Abstract
The multifunctional nuclear protein positive cofactor 4 (PC4) is involved in various cellular processes including transcription, replication, and chromatin organization. Recently, PC4 has been identified as a suppressor of oxidative mutagenesis in Escherichia coli and Saccharomyces cerevisiae. To investigate a potential role of PC4 in mammalian DNA repair, we used a combination of live cell microscopy, microirradiation, and fluorescence recovery after photobleaching analysis. We found a clear accumulation of endogenous PC4 at DNA damage sites introduced by either chemical agents or laser microirradiation. Using fluorescent fusion proteins and specific mutants, we demonstrated that the rapid recruitment of PC4 to laser-induced DNA damage sites is independent of poly(ADP-ribosyl)ation and γH2AX but depends on its single strand binding capacity. Furthermore, PC4 showed a high turnover at DNA damages sites compared with the repair factors replication protein A and proliferating cell nuclear antigen. We propose that PC4 plays a role in the early response to DNA damage by recognizing single-stranded DNA and may thus initiate or facilitate the subsequent steps of DNA repair.
Introduction
The human positive cofactor 4 (PC4) is an abundant nuclear protein that plays an important role in various cellular processes including transcription, replication, chromatin organization, and cell cycle progression (Ge and Roeder, 1994; Kretzschmar et al., 1994; Pan et al., 1996; Wang et al., 2004; Das et al., 2006). PC4 was originally identified as a transcription cofactor that was minimally needed, in addition to the basal transcription machinery consisting of TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH, to mediate the response of RNA polymerase II to transcriptional activators (Meisterernst et al., 1991; Ge and Roeder, 1994; Kretzschmar et al., 1994).
PC4 is thought to facilitate the formation of the preinitiation complex at the level of TFIID-TFIIIA binding as well as during promoter opening and the escape of RNA polymerase II through interaction with TFIIH (Kaiser et al., 1995; Fukuda et al., 2004). In addition to its cofactor function, PC4 represses transcription through interaction with single-stranded DNA (ssDNA) at open promoter regions (Werten et al., 1998; Wu and Chiang, 1998). Interestingly, PC4 was found to interact genetically and physically with a component of the polyadenylation complex CtsF-64–Rna15p, which indirectly supported the hypothesis that transcription, polyadenylation, and termination may be closely linked (Calvo and Manley, 2001).
The 127–amino acid protein PC4 consists of two major domains that are critical for distinct functions. The lysine-rich N-terminal regulatory domain (amino acid residues 1–62) is required for protein–protein interactions and is essential for coactivator function in vitro (Kretzschmar et al., 1994; Kaiser et al., 1995). The C-terminal domain (CTD), comprising amino acid residues 63–127, allows binding to ssDNA and double stranded DNA in a sequence-independent manner, mediating both transcriptional repression and coactivation (Kaiser et al., 1995; Werten et al., 1998). Structural analysis of the CTD revealed that PC4 dimerizes and binds ssDNA through the CTD (Brandsen et al., 1997; Werten and Moras, 2006). Mutation of critical amino acid residues within the CTD of PC4, predicted to be essential for ssDNA binding based on structural comparison analyses using the replication protein A (RPA)–ssDNA cocrystal structure (Bochkarev et al., 1997), resulted in the loss of its ability to bind to ssDNA and to repress transcription (Werten et al., 1998). Within its N-terminal regulatory domain, PC4 contains the so-called SEAC motif, which is rich in serine and acidic residues and was shown to be a target of casein kinase II (CK2) phosphorylation (Kretzschmar et al., 1994), regulating the activity of PC4 in mammalian cells (Ge et al., 1994). In proliferating mammalian cells, ∼95% of PC4 was shown to be phosphorylated, which affects its DNA-binding properties. Phosphorylated PC4 was shown to lose its coactivator and double stranded DNA–binding activities, but maintained its ability to bind to ssDNA mediating transcriptional repression (Ge et al., 1994; Werten et al., 1998).
Recently, it has been shown that the ssDNA-binding capacity of PC4 is required for resistance to hydrogen peroxide (H2O2) and prevents mutagenesis by oxidative DNA damage in Escherichia coli and Saccharomyces cerevisiae (Wang et al., 2004). Although these genetic studies argue for a role of PC4 in DNA repair, the direct involvement of PC4 in the DNA damage response of mammalian cells remains elusive. We used a combination of live cell microscopy, laser microirradiation, and FRAP analysis to study the recruitment of PC4 to DNA damage sites in vivo. We found a very rapid and transient accumulation of PC4 at DNA damage sites, which was independent of poly(ADP-ribosyl)ation and phosphorylation of H2AX but depended on its ability to bind ssDNA. These results argue for a role of this multifunctional cofactor in the very early steps of DNA repair.
