Abstract
pUO-StVR2 is a virulence-resistance plasmid which originated from pSLT of Salmonella enterica serovar Typhimurium through acquisition of a complex resistance island, flanked by regions that provide a toxin-antitoxin system and an iron uptake system. The presence of resistance and virulence determinants on the same plasmid allows coselection of both properties, potentially increasing health risks.
pUO-StVR2 is a hybrid virulence-resistance plasmid that is widely distributed among isolates of Salmonella enterica serovar Typhimurium, recovered in Spain not only from clinical samples but also from food and food-producing animals (2, 9, 11, 12). Similar isolates have recently been detected in the United Kingdom (A. Herrero, unpublished results), and indirect evidence suggests their presence in other European countries (1, 14). pUO-StVR2 confers resistance to ampicillin, chloramphenicol, streptomycin-spectinomycin, sulfonamides, and tetracycline, encoded by the blaOXA-1 (also called blaOXA-30) (3), catA1, aadA1, sul1, and tetA(B) genes, with two of them (blaOXA-1 and aadA1) located in the variable region of an integron termed InH. It also contains the spvC, rck, samA, traT, traX, repA, and parA/B genes of pSLT, the virulence plasmid specific for S. enterica serovar Typhimurium (17, 18), but it lacks the InFIB/repA2 replication region, including rsk (a 66-bp sequence that corresponds to the B, C, and D iterons of the replicon), and most of the pef operon (9, 11, 12).
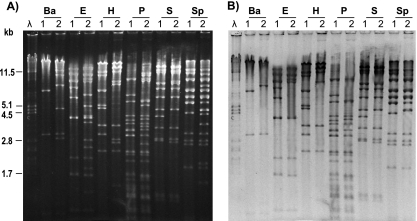
The possibility that pUO-StVR2 originated from pSLT was investigated by hybridization experiments. For these experiments, pSLT and pUO-StVR2 were extracted from S. enterica serovar Typhimurium LT2 and S. enterica serovar Typhimurium LSP 146/02, respectively (11), and digested with different endonucleases (Fig. 1). After separation by conventional agarose gel electrophoresis, fragments were transferred onto a nylon membrane and hybridized with a probe comprising the entire pSLT (19). As shown in Fig. 1, many fragments of pUO-StVR2 have a counterpart in pSLT, as revealed by both coincidence in size and hybridization, hence demonstrating the close relationship between the two plasmids. Other fragments generated from pUO-StVR2 were absent in the lanes corresponding to pSLT and failed to hybridize with the probe. They could be internal to DNA newly gained by pSLT, and this was in fact confirmed by identification of the junction regions as the fragments generated from pUO-StVR2 which hybridized with the probe but lacked a pSLT counterpart.
FIG. 1.
Relationship between pSLT and pUO-StVR2. (A) Restriction analysis of pSLT and pUO-StVR2. (B) Hybridization with the entire pSLT plasmid. Ba, BamHI; E, EcoRI; H, HindIII; P, PstI; S, SalI; Sp, SphI; λ, DNA of phage lambda digested with PstI, used as a size standard; lanes 1 and 2, pSLT and pUO-StVR2, respectively.
The genetic organization of the DNA acquired by pSLT to build pUO-StVR2 was further established by cloning and sequencing of the differential DNA and the junction regions. The hybrid plasmid was subjected to single and double digestions with the endonucleases used in the hybridization experiment (described above). The generated fragments were ligated into the corresponding sites of pUK1921, a cloning vector that contains a kanamycin resistance gene as a selectable marker (10). The obtained libraries were transformed into Escherichia coli DH5α (Invitrogen) (19). Recombinant plasmids were selected for acquisition of resistance properties associated with pUO-StVR2 or for the presence of fragments with sizes matching internal or junction regions of the differential DNA (described above). Relevant insertions were sequenced at the Servicio de Secuenciación de DNA, Centro de Investigaciones Biológicas (CSIC [Madrid, Spain]). Overlapping fragments were assembled to generate a contiguous sequence of 49,507 bp, which was analyzed online at the European Bioinformatics Institute web site (http://www.ebi.ac.uk/).
