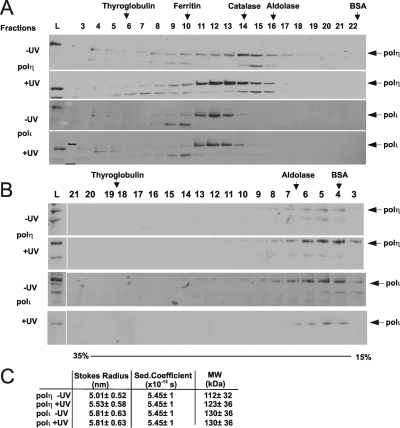
To explore further the relationship between polη and polι inside cells, we have fractionated cell lysates by both gel filtration and glycerol gradient centrifugation and analyzed the fractions for the polymerases by immunoblotting. Gel filtration separates proteins on the basis of their size and shape, whereas glycerol gradient fractionates on the basis of sedimentation coefficient, which is determined by mass, size, and shape (see Materials and Methods). Using gel filtration (Figure 4A), we found that polη and polι were associated with complexes of different Stokes radii, and interestingly the exclusion of polη increased following UV-irradiation. On the glycerol gradients (Figure 4B), both polymerases sedimented at approximately the same rate and this was independent of UV-irradiation. Putting these data together (Figure 4C) suggests that polη and ι are in complexes of 112 and 130 kDa, respectively, somewhat greater than the molecular weights of the polymerases themselves (78 kDa).
Figure 4.
Fractionation of polη and polι from cell lysates. (A) Lysates from unirradiated or UV-irradiated MRC5 cells were fractionated on a Superdex 200 gel filtration column, and fractions were analyzed by immunoblotting for polη and polι. L, load. (B) Equivalent lysates were centrifuged on glycerol gradients. (C) Molecular weight calculations from data obtained in A and B.
Combining the biochemical with the cell biological data, we conclude that the majority of polη and ι molecules diffuse independently in the cell, possibly complexed with other proteins, but the major difference in their mobilities results from the larger fraction of transiently immobilized polη than polι.
Effect of Chromatin Structure on Mobility of Polymerases
To gain further insight into factors affecting the intracellular mobilities of the polymerases, we looked for ways of disrupting chromatin structure to expose the DNA. We made use of the intercalating agent DRAQ5, which binds to DNA with selectivity for A-T base pairs (Njoh et al., 2006). DRAQ5 has recently been shown to disrupt chromatin structure (Wojcik and Dobrucki, 2008), and we have shown that the immobile fraction of transcription factor TFIIH becomes mobilized on treatment of cells with DRAQ5 (Giglia-Mari and Vermeulen, unpublished data). We measured the effect of DRAQ5 on the mobility of the core histone H2B. Histones are normally completely immobile in chromatin, but remarkably, 20% of H2B became mobile within minutes of DRAQ5 treatment (Figure 5A). This result is consistent with findings of Wojcik and Dobrucki (2008). After 1 h in DRAQ5, the original immobility was restored (data not shown). These data suggest that DRAQ5 causes a temporary opening up of the chromatin structure. We next exposed cells to DRAQ5 and measured the effects on the mobilities of polη and ι. Strikingly, we found that treatment of cells in which polη is uniformly distributed resulted in a long-lasting immobilization of 25% of the total polη population within 3 min (Figure 5A). In contrast, the effect on the mobility of polι was much smaller (Figure 5A), with just a slightly reduced mobility and <5% increase in the long-lasting immobile fraction. The effect of DRAQ5 on polη was temporary, and normal mobility was restored within 1 h (data not shown), consistent with the reimmobilization of H2B. We interpret these data as follows: DRAQ5 loosens chromatin structure resulting in release of histones and exposure of the DNA to nucleoplasmic proteins. Polη is then able to bind to DNA and becomes immobilized for a long time (in contrast to the very transient immobilization seen under normal conditions). We can exclude the possibility that DRAQ5 generates a DNA damage response that somehow accounts for the observed changes in mobility, because DRAQ5 treatment does not result in either ubiquitination of PCNA or activation of a DNA damage checkpoint (Verbiest, Mari, Gourdin, Sabbioneda, Wijgers, Dinant, Lehmann, Vermeulen, and Giglia-Mari, unpublished data).
Figure 5.
Effects of DRAQ5 on the mobilities of polη and ι. (A) Effect of DRAQ5 on the mobility of eGFP-histone H2B, eGFP-pol η, and eGFP-polι. Cells were treated with or without DRAQ5 for 3 min and then subjected to FRAP analysis. (B) MRC5 cells transfected with either eGFP-polη or eGFP-polι were UV irradiated, incubated for 6 h, and either fixed immediately or extracted with Triton X-100 before analysis by epifluorescence.
