Abstract
The metamorphic Mad2 protein acts as a molecular switch in the checkpoint mechanism that monitors proper chromosome attachment to spindle microtubules during cell division. The remarkably slow spontaneous rate of Mad2 switching between its checkpoint inactive and active forms is catalyzed onto a physiologically relevant time scale by a self–self interaction between its two forms, culminating in a large pool of active Mad2. Recent structural, biochemical, and cell biological advances suggest that the catalyzed conversion of Mad2 requires a major structural rearrangement that transits through a partially unfolded intermediate.
Introduction
The essential goal of mitosis is the equal distribution of sister chromatids into genetically identical daughter cells (Cleveland et al., 2003; Walczak and Heald, 2008). Chromosome segregation is directed by the centromere, a locus epigenetically defined by a specialized chromatin domain marked by nucleosomes in which the histone variant CENP-A (centromere protein A) replaces H3 (Black and Bassett, 2008). The kinetochore, an enormous protein assembly consisting of >80 known proteins, assembles upon the centromere of each chromatid and connects to microtubule-based fibers that extend from opposite poles of the mitotic spindle. Accurate kinetochore attachment to the spindle is monitored by a diffusible checkpoint signal termed the mitotic checkpoint (also referred to as the spindle assembly checkpoint; Musacchio and Salmon, 2007; Yu, 2007). The checkpoint inhibits mitosis, halting progression to anaphase until all chromosomes are aligned on the metaphase plate and every kinetochore is properly attached to the spindle (Fig. 1 A).
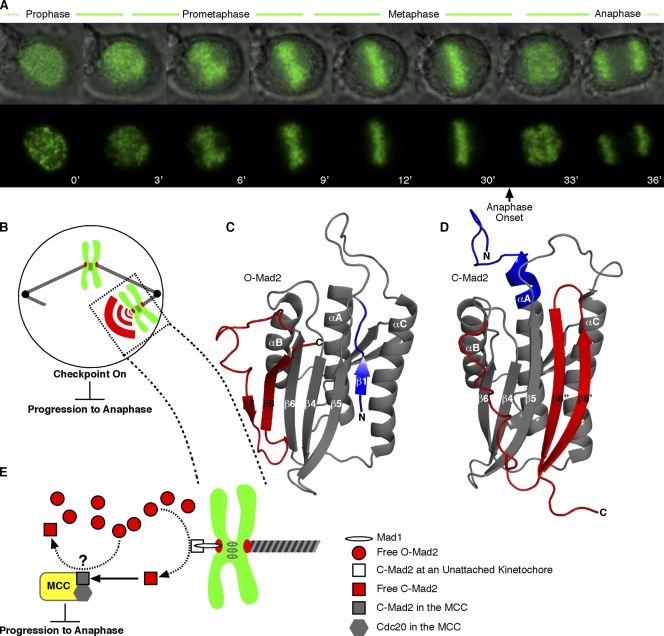
Figure 1.
The mitotic checkpoint ensures equal partitioning of chromosomes in anaphase. (A) A human tissue culture cell progressing through mitosis with time indicated in minutes. In the top row, chromosomes (green) are overlaid with a differential interference contrast image of the entire cell. Sister chromatids align at the metaphase plate early in mitosis and wait for ∼20 min before chromatid separation in anaphase. Upon final chromosome alignment, the mitotic checkpoint signal decays, allowing the cell to enter anaphase and initiate simultaneous separation of sister chromatids. (B) The mitotic checkpoint signal, comprised in part by a diffusible pool of C-Mad2, emanates from kinetochores that have not yet properly engaged the microtubule-based spindle. A single unattached chromosome is sufficient to generate a checkpoint signal that arrests mitosis before anaphase. (C and D) Interconversion between inactive O-Mad2 (PDB 1DUJ; Luo et al., 2000) and checkpoint-active C-Mad2 (PDB 1S2H; Luo et al., 2004) involves a major secondary and tertiary structural reorganization of N-terminal (blue) and C-terminal (red) segments. (E) Unattached kinetochores contain the checkpoint protein Mad1, which recruits C-Mad2, providing a catalytic surface for the conversion of the soluble pool of inactive O-Mad2 to active C-Mad2. C-Mad2 is able to bind and inhibit Cdc20 within the MCC, halting progression to anaphase. The Cdc20–C-Mad2 complex may also act to catalyze conversion of the O-Mad2 pool, although this aspect of Mad2 signaling remains controversial (Yu, 2006; Musacchio and Salmon, 2007). (B and E) Chromosomes are drawn in green with their kinetochores drawn in red.
