Abstract
Newly generated actin free barbed ends at the front of motile cells provide sites for actin filament assembly driving membrane protrusion. Growth factors induce a rapid biphasic increase in actin free barbed ends, and we found both phases absent in fibroblasts lacking H+ efflux by the Na-H exchanger NHE1. The first phase is restored by expression of mutant cofilin-H133A but not unphosphorylated cofilin-S3A. Constant pH molecular dynamics simulations and nuclear magnetic resonance (NMR) reveal pH-sensitive structural changes in the cofilin C-terminal filamentous actin binding site dependent on His133. However, cofilin-H133A retains pH-sensitive changes in NMR spectra and severing activity in vitro, which suggests that it has a more complex behavior in cells. Cofilin activity is inhibited by phosphoinositide binding, and we found that phosphoinositide binding is pH-dependent for wild-type cofilin, with decreased binding at a higher pH. In contrast, phosphoinositide binding by cofilin-H133A is attenuated and pH insensitive. These data suggest a molecular mechanism whereby cofilin acts as a pH sensor to mediate a pH-dependent actin filament dynamics.
Introduction
At the leading edge of motile cells, increased assembly of a branched actin filament network is a driving force for membrane protrusion (Pollard and Borisy, 2003). Key events acting synergistically to generate this actin network are filament severing to increase the abundance of actin free barbed ends (Carlsson, 2006), and filament nucleation and branching by the Arp2/3 complex (Pollard, 2007). Regulation of Arp2/3 complex nucleating activity by Rho family GTPases and nucleation-promoting factors of the Wiskott-Aldrich syndrome protein (WASP) family has been extensively studied (Goley and Welch, 2006; Stradal and Scita, 2006). Less is known about control of actin filament severing in motile cells. Although actin free barbed ends act as nuclei for filament assembly and can be generated by Arp2/3 complex nucleating activity and by uncapping barbed ends of preexisting filaments (Condeelis, 2001), filament severing by the actin-binding protein cofilin generates a rapid increase in free barbed ends in motile cells and is critical for membrane protrusion (Chan et al., 2000; Ghosh et al., 2004; Mouneimne et al., 2004).
All eukaryotes express one or more members of the actin-depolymerizing factor (ADF)/cofilin family, including three isoforms in mammals: ADF, nonmuscle cofilin, and muscle cofilin. In motile cells, cofilin promotes actin filament dynamics by increasing filament disassembly at the rear of actin networks, presumably to recycle actin monomers (Maciver et al., 1998; Blanchoin et al., 2000), and by severing filaments at the leading edge to generate new free barbed ends for nucleation by the Arp2/3 complex (Ichetovkin et al., 2002; van Rheenen et al., 2007). To sever filaments, cofilin binds F-actin at two sites, an N-terminal “G-site” and a C-terminal “F-site” (Pope et al., 2000). Dephosphorylation of a conserved Ser3 in cofilin by the phosphatases slingshot (Niwa et al., 2002) or chronophin (Gohla et al., 2005) promotes actin binding at the G-site (for review see Bamburg and Wiggan, 2002). Although dephosphorylation of Ser3 is necessary for cofilin activity, it is not sufficient (Song et al., 2006). Additional mechanisms, including dissociation of bound phosphotidylinositol-4,5-bisphosphate (PI(4,5)P2) (Yonezawa et al., 1990; Ojala et al., 2001; Gorbatyuk et al., 2006; van Rheenen et al., 2007) or an increase in intracellular pH (pHi), presumably by deprotonation of His133 in the F-site (Pope et al., 2004), may be necessary, which suggests that cofilin acts as a coincidence detector with its activation, requiring several independent regulatory events. The activity of cofilin in most species is recognized to be pH sensitive. Cofilin activity in vitro increases at neutral and higher pH (Hawkins et al., 1993; Maciver et al., 1998; Chen et al., 2004), and in wounded fibroblasts, increased pHi is necessary for ADF- and cofilin-regulated actin dynamics (Bernstein et al., 2000). H+ efflux mechanisms at the leading edge of motile cells have been speculated (Bailly and Jones, 2003; Bernstein and Bamburg, 2004) but have not been confirmed to spatially regulate cofilin activity.
We find here that H+ efflux by the mammalian Na-H exchanger NHE1 promotes a cofilin-dependent increase in actin free barbed ends in response to migratory cues. NHE1 catalyzes an electroneutral exchange of extracellular Na+ for intracellular H+, and its activity increases in response to migratory cues, including monolayer wounding (Frantz et al., 2007), growth factors (Putney et al., 2002; Frantz et al., 2007), and integrin engagement (Schwartz et al., 1991; Tominaga and Barber, 1998). In motile fibroblasts (Denker and Barber, 2002) and Dictyostelium discoideum cells (Patel and Barber, 2005), NHE1 localizes at the distal margin of membrane protrusions, and its H+ efflux is necessary for directed migration of mammalian fibroblasts (Denker and Barber, 2002), leukocytes (Ritter et al., 1998), and epithelial (Klein et al., 2000; Reshkin et al., 2000) and melanoma cells (Stock et al., 2005), and for chemotaxis of Dictyostelium cells (Patel and Barber, 2005). In Dictyostelium cells with a targeted deletion of nhe1, actin filament assembly in response to a chemoattractant is attenuated (Patel and Barber, 2005).
Our current findings indicate that motile fibroblasts expressing a mutant NHE1 lacking H+ efflux have attenuated de novo actin filament assembly and no increase in actin free barbed ends compared with fibroblasts expressing wild-type (WT) NHE1. A rapid first phase of actin free barbed end formation was restored by expression of pH-insensitive Acanthamoeba castellanii actophorin or mutant cofilin-H133A but not by unphosphorylated cofilin-S3A. Computational modeling, nuclear magnetic resonance (NMR) spectroscopy, and functional studies revealed the significance of His133 in pH-sensitive cofilin activity and actin free barbed end formation. However, purified cofilin-H133A retained pH-sensitive conformational changes and severing activity. We found that PI(4,5)P2 binding to WT cofilin is pH sensitive, with increased binding at pH 7.5, compared with pH 6.5, but PI(4,5)P2 binding to cofilin-H133A is pH-insensitive, with binding similar to WT at pH 7.5. Although pH and PI(4,5)P2 regulate cofilin activity, our data indicate that these two controls are related and suggest that pH-dependent PI(4,5)P2 binding by cofilin regulates actin-severing activity. Additionally, our findings support an emerging (Lee et al., 2005; Frantz et al., 2007) role for pH-sensitive His switches in phosphoinositide binding.
Results
H+ efflux by NHE1 is necessary for increased actin filament assembly in response to migratory cues
In mammalian fibroblasts (Denker and Barber, 2002) and in Dictyostelium cells (Patel and Barber, 2005), H+ efflux by NHE1 is necessary for efficient directed migration. In chemotaxing Dictyostelium cells, NHE1 also is necessary to suppress lateral pseudopods and to promote de novo actin assembly at the cell front (Patel and Barber, 2005). We asked whether NHE1 regulates actin filament assembly in motile fibroblasts by using NHE1-deficient cells stably expressing WT NHE1 (WT cells) or a mutant NHE1 containing an isoleucine substitution for glutamine 266 that lacks H+ efflux (E266I cells; Denker et al., 2000). WT and E266I cells at the edge of a wounded monolayer had a time-dependent increase in cortical F-actin after wounding; however, at all time points, the abundance of F-actin was significantly lower in E266I cells compared with WT cells (Fig. 1, A and B). Conversely, the abundance of cortical F-actin before wounding (not depicted) and immediately after wounding (Fig. 1 A) was similar in E266I and WT cells. De novo increases in F-actin in quiescent subconfluent cells treated with 50 ng/ml PDGF were also attenuated in E266I cells compared with WT cells (Fig. 1 C). With PDGF, pHi increases in WT cells from 7.15 ± 0.03 at quiescence to 7.47 ± 0.05, but pHi in E266I cells does not change and is 7.03 ± 0.05 in the absence and presence of PDGF (Frantz et al., 2007). Like confluent cells, the abundance of F-actin in quiescent E266I cells was similar to WT cells. However, with 50 ng/ml PDGF, total F-actin increased significantly in WT cells at 1 min (P < 0.05), 3 min (P < 0.05), and 6 min (P < 0.01) but not in E266I cells (P > 0.1 at 1, 3, and 6 min; Fig. 1 C). These data suggest that H+ efflux by NHE1 is not necessary for steady-state F-actin abundance but is required for a rapid increase in actin filament assembly in response to migratory cues.
Figure 1.
H+ efflux by NHE1 is necessary for de novo actin filament assembly. (A) Cortical F-actin at the leading edge of migrating WT and E266I cells after wounding of a confluent monolayer. At the indicated times, cells were fixed and labeled with rhodamine-phalloidin, and F-actin at the leading-edge was quantified in acquired images by using NIH Image. Data are expressed relative to the abundance of F-actin in WT cells immediately after wounding and represent means ± SEM of 40 to 50 cells for each condition analyzed in two cell preparations. Significant differences in F-actin abundance in WT and E266I cells are indicated for each time point. (B) Representative images of WT and E266I cells labeled with rhodamine-phalloidin at the indicated times after wounding. Bar, 7.5 μm. (C) Total F-actin in WT and E266I cells in the absence and presence of 50 ng/ml PDGF for the indicated times. Data are expressed relative to the abundance of F-actin in quiescent WT cells (t = 0) and represent means ± SEM of three separate cell preparations.
