Abstract
The aim of this study was to identify human immunodeficiency virus (HIV) protease mutations associated with virological response (VR) to fosamprenavir-ritonavir (FPV/r) in 113 protease inhibitor (PI)-experienced patients randomized in both CONTEXT and TRIAD clinical trials and receiving the same dose (700/100 mg twice daily) of FPV/r. The impact of each protease mutation on the VR to FPV/r, defined as the decrease in HIV RNA at week 12, was investigated with nonparametric analyses. A step-by-step procedure was done using a Jonckheere-Terpstra (JT) test that retains the group of mutations most strongly associated with the VR. Mutations at the following 14 codons were associated with a reduced VR to FPV/r: 10, 15, 33, 46, 54, 60, 62, 63, 72, 73, 82, 84, 89, and 90. The JT procedure led to selecting the CONTEXT/TRIAD genotypic set of mutations, I15V, M46I/L, I54L/M/V, D60E, L63P/T, and I84V, as providing the strongest association with the VR (P = 1.45 × 10−11). In the nine patients with zero mutations within this set, the median decrease in HIV RNA was −2.63 log copies/ml, and was −2.22 (n = 45), −1.50 (n = 26), −0.58 (n = 23), −0.47 (n = 6), −0.13 (n = 3), and 0.04 (n = 1) log copies/ml in those with one, two, three, four, five, and six mutations, respectively. This study identified six mutations associated with VR to FPV/r. Some of these mutations are shared with the current FPV/r Agence Nationale de Recherches sur le SIDA (ANRS) resistance score, which has been cross-validated in the CONTEXT/TRIAD data set, suggesting that the current ANRS FPV/r score is a useful tool for the prediction of VR to FPV/r in PI-experienced patients.
The use of antiretroviral therapy (ART) can sharply lower the level of human immunodeficiency virus type 1 (HIV-1) in plasma and delay the onset of clinical disease and death. Resistance to these therapies, however, can reduce or eliminate their usefulness. Drug resistance testing has proven its use in guiding treatment decisions for HIV-1-infected patients and is a recommended component of several international ART treatment guidelines. In this setting, rule-based algorithms are often proposed on the basis of the correlation between genotype and virological response (Agence Nationale de Recherches sur le SIDA [ANRS; http://www.hivfrenchresistance.org/], Stanford HIV Drug Resistance Database [http://hivdb.stanford.edu/], and the Rega Institute [http://www.rega.kuleuven.be/cev/]).
Fosamprenavir (FPV) is a prodrug of the protease inhibitor (PI) amprenavir and has shown, when boosted with ritonavir (FPV/r), safety and efficacy in antiretroviral-naïve and -experienced patients (1, 5, 8, 21). The protease mutations selected in vitro or in vivo in the presence of amprenavir (I50V, V32I plus I47V, I54LM/M, and I84V) (13) are rarely selected in patients treated by first-line FPV-containing regimens, particularly when boosted with ritonavir (RTV) (12, 22). In PI-experienced patients, few data are available concerning the prediction of virological response to FPV/r by genotypic resistance analysis (2, 17, 20). Recently, the ANRS proposed a genotypic interpretation algorithm for FPV/r including a genotypic score (L10F/I/V, L33F, M36I, I54L/M/V/A/T/S, I62V, V82A/F/C/G, I84V, and L90M) and the presence or absence of the mutations I50V or V32I plus I47V/A as an independent predictor of virological response in 73 PI-experienced patients (17). Thus, isolates with zero to three mutations among this score are considered not resistant to FPV/r, and isolates with more than three mutations among the score or with the presence of I50V or V32I plus I47V/A are considered resistant to FPV/r (http://www.hivfrenchresistance.org/; version 16). Another study identified 12 amino acid substitutions among those on the International AIDS Society (IAS) list (L10I/F/R/V, L33F, M36I, M46I/L, I54L/M/T/V, I62V, L63P, A71I/L/V/T, G73A/C/F/T, V82A/F/S/T, I84V, and L90M) and four polymorphism mutations (I13V, L19I, K55R, and L89M) that were associated with failure of FPV/r therapy (20). Failure was associated with the number of mutations, and a clinical cutoff of at least four mutations yielded a significantly poorer response. The most important difference between this study and the ANRS FPV/r genotypic interpretation was the absence of a primary mutation at codon 46 in the ANRS score. This mutation was also retained in the mutation set defined by Elston et al. in the CONTEXT study as associated with decreased virological response to FPV/r at week 12 (2). However, the differences between these mutation sets may be the consequence of using different patient populations and hence different baseline viruses with different levels and ranges of resistance to FPV/r and statistical approaches.
As it is now widely recognized that correlation studies analyzing the virological response in treatment-experienced patients according to the genotypic profile at baseline provide relevant information for establishing resistance algorithms, we report here the identification of baseline protease mutations associated with the virological response to FPV/r (700/100 mg twice daily [b.i.d.]), with a simple and previously described methodology, in PI-experienced patients enrolled in the CONTEXT and TRIAD clinical trials.
