Abstract
A conditional expression system has been developed using the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter to validate essential genes of Staphylococcus aureus in vivo. The system has been applied to prove the essentiality of ligA and to evaluate the function of tarI, which was found to be essential in vitro but not in vivo.
The current increase of antimicrobial resistance in pathogenic staphylococci due to the spread of multiply resistant strains has encouraged the search for alternative targets for antimicrobial chemotherapy. Potential target genes should be essential for bacterial viability or pathogenesis (3). However, gene essentiality can vary depending on test conditions, particularly in vitro versus in vivo situations (1). Therefore, the use of in vivo expression systems to validate putative target candidates is critical (12). In this study, a conditional expression system based on a regulatable Pspac-lacI promoter element has been evaluated for in vivo use. This regulatable gene expression system was originally developed by Jana et al. for the analysis of essential genes in vitro but has not been applied for target validation in vivo before (4). The system consists of a Pspac-lacI promoter element that regulates the expression of genes of interest depending on the concentration of the inducer isopropyl-β-d-thiogalactopyranoside (IPTG). Repression of the target gene is achieved by production of the repressor LacI, which binds to the Pspac-lacI promoter element. The production of sufficient amounts of the repressor LacI is ensured by the introduction of the multicopy plasmid pMJ8426, carrying lacI behind the constitutively expressed penicillinase (pcn) promoter, into the promoter fusion strain.
The strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F−endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 λ− Δ(argF-lac)U169 φ80dlacZΔM15 | Bethesda Research Laboratories |
| S. aureus | ||
| 8325 | NCTC 8325; laboratory strain | 10 |
| RN4220 | Restriction minus derivative of NCTC 8325-4 | 8 |
| Xen29 | S. aureus strain ATCC 12600 (a pleural fluid isolate) with a chromosomally integrated lux operon (Tn4001 luxABCDE Kmr) | 6, 7 |
| RN4220 Pspac::ligA | Conditional mutant of ligA; Cmr; with repressor plasmid pMJ8426 (lacI Tetr) | This study |
| RN4220 Pspac::tarI | Conditional mutant of tarI (SA0245); Cmr; with repressor plasmid pMJ8426 (lacI Tetr) | This study |
| 8325 Pspac::tarI | Conditional mutant of tarI (SA0245); Cmr; with repressor plasmid pMJ8426 (lacI Tetr) | This study |
| Xen29 Pspac::tarI | Conditional mutant of tarI (SA0245); Cmr; with repressor plasmid pMJ8426 (lacI Tetr) | This study |
| Plasmids | ||
| pLL30 | Shuttle vector; Spcr in E. coli; Tetr Cmr in S. aureus | 4 |
| pMJ8426 | Shuttle vector; lacI Spcr in E. coli; Tetr in S. aureus | 4 |
| pKSLligA | pLL30 with upstream and 5′ regions of the ligA gene for conditional mutation of ligA | This study |
| pKSLtarI | pLL30 with upstream and 5′ regions of the tarI gene for conditional mutation of tarI (SA0245) | This study |
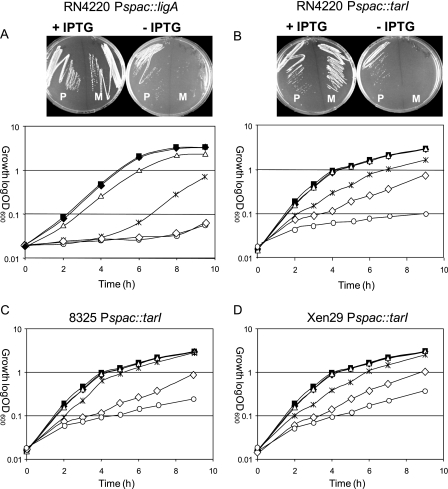
In a first step, the in vivo applicability of this conditional expression system was tested. A conditionally lethal mutant expressing the essential gene ligA, encoding DNA ligase, in an IPTG-dependent manner was constructed (5). The target gene was placed behind the Pspac promoter and integrated into the chromosome by a double-crossover event as described by Jana et al. (4). The upstream fragment was amplified by using primers 5′-CGG AAT TCA AGG AGG ATT AAG GGA TGG C-3′ and CGG GAT CCG CTC TTG ATA CAG TTG TAC C-3′, and the downstream fragment was amplified by using primers 5′-CCG CTC GAG CCA TAC ACA ATG GTT GGT GG-3′ and 