H+ efflux by NHE1 is necessary for biphasic actin free barbed end formation
The assembly of new actin filaments is enhanced by severing of existing filaments to increase the abundance of actin free barbed ends (Falet et al., 2002; Mouneimne et al., 2004; Carlsson, 2006). In motile epithelial cells (Mouneimne et al., 2004), macrophages (Cox et al., 1996), and Dictyostelium cells (Hall et al., 1989), the generation of new free barbed ends is biphasic. WT fibroblasts treated with PDGF also had a biphasic increase in the number of actin free barbed ends (Fig. 2, A and B). A rapid and transient first phase was maximal at 1 min, with a twofold increase, and returned to near control levels at 2 min. A second phase included a maximal increase at 3 min that was smaller than the first phase, and at 4 min, the number of actin free barbed ends was at control levels. The maximal increase in free barbed end formation was at ∼0.5 μm of the submembranous region (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200804161/DC1). In quiescent cells, the number of actin free barbed ends was similar in WT and E266I cells. However, in E266I cells, there was no increase in free barbed ends with PDGF, and at 1 and 3 min, the abundance of free barbed ends was significantly less than in WT cells (P < 0.001 and P < 0.01, respectively; Fig. 2, A and B). Hence, H+ efflux is not necessary for the number of actin free barbed ends at steady state but is necessary for increased formation in response to PDGF.
Figure 2.
H+ efflux is necessary for biphasic actin free barbed end formation. (A) Actin free barbed ends in the zone between 0 and 0.66 μm inside the plasma membrane in the absence and presence of PDGF for the indicated times. Data are expressed relative to the number of free barbed ends in quiescent WT cells (t = 0; arbitrary units of fluorescence intensity) and represent means ± SEM of four cell preparations, with at least 15 cells scored at each time point for each cell preparation. The relative free barbed end number was significantly greater in WT cells compared with E266I cells at 30 and 90 s (P < 0.001) and at 180 s (P < 0.01). (B) Representative images of WT and E266I in the absence (0 min) and presence of PDGF (1 and 3 min) used for measuring actin free barbed ends. Bar, 10 μm. (C) Steady-state pHi in SN1 cells maintained for 4 h in glutamine-free DME supplemented with the indicated concentrations of glutamine. Data represent means ± SEM of three cell preparations, with significant differences in glutamine-dependent pHi indicated. (D) Relative barbed end number in SN1 cells maintained in the indicated concentrations of glutamine in the absence and presence of PDGF. Data represent the means ± SEM of three cell preparations of SN1 cells maintained in 400 and 25 μM glutamine, and the mean of two cell preparations of SN1 cells maintained in 1 μM glutamine. Indicated are significant differences in PDGF- and glutamine-dependent increases in free barbed ends.
We also found a pH-dependent regulation of actin free barbed ends in NHE1-deficient PS120 cells stably expressing the system N1 transporter (SN1 cells). SN1 is a plasma membrane amino acid transporter expressed in the central nervous system that couples uptake of extracellular glutamine with efflux of intracellular H+ efflux. When expressed in PS120 cells, SN1 is uniformly localized along the plasma membrane, and H+ efflux is dependent on the concentration of extracellular glutamine (Chaudhry et al., 1999). Although short-term (4 h) incubation with extracellular glutamine between 1 and 400 μM does not change the pHi of WT cells (Srivastava et al., 2008), it significantly increased the pHi of SN1 cells (Fig. 2 C). With 1 μM glutamine, pHi was 7.06 ± 0.03 and similar to E266I cells; with 25 μM glutamine, pHi was 7.18 ± 0.02 and similar to quiescent WT cells; and with 400 μM glutamine, pHi was 7.38 ± 0.04 and similar to PDGF-stimulated WT cells. In three SN1 cell preparations, PDGF induced a biphasic increase in actin free barbed ends with 400 μM glutamine, with significant increases at 1 min (P > 0.001) and 3 min (P > 0.01); with 25 μM glutamine, there was a significant increase at 1 min (P > 0.05) but not at 3 min (Fig. 2 D). In two SN1 cell preparations incubated with 1 μM glutamine, there was no increase in free barbed ends with PDGF (Fig. 2 D). However, decreasing extracellular glutamine from 400 to 1 μM had no effect on the relative number of free barbed ends in the absence of PDGF. In addition to confirming pH dependence, data with SN1 cells indicate that increasing pHi from ∼7 to ∼7.4 is necessary but not sufficient in the absence of PDGF to increase actin free barbed end formation. Moreover, because SN1 is not clustered at lamellipodia like NHE1, these data suggest that the abundance of cortical free barbed ends can be regulated by global increases in pHi and that local changes may not be necessary.
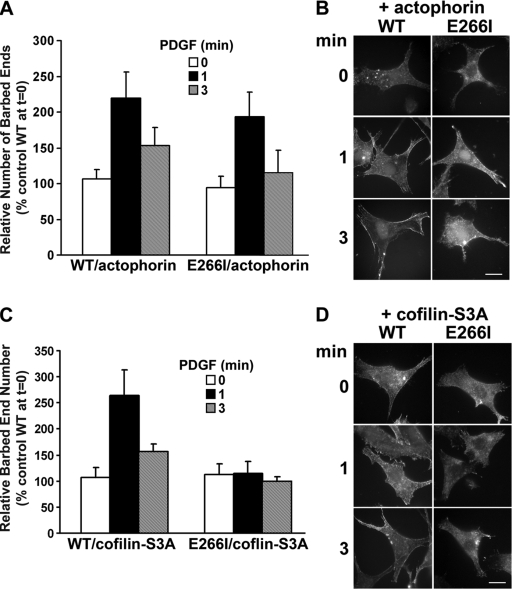
The first but not second phase of increased free barbed ends in epithelial cells treated with growth factors is generated predominantly by severing activity of cofilin (Mouneimne et al., 2004), which is recognized to be pH-dependent (Bamburg and Wiggan, 2002). Because the pHi of 7 in E266I cells treated with growth factors is significantly lower than the pHi of 7.5 in WT cells (Yan et al., 2001; Frantz et al., 2007), we speculated that cofilin-induced free barbed end formation might be inhibited at the lower pHi of E266I cells. Although most species of cofilin are activated by pH > 7, the activity of A. castellanii cofilin actophorin is pH-insensitive (Maciver et al., 1998). The number of free barbed ends in WT fibroblasts expressing actophorin was not significantly different in the absence of PDGF (10.42 ± 2.13 arbitrary units) compared with untransfected (control) WT cells (9.56 ± 1.85; Fig. 3, A and B; P > 0.5; n = 3). With PDGF, there was a twofold and 1.5-fold increase in barbed ends at 1 and 3 min, respectively, that was similar to increases in cells without actophorin (Fig. 3, A and B). In E266I cells, actophorin had no effect on the relative number of free barbed ends in the absence of PDGF (9.97 ± 1.90 and 8.42 ± 1.06 with and without actophorin, respectively) and nearly restored the number of free barbed ends in the first phase to values in WT cells (P > 0.1; E266I at 1 min compared with WT at 1 min, n = 3), but had no effect on attenuated free barbed end formation in the second phase (Fig. 3, A and B).
Figure 3.
Expression of actophorin but not cofilin-S3A restores the first phase of free barbed end formation in E266I cells. (A) Relative barbed end number in the absence and presence of PDGF for the indicated times in WT and E266I cells transiently expressing actophorin. Data are expressed relative to the abundance of barbed ends in quiescent untransfected WT cells and represent means ± SEM of three independent cell preparations, as described in Fig. 2. (B) Representative images of WT and E266I expressing actophorin used for measuring actin free barbed ends. (C) Relative barbed end number in the absence and presence of PDGF for the indicated times in WT and E266I cells transiently expressing cofilin-S3A. Data are expressed relative to the abundance of barbed ends in quiescent untransfected WT cells and represent means ± SEM of three cell preparations. (D) Representative images of WT and E266I expressing cofilin S3A used for measuring actin free barbed ends. Bars, 10 μm.
Cofilin activity also increases with dephosphorylation of Ser3 (Bamburg and Wiggan, 2002). Transient expression of human cofilin-S3A in WT cells had no effect on the number of free barbed ends in the absence of PDGF (11.15 ± 1.48 and 10.72 ± 0.89 with and without cofilin-S3A, respectively; P > 0.1; n = 3) and increased the number free barbed ends in the first phase but not the second phase compared with untransfected cells (Fig. 3, C and D). In E266I cells, cofilin-S3A also had no effect on the number of free barbed ends in the absence of PDGF or on attenuated first and second phases of free barbed end formation with PDGF (Fig. 3, C and D). Consistent with these findings, immunoblotting cell lysates indicated that total and phosphorylated cofilin in WT and E266I cells were similar in the absence of PDGF, and at 3 min of PDGF treatment, phosphorylated cofilin decreased ∼47% in WT cells and ∼50% in E266I cells (Fig. S1, B and C). Hence, H+ efflux by NHE1 is not necessary for cofilin expression or the regulated dephosphorylation of cofilin. Although cofilin-S3A has been found to be constitutively active in cells (Moriyama et al., 1996; Zebda et al., 2000; Ghosh et al., 2004), previous studies used cells with normal pHi homeostasis. Our data suggest a pH-dependent activation of cofilin for generating actin free barbed ends that is distinct from regulation by dephosphorylation of S3.
Structural models for regulation of cofilin by phosphorylation and by pH
To understand how phosphorylation and pH independently regulate cofilin-severing activity, we performed a series of molecular dynamics (MD) simulations. First, we performed explicit solvent MD on human cofilin, starting from the NMR structure (Pope et al., 2004), with Ser3 either phosphorylated or unphosphorylated. In the simulation of phosphorylated Ser3, the N-terminal portion interacted with the longest helix in the protein (α4), which forms part of the G-site implicated in binding to both G-actin and F-actin (Fig. 4 A, left). In particular, pSer3 forms salt-bridging interactions with Lys126 and Lys127 (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200804161/DC1). Other than this newly identified interaction, the overall structure of cofilin largely remains unperturbed. In contrast, the N terminus of unphosphorylated cofilin remained unstructured (Fig. 4 A, right), which is in agreement with the experimental structure. These results appear to be broadly consistent with existing functional data showing that phosphorylation of Ser3 abrogates binding of G-actin and with structural data from Pope et al. (2004) that indicate significant chemical shift perturbations in cofilin-S3D compared with WT cofilin. Gorbatyuk et al. (2006) also described chemical shift perturbations in the N terminus as well as α4 upon phosphorylation of chick cofilin by LIM kinase. These results are inconsistent, however, with data on actophorin (Blanchoin et al., 2000), where the phosphorylated residue is located at the immediate N terminus (Ser1). In that case, the protein structure was solved crystallographically in both the phosphorylated and unphosphorylated forms; pSer1 was not visualized in the electron density, and little conformational change was seen in the rest of the protein.