Results and discussion
PC4 accumulates at DNA damage sites
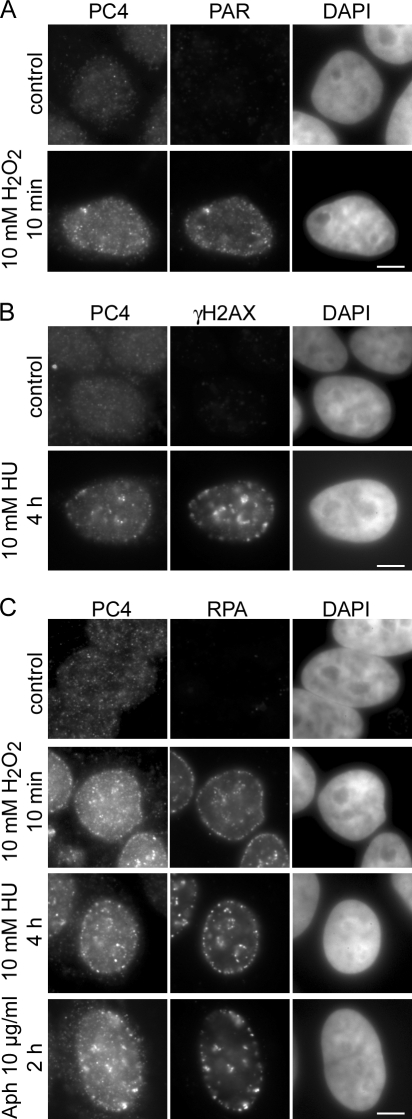
To investigate the role of PC4 in DNA repair we examined the redistribution of PC4 in response to DNA damage in human and mouse cells. After treatment with different chemical agents, which induce different types of DNA lesions, cells were in situ extracted and subsequently stained for endogenous PC4 and specific DNA damage markers. In untreated cells we found a diffuse distribution of PC4 in the nucleus. Upon treatment with H2O2 or Hydroxyurea (HU) PC4 accumulated at discrete subnuclear foci colocalizing with sites of DNA damage visualized by antibodies against poly(ADP-ribose) (PAR) and γH2AX, respectively (Fig. 1, A and B). Replication arrest with HU or aphidicolin (Aph), resulting in extended stretches of ssDNA bound by the single strand binding protein RPA (Gorisch et al., 2008), as well as treatment with H2O2, also led to a redistribution of PC4 into foci colocalizing with RPA (Fig. 1 C).
Figure 1.
PC4 accumulates at DNA damage sites. HeLa cells were treated with 10 mM HU, 10 mM H2O2, or 10 μg/ml Aph for the indicated time points and in situ extracted with 0.5% Triton X-100 for 30 s before fixation. (A and B) Widefield fluorescence images of human HeLa cells treated with H2O2 or HU show accumulation of PC4 at subnuclear sites colocalizing with the DNA damage markers PAR and γH2AX, respectively. (C) Replication arrest with HU or Aph, as well as DNA damage induction with H2O2 results in relocalization of PC4 to subnuclear foci colocalizing with the single strand binding protein RPA34. Bars, 5 μm.
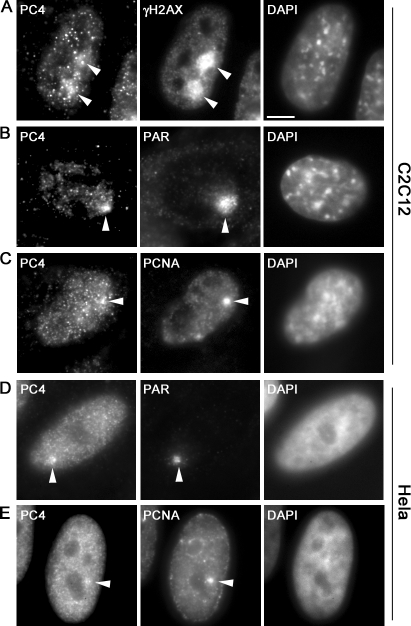
To locally introduce DNA lesions at preselected subnuclear sites we used microirradiation with a 405-nm diode laser as described previously (Mortusewicz et al., 2006, 2007). This treatment results in the generation of a mixture of different types of DNA damage, including single strand breaks and double strand breaks, which are substrates for different DNA repair pathways. Immunofluorescence stainings with specific antibodies revealed that endogenous PC4 accumulates at sites of DNA damage as early as 5 min after microirradiation in both human and mouse cells (Fig. 2, A, B, and D). Furthermore, we observed colocalization of PC4 with the replication and repair protein proliferating cell nuclear antigen (PCNA) at laser-induced DNA damage sites (Fig. 2, C and E). Collectively, these results show that PC4 accumulates at sites of DNA damage generated by chemical agents or laser microirradiation.
Figure 2.
PC4 accumulates at laser-induced DNA damage sites. Widefield fluorescence images of mouse C2C12 and human HeLa cells are shown. Fixation and immunostaining was performed ∼5 min after laser microirradiation. Arrowheads mark the sites of irradiation. Laser microirradiation results in local generation of DNA damage (A, B, and D) detected by antibodies against γH2AX and PAR, respectively. Endogenous PC4 accumulates at DNA damage sites in mouse (A and B) and human (D) cells and colocalizes with PCNA (C and E). Bar, 5 μm.