(This work was presented during the 14th Conference on Retroviruses and Opportunistic Infection, 25 to 28 February 2007 [16].)
MATERIALS AND METHODS
Patients and antiretroviral regimens.
The patients analyzed were enrolled in the CONTEXT and TRIAD trials. The CONTEXT (APV30003) trial was a phase III, randomized, multicenter, open-label study comparing the efficacy and safety of two dosing regimens of FPV/r (700 mg/100 mg twice daily or 1,400 mg/200 mg once daily) versus lopinavir-RTV (400 mg/100 mg twice daily) for 48 weeks in PI-experienced, HIV-infected adults experiencing virological failure (3). In this trial, there were three groups of patients as follows: group 1, FPV 700 mg b.i.d. plus RTV 100 mg b.i.d. plus two reverse transcriptase inhibitors (RTIs) (n = 107); group 2, FPV 1,400 mg once daily plus RTV 200 mg once daily plus two RTIs (n = 105); and group 3, lopinavir-RTV 400 mg/100 mg b.i.d. plus two RTIs (n = 103). Randomization was stratified according to the subject's plasma HIV-1 RNA level at the time of screening (1,000 to 10,000 copies/ml, >10,000 to 100,000 copies/ml, and >100,000 copies/ml). The TRIAD (APV102002) trial was a phase III, randomized, controlled, open-label, multicenter three-arm study in heavily PI-experienced subjects (defined as ≥2 prior PI-based regimens and a history of treatment with at least one ART from the three main available antiretroviral classes) comparing, in a 1:1:1 ratio, the following treatment groups: group A, 700 mg FPV/100 mg RTV b.i.d. plus optimized background treatment (OBT) (n = 21); group B, 1,400 mg FPV/100 mg RTV b.i.d. plus OBT (n = 21); and group C, 1,400 mg FPV/533 mg lopinavir/133 mg RTV b.i.d. plus OBT (n = 21) (http://ctr.gsk.co.uk/Summary/fosamprenavir/studylist.asp/).
Only data from patients enrolled in group 1 from CONTEXT and group A from TRIAD, i.e., patients receiving the approved dose of FPV/r, were pooled and analyzed in the present resistance analysis.
Genotypic resistance testing.
Genotypic resistance analysis was performed on plasma samples collected at baseline. Reported mutations were as listed at the IAS website (http://www.iasusa.org/) in September 2006 (9).
Statistical methods.
The end-point for the analysis was the decrease from baseline in HIV RNA at week 12.
First, the impact of the presence of each mutation along the protease gene (codons 1 to 99) on virological response was analyzed by comparing the change in plasma HIV-1 RNA in patients with and without the mutation by using the nonparametric Kruskal-Wallis test. Mutations present in at least 10% of patients and having P values lower than 0.10 in the univariate analysis described above were retained and then analyzed by using the removing procedure with a nonparametric test to select a final set of mutations best associated with the virological response. The removing procedure began with all k mutations retained from the univariate analysis. The nonparametric Jonckheere-Terpstra (JT) test for ordered alternatives that allows testing of a specific a priori sequence was used (6, 10, 15). From the initial set of k mutations, the first step was to remove one mutation. One by one, all combinations of k−1 mutations were investigated, and the combination providing the lower P value with the JT test was retained. In the second step, mutations were again removed one by one to compare the combinations of k−2 mutations, the combination providing the lower P value was again retained, and so on (6). The procedure ended when removing a mutation did not provide a lower P value than the previous P value.
To assess whether or not the genotypic score was an independent predictor of response, a linear multivariate regression was used, accounting for the baseline variables which were predictive of response in the univariate analysis (P < 0.10).
The statistical program used for analyses was SAS (version 9.0), and no correction for multiple testing was done.
RESULTS
Baseline patient characteristics.
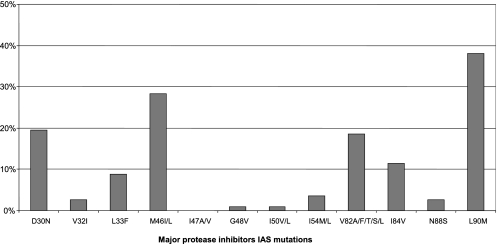
Among the 128 patients enrolled in group 1 from CONTEXT and group A from TRIAD, 113 patients were included in the present resistance analysis (CONTEXT, n = 96; TRIAD, n = 17). Fifteen out of 128 patients were excluded from this analysis because they either stopped treatment before week 12 or had missing week 12 HIV-1 RNA values. For the 113 patients retained in the analysis, baseline characteristics and details of the antiretroviral drugs associated with FPV/r are summarized in Table 1. None of the patients received nonnucleoside RTIs (NNRTIs) in the FPV/r-associated treatment. The prevalence of major PI resistance mutations at baseline defined by the IAS panel (http://www.iasusa.org/) is presented in Fig. 1 (9). The median number of major and minor IAS PI mutations (interquartile range [IQR]) was 2 (0 to 2) and 5 (3 to 7), respectively.