5′-ACG CGT CGA CAG CCA TCC CTT AAT CCT CC-3′ (restriction sites introduced to facilitate cloning are underlined). In the presence of plasmid pMJ8426 carrying lacI, the growth of Pspac::ligA depends on the presence of the inducer IPTG (Fig. 1). The conditional mutant strain RN4220 Pspac::ligA was not able to grow in vitro without IPTG in liquid culture and on agar plates. However, in the presence of 0.3 mM IPTG, the mutant strain showed the same growth rate as the parent strain, RN4220 (Fig. 1). These in vitro results confirm the essential function of ligA and the conditionally essential phenotype of the constructed mutant. Subsequently, the Pspac::ligA fusion strain was used in an experimental Staphylococcus aureus mouse abscess model based on collagen Gelfoam implants (Pharmacia & Upjohn, Kalamazoo, MI). The Gelfoam was cut into pieces of 1 cm2, incubated overnight in sterile phosphate-buffered saline, and implanted subcutaneously in the backs of female CFW1 mice. One group of mice drank water without and the other group with 25 mM IPTG ad libitum. Three days later, the mice were infected with the S. aureus parent strain, RN4220, or with a conditional mutant strain (104 CFU) injected into the implanted Gelfoam. For stable propagation of the plasmid, the animals were treated with tetracycline (due to the corresponding plasmid-encoded antibiotic resistance) immediately and 6 h after the infection of the Gelfoam implants (50 mg/kg of body weight, given intraperitoneally). One day after infection, Gelfoam implants were removed and homogenized, and bacterial cells were counted on Mueller-Hinton agar containing 0.3 mM IPTG alone or with 10 μg/ml tetracycline. Statistical analysis was performed using the nonparametric Mann-Whitney U test. For all comparisons, a P value of <0.05 was considered statistically significant. These experiments revealed that the growth of the Pspac::ligA mutant in the Gelfoam was dependent on the presence of IPTG, whereas the parent strain, RN4220, grew independently of IPTG (Fig. 2). Bacteria isolated from the Gelfoam were tetracycline resistant, indicating the stability of the repressor plasmid pMJ8426 in vivo under the test conditions.
FIG. 1.
In vitro growth of Pspac::ligA (A) and Pspac::tarI (B, C, and D) mutant strains depends on the inducer IPTG. Growth of all conditional mutants can be regulated by IPTG in the presence of the repressor plasmid pMJ8426. The parent strain RN4220 (P) and the Pspac mutants (M) in RN4220 were grown in solid medium either supplemented with 300 μM IPTG (+ IPTG) or without IPTG (− IPTG). Representative growth curves of strains RN4220, 8325, and Xen29 without IPTG (▪) or with 0.3 mM IPTG (♦) and of the Pspac mutants in strain RN4220 (B), 8325 (C), or Xen29 (D) without IPTG (○) or with IPTG at 0.3 mM (▵), 0.03 mM (*), or 0.003 mM (⋄) are shown.
FIG. 2.
Determination of CFU of the conditional S. aureus RN4220 Pspac::ligA strain (A) and the conditional RN4220 Pspac::tarI strain (C) 1 day after infection of the murine abscess (Gelfoam) model compared to CFU of strain RN4220 containing the repressor plasmid pMJ8426 (B and D). The number of CFU was determined on agar plates with (+) or without (−) tetracycline. The bacterial load in the Gelfoam homogenate of each individual mouse is indicated by a dot. Each horizontal line indicates the mean for all five mice per group. Asterisks indicate significant differences between the presence and absence of IPTG (P < 0.05 by the Mann-Whitney test). The growth of the Pspac::ligA mutant strain depends on the addition of IPTG (A), which does not affect the growth of the parent, RN4220 (B). The growth of the conditional Pspac::tarI strain is not dependent on the presence or absence of IPTG (C).