Recruitment kinetics and mobility of PC4 at DNA repair sites
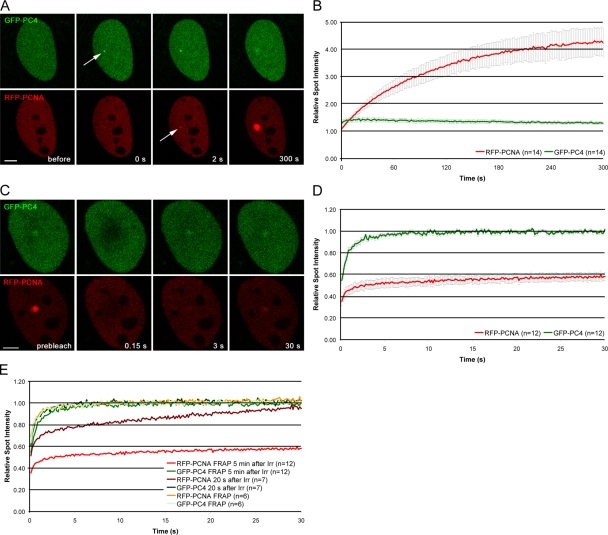
Having shown that endogenous PC4 accumulates at DNA damage sites, we generated GFP- and RFP-tagged fusion proteins to study the recruitment of PC4 in living cells (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200808097/DC1). As a positive control we chose the proccessivity factor PCNA, which is involved in various DNA repair pathways including nucleotide excision repair (Shivji et al., 1992), base excision repair (Gary et al., 1999; Levin et al., 2000), mismatch repair (Johnson et al., 1996; Umar et al., 1996; Jiricny, 2006), and repair of double strand breaks. Using a combination of microirradiation and time-lapse analysis we followed the spatiotemporal accumulation of GFP-PC4 and RFP-PCNA in vivo. For quantification, the fluorescence intensity at the irradiated sites were measured, corrected for background and total nuclear loss of fluorescence over the time course, and normalized to the preirradiation value as described previously (Mortusewicz et al., 2006, 2007). We found that GFP-PC4 accumulated at DNA damage sites immediately after microirradiation, preceding recruitment of RFP-PCNA (Fig. 3 A). Although RFP-PCNA showed a slow and constant increase of accumulation at repair sites during the observation period of 5 min, fluorescence intensities of GFP-PC4 declined after reaching a maximum around 20–40 s after microirradiation (Fig. 3 B). To determine whether the recruitment of PC4 to DNA damage sites is cell cycle dependent, we microirradiated cells in different S phase stages using RFP-PCNA as a cell cycle marker. We found that PC4 accumulates at laser-induced DNA damage sites in early, mid, and late S phase cells (Fig. S2).
Figure 3.
Recruitment and mobility of PC4 and PCNA at DNA damage sites in living cells. (A) Live cell imaging of a microirradiated C2C12 cell coexpressing GFP-PC4 and RFP-PCNA. Accumulation of GFP-PC4 can be observed immediately after microirradiation, whereas RFP-PCNA accumulates with a short delay of ∼2–10 s. Arrows mark the site of microirradiation. (B) Quantitative evaluation of recruitment kinetics showing mean curves. (C) To analyze the mobility of PC4 and PCNA at DNA damage sites, the microirradiated region was bleached 5 min after microirradiation and the fluorescence recovery was measured. (D) Quantitative evaluation of FRAP data showing mean curves. Error bars represent the standard error of the mean. (E) To analyze the mobility of GFP-PC4 and RFP-PCNA at the peak of GFP-PC4 accumulation, the microirradiated region was bleached 20 s after microirradiation and the fluorescence recovery was measured. As a control, a similar sized region in nonirradiated cells cotransfected with GFP-PC4 and RFP-PCNA was bleached and the fluorescence recovery was measured. The recovery curves obtained from FRAP analysis 5 min after microirradiation are also displayed as a reference. Quantitative evaluation of FRAP data showing mean curves. For clarity, error bars are not shown. Bars, 5 μm.
To determine the mobility of PC4 at laser-induced DNA damage sites, we performed FRAP analysis 5 min after microirradiation. The irradiated region was bleached with a high energy laser pulse for 300 ms and the fluorescence recovery was determined. After bleaching of the repair foci, we observed complete recovery of the PC4 signal within 5 s, indicating a high mobility of PC4 at repair sites (Fig. 3, C and D). In contrast, no recovery of PCNA at repair sites could be observed within the observation period, which is in good agreement with previous studies where DNA damage was induced by chemical agents or irradiation with a UV lamp (Solomon et al., 2004; Essers et al., 2005). As the fluorescence intensity of PC4 already begins to decline during the observation period of 5 min, we also performed FRAP analysis 20 s after microirradiation to determine the mobility of PC4 at the peak of accumulation. We could not detect any differences in the mobility of PC4 20 s or 5 min after microirradiation (Fig. 3 E). The constant increase in RFP-PCNA fluorescence observed when FRAP analysis was performed 20 s after microirradiation can be explained by new RFP-PCNA molecules being recruited during the time course of the FRAP experiment. Collectively, these results show an early and transient binding of PC4 at DNA damage sites, suggesting a role for PC4 in the early steps of DNA repair, like damage recognition and/or signaling.