TABLE 1.
Baseline patient characteristics (n = 113)
| Characteristic | Valuea |
|---|---|
| Median plasma HIV-1 RNA log copies/ml (IQR) | 4.24 (3.7-4.7) |
| Median CD4 cell count/mm3 (IQR) | 300 (173-424) |
| Median age (yr) (IQR) | 40 (37-44) |
| Gender | |
| Male | 94 (83) |
| Female | 19 (17) |
| Ethnic origin | |
| American-Hispanic | 9 (8) |
| Asian | 1 (1) |
| Black | 18 (16) |
| White (Caucasian/Arabic/North African) | 85 (75) |
| Subjects with CDC classification of HIV | |
| A | 45 (40) |
| B | 31 (27) |
| C | 37 (33) |
| Subjects with prior antiretroviral therapy | |
| NRTIs | 112 (99) |
| NNRTIs | 69 (61) |
| PIs | 113 (100) |
| Subjects who had previously received PIs | |
| 1 | 61 (54) |
| 2 | 37 (33) |
| 3 | 6 (5) |
| 4 | 7 (6) |
| 5 | 2 (2) |
| Subjects with antiretrovirals administered concomitantly with FPV/r | |
| 1 NRTI | 20 (18) |
| 2 NRTIs | 80 (71) |
| 3 NRTIs | 10 (9) |
| 4 NRTIs | 1 (0.8) |
| NRTIs + ENFb | 2 (1.2) |
All values are number (%) of subjects except where indicated otherwise.
ENF, enfuvirtide.
FIG. 1.
Baseline genotypic characteristics; prevalence of major protease inhibitors resistance mutations defined by the IAS panel.
Impact of PI mutations on the virological response to FPV/r.
Overall, the median virological response at week 12 was −1.86 (range, −3.59 to 1.08) log copies/ml.
For univariate analysis, the following 14 positions on the protease gene were found to be associated with a reduced virological response to FPV/r (P < 0.1): L10F/I/V/Y, I15V, L33F, M46L, I54A/L/V/M/S, D60E/N, I62V, L63A/S/T/P/Q/V, I72E/L/M/T/V, G73C/S/T, V82A/F/I/T, I84C/K/V, L89I/M/V, and L90M. Table 2 shows the univariate analysis of the virological response according to the presence of mutated or wild-type codons at specific sites of the protease gene.
TABLE 2.
Univariate analysis of the virological response to FPV/r according to the presence of mutated or wild-type codons at specific sites of the protease gene
| Position | Amino acid | n | Median HIV-1 RNA reduction at wk 12 | P value |
|---|---|---|---|---|
| 10 | L (wild type) | 63 | −2.1 | 0.0001 |
| F/I/V/Y | 50 | −1.21 | 0.0001 | |
| 15 | I (wild type) | 89 | −1.94 | 0.0073 |
| V | 24 | −0.93 | 0.0073 | |
| 33 | L (wild type) | 100 | −1.89 | 0.0068 |
| F/I | 13 | −0.36 | 0.0068 | |
| 46 | M (wild type) | 81 | −2.09 | 0.0002 |
| I/L | 32 | −0.98 | 0.0002 | |
| 54 | I (wild type) | 89 | −2.02 | <0.0001 |
| A/L/V/M/S | 24 | −0.39 | <0.0001 | |
| 60 | D (wild type) | 99 | −1.89 | 0.0038 |
| E/N | 14 | −0.34 | 0.0038 | |
| 62 | I (wild type) | 74 | −1.94 | 0.039 |
| V | 39 | −1.12 | 0.039 | |
| 63 | L (wild type) | 11 | −2.58 | 0.012 |
| A/S/T/P/Q/V | 102 | −1.7 | 0.012 | |
| 72 | I (wild type) | 83 | −1.89 | 0.065 |
| E/L/M/T/V | 30 | −1.38 | 0.065 | |
| 73 | G (wild type) | 101 | −1.89 | 0.008 |
| C/S/T | 12 | −0.71 | 0.008 | |
| 82 | V (wild type) | 92 | −1.95 | 0.002 |
| A/F/I/T | 21 | −0.75 | 0.002 | |
| 84 | I (wild type) | 98 | −1.91 | 0.0003 |
| C/K/V | 15 | −0.31 | 0.0003 | |
| 89 | L (wild type) | 102 | −1.88 | 0.034 |
| I/M/V | 11 | −0.31 | 0.034 | |
| 90 | L (wild type) | 70 | −1.95 | 0.016 |
| M | 43 | −1.38 | 0.016 |
Boosted FPV/r genotypic score. (i) Removing procedure.