Polι has a lower affinity for DNA than polη and remains mobile. Consistent with the idea that polι is more loosely associated with nuclear structures than polη, we confirmed our earlier findings (Kannouche and Lehmann, 2004) that polη localized in foci was resistant to extraction with triton, whereas polι was quantitatively extracted under identical conditions (Figure 5B).
DISCUSSION
Our data show that 1) Polη is highly mobile in nuclei of human fibroblasts; 2) even when localized in replication factories, it remains very mobile, albeit somewhat less so than when uniformly distributed in the nuclei, and this mobility in foci is similar during a normal S phase or in cells treated with UV light or hydroxyurea; 3) although ubiquitination of PCNA is not required for the localization of polη in replication foci, it results in an increased residence time in foci; 4) polι is even more mobile than polη, both when uniformly distributed and when localized in factories; and 5) treatment of cells with DRAQ5, which seems to result in the transient opening of the chromatin structure, causes a dramatic immobilization of polη but not polι.
The high mobility of polη in human cells, both uniformly distributed and in foci, agrees with the observations of Solovjeva et al. (2005) using Chinese hamster cells, and emphasizes that even though visible in fluorescent replication structures, proteins may still interact there very transiently. Our biochemical data suggest that polη may be associated with another protein in a complex of total molecular mass of 112 kDa. Rad18 has been shown to interact with polη both in cell lysates and as recombinant proteins (Watanabe et al., 2004; Yuasa et al., 2006). However in cells depleted of Rad18 the mobility of diffusely localized polη is hardly affected (data not shown), ruling out the possibility that binding to Rad18 is responsible for the reduced mobility of polη inside cells.
Our modeling shows that the principal factor responsible for the reduced mobility of polη relative to polι is the greater proportion of transiently immobilized polη molecules. We hypothesize that this immobilization represents polη transiently probing either the DNA itself or proteins associated with the DNA. Our data are consistent with a model in which polη has a weak affinity for DNA (Kusumoto et al., 2004) and is continually probing the chromatin. Outside S phase, the DNA is almost inaccessible inside chromatin, so polη is only retarded very briefly. During S phase, DNA is exposed at the replication forks, polη probes the exposed DNA for suitable substrates and its residence in the foci is increased by binding to the exposed DNA and by interaction with PCNA, especially when PCNA is ubiquitinated. However even under these circumstances, binding is weak and the polymerase remains at the fork for <1 s. Only when the fork is blocked is a substrate available for polη to engage and carry out TLS. This is likely to render the engaged polη molecule immobile for a relatively long period (compared with the transient immobilization discussed above). We have calculated that there are ∼80,000 molecules of polη in MRC5 cells (and a similar number of polι molecules) (Supplemental Figure S3), and it is likely that only a small fraction of these are engaged in TLS at any one time. This explains why we detect only a small long-lasting immobile fraction, even in UV-irradiated cells. In the pol dead mutant, we interpret the increased long-lasting immobile fraction as indicating that on engagement, the polymerase becomes temporarily trapped with substrate in its active site.
Relationship between polη and polι
In a previous study, we showed that polη and ι colocalize in replication foci and that the localization of polι in foci is dependent on polη. The two polymerases are able to interact physically, as demonstrated by Far Western blotting, yeast two-hybrid analysis, and coimmunoprecipitation in insect cells (Kannouche et al., 2003). However, three observations suggest that polι binds less strongly to chromatin than polη inside cells. First, our modeling data suggest that less polι is transiently immobile (Table 1). Second, polι is less tightly bound in replication foci than polη (Figure 5B). And third, polη is temporarily immobilized after treatment with DRAQ5, whereas polι is not (Figure 5A). Taking our previous and present observations together, we conclude that interactions between polη and ι must be transient or unstable, that polη helps polι to accumulate in foci, but that polι dissociates from foci more rapidly than polη. Our finding of polη and ι in different complexes on gel filtration also suggests that interactions between them are likely to be transient.
Ubiquitination of PCNA and Localization of polη in Foci
Our finding that PCNA ubiquitination is not required for polη to localize in foci is at first sight surprising, because focal localization is dependent on the UBZ ubiquitin-binding motif of polη (Bienko et al., 2005). However, polη localization in undamaged S phase cells is also dependent on the UBZ motif, even though there seems to be minimal ubiquitination of PCNA under these conditions. We conclude that ubiquitinated PCNA is not the only ubiquitinated target that drives polη into foci. However, once localized in foci, our data are consistent with the idea that ubiquitinated PCNA increases the residence time of polη, presumably by binding to polη via its UBZ motif at sites of stalled replication forks.