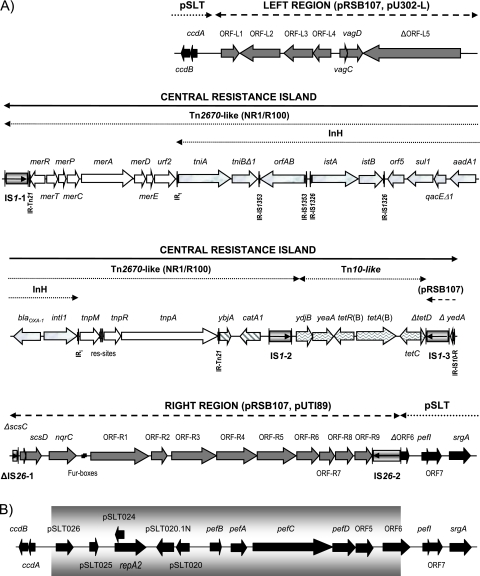
The sequence obtained (Fig. 2A) consists of 47,606 bp of foreign DNA and 1,901 bp of flanking pSLT DNA (525 and 1,376 bp at the conventional left and right ends, respectively). Within the latter, the new DNA is located between the noncoding region upstream of the ccdA gene and the 3′ end of open reading frame 6 (ORF6) upstream of pefI. The intermediate segment of pSLT (ca. 12 kb), which includes IncFIB/repA2, the overlapping rsk sequence (described above), and most of the pef operon (pefBACD), appears to have been replaced by the acquired DNA (Fig. 2B).
FIG. 2.
(A) Schematic representation of the DNA acquired by pSLT to compose pUO-StVR2. Solid and dashed horizontal lines above the scheme are used to point out the distinct components of the represented DNA. Plasmids showing the highest similarity to the main regions are indicated in parentheses. (B) pSLT region where the foreign DNA was inserted (based on accession no. AE006471). The segment of pSLT absent in pUO-StVR2 is shown within a gray box.
The foreign DNA has a mosaic structure and consists of a central region containing all of the resistance genes previously identified in pUO-StVR2 clustered within an antimicrobial resistance island of 28,756 bp and two flanking regions where additional virulence genes were identified (Fig. 2A). The left-hand region, at the ccdA end, comprises 7,102 bp with a GC content of 46.9%, which is nearly identical (99%) to sequences present in two multidrug-resistant plasmids, pRSB107 isolated from an uncultured bacterium in a sewage treatment plant (20) and pU302L from S. enterica serovar Typhimurium (5). It contains several ORFs of unknown function (here termed ORFL1 to ORFL5), and the vagC and vagD genes that encode a toxin-antitoxin system. The latter were originally detected in pSDV, the virulence plasmid specific for S. enterica serovar Dublin, and shown to be important for virulence (16), although the effect is now considered to be indirect, due to an enhancement of plasmid stability (8). In pUO-StVR2, the capture of a second toxin-antitoxin system, in addition to that encoded by the ccdA/ccdB genes already present in pSLT (8, 18), provides an extra level of security, further ensuring plasmid stability and therefore persistence of the resistance and virulence functions it confers.
The central resistance cluster includes a composite transposon almost identical to Tn2670, harbored by the NR1 (R100) plasmid of Shigella flexneri, which consists of Tn21 inserted within Tn9 and is thus delineated by the two copies of IS1 in direct orientation (13). However, the InH integron carried by the Tn21-like transposon of pUO-StVR2 differs from the In2 integron of Tn21 by insertion of the blaOXA-1 gene cassette upstream of aadA1. A defective Tn10 completes the resistance island of the hybrid plasmid. It carries ydjB, yeaA, tetR(B), tetA(B), tetC, and a truncated tetD. Yet, IS10-L, ydhA and ydjA are missing at one end, while most of tetD and part of yedA (which encodes the transposase of IS10-R) are replaced by a copy of IS1 at the other end. The resulting structure comprises the tetR(B)-tetA(B) genes between two oppositely oriented copies of IS1 and could therefore represent a new composite tetracycline transposon. It should further be noted that colinearity between a Tn2670-like transposon and a defective Tn10 has also been found in pRSB107 as well as in a pathogenicity island of Shigella flexneri 2a (15). However, the complexity of the Tn2670 derivative and/or the extent of the deletions affecting Tn10 differ in the three resistance clusters.