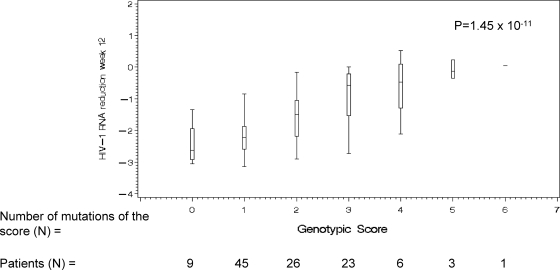
From the univariate analysis which retained 14 PI mutations (L10F/I/V/Y, I15V, L33F, M46L, I54A/L/V/M/S, D60E/N, I62V, L63A/S/T/P/Q/V, I72E/L/M/T/V, G73C/S/T, V82A/F/I/T, I84C/K/V, L89I/M/V, and L90M), the removing procedure using the JT test did not retain mutations at codons 10, 33, 62, 72, 73, 82, 89, and 90 and led to selecting the following genotypic score, named CONTEXT/TRIAD FPV/r score: I15V plus M46I/L plus I54L/M/V plus D60E plus L63P/T plus I84V; this provided the strongest association with virological response (P = 1.45 × 10−11). Figure 2 shows the median decrease in viral load as a function of the number of mutations identified in the CONTEXT/TRIAD data set providing the strongest association with virological response to FPV/r. In the 9 patients with zero mutations within this set, the median decrease in HIV RNA was −2.63 log copies/ml, and it was −2.22 (n = 45), −1.50 (n = 26), −0.58 (n = 23), −0.47 (n = 6), −0.13 (n = 3), and 0.04 (n = 1) log copies/ml in those with one, two, three, four, five, and six mutations, respectively (the median decrease in HIV RNA was −0.16 log copies/ml for the 10 patients with four or more mutations).
FIG. 2.
Median decrease in HIV RNA as a function of the number of mutations identified in the CONTEXT/TRIAD data set providing the strongest association with virological response to FPV/r (I15V plus M46I/L plus I54L/M/V plus D60E plus L63P/T plus I84V).
(ii) Multivariate analysis.
A series of univariate regression models were fitted to the data to search for predictive factors of the viral load reduction at week 12. Among the variables investigated (sex; age; CDC stage; baseline CD4; baseline HIV RNA; number of prior nucleoside RTIs [NRTIs], NNRTIs, and PIs; total number of NRTIs associated with FPV/r; use of abacavir, lamivudine, zidovudine, didanosine, stavudine, tenofir disoproxil fumarate, or T20 associated with FPV/r; and the CONTEXT/TRIAD-derived FPV/r genotypic score as a continuous variable), the following three variables were associated with a decreased virological response at week 12 (P < 0.10): number of prior NRTIs (P = 0.004), number of prior PIs (P = 0.006), and CONTEXT/TRIAD FPV/r genotypic score (P < 0.0001). From these three variables, the final multivariate model retained only the CONTEXT/TRIAD FPV/r genotypic score as being significantly associated with the virological response (P < 0.0001).
(iii) Impact of the FPV/r ANRS genotypic score on the virological response at week 12.
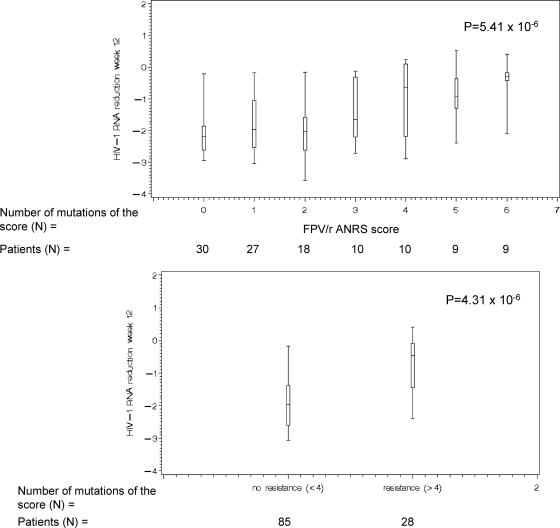
Another FPV/r genotypic score has been previously described, which is currently recommended by the ANRS (17). The ANRS FPV/r classifies strains as resistant to FPV/r if there are at least four mutations among the following combination of mutations (L10F/I/V plus L33F plus M36I plus I54A/L/M/S/T/V plus I62V plus V82A/C/F/G plus I84V plus L90M) or the presence of I50V or 32I plus 47V and classifies strains as susceptible to FPV/r in cases of three or fewer mutations among this score (http://www.hivfrenchresistance.org/; version 16). In order to evaluate the accuracy of the FPV/r ANRS genotypic score, we applied this score in the CONTEXT/TRIAD data set. Figure 3 shows the median decrease in HIV RNA in the CONTEXT/TRIAD data set according to the FPV/r ANRS genotypic score as a function of the number of mutations and having <4 or ≥4 mutations as it is defined in the ANRS rules (P = 5.41 × 10−6 and P = 4.31 × 10−6, respectively).