On and off switching of the mitotic checkpoint must be fast and definitive because either a weak checkpoint or an asynchronous metaphase to anaphase transition leads to irreversible missegregation of one or more chromosomes. The checkpoint must be active upon entry into mitosis and sufficiently robust so that checkpoint activation is maintained if even a single kinetochore remains unattached to the spindle (Fig. 1 B). After proper spindle attachment to all kinetochores, the checkpoint rapidly inactivates to allow for the destruction of mitotic targets (e.g., cyclin B and securin), which leads to synchronous chromosome separation and segregation. Inappropriate early inactivation of the checkpoint produces lethal chromosomal missegregation (Kops et al., 2004; Michel et al., 2004). However, a functional mitotic checkpoint is required for tumor cell death resulting from treatment with microtubule toxins such as taxol that are widely used in the clinic (Gascoigne and Taylor, 2008).
The Mad2 protein is a centrally important regulator of the mitotic checkpoint machinery. Its activity is controlled by switching between its two different native conformations, open Mad2 (O-Mad2; also referred to as Mad2N1; Fig. 1 C) and closed Mad2 (C-Mad2; also referrred to as Mad2N2; Fig. 1 D; Luo et al., 2004; De Antoni et al., 2005). Before checkpoint activation, freely diffusible monomeric Mad2 is thought to exist largely as O-Mad2, its inactive conformation, as is common for many regulatory proteins. Conformational conversion from inactive free O-Mad2 to active free C-Mad2 is catalyzed by a self–self interaction, namely by binding to the C-Mad2 subunit of a Mad1–C-Mad2 complex (Luo et al., 2000; Sironi et al., 2002; Vink et al., 2006) anchored at kinetochores that are not yet properly engaged with a spindle (Fig. 1 E; Chen et al., 1998, 1999; Waters et al., 1998). Although a direct physical demonstration that Mad2 structural conversion is catalyzed by unattached kinetochores is currently lacking, purified Mad1-bound Mad2 is known to catalyze the O-Mad2→C-Mad2 transition in the absence of any other effector molecules (Yang et al., 2008). Newly converted Mad2 releases from the kinetochore and blocks premature progression to anaphase by binding to and deactivating Cdc20 in conjunction with other essential checkpoint proteins (including BubR1 kinase and Bub3) as part of a mitotic checkpoint complex (MCC; Fig. 1 E; Hardwick et al., 2000; Sudakin et al., 2001). Although the checkpoint remains active, the inhibition of Cdc20 by C-Mad2 serves to restrain an E3 ubiquitin ligase known as the anaphase-promoting complex/cyclosome (APC; Fig. 1 E; Musacchio and Salmon, 2007; Yu, 2007). Once all kinetochores have properly attached to the spindle, Mad2 deactivates and releases Cdc20, allowing it to bind and activate the APC. APC–Cdc20 ubiquitinates several key mitotic substrates, including securin and cyclin B, leading to their removal by the proteasome and initiation of the metaphase to anaphase transition.
Mad2 differs strikingly from most regulatory proteins. Other proteins that change structure drastically, known as metamorphic proteins (Murzin, 2008), require the selective stabilization of their intrinsically less stable active form through substrate binding, chemical modification, or environmental change. For Mad2, the structural changes from the inactive form to the active form are unusually large (Fig. 1, C and D) and remarkably slow (Luo et al., 2004). Furthermore, Mad2 is found initially out of equilibrium in its inactive form (O-Mad2) even though its active form (C-Mad2) is the more stable conformation (Luo et al., 2004). Thus, checkpoint activation simply requires Mad2 to reach its equilibrium distribution. These properties raise key questions about the mechanism of mitotic checkpoint regulation. How do effector molecules modulate the rate of Mad2 interconversion? Could Mad2 regulation involve kinetic trapping in one of its two conformational states? Do transient conformational intermediates play a functional role?
The Mad2 structural rearrangement
The spontaneous Mad2 activation reaction, O-Mad2→C-Mad2, proceeds with a lifetime of 9 h; the reverse reaction is sixfold slower (Luo et al., 2004)! These unusually slow interconversion rates stem from the magnitude of the structure change, which involves a complete rearrangement of the secondary and tertiary structure of 60 out of 205 amino acids. In O-Mad2, the N-terminal segment forms a long loop and a short β strand (β1) that connects to the static core (Fig. 1 C). In the transition to C-Mad2, this segment loses its β conformation and reconfigures, adding two more turns to the αA helix (Fig. 1 D; Luo et al., 2002, 2004; Sironi et al., 2002). The C terminus undergoes an even more dramatic change. In O-Mad2, the C-terminal segment forms strands β7 and β8 (and connecting loops) that dock onto the static core β6 strand. In C-Mad2, the whole segment moves to the opposite side of the major β sheet and forms two new strands, β8′ and β8″, with a completely different hydrogen-bonding network. Overall, the transition to the C-Mad2 conformer relocates the N-terminal segment to make room for the incoming C-terminal segment, the displacement of which exposes an extended active site that is occluded in O-Mad2.