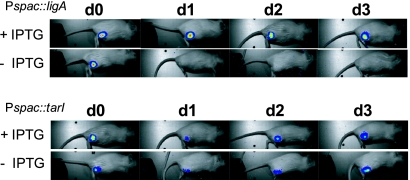
Moreover, the conditional essentiality of Pspac::ligA in vivo has also been confirmed in a second animal model. For this purpose, a conditional Pspac::ligA mutant was constructed in strain Xen29, a clinical isolate that has been used for in vivo imaging of staphylococcal infections. The growth of this strain can be visualized by means of bioluminescent imaging due to expression of luxABCED (6, 7). We injected 5 × 107 Xen29 Pspac::ligA CFU intramuscularly into the musculus tibialis anterior of female BALB/c mice. Bioluminescence was measured for three consecutive days, as shown in Fig. 3. As in the Gelfoam model, IPTG was given via drinking water (25 mM) to one group from day −4, and tetracycline was injected intraperitoneally at 30 mg/kg daily starting on day zero. These studies showed a significant decline of bioluminescence in mice that drank water without IPTG (Fig. 3, upper panels). Consequently, no bioluminescent signal was detectable after 24 h (day 1) of infection. In contrast, bioluminescent bacteria were visible until day 3 postinfection in a control group of mice that received IPTG via drinking water (Fig. 3).
FIG. 3.
Bioluminescence of strain Xen29 Pspac::ligA (upper panels) and Xen29 Pspac::tarI (lower panels) in the muscular infection model in the presence or absence of IPTG. The time course of bioluminescence levels was monitored for 3 consecutive days (day zero to day 3 [d0 to d3]) after intramuscular infection of the musculus tibialis anterior of mice with S. aureus Xen29 Pspac::ligA or Xen29 Pspac::tarI. Images of representative mice are presented. Signal intensity is indicated by a pseudocolor scale superimposed on the grayscale image to produce a composite image of photon emission in relation to the subject. Pixels with low photon counts appear blue, and magenta represents the most intense light emission. The signals were obtained using a Night Owl LB 981 luminograph (Berthold Technologies, Bad Wildbad, Germany) by integrating photons over 1 min.
In a different approach, we fused the tarI gene behind Pspac and integrated the construct into the chromosomes of three S. aureus strains: RN4220, 8325, and Xen29. Originally, tarI was annotated in strain N315 as ispD (SA0245) due to its homology to essential 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferases of Escherichia coli and Bacillus subtilis, which belong to the 1-deoxy-d-xylulose 5-phosphate pathway (DXP pathway) for isoprenoid biosynthesis (9). However, S. aureus—like other gram-positive cocci, such as Streptococcus pneumoniae and Enterococcus faecalis—does not possess the DXP pathway for isoprenoid biosynthesis. Instead, these species use the mevalonate pathway (13, 14). Meanwhile, there is increasing evidence that tarI is involved in teichoic acid synthesis in S. aureus (2, 11). Interestingly, S. aureus possesses two homologs of the tarI gene (SA0241 and SA0245 in strain N315) in the genome that are about 5,000 bp apart and share 75% identity on the amino acid sequence level. Both genes might encode d-ribitol-5-phosphate cytidylyltransferases. Reverse transcriptase PCR analyses revealed that both genes are transcribed in S. aureus grown to mid-log phase (data not shown). We could delete the SA0241 homolog, showing that this gene is clearly nonessential in S. aureus (data not shown). However, we were not able to delete the SA0245 gene. This observation differs from the results of a previous report by Zalacain et al. (15). Zalacain et al. could make a single mutant of either tarI homologous gene (yacM according to Bacillus subtilis nomenclature) in S. aureus RN4220, but they were not able to delete the second gene from either mutant. Interestingly, in a more recent study, the tarI-homologous gene SACOL0240 in S. aureus strain COL was essential for viability in vitro, as long as the tarI homolog tarO was not knocked out (2).