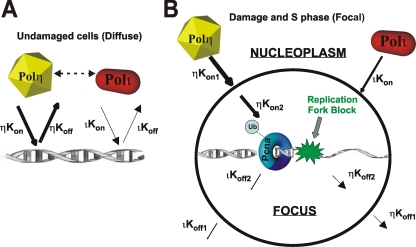
A schematic diagram to account for our data is indicated in Figure 6. Outside of S phase, the polymerases are probing the chromatin with Kon-/Koff for polη greater than that for polι. In S phase cells exposed to HU or DNA damage, there are two steps, namely, accumulation into foci and binding at the fork. For the first step, accumulation of polη in foci (Kon1) is independent of PCNA ubiqitination. The second step is facilitated by PCNA ubiquitination, which stabilizes the presence of polη and polι at the stalled replication fork. This results in an increase in the overall Kon-/Koff for both polymerases with consequent decreased mobility.
Figure 6.
Model for dynamics of polη and ι. (A) In undamaged cells, polη and polι probe the chromatin, but the residence time of polη is greater than that of polι, implying either a higher Kon or lower Koff rate. The double-headed arrow signifies weak interaction between the two polymerases. (B) In damaged S phase cells, where there is a replication fork blocked by damage and resulting ubiquitination of PCNA, there are two dynamic processes, transport into the focus and association with the blocked fork.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Roger Woodgate, Tony Huang, Cristina Cardoso, and Sally Wheatley for reagents, and to Roger Phillips for assistance with the microscopy. This work was supported by grants from ESF Eurodyna program, the UK Medical Research Council, the Dutch Science organization (NWO) for medical Sciences (ZonMW) VIDI grants, NWO Molecule to Cell program, and an European Union research training network and integrated project on DNA Repair.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-07-0724) on September 17, 2008.
REFERENCES
- Bienko M., Green C. M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A. R., Hofmann K., Dikic I. Ubiquitin-binding domains in translesion synthesis polymerases. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- Dantuma N. P., Groothuis T. A., Salomons F. A., Neefjes J. A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J. Cell Biol. 2006;173:19–26. doi: 10.1083/jcb.200510071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A. A., Huttner D., Daigaku Y., Chen S., Ulrich H. D. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol. Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J., Theil A. F., Baldeyron C., van Cappellen W. A., Houtsmuller A. B., Kanaar R., Vermeulen W. Nuclear dynamics of PCNA in DNA replication and repair. Mol. Cell. Biol. 2005;25:9350–9359. doi: 10.1128/MCB.25.21.9350-9359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C., Lehmann A. R., Fuchs R. P. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Haracska L., Johnson R. E., Unk I., Phillips B., Hurwitz J., Prakash L., Prakash S. Physical and functional interactions of human DNA polymerase η with PCNA. Mol. Cell. Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G.-L., Pyrolowakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Houtsmuller A. B., Vermeulen W. Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching. Histochem. Cell Biol. 2001;115:13–21. doi: 10.1007/s004180000234. [DOI] [PubMed] [Google Scholar]
- Huang T. T., Nijman S. M., Mirchandani K. D., Galardy P. J., Cohn M. A., Haas W., Gygi S. P., Ploegh H. L., Bernards R., D'Andrea A. D. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 2006;8:341–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- Johnson R. E., Kondratick C. M., Prakash S., Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999a;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- Johnson R. E., Prakash S., Prakash L. Requirement of DNA polymerase activity of yeast Rad30 protein for its biological function. J. Biol. Chem. 1999b;274:15975–15977. doi: 10.1074/jbc.274.23.15975. [DOI] [PubMed] [Google Scholar]
- Kannouche P., Broughton B. C., Volker M., Hanaoka F., Mullenders L.H.F., Lehmann A. R. Domain structure, localization and function of DNA polymerase η, defective in xeroderma pigmentosum variant cells. Genes Dev. 2001;15:158–172. doi: 10.1101/gad.187501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche P., Fernandez de Henestrosa A. R., Coull B., Vidal A. E., Gray C., Zicha D., Woodgate R., Lehmann A. R. Localization of DNA polymerases η and ι to the replication machinery is tightly co-ordinated in human cells. EMBO J. 2003;22:1223–1233. doi: 10.1093/emboj/cdf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche P., Lehmann A. Localization of Y-family polymerases and the DNA polymerase switch in mammalian cells. Methods Enzymol. 2006;408:407–415. doi: 10.1016/S0076-6879(06)08025-6. [DOI] [PubMed] [Google Scholar]