The active site of Mad2 is tailored, remarkably, to interact with both its upstream activator Mad1 (Fig. 2 A) and its downstream target Cdc20 (Luo et al., 2002; Sironi et al., 2002). Although Mad1 and Cdc20 appear to be otherwise unrelated, their Mad2-interacting regions are highly homologous and can be mimicked by a synthetic 12-residue consensus sequence peptide (Mad2-binding peptide 1 [MBP1]; Fig. 2 B; Luo et al., 2002). These partners bind by incorporating into the major Mad2 β sheet as a single β strand, interacting with the β6 strand and a new β7′ strand that forms upon the binding (Luo et al., 2000, 2002; Sironi et al., 2002). As shown in Fig. 2 (A and B), they actually thread through the C-Mad2 sheet like links in a concatenated chain. Once Mad1 binds to Mad2, it forms a very stable complex with no detectable turnover in 4 min, as detected with purified components by FRAP (Vink et al., 2006), correlating with earlier cell-based FRAP measurements of the hyperstable pool of kinetochore-bound C-Mad2 that is presumably bound to Mad1 (Shah et al., 2004).
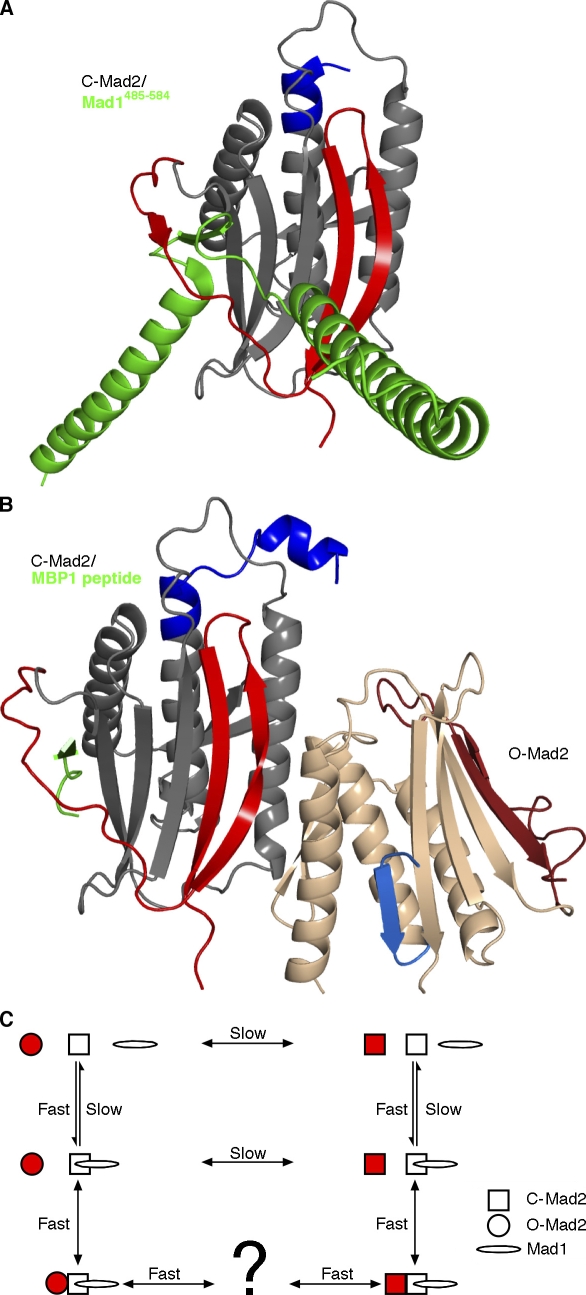
Figure 2.
Mad2-containing complexes. (A and B) The displacement of the C-terminal segments in C-Mad2 exposes a new β sheet edge that can incorporate Mad1 (A; PDB 1GO4; Sironi et al., 2002), Cdc20, or the synthetic peptide MBP1 (B; PDB 2V64; Mapelli et al., 2007) between newly exposed β6 and newly formed β7′. In the crystal structure of the O-Mad2–C-Mad2 dimer (B), asymmetrical dimerization occurs mainly through the unaltered core of Mad2 (gray to tan) but also includes the β8′ strand that is unique to C-Mad2 (coloring as in Fig. 1 C). O-Mad2, the form undergoing conversion (tan), interacts with C-Mad2 only through its unchanging core. (C) A reaction scheme for Mad2 catalysis. Mad2 (red) represents the molecule undergoing conversion. Uncatalyzed Mad2 interconversion proceeds far more slowly (lifetime >9 h; Luo et al., 2004) than the duration of metaphase (∼20 min). The Mad2 structural rearrangement is catalyzed by binding to the Mad1–C-Mad2 complex. In this reaction scheme, catalysis by induced fit would increase the forward O-Mad2→C-Mad2 rate, whereas the conformational selection of C-Mad2 would reduce the reverse O-Mad2←C-Mad2 rate. It is unknown whether Mad2 releases from the Mad1–C-Mad2 dimer as fully folded C-Mad2 or as a partially unfolded intermediate.