FIG. 3.
Median decrease in HIV RNA as a function of the number of mutations defined by the ANRS FPV/r genotypic score (L10F/I/V plus L33F plus M36I plus I54A/L/M/S/T/V plus I62V plus V82A/C/F/G plus I84V plus L90M).
DISCUSSION
Generation and clinical validation of genotypic resistance scores are based on correlation analyses between the genotypic profile at baseline and virological response. This provides objective information for the guidance of drug selection based on genotypes, in particular for treatment-experienced patients (11, 14, 23). Here, we have identified a list of protease mutations that impair the virological response to boosted FPV in PI-experienced patients using stepwise methodologies as previously proposed (6, 14, 15, 23). We described the mutations associated with FPV/r virological response at week 12 based on virological data from a pooled analysis of FPV/r 700/100 mg b.i.d. arms of two different clinical trials conducted in PI-experienced patients.
Among the mutations identified in the CONTEXT/TRIAD genotypic score (I15V plus M46I/L plus I54L/M/V plus D60E plus L63P/T plus I84V) associated with FPV/r virological response at week 12, some of them have been already identified as amprenavir- and FPV/r-resistant mutations in clinical trials using amprenavir or FPV/r and are already considered FPV/r IAS mutations (i.e., M46I/L, I54L/M/V, and I84V). Indeed, mutations at position 46 have already been identified in other FPV/r resistance scores (2, 20). Moreover, mutations at positions 46, 54, and 84 are selected in vitro and in vivo in the presence of amprenavir or FPV/r and have also been identified in other FPV/r resistance scores (2, 17, 20). This study also highlights the possible importance of substitutions of two new amino acid residues (I15V and D60E) in virological response to FPV/r therapy that will need to be independently verified. However, both mutations have already been implicated in decreasing virological response to other PIs, such as saquinavir for I15V and atazanavir for D60E (14, 23). The differences of PI resistance mutations highlighted in different genotypic scores might be due in part to higher trough exposure in patients receiving FPV/r compared to amprenavir. It has been shown that plasma trough levels drive the selection of mutation pathways during development of amprenavir resistance (4).
A linear, gradual relationship was observed between the week 12 change from baseline in plasma HIV-1 RNA and the increasing number of mutations. However, the number of patients with no virological response (less than −0.5 log10 plasma HIV-1 RNA decrease from baseline) and with four or more mutations from the identified score in this data set was low (n = 10). Hence, no clear clinical cutoff could be derived from the analysis and no categorical grouping (susceptible or resistant) could be defined. When applying the current ANRS FPV/r mutation set in the CONTEXT/TRIAD data set, the baseline resistance range equally illustrates the limited representation of patients with nonresponse and ≥4 baseline mutations (n = 28) and therefore potentially makes the current ANRS FPV/r score not applicable to a broad range of PI-experienced patients with viruses more likely to show reduced susceptibility to FPV/r.
However, the current ANRS FPV/r score (version 16) was highly predictive (P = 5.41 × 10−6) of the virological response in the CONTEXT/TRIAD data set, providing an external cross-validation of this score in an independent data set (17). It is not surprising that a score derived from another data set predicts the virological response less efficiently than a score derived from this data set (P = 1.45 × 10−11). This may be the consequence of using different patient and virus populations. Of note, although the current ANRS FPV/r score was derived from a smaller data set (n = 73) than the present study, the number of patients with lower susceptibilities and who are less responsive to FPV/r (less than −0.5 log10 change from baseline in plasma HIV-1 RNA) was greater than in this study (n = 39 and n = 28, respectively) and therefore more applicable to a wide population of PI-experienced patients. Although the statistical methodology used to determine FPV/r genotypic score and the median number of major and minor PI resistance mutations were similar in the present study and in the ANRS study, genotypic scores were not strictly identical, probably because of different patterns of mutations at baseline in the two different data sets. This may reflect the multiplicity and complexity of mutational interactions in determining genotypic resistance, the various therapeutic histories of the patient populations studied, and limitations of the study population sizes in view of that complexity. The best algorithm for predicting virological responses to FPV/RTV might be somehow derived by a meta-analysis combining the data from the present study and other studies (17, 20). However, the presence of I54L/M/V and I84V in both scores probably reflects a large weight of these mutations on the response to FPV/r. This is highlighted in the Stanford HIV Drug Resistance Database (http://hivdb.stanford.edu/), where each drug resistance mutation is assigned a drug penalty score. For example, for FPV, mutations I54V, I54M, and I54L have penalty scores of 10, 20, and 15, respectively, and mutation I84V has a penalty score of 35. In addition, by themselves, I54M/L and I84V each have a strong effect on amprenavir susceptibility, reducing it by about fourfold (18, 19). The present study and the ANRS studies did not highlight the impact of either I50V or V32I plus I47V that are known to be selected by amprenavir and FPV in patients failing a first-line regimen; however, this is explained simply by the fact that patients analyzed in both studies had no prior experience with either amprenavir or FPV and therefore viruses harbored a prevalence of these mutations that was too low to detect any association with virological response to FPV/r (e.g., no I50V or I47V in the CONTEXT/TRIAD data set). However, in the current ANRS algorithm (version 16), the presence of I50V and/or V32I plus I47V is even considered a resistance to FPV/r, based on the known specific impact of these mutations on phenotypic resistance to amprenavir and the ability of the drug to select for them upon first-line and further lines of therapies.