Figure 4.
Structural models of cofilin regulation. The structures shown are representative snapshots from MD simulations. (A, left) When Ser3 is phosphorylated, it is predicted to form strong ionic interactions with Lys126 and Lys127, which orders the N terminus and partially occludes the G-site for interacting with actin. (A, right) When Ser3 is not phosphorylated, the N terminus remains unstructured and flexible. (B) Varying pH between 6.5 and 8 does not introduce any major perturbations in the tertiary structure. However, His133 is predicted to be protonated at lower pH, forming a salt bridge with Asp98 (left). At a higher pH (8), His133 is unprotonated, and Asp98 instead interacts with Lys96 (right). At intermediate values of pH, the dynamics smoothly interpolate between these two extremes (not depicted).
To understand how increased pH might modulate cofilin activity independent of unphosphorylated Ser3, constant pH MD (CpHMD) simulations were performed on human cofilin at pH values between 6 and 8. The solution structure of human cofilin determined by NMR spectroscopy suggests that at pH 6, a solvent-exposed salt bridge forms between His133 and Asp98 (Pope et al., 2004). Our data corroborated this finding (Fig. 4 B, left) and also showed that at higher pH values, deprotonation of His133, the only histidine in human cofilin, weakens the interaction with Asp98, which instead tends to interact with Lys96 (Fig. 4 B, right). In the absence of a structure for cofilin bound to F-actin, the relationship of this pH-dependent conformational change to pH-dependent binding to F-actin is uncertain, but the His133 protonation state change and accompanying conformational changes could in principle modulate binding affinity at the F-site. Other conformational changes accompanying the change in pH are relatively minor, occurring primarily in the immediate vicinity of His133, as well as α1 and α4.
We also used NMR to monitor pH-dependent changes in cofilin. Comparison of 2D 15N heteronuclear single quantum coherence (HSQC) spectra of recombinant human cofilin at pH 6.5 and 7.5 showed chemical shift changes for residues in the vicinity of His133, which indicates changes in their chemical environments (Fig. S2 B). These findings are again consistent with chemical shift perturbations reported previously (Pope et al., 2004). Collectively, our simulation and NMR results, in combination with prior structural and biochemical data, support a model in which phosphorylation of Ser3 blocks binding to actin at the G-site, whereas protonation of His133 modulates binding at the F-site.
Cofilin H133A restores the first phase of free barbed end formation in E266I cells but retains pH dependence
On the basis of structural data, we speculated that deprotonation of His133 at the pHi of 7.5 in WT cells but not at the pHi of 7 in E266I cells may be necessary for the first phase of free barbed end formation with PDGF. Additionally, Pope et al. (2004) predict that His133 has an upshifted pKa of 7.4, although this could not be measured directly because of absence of signals from His133 in NMR spectra. We confirmed that expression of a mutant cofilin-H133A in E266I cells restored the first but not second phase of free barbed end formation. Compared with vector controls, WT cells expressing WT cofilin and cofilin-H133A had similar biphasic increases in the number of free barbed ends with PDGF, although the relative increase with WT cofilin was significantly greater than with cofilin-H133A (Fig. 5, A and B; P < 0.05; n = 3). Expressing WT cofilin in E266I cells did not restore free barbed end formation in either first or second phases, and the number of free barbed ends in both phases was not significantly different compared with quiescent cells (P > 0.1; n = 3; Fig. 5 A). However, in E266I cells expressing cofilin-H133A, the number of free barbed ends in the first phase was restored to that of WT cells expressing WT cofilin or cofilin-H133A, but the number of free barbed ends at quiescence and in the second phase were unchanged compared with untransfected cells (P > 0.1; n = 3). Additionally, cofilin-H133A but not WT cofilin partially restored an increase in cortical F-actin in E266I cells at the edge of a wounded monolayer (Fig. 5 C).
Figure 5.
Cofilin H133A restores the first phase of free barbed formation and de novo actin filament assembly in E266I cells. (A) Relative abundance of barbed ends in the absence and presence of PDGF for the indicated times in WT and E266I cells transiently expressing WT cofilin or cofilin-H133A. Data are expressed relative to the abundance of barbed ends in quiescent vector-transfected WT cells and represent means ± SEM of three independent transfections, with at least 15 cells scored at each time point for each cell preparation. (B) Representative images of WT and E266I expressing cofilin-H133A used for measuring actin free barbed ends. Bar, 10 μm. (C) Cortical F-actin at the leading-edge of wounded WT and E266I cells transfected with vector, WT cofilin, or cofilin-H133A. Cells were fixed at the indicated times and labeled with rhodamine-phalloidin, and F-actin at the leading-edge was quantified using National Institutes of Health Image. Data are expressed relative to the abundance of F-actin in vector-transfected WT cells immediately after wounding (initial) and represent means ± SEM of 20 to 30 cells for from two representative cell preparations.
NMR was used to follow pH-dependent changes in the chemical environments of residues in H133A cofilin. 2D 15N-HSQC spectra of H133A cofilin at pH 6.5 and 7.5 showed that the mutant adopts a fold very similar to WT (Fig. 6, A and B). MD simulations of cofilin-H133A are consistent with this observation, with major conformational differences largely confined to the flexible loops (Fig. 6 C). For H133A, similar pH-dependent chemical shift changes or broadening of NMR signals, as seen in the WT protein, were also observed for residues in the vicinity of Ala133 (His133). These changes indicate the presence of one or more residues, probably in addition to His133, that titrate in a physiological range.
Figure 6.
NMR spectra and severing activity for WT and H133A cofilin. (A) Comparison of 15N-HSQC spectra for the H133A mutant at pH 6.5 (red) and 7.5 (blue). Selected residue assignments are shown. These spectra show that peaks corresponding to several residues show chemical shift changes on changing the pH, which suggests that one or more side chains titrating in a physiological range are still present in the mutant. (B) Comparison of 15N-HSQC spectra for WT and H133A at pH 6 (selected residue assignments are shown). The H133A mutation introduces some structural changes but the protein remains well folded. (C) Superposition of MD snapshots for WT (green) and H133A (pink) cofilin at pH 6.5. (D, top) Quantification of F-actin severing by WT cofilin and cofilin H133A at indicated pH. Each bar represents the percent increase in the number of filaments 4 min after treatment with cofilin, expressed as means ± SEM. (bottom) Sample images of F-actin severing by WT cofilin and cofilin H133A at indicated pH. The filaments were observed before (as 0 min) and 4 min after incubation with cofilin. Bar, 2 μm.
The NMR results suggested that cofilin-H133A may have a similar pH-dependent activity toward F-actin compared with WT. To determine whether cofilin-H133A retains pH-sensitive F-actin severing activity similar to the WT protein, a light microscope severing assay was used that allows direct observation of severing in vitro (Ichetovkin, et al., 2000, 2002). The results clearly indicate that cofilin-H133A exhibits pH-sensitive severing of F-actin that is indistinguishable from WT (Fig. 6 D). H133A had minimal severing activity at pH 6 but a 25- to 30-fold increased activity at pH 7.
Deprotonation of His133 attenuates PI(4,5)P2 binding
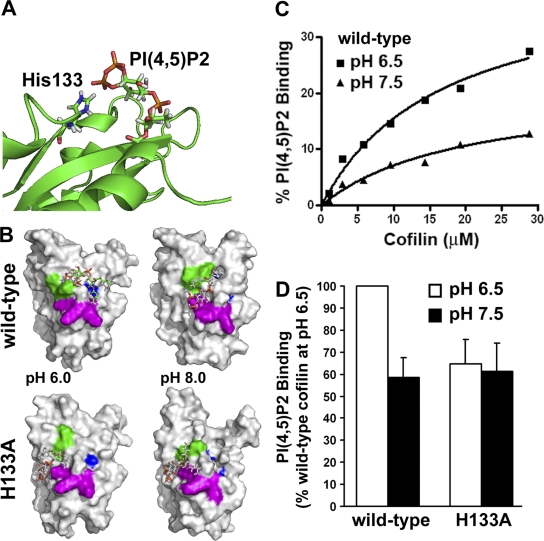
In vitro, PI(4,5)P2 and F-actin competitively bind to cofilin (Yonezawa et al., 1990) and actophorin (Van Troys et al., 2000). In epithelial cells, activation of phospholipase Cγ, which hydrolyzes PI(4,5)P2, is necessary for the cofilin-dependent first phase of actin free barbed end formation (Mouneimne et al., 2004), and in fibroblasts, PDGF induces an increase in phospholipase Cγ activity (Margolis et al., 1990) and hydrolysis of PI(4,5)P2 (McNamee et al., 1993). In cells, Förster resonance energy transfer analysis of the interaction between cofilin and PI(4,5)P2 demonstrates that cofilin is bound to PI(4,5)P2 and is released and activated in response to EGF stimulation by PI(4,5)P2 hydrolysis (van Rheenen et al., 2007). Although NMR experiments implicate His133 in binding the PI(4,5)P2 head group (Gorbatyuk et al., 2006), whether the protonation state of His133 regulates PI(4,5)P2 binding has not been reported. We used computational docking experiments to suggest a plausible model of interaction between PI(4,5)P2 and cofilin. When the head group of PI(4,5)P2 is docked to this site with His133 doubly protonated, the terminal phosphates interact closely with the His133 side chain (Fig. 7 A). When His133 is neutral, the head group does not dock in this pose and instead interacts with Lys125 (Fig. S3 B). This residue is part of a small group of residues (Ile124-Lys125-Lys126-Lys127-Leu128-Thr129) that had observed NMR spectral perturbations when bound to PI(4,5)P2 for a K132A/H133A double mutant (Gorbatyuk et al., 2006). The three cationic residues (Lys125, Lys126, and Lys127) may represent a secondary interaction site for the PI(4,5)P2 head group. Similarly, PI(4,5)P2 binds to yeast cofilin, where Lys132 and His133 are absent, but residues Arg109 and Arg110, which are equivalent to Lys125 and Lys126, are important for binding PI(4,5)P2 (Ojala et al., 2001). These results, in combination with the previous NMR studies with short-chain PI(4,5)P2 constructs (Gorbatyuk et al., 2006), suggest that deprotonation of His133 might decrease binding but not abolish it entirely.