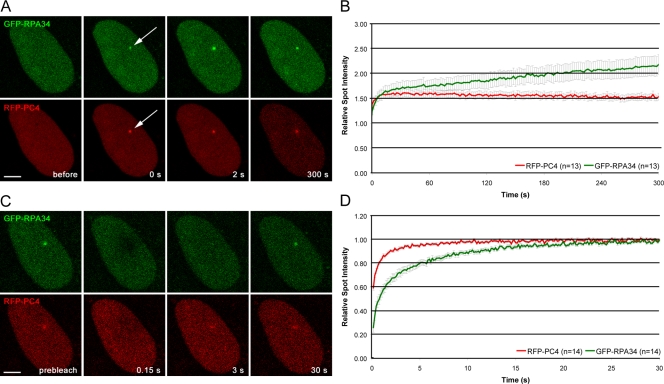
This raises the question of how PC4 gets recruited to DNA lesions. Given that the single strand binding capacity of PC4 is needed for resistance against H2O2 in repairing deficient E. coli (Wang et al., 2004), it was tempting to speculate that PC4 is recruited by binding to ssDNA generated at microirradiated sites. In addition, the crystal structure of PC4 shows high similarity to the single strand binding domains of RPA70 and RPA34 (Fig. S1 B; Bochkarev et al., 1997, 1999; Brandsen et al., 1997). Therefore, we directly compared the recruitment kinetics and the mobility of RFP-PC4 with GFP-RPA34. We found that both PC4 and RPA34 were recruited immediately after microirradiation, with PC4 accumulating slightly faster than RPA34 (Fig. 4 A). Like PCNA, RPA34 showed a slow and constant increase in fluorescence intensity at the irradiated site, whereas the intensity of PC4 gradually declined after reaching a maximum (Fig. 4 B). FRAP analysis revealed distinct recovery rates, indicating that PC4 exhibits a higher mobility at DNA damage sites than RPA34 (Fig. 4, C and D). Collectively, we could demonstrate that in comparison to the single strand binding protein RPA34, PC4 shows distinct recruitment and binding properties at laser-induced DNA damage sites.
Figure 4.
Comparison of recruitment and binding capacities of PC4 with RPA34 in living cells. (A) Live cell imaging of a microirradiated C2C12 cell coexpressing GFP-RPA34 and RFP-PC4. Both GFP-RPA34 and RFP-PC4 accumulate immediately after microirradiation (at sites of DNA damage). Arrows mark the site of microirradiation. (B) Quantitative evaluation of recruitment kinetics showing mean curves. (C) Mobility of PC4 and RPA34 at DNA damage sites. (D) Quantitative evaluation of FRAP data showing mean curves. The Error bars represent the standard error of the mean.
The C-terminal single strand binding domain of PC4 mediates recruitment to DNA damage sites
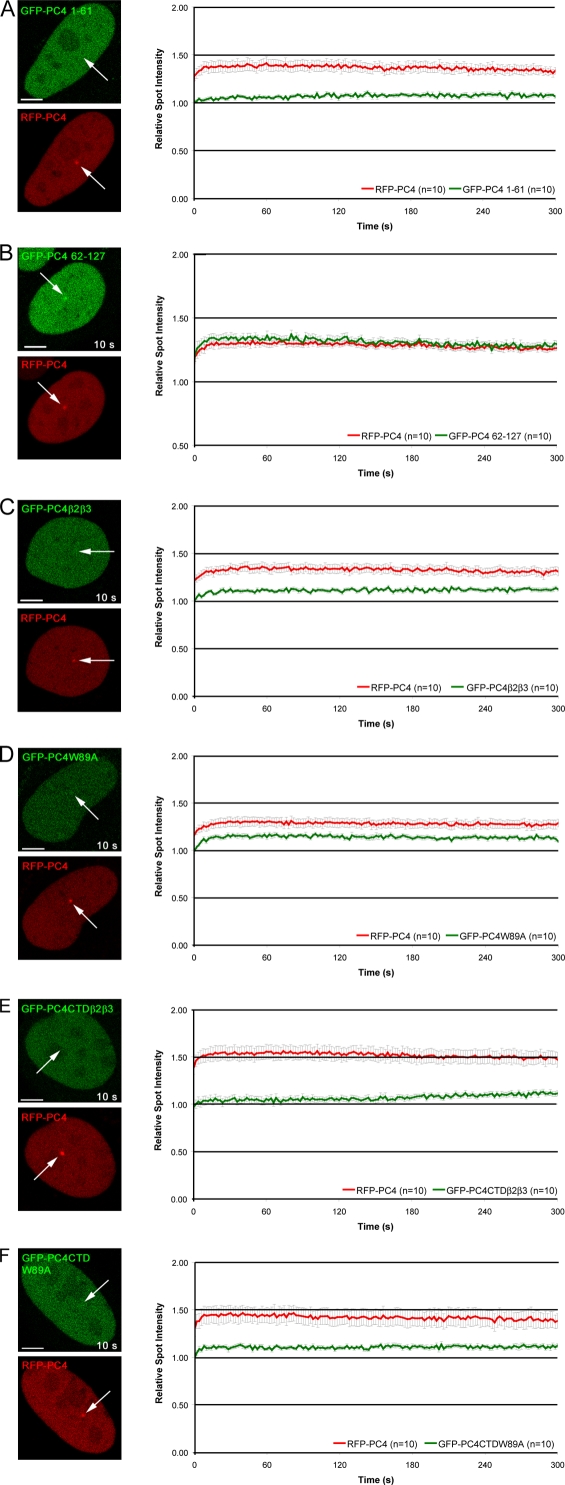
The fact that PC4 and RPA show different recruitment kinetics and turnover rates at DNA repair sites raises the question of whether PC4 indeed recognizes ssDNA generated after microirradiation. Earlier studies revealed a bipartite structure of PC4 comprising an N-terminal regulatory domain (amino acids 1–62) and a C-terminal ssDNA binding and dimerization domain (amino acids 63–127; Kretzschmar et al., 1994). It has also been shown that the ssDNA binding activity is required for transcription repression but is not needed for the activator-dependent stimulatory activity of PC4 (Werten et al., 1998). To investigate the mechanisms mediating the recruitment of PC4 to DNA damage sites we generated GFP fusion constructs comprising either the N-terminal regulatory domain (GFP-PC4 1–61) or the CTD (GFP-PC4 62–127). For direct comparison, we cotransfected the N-terminal domain and the CTD together with the full-length PC4. We found only a minor accumulation of GFP-PC4 1–61 at microirradiated sites (Fig. 5 A). In contrast, GFP-PC4 62–127 showed the same recruitment kinetics as the full-length protein (Fig. 5 B). In addition, we analyzed the recruitment of a fusion protein lacking the SEAC motif within the first 22 amino acids of PC4. This serine and acidic amino acids–rich motif is phosphorylated by CK2 (Kretzschmar et al., 1994), which has recently been implicated in the DNA damage response (Ayoub et al., 2008; Spycher et al., 2008). However, deleting this N-terminal domain did not significantly affect the recruitment of PC4 to DNA damage sites (Fig. S3, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200808097/DC1). We conclude that the CTD of PC4 is necessary and sufficient for recruitment to DNA damage sites and that PC4 recruitment does not depend on its N-terminal CK2 phosphorylation sites.