The remaining DNA incorporated by pSLT (11,748 bp [54.9% GC]) is 100% identical to another region of pRSB107. It includes truncated and intact copies of IS26 which flank the scsC (incomplete) and scsD genes (for suppression of copper sensitivity), an nqrC-like gene (for Na+ transport), and several ORFs designated here as ORF-R1 to ORF-R9. At least two of the latter, ORF-R1 and ORF-R2, which encode an Fe2+/Pb2+ permease and an Fe2+ transport protein, respectively, and which are preceded by two potential Fur boxes, could be involved in iron uptake (20). The similarity of the right-hand region of pUO-StVR2 to the corresponding DNA of pRSB107 extends to include the entire IS1 downstream of the truncated tetD, which is here considered as part of the central resistance island. The sequence from scsC to ORF-R9 is also found in pUTI89, a plasmid from the uropathogenic E. coli strain UTI89 (99% similarity) (6), as well as in the chromosome of Citrobacter koseri ATCC BAA-895 (accession no. CP000822 [98% similarity]). In the former, a copy of IS26 is placed at the right-hand end but not at the scsC end, while no IS26 flanked the equivalent segment of the C. koseri chromosome. Interestingly, the ORFs encoding the putative iron acquisition system are also present in a pathogenicity island of the highly invasive organism Yersinia pestis (accession no. AL031866).
As for the origin of pUO-StVR2, it is not known whether the distinct components of the foreign DNA have been sequentially acquired by pSLT or if the tripartite structure has been previously assembled in an as-yet-unidentified precursor plasmid. Nevertheless, the simultaneous presence of resistance and virulence determinants on the same extrachromosomal element will allow coselection of both properties, yielding potentially more dangerous strains that represent a hazard to human and animal health (7). The capture of an iron acquisition system by pSLT is of particular relevance, since successful competition for this essential nutrient, which is not freely available in the host, is of crucial importance for pathogens to establish infection (4).
Nucleotide sequence accession number.
The sequence generated in this work has been deposited in the EMBL database under accession no. AM991977.
Acknowledgments
We thank Jürgen Heinisch (Fachbereich Biologie/Chemie, AG Genetik Universität Osnabrück, Osnabrück, Germany) for critical reading of the manuscript.
A. Herrero was the recipient of a grant from the Fundación para el Fomento en Asturias de la Investigación Científica Aplicada y la Tecnología (FICYT, reference no. BP03-014). This work has been supported by projects PI02-0172 and PI05-2489 of the Fondo de Investigación Sanitaria, Ministerio de Sanidad y Consumo, Spain.
Footnotes
Published ahead of print on 13 October 2008.