Figure 7.
Binding of PI(4,5)P2 to cofilin is pH dependent and mediated by His133 protonation. (A) Docking model of potential mode of interaction between the PI(4,5)P2 head group and cofilin, when His133 is doubly protonated. (B) Docking models for PI(4,5)P2 and WT (top) and H133A (bottom) cofilin at pH 6.5 and 8. Residues Lys132 and His133 are shown in green; residues Lys125, Lys126, and Lys127 are shown in magenta, considered an alternative PIP2 binding site; and residues Phe15 and Leu99 are shown in blue. (C) The percentage of specific binding of lipid micelles containing 20 μM PI(4,5)P2 to the indicated concentrations of WT cofilin at pH 6.5 and 7.5. The abundance of cofilin in centrifuged pellets was normalized to nonspecific binding with lipid micelles in the absence of PI(4,5)P2 and calculated relative to the sum of cofilin in supernatant and pellet. Data are representative of two binding preparations. Kd values were relatively pH-independent but Bmax was twofold greater at pH 6.5 compared with pH 7.5. (D) Binding of lipid micelles containing 20 μM PI(4,5)P2 to 3 μM WT cofilin or cofilin-H133A at the indicated pH values. Data are expressed as percent of PI(4,5)P2 binding at pH 6.5 and represent means ± SEM of three binding preparations.
We used liposome sedimentation with purified recombinant rat cofilin to show pH-dependent PI(4,5)P2 binding to WT cofilin but not cofilin-H133A (Fig. 7, B and C). For WT cofilin, the dissociation constant (Kd) at pH 6.5 was 14.87 μM and relatively similar to that of 19.87 μM at pH 7.5. However, maximum specific binding (Bmax) was 2.5-fold greater at pH 6.5 (32.94%) compared with pH 7.5 (12.87%). Consistent with our docking model, which indicates that His133 interacts with the head group of PI(4,5)P2, PI(4,5)P2 binding by cofilin-H133A at pH 6.5 was not significantly different compared with binding by WT cofilin at pH 7.5, and was pH-insensitive (Fig. 7 D). For H133A, Bmax was 21.65% and 18.13% at pH 6.5 and 7.5, respectively, although Kd values were relatively similar, with 21.65 μM at pH 6.5 and 18.13 μM at pH 7.5. These data indicate that higher maximum PI(4,5)P2 binding is conferred by protonation of His133, and they suggest that pH-dependent cofilin activity in cells is determined in part by PI(4,5)P2 binding.
Discussion
Increased pHi is an evolutionarily conserved but poorly understood mechanism promoting cytoskeleton assemblies and cell movement. Earlier work on the fertilization of sea urchin eggs (Begg and Rebhun, 1979), the acrosomal reaction in echinoderm sperm (Tilney et al., 1978), and the motility of nematode sperm cells (King et al., 1994; Italiano et al., 1999) suggests that transient increases in pHi are necessary for de novo assembly of cytoskeletal filaments. A necessary role for increased pHi in directed migration has been shown in Dictyostelium cells (Van Duijn and Inouye, 1991) and in different mammalian cell types (Ritter et al., 1998; Klein et al., 2000; Reshkin et al., 2000; Denker and Barber, 2002; Stock et al., 2005). In growing plant pollen tubes, increased pHi at the distal cortex also promotes F-actin assembly at the tube tip (Lovy-Wheeler et al., 2006). Our understanding of how pHi regulates cytoskeleton dynamics and cell movement, however, is limited.
Our current data indicate that H+ efflux by NHE1 is necessary for increased assembly of actin filaments in response to monolayer wounding and growth factors. We previously found that increased actin filament assembly in response to chemoattractant is also attenuated in Ddnhe1-null Dictyostelium cells (Patel and Barber, 2005). In activated platelets (Falet et al., 2005) and in motile neutrophils (Norgauer et al., 1994), epithelial cells (Chan et al., 1998), macrophages (Cox et al., 1996), and Dictyostelium cells (Hall et al., 1989; Chen et al., 2003; Postma et al., 2003), de novo actin filament assembly is biphasic, although the regulation and function of each phase is not completely understood. The rapid and transient first phase is necessary for spatially localizing where membrane protrusions occur in tumor cells (Mouneimne et al., 2006), although this has not been experimentally confirmed in other cell types. In Dictyostelium cells, the first phase of F-actin assembly is attenuated but not eliminated by inhibition of Rac1B (Park et al., 2004), RasG (Sasaki et al., 2004), and DdNHE1 (Patel and Barber, 2005). The second phase is thought to drive membrane protrusion. In Dictyostelium cells, the second phase is also dependent on DdNHE1 and on increased abundance of PI(3,4,5)P3 at the cell front, which is regulated by phosphoinositide 3-kinases (PI3-kinases) and the PI3-phosphatase PTEN. Inhibiting PI3-kinases suppresses the second phase of actin polymerization, but the first phase is retained (Funamoto et al., 2001; Chen et al., 2003). However, several findings challenge the role of increased PI(3,4,5)P3 in regulating actin kinetics. First, a gene knockout strain in Dictyostelium lacking PTEN and all five type-1 PI3-kinases retains actin assembly (Hoeller and Kay, 2007). Second, a redundant phospholipase A2 pathway regulates actin kinetics independent of PI3-kinase activity (Chen et al., 2007; van Haastert et al., 2007). Third, mutant cells that lack a second phase of actin kinetics have decreased (Denker and Barber, 2002) or increased (Patel and Barber, 2005) PI(3,4,5)P3.
The generation of new free barbed ends is necessary for increased actin filament assembly (Condeelis, 2001; Falet et al., 2002; Carlsson, 2006) and has biphasic kinetics in response to migratory cues (Mouneimne et al., 2004). We used different fibroblast models expressing an inactive NHE1-E266I or the SN1 glutamine-H+ transporter to show that H+ efflux is necessary for both phases of free barbed end formation in fibroblasts, with the first but not the second phase being dependent on pH-sensitive cofilin activity. In epithelial cells, cofilin activity is also necessary for the first phase of free barbed end formation, but the second phase is dependent on PI3-kinase activity (Mouneimne et al., 2004), like the second phase of actin assembly in amoeboid cells. Although migrating E266I cells have decreased and mislocalized PI(3,4,5)P3 (Denker and Barber, 2002), whether this contributes to the absence of the second phase of free barbed end formation is uncertain. In Ddnhe1-null Dictyostelium cells, which lack a second phase of actin filament assembly, the abundance of PI(3,4,5)P3 is increased, which suggests that NHE1 regulation of the second phase may be mediated by mechanisms independent of PI(3,4,5)P3.
The ability of cofilin-H133A but not cofilin-S3A to restore the first phase of actin free barbed end formation in E266I cells indicates the importance of coincidence regulation for cofilin activity that requires S3 dephosphorylation and increased pHi. Coincidence regulation has also been suggested by previous work on the solution structure of cofilin (Pope et al., 2004) and by our computational modeling and NMR data. These data indicate distinct mechanisms for modulating actin binding that are determined by Ser3 dephosphorylation in the N terminus and by pH sensing in the C terminus. Changes in pH have little effect on the overall structure and dynamics of cofilin. Rather, lowered pH likely inhibits interactions with actin at the F-site by directly modulating the binding affinity at that site through the change in His133 charge, and through localized conformational changes. In contrast, we propose that phosphorylation of Ser3 inhibits binding to the G-site by steric occlusion, specifically by ionic interactions between pSer3 and Lys126, and Lys127. Little conformational change is predicted in the remainder of the structure, which is consistent with previous NMR work on human cofilin-S3D (Pope et al., 2004) and chick cofilin-pSer3 (Gorbatyuk et al., 2006).
Coincidence regulation by protons suggests that global changes in pHi may be sufficient for spatially restricted regulation, and that local pHi changes may not be necessary. Our data with SN1 cells support this possibility because SN1 is not localized at membrane protrusions like NHE1. In activated fibroblasts (Dawe et al., 2003) and platelets (Falet et al., 2005), dephosphorylated cofilin is restricted to the distal cortex, which would spatially limit a second activation step by increased pHi. Additionally, because PI(4,5)P2 and F-actin competitively bind cofilin (Van Troys et al., 2000; Yonezawa et al., 1990), dephosphorylated cofilin at the plasma membrane would be inactive when bound to PI(4,5)P2. Moreover, cofilin can only bind to tropomyosin-free actin filaments, which are only present at the plasma membrane interface with the cytoplasm, further restricting the location of F-actin severing by cofilin (DesMarais et al., 2002).
Our data support a mechanism whereby pH-dependent cofilin activity in cells is determined primarily by regulating cofilin binding to PI(4,5)P2. We predict that, at the lower pHi of resting cells, cofilin activity is inhibited by greater maximum binding to PI(4,5)P2. Increased pHi with migratory cues lowers the abundance of PI(4,5)P2 bound to cofilin, and, with dephosphorylation of S3, increases cofilin activity. Our finding that protonation of cofilin His133 increases PI(4,5)P2 binding is significant for two reasons. First, for activation of cofilin, it suggests that increased pH and release of PI(4,5)P2 are biochemically linked. Although pH and PI(4,5)P2 are recognized regulators of cofilin activity (Bailly and Jones, 2003), pH-dependent binding of PI(4,5)P2 to cofilin has not been shown. However, we speculate that pH-dependent release of PI(4,5)P2 would not be sufficient as a coincidence activator of cofilin without dephosphorylation of S3. In SN1 cells, increased actin free barbed ends required a higher pH and growth factor.