Figure 5.
The single strand binding capacity of PC4 is needed for efficient recruitment to DNA damage sites. Deletion proteins containing either the N-terminal domain (1–61) or the CTD (62–127) of PC4 were tested for in vivo recruitment to DNA damage sites. Whereas the N-terminal domain shows only a minor accumulation at microirradiated sites (A), the CTD is recruited with similar kinetics like the full-length PC4 (B). For further analysis, key residues essential for single strand binding within the full length or CTD of PC4 were mutated (outlined in Fig. 3 A). Recruitment of mutated fusion proteins to DNA damage sites is greatly reduced (C–F). Arrows mark the site of microirradiation. Bars, 5 μm.
In addition to phosphorylation of DNA repair factors by CK2, two other posttranslational modifications, poly(ADP-ribosyl)ation and phosphorylation of H2AX, have been shown to play a central role in the recruitment and/or retention of factors involved in later stages of the DNA repair process. We determined the recruitment kinetics of PC4 to laser-induced DNA damage sites in wild-type mouse embryonic fibroblasts (MEFs) in comparison with PARP-1−/− and H2AX−/− MEFs, which are largely devoid of DNA damage–induced poly(ADP-ribosyl)ation and phosphorylation of H2AX, respectively (Trucco et al., 1998; Celeste et al., 2002). Interestingly, recruitment of PC4 did not depend on any of these modifications (Fig. S3, C and D), which is another indication for the involvement of PC4 in the very early steps of the DNA damage response.
To further characterize the recruitment of PC4 to DNA damage sites, we generated mutants in the context of the full length and the CTD of PC4. We introduced a point mutation at position 89 replacing Trp by Ala (GFP-PC4W89A and GFP-PC4CTDW89A) and a triple mutation at positions 77, 78, and 80 (GFP-PC4β2β3 and GFP-PC4CTDβ2β3), which were previously described to be essential for ssDNA binding of PC4 (Werten et al., 1998). The results of the microirradiation analysis of the PC4 mutants are summarized in Fig. S1 A and shown in detail in Fig. 5. Both mutations led to a reduced accumulation of PC4 at microirradiated sites in the context of the full length and the CTD of PC4 (Fig. 5, C–F), indicating that the single strand binding capacity of PC4 is needed for efficient recruitment of PC4 to DNA repair sites in living cells.
The fast and transient binding of the transcriptional cofactor PC4 at DNA damage sites identified in this study raises several interesting questions concerning potential roles in DNA repair and connections to transcriptional regulation. The observation that the recruitment of PC4 depends on its single strand binding capacity suggests that PC4 might fulfill similar roles in DNA repair as RPA. However, the different binding kinetics and mobility of PC4 and RPA at DNA damage sites would argue for distinct functions in DNA repair.
As PC4 has been implicated in the regulation of DNA replication (Pan et al., 1996), it could stop DNA replication near DNA lesions. Similarly, PC4 might also stop transcription as a response to DNA damage, which is supported by the fact that PC4 is a potent repressor of transcription at specific DNA structures such as ssDNA, DNA ends, and heteroduplex DNA, which are generated during DNA repair (Werten et al., 1998). Moreover, PC4 could have a helicase-like function (Werten et al., 1998; Werten and Moras, 2006), which through binding and multimerization along ssDNA is predicted to enable ATP-independent unwinding of duplex DNA. In this regard, one could also envision a protective role of PC4 in preventing degradation of ssDNA by nucleases.
The crystallization of PC4 in complex with ssDNA revealed that the subunits of the PC4 homodimer cooperate in the sequence-independent binding (Ballard et al., 1988) of two opposing DNA backbones, exposing the DNA bases to the surrounding environment (Werten and Moras, 2006). These observations, together with the rapid recruitment of PC4 to DNA damage sites, argue for a role of PC4 in the detection and/or exposure of DNA damages. During the subsequent repair process PC4 may be displaced, as suggested by the observed transient binding at damaged sites.