For the sake of simplicity, it is often stated that C-Mad2 itself is competent to bind Mad1 or Cdc20. However, in this binding reaction, Mad2 must expose a binding site, load its binding partner, and lock it in place. This implies that binding to either Mad1 or Cdc20 requires a substantial local rearrangement of Mad2 structure (Mapelli et al., 2007; Yang et al., 2008). Although the possibility exists that Mad1 and Cdc20 may themselves unfold, thread through the Mad2-binding loop, and refold, partial unfolding of Mad2 itself seems more likely, especially because the O-Mad2→C-Mad2 conversion appears to require a similar partial unfolding. In this view, a partially unfolded intermediate form of Mad2 would be required for Cdc20 binding and APC inhibition.
Conformational switching models
On time scales relevant to cell biology, the great majority of biomolecules assume their equilibrium distribution among alternative conformations, and their rates of interconversion can be safely ignored. However, the spontaneous O-Mad2→C-Mad2 conversion rate (many hours) is clearly inadequate for the rapid checkpoint activation required to inhibit anaphase immediately upon mitotic entry. The conversion of freely diffusible O-Mad2 is catalyzed by its self-interaction with the C-Mad2 partner of the kinetochore-bound Mad1–C-Mad2 complex (Fig. 2 C). How is this catalytic event accomplished? Thermodynamic principles dictate that molecular binding partners promote structure change in allosteric proteins by binding more strongly to the favored form. Two common structure change models exist. Association may promote the structure change by sacrificing some of its binding energy to forcefully distort the protein conformation (induced fit model), or selection may occur among preexisting dynamically cycling protein conformations by more strongly binding to and thereby trapping the preferred partner (conformational selection model).
If the O-Mad2→C-Mad2 conversion is catalyzed by induced fit, the structure of Mad2 in the catalytic complex should display the activation mechanism. Mapelli et al. (2007) crystallized a valid replica of the Mad1–C-Mad2–O-Mad2 catalytic complex (Fig. 2 B). The O-Mad2 subunit was trapped in the open conformation by shortening the loop connecting the β5 strand to the αC helix. The O-Mad2 loopless mutant (O-Mad2LL) was dimerized with a C-Mad2 molecule that was bound in turn to the synthetic activation peptide MBP1 to make a stable MBP1–C-Mad2–O-Mad2LL complex. The crystal structure of the complex reveals that the dimerization surface of O-Mad2, the form undergoing conformational change, only involves segments that are not substantially altered upon the O-Mad2→C-Mad2 switch. Thus, it does not appear that the interaction would serve to forcefully induce the transition, providing evidence against an induced fit mechanism.
In the case of a conformational selection mechanism, one can expect that the catalyzing kinetochore-bound Mad1–C-Mad2 complex would favor the closed form of the substrate Mad2 molecule by binding to sites that are specific for C-Mad2. In fact, the C-Mad2–C-Mad2 complex does involve some of those sites (Yang et al., 2008). However, conformational selection alone does not increase the rate of conversion to the target structure; rather, it stabilizes the selected form by decreasing the reverse rate. Therefore, conformational selection of C-Mad2 can be effective only if conformer sampling (O-Mad2→C-Mad2) is appropriately rapid. If rapid conformational sampling occurred naturally, catalysis would not be necessary because the target C-Mad2 is actually the more stable form (Luo et al., 2004), ruling out conformational selection of C-Mad2 (Fig. 2 C). In summary, recent structures elegantly display the static Mad2 dimerization interface, but they do not suggest a mechanism to explain how the C-Mad2–O-Mad2 interaction catalyzes the O-Mad2→C-Mad2 transition.
An unfolding model for Mad2 conformational change
It is hard to envision how any kind of straightforward conformational conversion (e.g., by a hinging or rigid body motion) could accomplish the major structural rearrangement between the two natively folded Mad2 forms. Rather, the conformational rearrangement is so extensive that it seems to require a significant unfolding of Mad2 to some transient high energy intermediate followed by kinetic partitioning between the two alternative forms upon refolding. Similarly, the fact that the main chain of Mad1 and Cdc20 actually threads through the major β sheet of C-Mad2 seems to require some transitional partially unfolded intermediate from which the C terminus could refold around the ligand upon binding.
A precedent for conformational change through partial unfolding can be found in the much smaller cytochrome c alkaline transition. At an elevated pH, the residue ligated to heme is switched from Met80 to the neighboring Lys79. Rather than simply shifting over by one amino acid residue, the transition involves the unfolding and refolding of a 15-residue loop that contains the two critical residues. The loop has been shown to unfold and refold repeatedly under native conditions as a cooperative unit known as a foldon. The stability of the loop foldon determines the equilibrium between the Met80-liganded and Lys79-liganded forms (Maity et al., 2006), and the foldon unfolding rate limits the kinetics of the transition (Hoang et al., 2003). More generally, recent work indicates that many proteins act as accretions of foldon units that repeatedly unfold and refold under native conditions. It now appears that cooperative foldons can account for the unit steps in protein folding pathways, and, having reached the native state, their continuing dynamic unfolding and refolding behavior can be exploited to control ligand on and off rates (Englander et al., 2007) and even allosteric communication (Hilser and Thompson, 2007).