To clarify whether or not tarI (SA0245) is essential in S. aureus, we constructed a conditional mutant of this gene by placing tarI behind the conditional Pspac promoter using primers 5′-CGG AAT TCG ATA TAG TTG AAT GGA GGA AG-3′ and 5′-CGG GAT CCA ATA CGA CCT CAC CAA CAC C-3′ (upstream fragment) and primers 5′-ACG CGT CGA CTT CCT CCA TTC AAC TAT ATC-3′ and 5′-CCG CTC GAG TAT CTA GTG TCA CCT AAT CC-3′ (downstream fragment). As shown in Fig. 1B, the growth of the conditional Pspac::tarI mutant in strain RN4220 was dependent on IPTG in vitro, indicating an essential function of tarI in S. aureus. In addition to strain RN4220, conditional Pspac::tarI mutants were also constructed in strain 8325 and Xen29. Conditional growth experiments using different IPTG concentrations revealed that in the absence of IPTG, growth inhibition was less pronounced in Xen29 and 8325 than in RN4220 (Fig. 1B to D). Consequently, we employed the Pspac::tarI mutant of strain RN4220 in the in vivo implant model. For this purpose, both RN4220 and the mutant strain were used in the same manner as that described above for the ligA conditional mutant. Surprisingly, these experiments revealed that the mutant strain showed the same capability to grow in vivo as the parent strain (Fig. 2). Obviously, tarI (SA0245) is important for the growth of S. aureus in vitro, which has been demonstrated in three strains (RN4220, 8325, and Xen29). However, the gene product seems to be dispensable for in vivo growth. To rule out the possibility that the in vivo essentiality of tarI is related to the strain background, conditional tarI expression was also investigated in the muscle model using the bioluminescent strain Xen29 Pspac::tarI. Following challenge with 5 × 107 Xen29 Pspac::tarI CFU in the musculus tibialis anterior, no significant difference in bioluminescence between mice that drank water with IPTG and mice that drank water without IPTG could be detected (Fig. 3, lower panels). Furthermore, the musculus tibialis anterior was removed after 3 days of infection and homogenized, and CFU counts were determined, revealing that there was no significant difference between the groups with and without IPTG (data not shown). In addition, to test if the conditional phenotype of the Pspac::tarI mutant strain was lost during in vivo passage, in vitro essentiality was reevaluated by selecting representative colonies from the agar plates that had been used for calculation of bacterial loads in infected muscles. Because these colonies maintained their conditionally essential phenotype in vitro, it can be concluded that tarI is not essential in vivo.
These data indicate consistently that tarI is essential for growth only under specific laboratory conditions. Moreover, the function of the tarI (SA0245)-homologous gene SA0241 must be elucidated, since it might complement the functions of tarI under certain conditions. Nevertheless, the results of the study highlight the importance of in vivo validation of in vitro data, especially if new targets for antibiotic development are selected.
In summary, the conditional in vivo systems presented here are suitable for testing in vivo essentiality. The application of IPTG via drinking water was a suitable way of providing sufficient concentrations of the inducer in the staphylococcal cells. Thus, the Pspac expression system has proven to be applicable as a conditional expression system for investigating the essentiality of a particular target gene in vivo during S. aureus infection.
Acknowledgments
This study was supported by a grant of the Bavarian Science Foundation (FORINGEN) and the Deutsche Forschungsgemeinschaft (SFB-TR34 and SFB630).
We thank U. Wallner, K. Merfort, S. Obertegger, M. Haas, and Martin Eckart for technical assistance. We thank C. Ladel for support in animal experiments.