As PC4 is a cofactor of RNA polymerase II and also interacts with p53 (Banerjee et al., 2004), a central regulator of the cellular DNA damage response, it is tempting to speculate that binding to ssDNA may lead to a transient depletion of nuclear levels of free PC4 that affects the stoichiometry and/or activity of regulatory complexes and thereby contributes to the sensing and signaling of DNA damage.
Materials and methods
Cell culture and transfection
Human HeLa, wild-type MEFs, PARP1−/− MEFs (Trucco et al., 1998), H2AX−/− MEFs (Celeste et al., 2002), and mouse C2C12 cells were cultured in DME containing 50 μg/ml gentamicin supplemented with 10 and 20% FCS, respectively. PARP1−/− and H2AX−/− MEFs were provided by V. Schreiber (Ecole Supérieure de Biotechnologie de Strasbourg, Strasbourg, France) and A. Nussenzweig (National Cancer Institute, National Institutes of Health, Bethesda, MD), respectively.
Cells grown on μ-slides (Ibidi) or on gridded coverslips were cotransfected with jetPEI (PolyPlus Transfection) according to the manufacturer's instructions. For microirradiation experiments, cells were sensitized by incubation in medium containing 10 μg/ml BrdU for 24–48 h. HU, H2O2, and Aph were obtained from Sigma-Aldrich.
Expression plasmids
The generation of PC4 deletion and point mutants was previously described (Kretzschmar et al., 1994; Werten et al., 1998). Corresponding GFP-PC4 fusion constructs were constructed by ligation of either restriction fragments (NdeI–ClaI for GFP-PC4; EcoRI–ClaI for the constructs GFP-PC4β2β3, GFP-PC4W89A, GFP-PC4 22–127, GFP-PC4 CTD β2β3, and GFP-PC4 CTDW89A; and XhoI–PstI for GFP-PC4 62–127) or PCR products (forward primer, 5′ GAAGATCTCCGGTTATTCTTCATATGCC 3′; reverse primer, 5′ TGGAATTCTCAATCATCTCTG 3′; BglII–EcoRI cloning for GFP-PC4 1–61) into matching restriction sites of pEGFP-C1 (Clontech Laboratories, Inc.). GFP-PC4 fusion constructs were verified by sequencing and tested by expression in HeLa cells followed by Western blot analysis. A red variant of PC4 was generated by replacing GFP with RFP (Campbell et al., 2002) and termed RFP-PC4. The mRFP1 expression vector was supplied by R. Tsien (University of California, San Diego, La Jolla, CA).
Mammalian expression constructs encoding translational fusions of human RPA34 and PCNA with either GFP or RFP were previously described (Sporbert et al., 2005). In all cases, expression was under the control of the cytomegalovirus promoter and correct expression of fusion proteins was verified by Western blot analysis.
Immunofluorescence and detergent extraction
Cells were fixed in 3.7% formaldehyde for 10 min and permeabilized with 0.5% Triton X-100 or ice-cold methanol for 5 min. The following primary antibodies (diluted in PBS containing 4% BSA) were used: anti-γH2AX (Ser139) mouse monoclonal antibodies (Millipore), anti-PAR mouse monoclonal antibodies (Trevigen), anti-RPA34 mouse monoclonal antibodies (EMD), anti-PC4 rabbit polyclonal antibodies (SA2249; generated by standard techniques; Eurogentech), and anti-PCNA rat monoclonal antibodies (Spada et al., 2007). Primary antibodies were detected using secondary antibodies (diluted 1:200 in PBS containing 4% BSA) conjugated to AlexaFluor 488 or 555 (Invitrogen). Cells were counterstained with DAPI and mounted in Vectashield (Vector Laboratories). For in situ extraction, cells were permeabilized for 30 s with 0.5% Triton X-100 in PBS before fixation.
Live cell microscopy, microirradiation, and photobleaching experiments
Live cell imaging, microirradiation, and photobleaching experiments were performed with a confocal laser scanning microscope (TCS SP2/AOBS or SP5/AOBS; Leica), each equipped with a UV-transmitting HCX PL 63x/1.4 oil objective. GFP and RFP were excited with a 488-nm Ar laser line and a 561-nm diode pumped solid state laser line (Leica), respectively. The microscopes were equipped with a heated environmental chamber set to 37°C. Confocal image series were typically recorded with a frame size of 256 × 256 pixels and a pixel size of 90 nm.
Microirradiation was performed as previously described (Mortusewicz et al., 2006, 2007). In brief, a preselected spot ∼1 μm in diameter within the nucleus was microirradiated for 1 s with a 405-nm diode laser (Leica) set to 50–80 μW. The laser power was measured after passing through the objective lens with a laser power meter (Coherent). Before and after microirradiation, confocal image series of one mid z section were recorded at 2-s time intervals (typically 6 pre- and 150 postirradiation frames). For evaluation of the recruitment kinetics, fluorescence intensities at the irradiated region were corrected for background and for total nuclear loss of fluorescence over the time course and normalized to the preirradiation value.