Second, of general significance is an emerging theme of pH-dependent phosphoinositide binding at sites containing histidine residues. Phosphoinositide binding to FYVE domains is pH-sensitive and regulated by a histidine switch (Lee et al., 2005). Additionally, we recently showed pH-sensitive PI(4,5)P2 binding to the guanine nucleotide exchange factor Dbs (Dbl's big sister), which contains a His (H843) in the predicted phosphoinositide-binding site (Frantz et al., 2007). In contrast, PI(4,5)P2 binding to the guanine nucleotide exchange factor intersectin, which binds phosphoinositides at a charged Lys/Arg cluster, is pH-insensitive. The PI(4,5)P2 binding site in actophorin lacks a His (Van Troys et al., 2000), which could contribute to the pH-insensitive activity of A. castellanii cofilin. A review of phosphoinositide-binding domains in the PDB indicates that although binding sites containing charged Lys/Arg-rich clusters are the most prevalent, many of these domains contain a His within predicted phosphoinositide-binding sites. As recently suggested (Gorbatyuk et al., 2006), phosphoinositide binding is a likely negative regulator of most proteins that cap or sever actin filaments. We predict that if phosphoinositides bind at His residues in these proteins, binding and protein function may be pH-sensitive. Additionally, the hydrolysis of PI(4,5)P2 by PLC, which is required for the full release and activation of cofilin in vivo (van Rheenen et al., 2007), would be facilitated by the weakened binding of cofilin to PIP2 resulting from a local increase in pH in the presence of activated NHE1.
Materials and methods
Cell culture
NHE1-deficient PS120 fibroblasts stably expressing WT NHE1 (WT cells), or a mutant NHE1 lacking ion translocation (E266I cells; Denker et al., 2000), or stably expressing SN1 (Chaudhry et al., 1999) have been described previously. Cells were maintained in DME-H21 medium supplemented with 5% FBS and penicillin-streptomycin (growth medium) at 5% CO2. For experiments with wounded cells, fibroblasts plated on glass coverslips were grown to confluence, wounded with a P1000 pipette tip, and maintained at 5% CO2 in growth medium for 15 min. For experiments with subconfluent cells, cells were plated on glass coverslips at ∼75% confluence, maintained for 24 h in DME containing 0.2% FBS (quiescent cells), and treated for the indicated times with 50 ng/ml PDGF-BB (Roche). For expression of WT and mutant cofilin or actophorin, cells were cotransfected with cherry-red histone pJAG 285 by electroporation (Amaxa Biosystems Nucleofector kit; Lonza), plated on glass coverslips in growth medium for 24 h, then transferred to DME containing 0.2% FBS for 24 h before using. SN1 cells were maintained in glutamine-free DME in the absence of FBS and supplemented with the indicated concentrations of glutamine 4 h before use.
F-actin abundance
Wounded monolayers plated on glass coverslips were fixed in 3.7% formaldehyde for 20 min, permeabilized in 0.1% Triton X-100, incubated with rhodamine-phalloidin (1:500; Invitrogen) in PBS for 30 min, and processed for imaging with mounting medium containing 50% glycerol and N-propyl gallate. Images were collected at room temperature using a microscope (Axiophot; Carl Zeiss, Inc.) and a Plan-neofluar 100× objective with 1.3 numerical aperture (Carl Zeiss, Inc.) adapted with a computer-driven cooled charge-coupled device Spot camera (RT slider model 2.3.0) and Spot advanced 4.1.5 acquisition software (Diagnostic Instruments, Inc.). Images were acquired in the linear range of the detector's response at a sensitivity such that none of the pixels in the image were saturated. F-actin at the leading edge was quantified using National Institutes of Health (NIH) Image software. For each cell, a line perpendicular to the wound edge was drawn bisecting the nucleus and extending to the leading-edge membrane. Fluorescence intensity was determined in an area from the membrane to 5 μm within the cortex, and four pixels to the left and right of the perpendicular line. For total F-actin, untransfected subconfluent cells in the absence and presence of PDGF were fixed and labeled with rhodamine-phalloidin as described previously. After washing, the rhodamine dye was extracted in methanol at −20°C, and fluorescence intensity was measured at Ex 554 and Em 575 using a SpectraMax M5 (MDS Analytical Devices). Cortical F-actin in transfected cells was determined as described for wound-edge cells.
Actin free barbed ends assay
The number of actin free barbed ends was determined by a modification of previously described methods (Chan et al., 1998). Quiescent cells plated in MatTek tissues culture dishes (MatTek Corporation) were treated with 50 ng/ml PDGF for the indicated times and permeabilized in buffer containing biotin–G-actin. 20 μg biotin–G-actin (Cytoskeleton, Inc.) in 100 μl of dilution buffer (1 mM Hepes, pH 7.5, 0.2 mM MgCL2, and 0.2 mM ATP) was centrifuged at 100,000 g for 20 min to remove aggregates. The mixture was diluted to a final concentration of 0.02 g/liter in permeabilizing buffer (5 mM KCl, 137 mM NaCl, 4 mM NaHCO3, 0.4 mM KH2PO4, 1.1 mM Na2HPO4, 2 mM MgCl2, 5 mM Pipes, pH 7.2, 2 mM EGTA, and 5.5 mM glucose) containing 0.04 g/liter saponin and 1% BSA. After permeabilizing cells for 45 s at 37°C in the presence of biotin–G-actin, the reaction was stopped by adding permeabilizing buffer without saponin and BSA, and cells were fixed for 5 min with 3.7% formaldehyde in permeabilizing buffer. Cells were then incubated for 10 min with permeabilizing buffer containing 0.1 M glycine; blocked for 20 min in TBS buffer containing 1% BSA, 1% FBS, and 5 μg/ml phalloidin; and incubated for 1 h with FITC- or rhodamine-conjugated anti-biotin antibody (1:50; Jackson ImmunoResearch Laboratories) and washed with TBS buffer containing 1% BSA. Coverslips from MatTek dishes were mounted with 0.1 M N-propyl gallate in 50% glycerol in TBS, pH 7.5, for imaging. Images for all experiments were collected using identical settings within the linear range of the detector's response and at a sensitivity such that none of the pixels in the image were saturated. Digital images were then linearly converted in NIH Image and analyzed using macro analysis as described previously (Mouneimne et al., 2004). In brief, the software averages the fluorescence intensity in 29 consecutive annuli of 0.22 μm, beginning 1.1 μm outside the cell periphery and extending inside the cell. Fluorescence intensity versus distance from the cell periphery was used to determine the number of cortical free barbed ends. Mean intensities of three annuli (0–0.22, 0.22–0.44, and 0.44–0.66 μm inside the cell were used to determine the number of free barbed ends.
pHi
pHi was determined in cells loaded with the fluorescent H+-sensitive dye 2,7-biscarboxyethyl-5(6)-carboxyfluorescein (BCECF; Invitrogen) as described previously (Denker et al., 2000). Ratios of BCECF fluorescence at Ex 490/Em 530 and Ex 440/Em 530 were converted to pHi by calibrating with 10 μM nigericin in 105 mM KCl.
Cofilin immunoblotting
Quiescent cells in 6-well plates were treated with 50 ng/ml PDGF for the indicated times and lysed in a modified RIPA buffer (50 mM Tris-HCl, 135 mM NaCl, 3 mM KCl, 1% NP-40, protease inhibitors, 1 mM EGTA, 5 mM NaF, 10 mM sodium pyrophosphate, and 1 mM sodium vanadate, pH 7.4), then lysates were centrifuged at 850 g for 5 min to obtain a postnuclear supernatant. Equal amounts of protein were separated by SDS-PAGE, transferred to PVDF membranes, and immunoblotted with antibodies to cofilin or phospho-cofilin (1:1,000; Cell Signaling Technology), or β-actin (C4; 1:10,000; Millipore). Immunoblots were analyzed by using NIH Image.
Recombinant cofilin purification
Mutant cofilin-H133A and cofilin-S3A were generated using the QuickChange Site-Directed Mutagenesis kit (Agilent Technologies) by using the following primers: forward 5′–3′: GAAACTGACAGGAATCAAGGCCGAATTACAAGCTAACTGC; reverse 5′–3′: GCAGTTAGCTTGTAATTCGGCCTTGATTCCTGTCAGTTTC. A C-terminal Myc tag was added by PCR using the following primers: forward 5′–3′: ATGGCCTCTGGTGTGGCTG for WT cofilin and ATGGCCGCTGGTGTGGCTG for S3A cofilin; reverse 5′–3′: TCACAGATCTTCTTCTGAGATGAGTTTTTGTTCCAAAGGCTTGCCCTCCAGGGA.
DNA subcloned into pET15b (EMD) was used to transform Rosetta BL21 DE3 bacteria cells, which were grown in Luria broth (LB) media with 100 μg/ml ampicillin and 35 μg/ml chloramphenicol at 30°C. For 15N-labeled protein, LB was replaced by minimal medium at pH 7.4 (1× M9 salts, 2 mM MgSO4, 100 μM CaCl2, 1× MEM vitamin mix [Invitrogen], 1 g/liter 15N-H4Cl [Sigma-Aldrich], 1 g/liter Isogrow 15N13C [Sigma-Aldrich], 0.5 mg/liter biotin, and 20 mg/liter FeCl2). Cells were grown at 30°C, and protein expression was induced by the addition of 1 mM IPTG. Cells were collected by centrifugation, suspended in buffer A (50 mM NaH2PO4, pH 8, 300 mM NaCl, and 5 mM imidazole, pH 8), and lysed using a microfluidizer (Microfluidics). Lysates were applied to Talon resin columns (BD), columns were washed in buffer A with 20 mM imidazole, and protein was eluted with buffer A containing 300 mM imidazole in fractions of 0.5 ml. 20 μl of each fraction was separated by SDS-PAGE and visualized by Coomassie staining, and fractions containing cofilin were pooled and buffer exchanged by dialysis against thrombin cleavage buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 2.5 mM CaCl2). The His tag of purified cofilin was cleaved using the Thrombin Cleavage Capture kit (EMD). Buffer was then exchange by dialysis to NMR storage buffer (10 mM sodium phosphate buffer, 25 mM NaCl, 1 mM DTT, 0.2 mM EGTA, and 1 mM NaN3, pH 6.0) or to freeze storage buffer (10 mM Tris, pH 7.5, 1 mM NaN3, 5 mM DTT, and 10% glycerol) for storage at −80°C. Protein was quantified by SDS-PAGE or on a spectrofluorometer.