Can the emerging foldon paradigm help to explain the Mad2 conformational switching mechanism? In addition to the aforementioned structural issues (e.g., massive rearrangement and threading), some other Mad2 folding–related observations are suggestive. Chemically denatured Mad2 spontaneously refolds into a nonequilibrium mixture of its two alternative conformations (C-Mad2 and O-Mad2 in a 2:1 ratio; Luo et al., 2004). The implication is that the refolding pathway contains some intermediate stage from which Mad2 partitions into its two different stable forms (Fig. 3, A and B). Spontaneous equilibration from this point is extremely slow. Thus, O-Mad2 is not itself a facile on-pathway precursor for generation of C-Mad2. Rather, O-Mad2 appears to transit to C-Mad2 by backtracking through a partially unfolded intermediate Mad2 (I-Mad2; Fig. 3 A) and redistributing between O-Mad2 and C-Mad2 over several equilibration cycles.
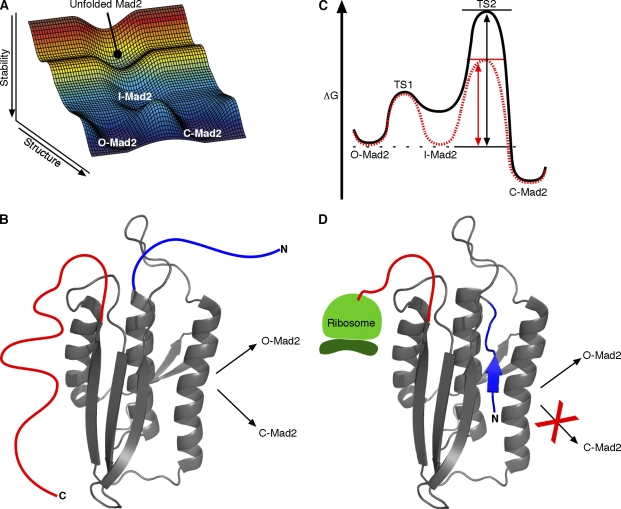
Figure 3.
Mad2 unfolding and refolding considerations. (A) Free energy reaction landscape for Mad2 interconversion through a partially folded intermediate that lies on the folding pathway (created with Matlab version R2007a; The MathWorks, Inc.). When chemically denatured Mad2 is refolded, it initially reaches a nonequilibrium O-Mad2–C-Mad2 mixture, suggesting that the folding pathway to reach either form passes through a common intermediate and kinetically partitions rather than passing through one form on the way to the other. We suggest that the catalyzed interconversion seems likely, on this and other grounds, to pass back through the same partially unfolded intermediate. (B) A notional structure for a Mad2-folding intermediate showing the common (gray) and variable (colored as in Fig. 1 C) segments. (C) Catalysis through intermediate stabilization. The conversion reaction of Mad2 is drawn with (red dashed line) or without (black solid line) dimerization with the kinetochore-bound C-Mad2–Mad1 complex. The measured C-Mad2/O-Mad2 equilibrium ratio is 8:1 (Luo et al., 2004), indicating that C-Mad2 is ∼1 kcal/mol more stable than O-Mad2. Conformational selection of I-Mad2 would equally stabilize I-Mad2 and the TS2 transition barrier relative to O-Mad2, effectively increasing the O-Mad2→C-Mad2 rate even though the energy difference between I-Mad2 and TS2 remains unchanged. The dashed black line indicates the energy state of O-Mad2, the black arrow (left) indicates increasing energy, the double-headed black arrow indicates the energy difference between O-Mad2 and TS2 without dimerization, and the doubled-headed red arrow indicates the energy difference between O-Mad2 and TS2 with dimerization. (D) As it emerges from the ribosome, Mad2 may preferentially fold to O-Mad2 because the last emerging C-terminal segment is required for forming C-Mad2, and protein folding is typically much faster than translation.
How can an unfolding-dependent binding model promote the rate of the Mad2 conformational transition? As noted before, selective binding to C-Mad2 itself would not be helpful. Instead, the Mad1–C-Mad2 complex needs only to selectively stabilize a partially unfolded intermediate on the O-Mad2 side of the rate-limiting transition barrier, such as the hypothetical I-Mad2 in Fig. 3 C. The stabilization of I-Mad2 would equally stabilize the rate-limiting transition state relative to O-Mad2 and therefore increase the rate of O-Mad2→C-Mad2 (it should be noted that I-Mad2 may be but is not necessarily the same as the intermediate that binds Mad1 and Cdc20 discussed in The Mad2 structural rearrangement section).