Footnotes
Published ahead of print on 22 September 2008.
REFERENCES
- 1.Becker, D., M. Selbach, C. Rollenhagen, M. Ballmaier, T. F. Meyer, M. Mann, and D. Bumann. 2006. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440:303-307. [DOI] [PubMed] [Google Scholar]
- 2.D'Elia, M. A., M. P. Pereira, Y. S. Chung, W. Zhao, A. Chau, T. J. Kenney, M. C. Sulavik, T. A. Black, and E. D. Brown. 2006. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J. Bacteriol. 188:4183-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Lara, J., M. Masalha, and S. J. Foster. 2005. Staphylococcus aureus: the search for novel targets. Drug Discov. Today 10:643-651. [DOI] [PubMed] [Google Scholar]
- 4.Jana, M., T. T. Luong, H. Komatsuzawa, M. Shigeta, and C. Y. Lee. 2000. A method for demonstrating gene essentiality in Staphylococcus aureus. Plasmid 44:100-104. [DOI] [PubMed] [Google Scholar]
- 5.Kaczmarek, F. S., R. P. Zaniewski, T. D. Gootz, D. E. Danley, M. N. Mansour, M. Griffor, A. V. Kamath, M. Cronan, J. Mueller, D. Sun, P. K. Martin, B. Benton, L. McDowell, D. Biek, and M. B. Schmid. 2001. Cloning and functional characterization of an NAD+-dependent DNA ligase from Staphylococcus aureus. J. Bacteriol. 183:3016-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadurugamuwa, J. L., L. V. Sin, E. Albert, J. Yu, K. P. Francis, M. DeBoer, M. Rubin, C. Bellinger-Kawahara, T. R. Parr, Jr., and P. R. Contag. 2003. Direct continuous method for monitoring biofilm infection in a mouse model. Infect. Immun. 71:882-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadurugamuwa, J. L., L. V. Sin, J. Yu, K. P. Francis, R. Kimura, T. Purchio, and P. R. Contag. 2003. Rapid direct method for monitoring antibiotics in a mouse model of bacterial biofilm infection. Antimicrob. Agents Chemother. 47:3130-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 9.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 10.Novick, R. P. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155-166. [DOI] [PubMed] [Google Scholar]
- 11.Qian, Z., Y. Yin, Y. Zhang, L. Lu, Y. Li, and Y. Jiang. 2006. Genomic characterization of ribitol teichoic acid synthesis in Staphylococcus aureus: genes, genomic organization and gene duplication. BMC Genomics 7:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomson, C. J., E. Power, H. Ruebsamen-Waigmann, and H. Labischinski. 2004. Antibacterial research and development in the 21st Century—an industry perspective of the challenges. Curr. Opin. Microbiol. 7:445-450. [DOI] [PubMed] [Google Scholar]
- 13.Voynova, N. E., S. E. Rios, and H. M. Miziorko. 2004. Staphylococcus aureus mevalonate kinase: isolation and characterization of an enzyme of the isoprenoid biosynthetic pathway. J. Bacteriol. 186:61-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilding, E. I., J. R. Brown, A. P. Bryant, A. F. Chalker, D. J. Holmes, K. A. Ingraham, S. Iordanescu, C. Y. So, M. Rosenberg, and M. N. Gwynn. 2000. Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. J. Bacteriol. 182:4319-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zalacain, M., S. Biswas, K. A. Ingraham, J. Ambrad, A. Bryant, A. F. Chalker, S. Iordanescu, J. Fan, F. Fan, R. D. Lunsford, K. O'Dwyer, L. M. Palmer, C. So, D. Sylvester, C. Volker, P. Warren, D. McDevitt, J. R. Brown, D. J. Holmes, and M. K. Burnham. 2003. A global approach to identify novel broad-spectrum antibacterial targets among proteins of unknown function. J. Mol. Microbiol. Biotechnol. 6:109-126. [DOI] [PubMed] [Google Scholar]