MD simulations
To study the effects of phosphorylation, we used explicit solvent MD. The 20 initial structures were taken from the NMR structure 1Q8G from the Protein Data Bank. For each one of these 20 structures of the cofilin, we performed two different simulations: with Ser3 unphosphorylated and with Ser3 phosphorylated. The calculations were done with GROMACS (version 3.2.1; GNU General Public License) using the GROMOS96 force field for pSer. All systems were solvated with the SPC water model in a periodic cubic cell. The solvated systems were subject to 2,500 steps of steepest descent minimization to prepare the system for the MD simulations. The equilibration of the systems was done in two parts: the first part consisted of 50 ps of dynamics in which the temperature was raised from 0 to 300 K. The second part of the equilibration consisted of 1 ns with an NPT ensemble. Once the systems were equilibrated, we calculated one additional nanosecond in the NPT ensemble at 300 K using a thermostat (Berendsen) and particle mesh Ewald (PME) for the Coulombic interactions. The results from the 20 simulations starting from different NMR models were very similar, and we show one representative result.
Constant pH MD simulations were performed using the AMBER 8 suite of programs (Case et al., 2005). The AMBER parm99 force field (Wang et al., 2000) and generalized Born solvation model (Onufriev et al., 2004) were used. First, the system was energy minimized using six consecutive rounds of 800 steps of the steepest descent algorithm followed by 1,200 steps of the conjugate gradient algorithm, giving a total of 12,000 steps. Harmonic restraints applied to the α carbons were slowly relaxed from 25 to 1 kcal/mol/Å2 by the end of the energy minimization step. To save computational time, a 15-Å cutoff for nonbonded interactions was used. The equilibration period in the MD simulations consisted of three stages. In the first one, the system was gradually heated from 100 K to 300 K for 30 ps at 100 K intervals, followed by 70 ps at 300 K. The remaining restraints were gradually reduced to zero in this stage. The second stage consisted of 50 ps of unrestrained equilibration. Finally, in the third stage, the constant pH MD (CpHMD) approach was applied (Mongan et al., 2004). The method implemented in AMBER addresses the dynamic dependence of pKa through Monte Carlo sampling of the Boltzmann distribution of protonation states concurrent with the MD simulations. The nature of the distribution is affected by the solvent pH, which is set as an external parameter. In the final equilibration stage, the pH variable was defined and the system was submitted to an MD run of 20 ps. The salt concentration was set at 0.1 M. The MD run was then continued for 10 ns at conditions of low (6.5) and high pH (8). Solute temperature was weakly coupled to a Berendsen temperature bath at 300 K, with a time constant of 2 ps. Bond lengths including hydrogens were constrained using the SHAKE algorithm. The time step was 1 fs. The center-of-mass motion was removed at regular intervals of 1,000 fs. The trajectories were saved every 1 ps. In the MD simulations, the cutoff for nonbonded interactions was 12 Å. Analyses were performed primarily with the ptraj program (AMBER; distributed by the University of California, San Francisco).
Docking
The NMR structures for WT cofilin (protein database ID No. 1Q8G, Model 1) and its H133A mutant were used in the docking studies of PI(4,5)P2. To generate the structure for the mutant, His133 in the NMR Model 1 was substituted by an alanine residue using Maestro (Schrödinger, LLC). The docking calculations were performed using the Induced Fit protocol (Sherman et al., 2006), a combination of Glide (Friesner et al., 2004; Halgren et al., 2004) and Prime (Jacobson et al., 2004) that accounts for both ligand and receptor side chain flexibility. Before the docking calculations, both WT and mutant proteins were submitted to a series of restrained, partial minimizations using the OPLS_2005 force field (Jorgensen et al., 1996; Kaminski et al., 2001). PI(4,5)P2 was submitted to a preminimization with the MMFF94 force field using a “4r” distance-dependent dielectric constant.
NMR
Experiments were performed on Avance DRX 500 and Avance 800 MHz spectrometers (Bruker) at 300 K. Data were processed using TopSpin 2.0 software (Bruker). Spectra were analyzed using ANSIG (Kraulis et al., 1994). Resonance assignments were transferred to spectra using values deposited in the Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu/) under accession code BMRB-6004 (Zierler-Gould et al., 2004).
F-actin severing assay
The ability of cofilin to sever F-actin was directly observed using an in vitro severing assay described previously (Ichetovkin et al., 2000; Ichetovkin et al., 2002). In brief, 2 μM actin containing 20% rhodamine-labeled and 10% biotin-labeled actin were polymerized at room temperature for 2 h in ISAP buffer (20 mM Pipes, pH 7, 1 mM ATP, 1 mM DTT, 50 mM KCl, 5 mM EGTA, and 2 mM MgCl2) containing 0.2 μM phalloidin. Coverslips (50, 3, and 24 mm, no. 1.5) were coated with 15 ml of 0.1% nitrocellulose in amyl acetate (Ernest F. Fullam, Inc.) and allowed to dry in a fume hood. Another coverslip (18 mm2, no. 1) was mounted onto the coated coverslip with two pieces of double sticky tape so as to form a perfusion chamber. The resulting chamber holds a volume of around 30 ml. Antibiotin antibody was diluted to 30 mg/ml with wash buffer (ISAP buffer containing 0.5 mg/ml BSA) perfused into the chamber, and incubated for 5 min before washing twice with wash buffer. Rhodamine/biotin-labeled F-actin was diluted 30-fold to 0.067 μM with antibleaching buffer A (ISAP containing 5 mg/ml BSA and antibleaching components: 0.036 mg/ml catalase, 0.2mg/ml glucose oxidase, 6 mg/ml glucose, and 100 mM DTT; Kron and Spudich, 1986; Kron et al., 1991) and perfused into the chamber. After a 5-min incubation, free actin was washed off with two 30-μl rinses of antibleaching buffer A and then with two 30-μl rinses of antibleaching buffer B (cofilin storage buffer containing 0.036 mg/ml catalase, 0.2 mg/ml glucose oxidase, 6 mg/ml glucose, and 100 mM DTT). Cofilin diluted in antibleaching buffer B to a final concentration of 5 or 16 μM was perfused into the chamber, and images of bound filaments were taken with a microscope (Olympus) equipped with a charge-coupled device camera and scored for severing as described previously (Ichetovkin et al., 2000).
Phospholipid binding
Lipid micelles were prepared as described previously (Lebensohn et al., 2006) by using a mini-extruder (Avanti Polar Lipids, Inc.), and they contained phosphatidyl ethanolamine/phosphatidyl choline/PI(4,5)P2 (71:25:4 molar ratio; Avanti Polar Lipids, Inc.). Vesicle suspensions were adjusted to the indicated pH with KOH or HCl. Quantitative binding and Kds were obtained by incubating 20 μM PI(4,5)P2 with the indicated concentrations of cofilin for 15 min at room temperature and then collected by centrifugation at 100,000 g for 60 min. Supernatants and pellets were analyzed by SDS-PAGE and Coomassie staining. The amount of protein on the gel was determined by densitometry analysis by using NIH Image. Specific binding was calculated as the abundance of protein bound to vesicles containing PI(4,5)P2 minus binding to vesicles in the absence of PI(4,5)P2. The abundance of protein bound to vesicles in the absence of PI(4,5)P2 was minimal and pH-independent. pH-dependent dissociation constants were calculated from transformations of binding curves using GraphPad Prism 5 software (GraphPad Prism). Binding at pH 6.5 and 7.5 was also determined by incubating 20 μM PI(4,5)P2 with 3 μM of recombinant WT cofilin or coflin-H133A. To correct for variations in lipid vesicle preparations, data were expressed relative to binding at pH 6.5 for each determination.
Statistical analysis
Data were analyzed with GraphPad Prism software using an unpaired t test with 95% confidence intervals. The sample size represented the number of separate cell preparations, except for analysis of F-actin after monolayer wounding, which included data obtained from 40–60 cells in two separate cell preparations.
Online supplemental material
Fig. S1 shows spatially localized cortical actin free barbed end formation and the time-dependent phosphorylation of cofilin in cells treated with PDGF. Fig. S2 shows predicted and measured structural changes in cofilin with increasing pH within the physiological range. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200804161/DC1.
Acknowledgments
This work was supported by National Institutes of Health grants GM38511 to J. Condeelis and GM58642 to D.L. Barber, by National Science Foundation grant no. MCB-0346399 to M.P. Jacobson, and by a grant from the Sandler Family Foundation to M.J.S. Kelly, M.P. Jacobson, and D.L. Barber. Work by C. Frantz and D.L. Barber was conducted in a facility constructed with support from Research Facilities Improvement Program grant no. C06 RR16490 from the National Center for Research Resources, National Institutes of Health. M.P. Jacobson is a consultant to Schrödinger, LLC.
Abbreviations used in this paper: ADF, actin-depolymerizing factor; HSQC, heteronuclear single quantum coherence; MD, molecular dynamics; NMR, nuclear magnetic resonance; pHi, intracellular pH; PI(4,5)P2, phosphotidylinositol-4,5-bisphosphate; PI3-kinase, phosphoinositide 3-kinase; WT, wild type.