Unfortunately, a Mad1–C-Mad2–I-Mad2 structure is not likely to be solved by x-ray crystallography because partially unfolded and dynamically interconverting structures are not conducive to crystal formation. Available crystal structures of pertinent dimers used Mad2 variants that would prevent I-Mad2 formation. The Mad2LL mutant used to obtain MBP1–C-Mad2–O-Mad2LL crystals prevents O-Mad2 from switching into the C-Mad2 conformation by restricting the conformational search space of the N terminus (Mapelli et al., 2007). The L13A mutation used to obtain C-Mad2–C-Mad2 crystals stabilizes the native closed conformation so that the alternative O-Mad2 or I-Mad2 forms would not significantly populate (Yang et al., 2008). The structure of I-Mad2 will have to be studied by methods more applicable to dynamic systems. Foldon-dependent unfolding behavior in other proteins has so far been studied successfully, not by static crystallography but by dynamic hydrogen exchange (Englander et al., 2007) and nuclear magnetic resonance relaxation dispersion (Korzhnev and Kay, 2008) methods.
The focus on folding models and kinetic trapping may also explain another puzzling observation. The in vitro equilibrium ratio of C-Mad2/O-Mad2 is 8:1 (Luo et al., 2004). Nevertheless, upon mitotic entry, a catalyzed conversion to the equilibrium C-Mad2 form is necessary. Apparently, nascent Mad2 polypeptide emerging from the ribosome folds preferentially to O-Mad2 (Fig. 3 D). This conclusion is supported by the observation that recombinant Mad2 expressed in Escherichia coli exists predominantly as O-Mad2 (when kept at low temperature to minimize interconversion) even though Mad2 refolded in solution favors C-Mad2 by 2:1 (Luo et al., 2004). The preferential nonequilibrium folding to O-Mad2 may occur before final ribosomal disengagement because the later emerging C terminus is required to form C-Mad2 but not O-Mad2 (a Mad2 mutant with 10 C-terminal residues truncated cannot form C-Mad2; Luo et al., 2004). Given that the time constant for the O-Mad2→C-Mad2 transition is >9 h, newly expressed Mad2 will be kinetically trapped as O-Mad2.
Protein synthesis alone appears to be sufficient to create a soluble pool of O-Mad2 poised for activation upon mitotic entry and interaction with the kinetochore-bound Mad1–C-Mad2 complex. It remains unknown whether or not nuclear pore-tethered Mad1–C-Mad2 in interphase (Campbell et al., 2001) and/or spindle pole–tethered Mad1–C-Mad2 after anaphase onset (Shah et al., 2004) are capable of catalyzing the O-Mad2→C-Mad2 conversion. In all of the likely models, Mad1–C-Mad2 at the kinetochore serves as an active catalytic platform for Mad2 conversion, providing a mechanism for rapid checkpoint activation at the onset of mitosis (De Antoni et al., 2005; Yu, 2006).
Comparison of autocatalyzed Mad2 conversion with prion propagation
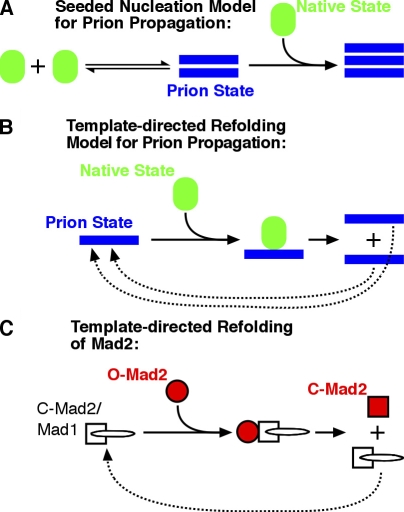
The O-Mad2→C-Mad2 conversion process has been referred to as prionlike (Mapelli et al., 2006) because it shares with prions the general property of structural conversion of one folded state to another via self–self interactions. Prions are proteins that can switch to self-perpetuating infectious conformations. The prions that have been extensively characterized to date, including PrP, Sup35, Ure2, HET-s, and Rnq1 (Shorter and Lindquist, 2005), are propagated as self-templating cross β-amyloid forms in a reaction, wherein the equilibrium between the native and prion states is dramatically shifted by interaction with the stable self-templating amyloid (Fig. 4 A). Obviously, O-Mad2→C-Mad2 conversion has little in common structurally with amyloid. For instance, amyloids do not release newly converted monomers (Carulla et al., 2005). However, before the importance of seeded nucleation by amyloid for prion propagation was realized, the prion field had generated several models for how proteins could self-perpetuate structural change. Curiously, the Mad2 switch is strikingly reminiscent of one of the early models proposed to explain prion propagation, termed template-directed refolding, in which newly converted prion conformers dissociate from the original template (Fig. 4, B and C; Prusiner, 1991; Aguzzi, 2004; Tuite and Koloteva-Levin, 2004). The main difference between Mad2 and this prion model is that the dissociation of equally infectious subunits would generate an explosive chain reaction (Fig. 4 B). In contrast, the rate of O-Mad2→C-Mad2 conversion (Fig. 4 C) is limited by the size of the static pool of kinetochore-bound Mad1–C-Mad2 complex (and perhaps also that of the MCC-bound Cdc20–C-Mad2 complex; De Antoni et al., 2005). That is, free C-Mad2 monomers do not catalyze further conversion of O-Mad2 (Yang et al., 2008). Despite this important distinction, we note that Mad2 is a stunning example of a protein that undergoes template-directed refolding as part of its adaptive cellular function (Fig. 4 C). It will be important to determine what properties of the Mad1-bound C-Mad2 conformation endow it with the ability to catalyze O-Mad2→C-Mad2 conversion. Earlier work on prions generated the prediction that self-perpetuating conformational switching may be used in various cell biological niches (Lindquist, 1997). Although distinct from the self-templating mechanism of amyloids, the template-driven refolding of Mad2 represents a clear example in which this is the case.