The nonparametric statistical methodology allows a comparison of groups of patients according to the number of mutations harbored by their viruses. The JT test with a specific alternative hypothesis is specifically adapted to the situation (6, 7). Indeed, when comparing groups of patients with distinct numbers of resistance mutations, one would expect that patients with no resistance mutations would have better virological responses than patients with one mutation, who in turn would have better virological responses than patients with two mutations, and so on. However, one limitation of the JT test is that no adjustment can be made. Indeed, adjustment can be done only with parametric models and not with nonparametric methods. Thus, in the statistical analysis, no adjustment has been made regarding the susceptibility to the NRTIs received in the background regimen. Consequently, this can potentially introduce a bias effect in the analysis, but in CONTEXT and TRIAD trials, where the choice of the NRTIs in the background regimen was guided by a resistance test for all patients, the importance of making an adjustment to susceptibility to NRTI is then reduced.
In conclusion, we identified a set of mutations associated with virological response to FPV/r in PI-experienced patients enrolled in CONTEXT and TRIAD clinical trials but no clear cutoff could be identified. However, this set of mutations shares some mutations with the current FPV/r ANRS resistance score, which has been cross-validated in the CONTEXT/TRIAD data set, confirming that FPV/r ANRS score (version 16) is a useful tool for the prediction of virological response to FPV/r in PI-experienced patients.
Acknowledgments
The APV30003 investigators are as follows: Mark Bloch, John Chuah, David Cooper, Robert Finlayson, Jennifer Hoy, Robert McFarlane, John Quin, Norm Roth, and Cassy Workman from Australia; Nathan Clumeck from Belgium; Kevin A. Gough, Patrice Junod, Donald Kilby, John MacLeod, Anita Rachlis, Walter F. Schlech III, Fiona Smaill, Benoit Trottier, and Christos Tsoukas from Canada; Carlos Beltran from Chile; Michelle Bentata, J. Francois Delfraissy, Gilles Force, Herve Gallais, Jean Albert Gastaut, Bruno Hoen, Marie-Aude Khuong Josses, Alain Lafeuillade, Jean Marie Lang, Jean-Michel Livrozet, Jean-Michel Molina, Willy Rozenbaum, Daniel Sereni, Pierre de Truchis, Daniel Vittecoq, and Patrick Yeni from France; Keikawus Arasteh, Gerd Faetkenheuer, Stefan Fenske, Hans Jaeger, Stefan Mauss, Andreas Plettenberg, Juergen Rockstroh, Rheinhold Schimdt, and Schlomo Staszewski from Germany; Adriano Lazzarin and Nicolo Piernsatelli from Italy; Antonio Diniz and Eugenio Teofilo from Portugal; Jorge L. Santana Bagur and Gabriel A. Martinez from Puerto Rico; Maria Jesus Perez Elias, Bonaventura Clotet-Sala, Vicente Estrada, Pompeyo Viciana Fernandez, Jose Maria Gatell-Artigas, Enrique Ortega, Jose Maria Pena, Federico Pulido, Rafael Rubio, and Agustin Munoz Sanz from Spain; Enos Bernasconi, Milos Opravil, and Amalio Telenti from Switzerland; Philippa J. Easterbrook, Brian George Gazzard, and Celia J. Skinner from the United Kingdom; and Bisher Akil, Stephen L. Becker, Nicolaos C. Bellos, Daniel S. Berger, Andre Brutus, Paul Cimoch, David V. Condoluci, Gregg O. Coodley, Timothy P. Cooley, Edwin DeJesus, Thomas M. File, Jr., Gervais Frechette, Joseph C. Gathe, Jr., Jose A. Giron, Eliot Godofsky, Mitchell Goldman, Stephen L. Green, Patrick G. Haggerty, Barbara Hanna, Kevin King, G. Steven Kooshian, Pardeep Kumari, Anthony LaMarca, Danny J. Lancaster, Christopher Lucasti, Alberto Mestre, Robert A. Myers, Jr., Jeffrey P. Nadler, Ronald G. Nahass, Cheryl L. Newman, Robert Orenstein, David A. Parks, Gerald Pierone, Jr., Arnaldo R. Quinones, Bruce S. Rashbaum, Frank S. Rhame, Gary Richmond, Allan E. Rodriguez, James H. Sampson, Michael G. Sension, Gail Skowron, Kimberly Y. Smith, Corklin Steinhart, Richard Stryker, Kimberly K. Summers, Karen T. Tashima, Nathan M. Thielman, Fehmida Visnegarwala, Barbara H. Wade, Charles M. Walworth, Lawrence J. Wheat, Michael Goldman, David P. Wright, Benjamin Young, and Christine A. Zurawski from the United States.