For FRAP analysis, a region of interest was selected and photobleached for 300 ms with all laser lines of the Ar laser and the 561-nm diode pumped solid state laser set to maximum power at 100% transmission. Before and after bleaching, confocal image series were recorded at 150-ms time intervals (typically 10 pre- and 200 postbleach frames). Mean fluorescence intensities of the bleached region were corrected for background and for total nuclear loss of fluorescence over the time course and normalized to the mean of the last four prebleach values.
For the quantitative evaluation of microirradiation and photobleaching experiments, data of at least nine nuclei were averaged and the mean curve and the standard error of the mean calculated and displayed using Excel software (Microsoft).
Images of fixed cells were taken with a widefield epifluorescence microscope (Axiophot 2; Carl Zeiss, Inc.) using a Plan Apochromat 63x/1.40 oil objective (Carl Zeiss, Inc.) and a cooled charge-coupled device camera (Visitron Systems).
Online supplemental material
Fig. S1 shows a schematic outline of fusion proteins used in this study and a comparison of the crystal structure of PC4 (Brandsen et al., 1997) with RPA70 (Bochkarev et al., 1997) and RPA34 (Bochkarev et al., 1999). Fig. S2 shows that recruitment of PC4 to laser-induced DNA damage sites occurs in all S phase stages. Fig. S3 shows that the recruitment of PC4 is independent of its N-terminal CK2 phosphorylation sites, poly(ADP-ribosyl)ation, and phosphorylation of H2AX. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200808097/DC1.
Supplementary Material
Acknowledgments
We are indebted to Dr. R. Tsien for providing the mRFP1 expression vector and to Dr. V. Schreiber and Dr. A. Nussenzweig for providing PARP-1−/− and H2AX−/− MEFs, respectively.
This work was supported by the Nanosystems Initiative Munich and by grants from the Deutsche Forschungsgemeinschaft to H. Leonhardt, M. Meisterernst, and M.C. Cardoso.
M. Meisterernst and H. Leonhardt contributed equally to this paper.
W. Roth's present address is Division of Cell Biochemistry, Institute of Physiological Chemistry, University of Bonn, 53115 Bonn, Germany.
Abbreviations used in this paper: Aph, aphidicolin; CK2, casein kinase II; CTD,C-terminal domain; HU, Hydroxyurea; MEF, mouse embryonic fibroblast; PAR, poly(ADP-ribose); PC4, positive cofactor 4; PCNA, proliferating cell nuclear antigen; RPA, replication protein A; ssDNA, single-stranded DNA.
References
- Ayoub, N., A.D. Jeyasekharan, J.A. Bernal, and A.R. Venkitaraman. 2008. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 453:682–686. [DOI] [PubMed] [Google Scholar]
- Ballard, D.W., W.M. Philbrick, and A.L. Bothwell. 1988. Identification of a novel 9-kDa polypeptide from nuclear extracts. DNA binding properties, primary structure, and in vitro expression. J. Biol. Chem. 263:8450–8457. [PubMed] [Google Scholar]
- Banerjee, S., B.R. Kumar, and T.K. Kundu. 2004. General transcriptional coactivator PC4 activates p53 function. Mol. Cell. Biol. 24:2052–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkarev, A., R.A. Pfuetzner, A.M. Edwards, and L. Frappier. 1997. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 385:176–181. [DOI] [PubMed] [Google Scholar]
- Bochkarev, A., E. Bochkareva, L. Frappier, and A.M. Edwards. 1999. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J. 18:4498–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsen, J., S. Werten, P.C. van der Vliet, M. Meisterernst, J. Kroon, and P. Gros. 1997. C-terminal domain of transcription cofactor PC4 reveals dimeric ssDNA binding site. Nat. Struct. Biol. 4:900–903. [DOI] [PubMed] [Google Scholar]
- Calvo, O., and J.L. Manley. 2001. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell. 7:1013–1023. [DOI] [PubMed] [Google Scholar]
- Campbell, R.E., O. Tour, A.E. Palmer, P.A. Steinbach, G.S. Baird, D.A. Zacharias, and R.Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 99:7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste, A., S. Petersen, P.J. Romanienko, O. Fernandez-Capetillo, H.T. Chen, O.A. Sedelnikova, B. Reina-San-Martin, V. Coppola, E. Meffre, M.J. Difilippantonio, et al. 2002. Genomic instability in mice lacking histone H2AX. Science. 296:922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, C., K. Hizume, K. Batta, B.R. Kumar, S.S. Gadad, S. Ganguly, S. Lorain, A. Verreault, P.P. Sadhale, K. Takeyasu, and T.K. Kundu. 2006. Transcriptional coactivator PC4, a chromatin-associated protein, induces chromatin condensation. Mol. Cell. Biol. 26:8303–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers, J., A.F. Theil, C. Baldeyron, W.A. van Cappellen, A.B. Houtsmuller, R. Kanaar, and W. Vermeulen. 2005. Nuclear dynamics of PCNA in DNA replication and repair. Mol. Cell. Biol. 25:9350–9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, A., T. Nakadai, M. Shimada, T. Tsukui, M. Matsumoto, Y. Nogi, M. Meisterernst, and K. Hisatake. 2004. Transcriptional coactivator PC4 stimulates promoter escape and facilitates transcriptional synergy by GAL4-VP16. Mol. Cell. Biol. 24:6525–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary, R., K. Kim, H.L. Cornelius, M.S. Park, and Y. Matsumoto. 1999. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Biol. Chem. 274:4354–4363. [DOI] [PubMed] [Google Scholar]