Figure 4.
Comparing the Mad2 conformational switching reaction to models for prion propagation. (A and B) Models for prion propagation adapted from Aguzzi (2004). (C) A simplified reaction scheme for Mad2 conversion catalyzed by self–self interactions.
Concluding remarks
The extensive secondary and tertiary structural reorganization that accomplishes Mad2 conformational switching is analogous to the major structural rearrangements that have now been seen for some other so-called metamorphic proteins (Murzin, 2008) but is distinguished structurally, kinetically, and in its self–self catalytic nature from any other known regulatory transition. In this review, we suggest a foldon-dependent molecular switching mechanism in which catalytic C-Mad2 selectively binds to a partially unfolded partner called I-Mad2. Rather than directly stabilizing the active conformation, the binding energy promotes equilibration that favors the more stable C-Mad2 form. The binding catalyzes the O-Mad2→C-Mad2 conversion by equivalently lowering the energy level of the transition state. This view relates Mad2 regulatory structure change to the foldon paradigm that emphasizes the role of the naturally occurring folding and unfolding behavior of nativelike foldon units in protein folding and function (Englander et al., 2007). Coherently, the same folding/unfolding picture can explain why newly synthesized Mad2 initially folds to and becomes trapped in a nonequilibrium conformational distribution, and it further suggests a mechanism that allows the tightly folded native C-Mad2 structure to be threaded by its binding partners (Mad1 or Cdc20).
The model suggested in this review represents the first description of cell cycle regulation in which a partial unfolding of the major signaling molecule and its refolding into an entirely different conformation directs distinct downstream biochemical outcomes. Future experiments designed to elucidate the unfolding and refolding events that appear to determine the cycle of Mad2 activation and silencing seem likely to provide important insight into the elegant but complex mechanisms that faithfully guard genome integrity at cell division.
Acknowledgments
We thank M. Lampson and J. Shah for their helpful comments on the manuscript, L. Mayne for helpful discussion, and D. Slochower for his assistance with Matlab.
This work was supported in part by the National Institutes of Health (research grants DP2OD002177 [J. Shorter], GM31847 [S.W. Englander], and GM82989 [B.E. Black]), by a seed money research grant through the Abramson Cancer Center (grant #IRG-78-002-30) from the American Cancer Society (B.E. Black), and by a career award in the biomedical sciences from the Burroughs Wellcome Fund (B.E. Black).
Abbreviations used in this paper: APC, anaphase-promoting complex/cyclosome; C-Mad2, closed Mad2; I-Mad2, intermediate Mad2; MBP1, Mad2-binding peptide 1; MCC, mitotic checkpoint complex; O-Mad2, open Mad2.
References
- Aguzzi, A. 2004. Understanding the diversity of prions. Nat. Cell Biol. 6:290–292. [DOI] [PubMed] [Google Scholar]
- Black, B.E., and E.A. Bassett. 2008. The histone variant CENP-A and centromere specification. Curr. Opin. Cell Biol. 20:91–100. [DOI] [PubMed] [Google Scholar]
- Campbell, M.S., G.K. Chan, and T.J. Yen. 2001. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 114:953–963. [DOI] [PubMed] [Google Scholar]
- Carulla, N., G.L. Caddy, D.R. Hall, J. Zurdo, M. Gairi, M. Feliz, E. Giralt, C.V. Robinson, and C.M. Dobson. 2005. Molecular recycling within amyloid fibrils. Nature. 436:554–558. [DOI] [PubMed] [Google Scholar]
- Chen, R.H., A. Shevchenko, M. Mann, and A.W. Murray. 1998. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 143:283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R.H., D.M. Brady, D. Smith, A.W. Murray, and K.G. Hardwick. 1999. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell. 10:2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland, D.W., Y. Mao, and K.F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 112:407–421. [DOI] [PubMed] [Google Scholar]
- De Antoni, A., C.G. Pearson, D. Cimini, J.C. Canman, V. Sala, L. Nezi, M. Mapelli, L. Sironi, M. Faretta, E.D. Salmon, and A. Musacchio. 2005. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 15:214–225. [DOI] [PubMed] [Google Scholar]
- Englander, S.W., L. Mayne, and M.M. Krishna. 2007. Protein folding and misfolding: mechanism and principles. Q. Rev. Biophys. 40:287–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne, K.E., and S.S. Taylor. 2008. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 14:111–122. [DOI] [PubMed] [Google Scholar]