The APV102002 investigators are as follows: Panagiotis Gargalianos-Kakolyris, George Chrysos, and Marios Lazanas from Greece; Pere Domingo, Vicente Estrada, Federico Pulido, Rafael Rubio, Vicente Soriano, Jose Alberto Terrón, Miguel Górgolas, Blai Coll, Vicente Abril, and Joaquín Portilla from Spain; Sylvie Trottier, Donald Kilby, Graham Smith, Jean-Guy Baril, Ken Logue, Brian Conway, Christos Tsoukas, and Fiona Smaill from Canada; Andreas Plattenberg from Germany; Maria-Louisa Partisani, Jean-Michel Molina, Daniel Sereni, Alain Lafeuillade, Thierry May, Jean-François Bergman, Marie-Aude Khuong-Josses, Olivier Bouchaud, Gilles Pialoux, and Félicia David-Ouaknine from France; Nathan Clumeck from Belgium; Laura Sighinolfi, Francesco Mazzotta, Francesco Leoncini, Paolo Sacchi, Roberto Cauda, Anna Maria Catellan, Giovanni Penco, and Pietro Caramello from Italy; Jonathon Anderson, David Cooper, and Cassy Workman from Australia; and Martin Fisher, Margaret Johnson, Mia Huengsberg, Chloe Orkin, Phillipa Easterbrook, Phillip Hay, Alan Winston, and Hiten Thaker from the United Kingdom.
Footnotes
Published ahead of print on 13 October 2008.
REFERENCES
- 1.DeJesus, E., A. Lamarca, M. Sension, C. Beltran, and P. Yeni. 2003. Abstr. 10th Conf. Retroviruses Opportun. Infect., abstr. 178.
- 2.Elston, R., S. White, P. Yates, F. Xu, N. Richards, S. Sharp, M. B. Wire, and M. Tisdale. 2004. Abstr. XIII Int. HIV Drug Resist. Workshop, abstr. 115.
- 3.Elston, R., P. Yates, M. Tisdale, N. Richards, S. White, and E. DeJesus. 2004. Abstr. XV Int. AIDS Conf., abstr. MoOrB1055.
- 4.Elston, R., S. Randall, F. Xu, W. Harris, V. Manohitharajah, M. Maguire, A. Rakik, M. Ait-Khaled, D. S. Stein, M. Tisdale, and W. Snowden. 2001. Abstr. 2nd Int. Workshop Clin. Pharmacol. HIV Ther., abstr. 5.
- 5.Eron, J., Jr., P. Yeni, J. Gathe, Jr., V. Estrada, E. DeJesus, S. Staszewski, P. Lackey, C. Katlama, B. Young, L. Yau, D. Sutherland-Phillips, P. Wannamaker, C. Vavro, L. Patel, J. Yeo, and M. Shaefer. 2006. The KLEAN study of fosamprenavir-ritonavir versus lopinavir-ritonavir, each in combination with abacavir-lamivudine, for initial treatment of HIV infection over 48 weeks: a randomised non-inferiority trial. Lancet 368:476-482. [DOI] [PubMed] [Google Scholar]
- 6.Flandre, P., A. G. Marcelin, J. Pavie, N. Shmidely, M. Wirden, O. Lada, M. C. Bernard, J. M. Molina, and V. Calvez. 2005. Comparison of tests and procedures to build clinically relevant genotypic scores: application to the Jaguar study. Antivir. Ther. 10:479-487. [PubMed] [Google Scholar]
- 7.Flandre, P., and J. O'Quigley. 2007. Predictive strength of Jonckheere's test for trend: an application to genotypic scores in HIV infection. Stat. Med. 26:4441-4454. [DOI] [PubMed] [Google Scholar]
- 8.Gathe, J. C., Jr., P. Ive, R. Wood, D. Schurmann, N. C. Bellos, E. DeJesus, A. Gladysz, C. Garris, and J. Yeo. 2004. SOLO: 48-week efficacy and safety comparison of once-daily fosamprenavir/ritonavir versus twice-daily nelfinavir in naive HIV-1-infected patients. AIDS 18:1529-1537. [DOI] [PubMed] [Google Scholar]
- 9.Johnson, V. A., F. Brun-Vezinet, B. Clotet, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2006. Update of the drug resistance mutations in HIV-1: fall 2006. Top. HIV Med. 14:125-130. [PubMed] [Google Scholar]
- 10.Jonckheere, A. R. 1954. A distribution-free k-sample test against ordered alternatives. Biometrika 41:133-145. [Google Scholar]
- 11.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, J. Sylte, B. Richards, B. Bernstein, R. Rode, and E. Sun. 2002. Analysis of the virological response with respect to baseline viral phenotype and genotype in protease inhibitor-experienced HIV-1-infected patients receiving lopinavir/ritonavir therapy. Antivir. Ther. 7:165-174. [PubMed] [Google Scholar]