References
- Bailly, M., and G.E. Jones. 2003. Polarised migration: cofilin holds the front. Curr. Biol. 13:R128–R130. [DOI] [PubMed] [Google Scholar]
- Bamburg, J.R., and O.P. Wiggan. 2002. ADF/cofilin and actin dynamics in disease. Trends Cell Biol. 12:598–605. [DOI] [PubMed] [Google Scholar]
- Begg, D.A., and L.I. Rebhun. 1979. pH regulates the polymerization of actin in the sea urchin egg cortex. J. Cell Biol. 83:241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, B.W., and J.R. Bamburg. 2004. A proposed mechanism for cell polarization with no external cues. Cell Motil. Cytoskeleton. 58:96–103. [DOI] [PubMed] [Google Scholar]
- Bernstein, B.W., W.B. Painter, H. Chen, L.S. Minamide, H. Abe, and J.R. Bamburg. 2000. Intracellular pH modulation of ADF/cofilin proteins. Cell Motil. Cytoskeleton. 47:319–336. [DOI] [PubMed] [Google Scholar]
- Blanchoin, L., T.D. Pollard, and R.D. Mullins. 2000. Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 10:1273–1282. [DOI] [PubMed] [Google Scholar]
- Carlsson, A.E. 2006. Stimulation of actin polymerization by filament severing. Biophys. J. 90:413–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case, D.A., T.E. Cheatham III, T. Darden, H. Gohlke, R. Luo, K.M. Merz Jr., A. Onufriev, C. Simmerling, B. Wang, and R.J. Woods. 2005. The Amber biomolecular simulation programs. J. Comput. Chem. 26:1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, A.Y., S. Raft, M. Bailly, J.B. Wyckoff, J.E. Segall, and J.S. Condeelis. 1998. EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J. Cell Sci. 111:199–211. [DOI] [PubMed] [Google Scholar]
- Chan, A.Y., M. Bailly, N. Zebda, J.E. Segall, and J.S. Condeelis. 2000. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J. Cell Biol. 148:531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry, F.A., R.J. Reimer, D. Krizaj, D. Barber, J. Storm-Mathisen, D.R. Copenhagen, and R.H. Edwards. 1999. Molecular analysis of system N suggests novel physiological roles in nitrogen metabolism and synaptic transmission. Cell. 99:769–780. [DOI] [PubMed] [Google Scholar]
- Chen, L., C. Janetopoulos, Y.E. Huang, M. Iijima, J. Borleis, and P.N. Devreotes. 2003. Two phases of actin polymerization display different dependencies on PI(3,4,5)P3 accumulation and have unique roles during chemotaxis. Mol. Biol. Cell. 14:5028–5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., B.W. Bernstein, J.M. Sneider, J.A. Boyle, L.S. Minamide, and J.R. Bamburg. 2004. In vitro activity differences between proteins of the ADF/cofilin family define two distinct subgroups. Biochemistry. 43:7127–7142. [DOI] [PubMed] [Google Scholar]
- Chen, L., M. Iijima, M. Tang, M.A. Landree, Y.E. Huang, Y. Xiong, P.A. Iglesias, and P.N. Devreotes. 2007. PLA2 and PI3K/PTEN pathways act in parallel to mediate chemotaxis. Dev. Cell. 12:603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis, J. 2001. How is actin polymerization nucleated in vivo? Trends Cell Biol. 11:288–293. [DOI] [PubMed] [Google Scholar]
- Cox, D., P. Chang, T. Kurosaki, and S. Greenberg. 1996. Syk tyrosine kinase is required for immunoreceptor tyrosine activation motif-dependent actin assembly. J. Biol. Chem. 271:16597–16602. [DOI] [PubMed] [Google Scholar]
- Dawe, H.R., L.S. Minamide, J.R. Bamburg, and L.P. Cramer. 2003. ADF/cofilin controls cell polarity during fibroblast migration. Curr. Biol. 13:252–257. [DOI] [PubMed] [Google Scholar]
- Denker, S.P., and D.L. Barber. 2002. Cell migration requires both ion translocation and cytoskeletal anchoring by the Na-H exchanger NHE1. J. Cell Biol. 159:1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denker, S.P., D.C. Huang, J. Orlowski, H. Furthmayr, and D.L. Barber. 2000. Direct binding of the Na–H exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H(+) translocation. Mol. Cell. 6:1425–1436. [DOI] [PubMed] [Google Scholar]
- DesMarais, V., I. Ichetovkin, J. Condeelis, and S.E. Hitchcock-DeGregori. 2002. Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. J. Cell Sci. 115:4649–4660. [DOI] [PubMed] [Google Scholar]
- Falet, H., K.M. Hoffmeister, R. Neujahr, J.E. Italiano Jr., T.P. Stossel, F.S. Southwick, and J.H. Hartwig. 2002. Importance of free actin filament barbed ends for Arp2/3 complex function in platelets and fibroblasts. Proc. Natl. Acad. Sci. USA. 99:16782–16787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falet, H., G. Chang, B. Brohard-Bohn, F. Rendu, and J.H. Hartwig. 2005. Integrin alpha(IIb)beta3 signals lead cofilin to accelerate platelet actin dynamics. Am. J. Physiol. Cell Physiol. 289:C819–C825. [DOI] [PubMed] [Google Scholar]
- Frantz, C., A. Karydis, P. Nalbant, K.M. Hahn, and D.L. Barber. 2007. Positive feedback between Cdc42 activity and H+ efflux by the Na-H exchanger NHE1 for polarity of migrating cells. J. Cell Biol. 179:403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner, R.A., J.L. Banks, R.B. Murphy, T.A. Halgren, J.J. Klicic, D.T. Mainz, M.P. Repasky, E.H. Knoll, M. Shelley, J.K. Perry, et al. 2004. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47:1739–1749. [DOI] [PubMed] [Google Scholar]
- Funamoto, S., K. Milan, R. Meili, and R.A. Firtel. 2001. Role of phosphatidylinositol 3′ kinase and a downstream pleckstrin homology domain-containing protein in controlling chemotaxis in Dictyostelium. J. Cell Biol. 153:795–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, M., X. Song, G. Mouneimne, M. Sidani, D.S. Lawrence, and J.S. Condeelis. 2004. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 304:743–746. [DOI] [PubMed] [Google Scholar]
- Gohla, A., J. Birkenfeld, and G.M. Bokoch. 2005. Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat. Cell Biol. 7:21–29. [DOI] [PubMed] [Google Scholar]
- Goley, E.D., and M.D. Welch. 2006. The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7:713–726. [DOI] [PubMed] [Google Scholar]
- Gorbatyuk, V.Y., N.J. Nosworthy, S.A. Robson, N.P. Bains, M.W. Maciejewski, C.G. Dos Remedios, and G.F. King. 2006. Mapping the phosphoinositide-binding site on chick cofilin explains how PIP2 regulates the cofilin-actin interaction. Mol. Cell. 24:511–522. [DOI] [PubMed] [Google Scholar]
- Halgren, T.A., R.B. Murphy, R.A. Friesner, H.S. Beard, L.L. Frye, W.T. Pollard, and J.L. Banks. 2004. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 47:1750–1759. [DOI] [PubMed] [Google Scholar]
- Hall, A.L., V. Warren, S. Dharmawardhane, and J. Condeelis. 1989. Identification of actin nucleation activity and polymerization inhibitor in ameboid cells: their regulation by chemotactic stimulation. J. Cell Biol. 109:2207–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins, M., B. Pope, S.K. Maciver, and A.G. Weeds. 1993. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry. 32:9985–9993. [DOI] [PubMed] [Google Scholar]
- Hoeller, O., and R.R. Kay. 2007. Chemotaxis in the absence of PIP3 gradients. Curr. Biol. 17:813–817. [DOI] [PubMed] [Google Scholar]
- Ichetovkin, I., J. Han, K.M. Pang, D.A. Knecht, and J.S. Condeelis. 2000. Actin filaments are severed by both native and recombinant dictyostelium cofilin but to different extents. Cell Motil. Cytoskeleton. 45:293–306. [DOI] [PubMed] [Google Scholar]
- Ichetovkin, I., W. Grant, and J. Condeelis. 2002. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 12:79–84. [DOI] [PubMed] [Google Scholar]
- Italiano, J.E. Jr., M. Stewart, and T.M. Roberts. 1999. Localized depolymerization of the major sperm protein cytoskeleton correlates with the forward movement of the cell body in the amoeboid movement of nematode sperm. J. Cell Biol. 146:1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, M.P., D.L. Pincus, C.S. Rapp, T.J. Day, B. Honig, D.E. Shaw, and R.A. Friesner. 2004. A hierarchical approach to all-atom protein loop prediction. Proteins. 55:351–367. [DOI] [PubMed] [Google Scholar]
- Jorgensen, W.L., D.S. Maxwell, and J. Tirado-Rives. 1996. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 118:11225–11236. [Google Scholar]
- Kaminski, G.A., R.A. Friesner, J. Tirado-Rives, and W.L. Jorgensen. 2001. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B. 105:6474–6487. [Google Scholar]
- King, K.L., J. Essig, T.M. Roberts, and T.S. Moerland. 1994. Regulation of the Ascaris major sperm protein (MSP) cytoskeleton by intracellular pH. Cell Motil. Cytoskeleton. 27:193–205. [DOI] [PubMed] [Google Scholar]
- Klein, M., P. Seeger, B. Schuricht, S.L. Alper, and A. Schwab. 2000. Polarization of Na(+)/H(+) and Cl(-)/HCO (3)(-) exchangers in migrating renal epithelial cells. J. Gen. Physiol. 115:599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis, P.J., P.J. Domaille, S.L. Campbell-Burk, T. Van Aken, and E.D. Laue. 1994. Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry. 33:3515–3531. [DOI] [PubMed] [Google Scholar]
- Kron, S.J., and J.A. Spudich. 1986. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc. Natl. Acad. Sci. USA. 83:6272–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron, S.J., Y.Y. Toyoshima, T.Q. Uyeda, and J.A. Spudich. 1991. Assays for actin sliding movement over myosin-coated surfaces. Methods Enzymol. 196:399–416. [DOI] [PubMed] [Google Scholar]
- Lebensohn, A.M., L. Ma, H.Y. Ho, and M.W. Kirschner. 2006. Cdc42 and PI(4,5)P2-induced actin assembly in Xenopus egg extracts. Methods Enzymol. 406:156–173. [DOI] [PubMed] [Google Scholar]