- Hardwick, K.G., R.C. Johnston, D.L. Smith, and A.W. Murray. 2000. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 148:871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilser, V.J., and E.B. Thompson. 2007. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc. Natl. Acad. Sci. USA. 104:8311–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang, L., H. Maity, M.M. Krishna, Y. Lin, and S.W. Englander. 2003. Folding units govern the cytochrome c alkaline transition. J. Mol. Biol. 331:37–43. [DOI] [PubMed] [Google Scholar]
- Kops, G.J., D.R. Foltz, and D.W. Cleveland. 2004. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl. Acad. Sci. USA. 101:8699–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzhnev, D.M., and L.E. Kay. 2008. Probing invisible, low-populated states of protein molecules by relaxation dispersion NMR spectroscopy: an application to protein folding. Acc. Chem. Res. 41:442–451. [DOI] [PubMed] [Google Scholar]
- Lindquist, S. 1997. Mad cows meet psi-chotic yeast: the expansion of the prion hypothesis. Cell. 89:495–498. [DOI] [PubMed] [Google Scholar]
- Luo, X., G. Fang, M. Coldiron, Y. Lin, H. Yu, M.W. Kirschner, and G. Wagner. 2000. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat. Struct. Biol. 7:224–229. [DOI] [PubMed] [Google Scholar]
- Luo, X., Z. Tang, J. Rizo, and H. Yu. 2002. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell. 9:59–71. [DOI] [PubMed] [Google Scholar]
- Luo, X., Z. Tang, G. Xia, K. Wassmann, T. Matsumoto, J. Rizo, and H. Yu. 2004. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat. Struct. Mol. Biol. 11:338–345. [DOI] [PubMed] [Google Scholar]
- Maity, H., J.N. Rumbley, and S.W. Englander. 2006. Functional role of a protein foldon: an omega-loop foldon controls the alkaline transition in ferricytochrome c. Proteins. 63:349–355. [DOI] [PubMed] [Google Scholar]
- Mapelli, M., F.V. Filipp, G. Rancati, L. Massimiliano, L. Nezi, G. Stier, R.S. Hagan, S. Confalonieri, S. Piatti, M. Sattler, and A. Musacchio. 2006. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 25:1273–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli, M., L. Massimiliano, S. Santaguida, and A. Musacchio. 2007. The mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell. 131:730–743. [DOI] [PubMed] [Google Scholar]
- Michel, L., E. Diaz-Rodriguez, G. Narayan, E. Hernando, V.V. Murty, and R. Benezra. 2004. Complete loss of the tumor suppressor MAD2 causes premature cyclin B degradation and mitotic failure in human somatic cells. Proc. Natl. Acad. Sci. USA. 101:4459–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin, A.G. 2008. Biochemistry. Metamorphic proteins. Science. 320:1725–1726. [DOI] [PubMed] [Google Scholar]
- Musacchio, A., and E.D. Salmon. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393. [DOI] [PubMed] [Google Scholar]
- Prusiner, S.B. 1991. Molecular biology of prion diseases. Science. 252:1515–1522. [DOI] [PubMed] [Google Scholar]
- Shah, J.V., E. Botvinick, Z. Bonday, F. Furnari, M. Berns, and D.W. Cleveland. 2004. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr. Biol. 14:942–952. [DOI] [PubMed] [Google Scholar]
- Shorter, J., and S. Lindquist. 2005. Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 6:435–450. [DOI] [PubMed] [Google Scholar]
- Sironi, L., M. Mapelli, S. Knapp, A. De Antoni, K.T. Jeang, and A. Musacchio. 2002. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 21:2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin, V., G.K. Chan, and T.J. Yen. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite, M.F., and N. Koloteva-Levin. 2004. Propagating prions in fungi and mammals. Mol. Cell. 14:541–552. [DOI] [PubMed] [Google Scholar]
- Vink, M., M. Simonetta, P. Transidico, K. Ferrari, M. Mapelli, A. De Antoni, L. Massimiliano, A. Ciliberto, M. Faretta, E.D. Salmon, and A. Musacchio. 2006. In vitro FRAP identifies the minimal requirements for Mad2 kinetochore dynamics. Curr. Biol. 16:755–766. [DOI] [PubMed] [Google Scholar]
- Walczak, C.E., and R. Heald. 2008. Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol. 265:111–158. [DOI] [PubMed] [Google Scholar]
- Waters, J.C., R.H. Chen, A.W. Murray, and E.D. Salmon. 1998. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 141:1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M., B. Li, C.J. Liu, D.R. Tomchick, M. Machius, J. Rizo, H. Yu, and X. Luo. 2008. Insights into mad2 regulation in the spindle checkpoint revealed by the crystal structure of the symmetric mad2 dimer. PLoS Biol. 6:e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H. 2006. Structural activation of Mad2 in the mitotic spindle checkpoint: the two-state Mad2 model versus the Mad2 template model. J. Cell Biol. 173:153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H. 2007. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol. Cell. 27:3–16. [DOI] [PubMed] [Google Scholar]