REFERENCES
- 1.Antunes, P., J. Machado, J. C. Sousa, and L. Peixe. 2004. Dissemination amongst humans and food products of animal origin of a Samonella typhimurium clone expressing an integron-borne OXA-30 β-lactamase. J. Antimicrob. Chemother. 54:429-434. [DOI] [PubMed] [Google Scholar]
- 2.Bances, M., A. Herrero, Y. González, M. R. Rodicio, and M. A. González-Hevia. 2007. Outbreak of gastroenteritis in a nursery school caused by a strain of Salmonella enterica serovar Typhimurium carrying the hybrid virulence-resistance plasmid pUO-StVR2. Enferm. Infecc. Microbiol. Clin. 25:376-381. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, D. A., and M. R. Mulvey. 2006. OXA-1 is OXA-30 is OXA-1. J. Antimicrob. Chemother. 58:224-225. [DOI] [PubMed] [Google Scholar]
- 4.Braun, V. 2001. Iron uptake mechanisms and their regulation in pathogenic bacteria. Int. J. Med. Microbiol. 291:67-79. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C. Y., G. W. Nace, B. Solow, and P. Fratamico. 2007. Complete nucleotide sequences of 84.5- and 3.2-kb plasmids in the multi-antibiotic resistant Salmonella enterica serovar Typhimurium U302 strain G8430. Plasmid 57:29-43. [DOI] [PubMed] [Google Scholar]
- 6.Chen, S. L., C.-S. Hung, J. Xu, C. S. Reigstad, V. Magrini, A. Sabo, D. Blasiar, T. Bieri, R. R. Meyer, P. Ozersky, J. R. Armstrong, R. S. Fulton, J. P. Latreille, J. Spieth, T. M. Hooton, E. R. Mardis, S. J. Hultgren, and J. I. Gordon. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach Proc. Natl. Acad. Sci. USA 103:5977-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fluit, A. C. 2005. Towards more virulent and antibiotic-resistant Salmonella? FEMS Immunol. Med. Microbiol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 8.Gerdes, K., S. K. Christensen, and A. Løbner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 9.Guerra, B., S. Soto, R. Helmuth, and M. C. Mendoza. 2002. Characterization of a self-transferable plasmid from Salmonella enterica serotype Typhimurium clinical isolates carrying two integron-borne gene cassettes together with virulence and drug resistance genes. Antimicrob. Agents Chemother. 46:2977-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinisch, J. J. 1993. PFK2, ISP42, ERG2 and RAD14 are located on the right arm of chromosome XIII. Yeast 9:1103-1105. [DOI] [PubMed] [Google Scholar]
- 11.Herrero, A., M. R. Rodicio, M. A. González-Hevia, and M. C. Mendoza. 2006. Molecular epidemiology of emergent multidrug-resistant Salmonella enterica serotype Typhimurium strains carrying the virulence resistance plasmid pUO-StVR2. J. Antimicrob. Chemother. 57:39-45. [DOI] [PubMed] [Google Scholar]
- 12.Herrero, A., M. R. Rodicio, M. A. Echeita, and M. C. Mendoza. 2008. Salmonella enterica serotype Typhimurium carrying hybrid virulence-resistance plasmids (pUO-StVR): a new multidrug-resistant group endemic in Spain. Int. J. Med. Microbiol. 298:253-261. [DOI] [PubMed] [Google Scholar]
- 13.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindstedt, B.-A., E. Heir, I. Nygård, and G. Kapperud. 2003. Characterization of class I integrons in clinical strains of Salmonella enterica subsp. enterica serovars Typhimurium and Enteritidis from Norwegian hospitals. J. Med. Microbiol. 52:141-149. [DOI] [PubMed] [Google Scholar]
- 15.Luck, S. N., S. A. Turner, K. Rajakumar, H. Sakellaris, and B. Adler. 2001. Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect. Immun. 69:6012-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pullinger, G. D., and A. J. Lax. 1992. A Salmonella dublin virulence plasmid locus that affects bacterial growth under nutrient-limited conditions. Mol. Microbiol. 6:1631-1643. [DOI] [PubMed] [Google Scholar]
- 17.Rotger, R., and J. Casadesús. 1999. The virulence plasmids of Salmonella. Int. Microbiol. 2:177-184. [PubMed] [Google Scholar]
- 18.Rychlik, I., D. Gregorova, and H. Hradecka. 2006. Distribution and function of plasmids in Salmonella enterica. Vet. Microbiol. 112:1-10. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Szczepanowski, R., S. Braun, V. Riedel, S. Schneiker, I. Krahn, A. Pühler, and A. Schlüter. 2005. The 120 592 bp IncF plasmid pRSB107 isolated from a sewage-treatment plant encodes nine different antibiotic-resistance determinants, two iron-acquisition systems and other putative virulence-associated functions. Microbiology 151:1095-1111. [DOI] [PubMed] [Google Scholar]