- Lee, S.A., R. Eyeson, M.L. Cheever, J. Geng, V.V. Verkhusha, C. Burd, M. Overduin, and T.G. Kutateladze. 2005. Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc. Natl. Acad. Sci. USA. 102:13052–13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovy-Wheeler, A., J.G. Kunkel, E.G. Allwood, P.J. Hussey, and P.K. Hepler. 2006. Oscillatory increases in alkalinity anticipate growth and may regulate actin dynamics in pollen tubes of lily. Plant Cell. 18:2182–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver, S.K., B.J. Pope, S. Whytock, and A.G. Weeds. 1998. The effect of two actin depolymerizing factors (ADF/cofilins) on actin filament turnover: pH sensitivity of F-actin binding by human ADF, but not of Acanthamoeba actophorin. Eur. J. Biochem. 256:388–397. [DOI] [PubMed] [Google Scholar]
- Margolis, B., A. Zilberstein, C. Franks, S. Felder, S. Kremer, A. Ullrich, S.G. Rhee, K. Skorecki, and J. Schlessinger. 1990. Effect of phospholipase C-gamma overexpression on PDGF-induced second messengers and mitogenesis. Science. 248:607–610. [DOI] [PubMed] [Google Scholar]
- McNamee, H.P., D.E. Ingber, and M.A. Schwartz. 1993. Adhesion to fibronectin stimulates inositol lipid synthesis and enhances PDGF-induced inositol lipid breakdown. J. Cell Biol. 121:673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongan, J., D.A. Case, and J.A. McCammon. 2004. Constant pH molecular dynamics in generalized Born implicit solvent. J. Comput. Chem. 25:2038–2048. [DOI] [PubMed] [Google Scholar]
- Moriyama, K., K. Iida, and I. Yahara. 1996. Phosphorylation of Ser-3 of cofilin regulates its essential function on actin. Genes Cells. 1:73–86. [DOI] [PubMed] [Google Scholar]
- Mouneimne, G., L. Soon, V. DesMarais, M. Sidani, X. Song, S.C. Yip, M. Ghosh, R. Eddy, J.M. Backer, and J. Condeelis. 2004. Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J. Cell Biol. 166:697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne, G., V. DesMarais, M. Sidani, E. Scemes, W. Wang, X. Song, R. Eddy, and J. Condeelis. 2006. Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr. Biol. 16:2193–2205. [DOI] [PubMed] [Google Scholar]
- Niwa, R., K. Nagata-Ohashi, M. Takeichi, K. Mizuno, and T. Uemura. 2002. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 108:233–246. [DOI] [PubMed] [Google Scholar]
- Norgauer, J., J. Krutmann, G.J. Dobos, A.E. Traynor-Kaplan, Z.G. Oades, and I.U. Schraufstatter. 1994. Actin polymerization, calcium-transients, and phospholipid metabolism in human neutrophils after stimulation with interleukin-8 and N-formyl peptide. J. Invest. Dermatol. 102:310–314. [DOI] [PubMed] [Google Scholar]
- Ojala, P.J., V. Paavilainen, and P. Lappalainen. 2001. Identification of yeast cofilin residues specific for actin monomer and PIP2 binding. Biochemistry. 40:15562–15569. [DOI] [PubMed] [Google Scholar]
- Onufriev, A., D. Bashford, and D.A. Case. 2004. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins. 55:383–394. [DOI] [PubMed] [Google Scholar]
- Park, K.C., R.F. Meili, S. Lee, F. Apone, and R.A. Firtel. 2004. Rac regulation of chemotaxis and morphogenesis in Dictyostelium. EMBO J. 23:4177–4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, H., and D.L. Barber. 2005. A developmentally regulated Na-H exchanger in Dictyostelium discoideum is necessary for cell polarity during chemotaxis. J. Cell Biol. 169:321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T.D. 2007. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 36:451–477. [DOI] [PubMed] [Google Scholar]
- Pollard, T.D., and G.G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. [DOI] [PubMed] [Google Scholar]
- Pope, B.J., S.M. Gonsior, S. Yeoh, A. McGough, and A.G. Weeds. 2000. Uncoupling actin filament fragmentation by cofilin from increased subunit turnover. J. Mol. Biol. 298:649–661. [DOI] [PubMed] [Google Scholar]
- Pope, B.J., K.M. Zierler-Gould, R. Kuhne, A.G. Weeds, and L.J. Ball. 2004. Solution structure of human cofilin: actin binding, pH sensitivity, and relationship to actin-depolymerizing factor. J. Biol. Chem. 279:4840–4848. [DOI] [PubMed] [Google Scholar]
- Postma, M., J. Roelofs, J. Goedhart, T.W. Gadella, A.J. Visser, and P.J. Van Haastert. 2003. Uniform cAMP stimulation of Dictyostelium cells induces localized patches of signal transduction and pseudopodia. Mol. Biol. Cell. 14:5019–5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney, L.K., S.P. Denker, and D.L. Barber. 2002. The changing face of the Na+/H+ exchanger, NHE1: structure, regulation, and cellular actions. Annu. Rev. Pharmacol. Toxicol. 42:527–552. [DOI] [PubMed] [Google Scholar]
- Reshkin, S.J., A. Bellizzi, S. Caldeira, V. Albarani, I. Malanchi, M. Poignee, M. Alunni-Fabbroni, V. Casavola, and M. Tommasino. 2000. Na+/H+ exchanger-dependent intracellular alkalinization is an early event in malignant transformation and plays an essential role in the development of subsequent transformation-associated phenotypes. FASEB J. 14:2185–2197. [DOI] [PubMed] [Google Scholar]
- Ritter, M., P. Schratzberger, H. Rossmann, E. Woll, K. Seiler, U. Seidler, N. Reinisch, C.M. Kahler, H. Zwierzina, H.J. Lang, et al. 1998. Effect of inhibitors of Na+/H+-exchange and gastric H+/K+ ATPase on cell volume, intracellular pH and migration of human polymorphonuclear leucocytes. Br. J. Pharmacol. 124:627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, A.T., C. Chun, K. Takeda, and R.A. Firtel. 2004. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J. Cell Biol. 167:505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, M.A., C. Lechene, and D.E. Ingber. 1991. Insoluble fibronectin activates the Na/H antiporter by clustering and immobilizing integrin alpha 5 beta 1, independent of cell shape. Proc. Natl. Acad. Sci. USA. 88:7849–7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, W., T. Day, M.P. Jacobson, R.A. Friesner, and R. Farid. 2006. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 49:534–553. [DOI] [PubMed] [Google Scholar]
- Song, X., X. Chen, H. Yamaguchi, G. Mouneimne, J.S. Condeelis, and R.J. Eddy. 2006. Initiation of cofilin activity in response to EGF is uncoupled from cofilin phosphorylation and dephosphorylation in carcinoma cells. J. Cell Sci. 119:2871–2881. [DOI] [PubMed] [Google Scholar]
- Srivastava, J., G. Barreiro, S. Groscurth, A.R. Gringas, B.T. Goult, D.R. Critchley, M.J.S. Kelly, M.P. Jacobson, and D.L. Barber. 2008. Structural model and functional significance of pH-dependent talin-actin binding for focal adhesion remodeling. Proc. Natl. Acad. Sci. USA. 105:14436–14441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock, C., B. Gassner, C.R. Hauck, H. Arnold, S. Mally, J.A. Eble, P. Dieterich, and A. Schwab. 2005. Migration of human melanoma cells depends on extracellular pH and Na+/H+ exchange. J. Physiol. 567:225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stradal, T.E., and G. Scita. 2006. Protein complexes regulating Arp2/3-mediated actin assembly. Curr. Opin. Cell Biol. 18:4–10. [DOI] [PubMed] [Google Scholar]
- Tilney, L.G., D.P. Kiehart, C. Sardet, and M. Tilney. 1978. Polymerization of actin. IV. Role of Ca++ and H+ in the assembly of actin and in membrane fusion in the acrosomal reaction of echinoderm sperm. J. Cell Biol. 77:536–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga, T., and D.L. Barber. 1998. Na-H exchange acts downstream of RhoA to regulate integrin-induced cell adhesion and spreading. Mol. Biol. Cell. 9:2287–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duijn, B., and K. Inouye. 1991. Regulation of movement speed by intracellular pH during Dictyostelium discoideum chemotaxis. Proc. Natl. Acad. Sci. USA. 88:4951–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haastert, P.J., I. Keizer-Gunnink, and A. Kortholt. 2007. Essential role of PI3-kinase and phospholipase A2 in Dictyostelium discoideum chemotaxis. J. Cell Biol. 177:809–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rheenen, J., X. Song, W. van Roosmalen, M. Cammer, X. Chen, V. Desmarais, S.C. Yip, J.M. Backer, R.J. Eddy, and J.S. Condeelis. 2007. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J. Cell Biol. 179:1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Troys, M., D. Dewitte, J.L. Verschelde, M. Goethals, J. Vandekerckhove, and C. Ampe. 2000. The competitive interaction of actin and PIP2 with actophorin is based on overlapping target sites: design of a gain-of-function mutant. Biochemistry. 39:12181–12189. [DOI] [PubMed] [Google Scholar]
- Wang, J., P. Cieplak, and P.A. Kollman. 2000. How well does a RESP (restrained electrostatic potential) model do in calculating the conformational energies of organic and biological molecules? J. Comput. Chem. 21:1049–1074. [Google Scholar]
- Yan, W., K. Nehrke, J. Choi, and D.L. Barber. 2001. The Nck-interacting kinase (NIK) phosphorylates the Na+-H+ exchanger NHE1 and regulates NHE1 activation by platelet-derived growth factor. J. Biol. Chem. 276:31349–31356. [DOI] [PubMed] [Google Scholar]
- Yonezawa, N., E. Nishida, K. Iida, I. Yahara, and H. Sakai. 1990. Inhibition of the interactions of cofilin, destrin, and deoxyribonuclease I with actin by phosphoinositides. J. Biol. Chem. 265:8382–8386. [PubMed] [Google Scholar]
- Zebda, N., O. Bernard, M. Bailly, S. Welti, D.S. Lawrence, and J.S. Condeelis. 2000. Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension. J. Cell Biol. 151:1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierler-Gould, K.M., B.J. Pope, A.G. Weeds, and L.J. Ball. 2004. Backbone and sidechain 1H, 13C and 15N resonance assignments of human cofilin. J. Biomol. NMR. 29:429–430. [DOI] [PubMed] [Google Scholar]