- Kannouche P. L., Lehmann A. R. Ubiquitination of PCNA and the polymerase switch in human cells. Cell Cycle. 2004;3:1011–1013. [PubMed] [Google Scholar]
- Kannouche P. L., Wing J., Lehmann A. R. Interaction of human DNA polymerase η with monoubiquitinated PCNA; A possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- Kusumoto R., Masutani C., Shimmyo S., Iwai S., Hanaoka F. DNA binding properties of human DNA polymerase eta: implications for fidelity and polymerase switching of translesion synthesis. Genes Cells. 2004;9:1139–1150. doi: 10.1111/j.1365-2443.2004.00797.x. [DOI] [PubMed] [Google Scholar]
- Li Y., Korolev S., Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher V. M., Ouellette L. M., Curren R. D., McCormick J. J. Frequency of ultraviolet light-induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature. 1976;261:593–595. doi: 10.1038/261593a0. [DOI] [PubMed] [Google Scholar]
- Masutani C., Araki M., Yamada A., Kusumoto R., Nogimori T., Maekawa T., Iwai S., Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C., Kusumoto R., Iwai S., Hanaoka F. Accurate translesion synthesis by human DNA polymerase η. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch S. D., Kokoska R. J., Masutani C., Iwai S., Hanaoka F., Kunkel T. A. Preferential cis-syn thymine dimer bypass by DNA polymerase η occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- Njoh K. L., et al. Spectral analysis of the DNA targeting bisalkylaminoanthraquinone DRAQ5 in intact living cells. Cytometry A. 2006;69:805–814. doi: 10.1002/cyto.a.20308. [DOI] [PubMed] [Google Scholar]
- Solovjeva L., Svetlova M., Sasina L., Tanaka K., Saijo M., Nazarov I., Bradbury M., Tomilin N. High mobility of flap endonuclease 1 and DNA polymerase eta associated with replication foci in mammalian S-phase nucleus. Mol. Biol. Cell. 2005;16:2518–2528. doi: 10.1091/mbc.E04-12-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporbert A., Gahl A., Ankerhold R., Leonhardt H., Cardoso M. C. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell. 2002;10:1355–1365. doi: 10.1016/s1097-2765(02)00729-3. [DOI] [PubMed] [Google Scholar]
- Tateishi S., Sakuraba Y., Masuyama S., Inoue H., Yamaizumi M. Dysfunction of human Rad18 results in defective postreplication repair and hypersensitivity to multiple mutagens. Proc. Natl. Acad. Sci. USA. 2000;97:7927–7932. doi: 10.1073/pnas.97.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A., McDonald J. P., Frank E. G., Woodgate R. Polι, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- Vidal A. E., Kannouche P. P., Podust V. N., Yang W., Lehmann A. R., Woodgate R. PCNA-dependent coordination of the biological functions of human DNA polymerase ι. J. Biol. Chem. 2004;279:48360–48368. doi: 10.1074/jbc.M406511200. [DOI] [PubMed] [Google Scholar]
- Volker M., Mone M. J., Karmakar P., van Hoffen A., Schul W., Vermeulen W., Hoeijmakers J. H., van Driel R., van Zeeland A. A., Mullenders L. H. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;28:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik K., Dobrucki J. W. Interaction of a DNA intercalator DRAQ5, and a minor groove binder SYTO17, with chromatin in live cells–influence on chromatin organization and histone-DNA interactions. Cytometry A. 2008;73:555–562. doi: 10.1002/cyto.a.20573. [DOI] [PubMed] [Google Scholar]
- Yang W., Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. USA. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa M. S., Masutani C., Hirano A., Cohn M. A., Yamaizumi M., Nakatani Y., Hanaoka F. A human DNA polymerase eta complex containing Rad18, Rad6 and Rev1; proteomic analysis and targeting of the complex to the chromatin-bound fraction of cells undergoing replication fork arrest. Genes Cells. 2006;11:731–744. doi: 10.1111/j.1365-2443.2006.00974.x. [DOI] [PubMed] [Google Scholar]
- Zhuang Z., Johnson R. E., Haracska L., Prakash L., Prakash S., Benkovic S. J. Regulation of polymerase exchange between Poleta and Poldelta by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proc. Natl. Acad. Sci. USA. 2008;105:5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.