- 12.MacManus, S., P. J. Yates. R. C. Elston. S. White, N. Richards, and W. Snowden. 2004. GW433908/ritonavir once daily in antiretroviral therapy-naive HIV-infected patients: absence of protease resistance at 48 weeks. AIDS 18:651-655. [DOI] [PubMed] [Google Scholar]
- 13.Maguire, M., D. Shortino, A. Klein, W. Harris, V. Manohitharajah, M. Tisdale, R. Elston, J. Yeo, S. Randall, F. Xu, H. Parker, J. May, and W. Snowden. 2002. Emergence of resistance to protease inhibitor amprenavir in human immunodeficiency virus type 1-infected patients: selection of four alternative viral protease genotypes and influence of viral susceptibility to coadministered reverse transcriptase nucleoside inhibitors. Antimicrob. Agents Chemother. 46:731-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcelin, A. G., P. Flandre, C. de Mendoza, B. Roquebert, G. Peytavin, L. Valer, M. Wirden, S. Abbas, C. Katlama, V. Soriano, and V. Calvez. 2007. Clinical validation of saquinavir/ritonavir genotypic resistance score in protease-inhibitor-experienced patients. Antivir. Ther. 12:247-252. [PubMed] [Google Scholar]
- 15.Marcelin, A.-G., P. Flandre, J. Pavie, N. Schmidely, M. Wirden, O. Lada, D. Chiche, J.-M. Molina, and V. Calvez. 2005. Clinically relevant genotype interpretation of resistance to didanosine. Antimicrob. Agents Chemother. 49:1739-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcelin, A.-G., P. Flandre, J.-M. Molina, C. Katlama, P. Yéni, F. Raffi, Z. Antoun, M. Ait-Khaled, and V. Calvez. 2007. Abstr. 14th Conf. Retroviruses Opportun. Infect., abstr. 608.
- 17.Masquelier, B., K. L. Assoumou, D. Descamps, L. Bocket, J. Cottalorda, A. Ruffault, A. G. Marcelin, L. Morand-Joubert, C. Tamalet, C. Charpentier, G. Peytavin, Z. Antoun, F. Brun-Vezinet, and D. Costagliola. 2008. Clinically validated mutation scores for HIV-1 resistance to fosamprenavir/ritonavir. J. Antimicrob. Chemother. 61:1362-1368. [DOI] [PubMed] [Google Scholar]
- 18.Murphy, M. D., G. I. Marousek, and S. Chou. 2004. HIV protease mutations associated with amprenavir resistance during salvage therapy: importance of I54M. J. Clin. Virol. 30:62-67. [DOI] [PubMed] [Google Scholar]
- 19.Parkin, N. T., C. Chappey, and C. J. Petropoulos. 2003. Improving lopinavir genotype algorithm through phenotype correlations: novel mutation patterns and amprenavir cross-resistance. AIDS 17:955-961. [DOI] [PubMed] [Google Scholar]
- 20.Pellegrin, I., D. Breilh, G. Coureau, S. Boucher, D. Neau, P. Merel, D. Lacoste, H. Fleury, M. C. Saux, J. L. Pellegrin, E. Lazaro, F. Dabis, and R. Thiebaut. 2007. Interpretation of genotype and pharmacokinetics for resistance to fosamprenavir-ritonavir-based regimens in antiretroviral-experienced patients. Antimicrob. Agents Chemother. 51:1473-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quercia, R., E. Garnier, V. Ferre, P. Morineau, B. Bonnet, C. Soulard, and F. Raffi. 2005. Salvage therapy with ritonavir-boosted amprenavir/fosamprenavir: virological and immunological response in two years follow-up. HIV Clin. Trials 6:73-80. [DOI] [PubMed] [Google Scholar]
- 22.Ross, L., C. Vavro, B. Wine, and Q. Liao. 2006. Fosamprenavir clinical study meta-analysis in ART-naive subjects: rare occurrence of virologic failure and selection of protease-associated mutations. HIV Clin. Trials 7:334-338. [DOI] [PubMed] [Google Scholar]
- 23.Vora, S., A. G. Marcelin, H. F. Gunthard, P. Flandre, H. H. Hirsch, B. Masquelier, A. Zinkernagel, G. Peytavin, V. Calvez, L. Perrin, and S. Yerly. 2006. Clinical validation of atazanavir/ritonavir genotypic resistance score in protease inhibitor-experienced patients. AIDS 20:35-40. [DOI] [PubMed] [Google Scholar]