- Ge, H., and R.G. Roeder. 1994. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 78:513–523. [DOI] [PubMed] [Google Scholar]
- Ge, H., Y. Zhao, B.T. Chait, and R.G. Roeder. 1994. Phosphorylation negatively regulates the function of coactivator PC4. Proc. Natl. Acad. Sci. USA. 91:12691–12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorisch, S.M., A. Sporbert, J.H. Stear, I. Grunewald, D. Nowak, E. Warbrick, H. Leonhardt, and M.C. Cardoso. 2008. Uncoupling the replication machinery: replication fork progression in the absence of processive DNA synthesis. Cell Cycle. 7:1983–1990. [DOI] [PubMed] [Google Scholar]
- Jiricny, J. 2006. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 7:335–346. [DOI] [PubMed] [Google Scholar]
- Johnson, R.E., G.K. Kovvali, S.N. Guzder, N.S. Amin, C. Holm, Y. Habraken, P. Sung, L. Prakash, and S. Prakash. 1996. Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J. Biol. Chem. 271:27987–27990. [DOI] [PubMed] [Google Scholar]
- Kaiser, K., G. Stelzer, and M. Meisterernst. 1995. The coactivator p15 (PC4) initiates transcriptional activation during TFIIA-TFIID-promoter complex formation. EMBO J. 14:3520–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar, M., K. Kaiser, F. Lottspeich, and M. Meisterernst. 1994. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell. 78:525–534. [DOI] [PubMed] [Google Scholar]
- Levin, D.S., A.E. McKenna, T.A. Motycka, Y. Matsumoto, and A.E. Tomkinson. 2000. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr. Biol. 10:919–922. [DOI] [PubMed] [Google Scholar]
- Meisterernst, M., A.L. Roy, H.M. Lieu, and R.G. Roeder. 1991. Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell. 66:981–993. [DOI] [PubMed] [Google Scholar]
- Mortusewicz, O., U. Rothbauer, M.C. Cardoso, and H. Leonhardt. 2006. Differential recruitment of DNA Ligase I and III to DNA repair sites. Nucleic Acids Res. 34:3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortusewicz, O., J.C. Ame, V. Schreiber, and H. Leonhardt. 2007. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 35:7665–7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Z.Q., H. Ge, A.A. Amin, and J. Hurwitz. 1996. Transcription-positive cofactor 4 forms complexes with HSSB (RPA) on single-stranded DNA and influences HSSB-dependent enzymatic synthesis of simian virus 40 DNA. J. Biol. Chem. 271:22111–22116. [DOI] [PubMed] [Google Scholar]
- Shivji, K.K., M.K. Kenny, and R.D. Wood. 1992. Proliferating cell nuclear antigen is required for DNA excision repair. Cell. 69:367–374. [DOI] [PubMed] [Google Scholar]
- Solomon, D.A., M.C. Cardoso, and E.S. Knudsen. 2004. Dynamic targeting of the replication machinery to sites of DNA damage. J. Cell Biol. 166:455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spada, F., A. Haemmer, D. Kuch, U. Rothbauer, L. Schermelleh, E. Kremmer, T. Carell, G. Langst, and H. Leonhardt. 2007. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J. Cell Biol. 176:565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporbert, A., P. Domaing, H. Leonhardt, and M.C. Cardoso. 2005. PCNA acts as a stationary loading platform for transiently interacting Okazaki fragment maturation proteins. Nucleic Acids Res. 33:3521–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spycher, C., E.S. Miller, K. Townsend, L. Pavic, N.A. Morrice, P. Janscak, G.S. Stewart, and M. Stucki. 2008. Constitutive phosphorylation of MDC1 physically links the MRE11–RAD50–NBS1 complex to damaged chromatin. J. Cell Biol. 181:227–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trucco, C., F.J. Oliver, G. de Murcia, and J. Menissier-de Murcia. 1998. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 26:2644–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar, A., A.B. Buermeyer, J.A. Simon, D.C. Thomas, A.B. Clark, R.M. Liskay, and T.A. Kunkel. 1996. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 87:65–73. [DOI] [PubMed] [Google Scholar]
- Wang, J.Y., A.H. Sarker, P.K. Cooper, and M.R. Volkert. 2004. The single-strand DNA binding activity of human PC4 prevents mutagenesis and killing by oxidative DNA damage. Mol. Cell. Biol. 24:6084–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werten, S., and D. Moras. 2006. A global transcription cofactor bound to juxtaposed strands of unwound DNA. Nat. Struct. Mol. Biol. 13:181–182. [DOI] [PubMed] [Google Scholar]
- Werten, S., G. Stelzer, A. Goppelt, F.M. Langen, P. Gros, H.T. Timmers, P.C. Van der Vliet, and M. Meisterernst. 1998. Interaction of PC4 with melted DNA inhibits transcription. EMBO J. 17:5103–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S.Y., and C.M. Chiang. 1998. Properties of PC4 and an RNA polymerase II complex in directing activated and basal transcription in vitro. J. Biol. Chem. 273:12492–12498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.