Abstract
Amphotericin B (AMB) is the most widely used polyene antifungal drug for the treatment of systemic fungal infections, including invasive aspergillosis. It has been our aim to understand the molecular targets of AMB in Aspergillus fumigatus by genomic and proteomic approaches. In transcriptomic analysis, a total of 295 genes were found to be differentially expressed (165 upregulated and 130 downregulated), including many involving the ergosterol pathway, cell stress proteins, cell wall proteins, transport proteins, and hypothetical proteins. Proteomic profiles of A. fumigatus alone or A. fumigatus treated with AMB showed differential expression levels for 85 proteins (76 upregulated and 9 downregulated). Forty-eight of them were identified with high confidence and belonged to the above-mentioned categories. Differential expression levels for Rho-GDP dissociation inhibitor (Rho-GDI), secretory-pathway GDI, clathrin, Sec 31 (a subunit of the exocyst complex), and RAB GTPase Ypt51 in response to an antifungal drug are reported here for the first time and may represent a specific response of A. fumigatus to AMB. The expression of some of these genes was validated by real-time reverse transcription-PCR. The AMB responsive genes/proteins observed to be differentially expressed in A. fumigatus may be further explored for novel drug development.
Invasive aspergillosis, caused primarily by Aspergillus fumigatus, has emerged as the leading cause of mortality among immunocompromised patients with underlying hematological diseases or bone marrow transplantation (7, 38). Amphotericin B (AMB), a broad-spectrum fungicidal agent, has been widely used to treat patients with invasive aspergillosis. AMB is reported to be fungicidal (MFC/MIC ≤ 4) against all A. fumigatus and Aspergillus flavus isolates but not on Aspergillus terreus isolates (25). Its therapeutic use is limited by its toxicity (nephrotoxicity, cytotoxicity, and hepatotoxicity, etc.) in the host (13) and development of resistance in fungal isolates (42). However, use of lipid formulations of AMB, administration by the inhalation route, and development of less toxic analogues have facilitated better therapeutic outcomes (35). In general, it is known that AMB intercalates with ergosterol of the fungal cell membrane and forms pores resulting in leakage of fungal cell components, which leads to death via osmotic collapse (15, 22). It also promotes oxidative damage to cell membranes through generation of reactive oxygen species (ROS) (43) and damage to DNA resulting in loss of cell viability, a characteristic of apoptosis (31).
Earlier efforts reported genome-wide expression analysis to understand the mechanism of action and specific effects of AMB and other antifungal drugs on nonfilamentous fungal species, such as Saccharomyces cerevisiae and Candida albicans (1, 24, 46). However, in an earlier study of interaction of A. fumigatus with voriconazole (11), decreased mRNA expression of ergosterol biosynthesis genes was observed, which was in contrast with previous reports of S. cerevisiae and C. albicans (1, 24) suggesting that A. fumigatus, being a filamentous fungus, might respond differently. Since AMB is effective against invasive aspergillosis, we studied its molecular targets in A. fumigatus by using proteomic and transcriptomic approaches.
MATERIALS AND METHODS
A. fumigatus strain.
A. fumigatus strain Af293, isolated from the lungs of an invasive-aspergillosis patient, was kindly provided by D. W. Denning (School of Medicine, University of Manchester, Manchester, United Kingdom). The Af293 strain was used for a whole-genome sequencing project by the Sanger Institute (Hinxton, Cambridge, United Kingdom) and the J. Craig Venter Institute (Rockville, MD).
Spore harvesting.
A. fumigatus culture was maintained on Sabouraud dextrose agar (SDA) (4.7 g/liter; Himedia, Mumbai, India) slants. Spores were harvested from A. fumigatus culture (after 72 h) growing on SDA by using phosphate-buffered saline (PBS) with 0.05% Tween 20 (PBST), centrifuged at 10,000 rpm for 10 min at 4°C, and washed with PBS twice. Conidia were then allowed to swell in PBS containing Ca2+ and Mg2+ for 1 h, followed by a viability count (numbers of CFU/ml) on SDA plates by a hemocytometer.
Determination of MIC50 of AMB.
A. fumigatus spores (1 × 106 spores/ml) were incubated in RPMI 1640 medium (with l-glutamine and sodium bicarbonate), pH 7.4 (Sigma, St. Louis, MO), alone or along with AMB (0.03 to 32 μg/ml) (Glaxo pharmaceuticals, Delhi, India) at 37°C for 24 h in a radiation-sterilized 96-well flat-bottom microtiter plate (Tarsons, Kolkata, India) (3). One hundred microliters of the diluted (two times) conidial suspension (the final volume in each well was 200 μl) was inoculated with each microdilution well, containing 100 μl of the diluted (two times) drug concentrations. Medium containing AMB was replaced with fresh medium (200 μl) in all the wells, and 10 μl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; 5 mg/ml) (Sigma, St. Louis, MO) was added to each well, which was again incubated at 37°C for 3 h. The plate was centrifuged at 3,000 rpm, and supernatant was removed. A. fumigatus (comprising spores and hyphae) was lysed with acid-isopropanol (0.4 N HCl in isopropanol). The plate was centrifuged at 3,000 rpm, and supernatant was transferred to the fresh plate. The percentage of MTT conversion to its formazan derivative for each well was calculated by comparing the optical densities at 570 nm of the wells with that of the AMB-free control, and the concentration of AMB (MIC50) for Af293 was determined from a standard curve (23). The experiment was performed thrice, and the mean MIC50 value was considered.
Large-scale culture for total RNA and protein extraction.
A. fumigatus spores (1 × 106/ml) were inoculated in 100 ml of RPMI 1640 medium (with l-glutamine and sodium bicarbonate) (Sigma, St. Louis, MO) alone or with AMB (at the MIC50, i.e., 0.125 μg/ml, as determined above) in culture flasks, followed by incubation at 37°C for 24 h. A. fumigatus alone (comprising spores and hyphae) or A. fumigatus treated with AMB (comprising spores and hyphae) (as observed under a microscope) was scraped from the culture flask and pelleted at 4°C by centrifugation at 18,000 rpm for 15 min and kept at −70°C. Microarray and proteomic analyses were carried out with each of two biological replicates obtained from the above-mentioned experiments.
Proteomic analysis. (i) Protein extraction.
A. fumigatus alone or A. fumigatus treated with AMB (at the MIC50) was ground in liquid nitrogen (1.5 g of the wet mat) separately, and total protein was extracted at 4°C in cold 50 mM sodium phosphate buffer, pH 7.0, with 1 mM phenylmethylsulfonyl fluoride, 2 mM EDTA, and 0.2 mM dithiothreitol (DTT) for 3 h with constant stirring. The protein extract was centrifuged at 12,000 × g for 20 min at 4°C, and supernatant was collected to give a final concentration of 5% trichloroacetic acid. The precipitate was washed with cold acetone, lyophilized, and stored at −70°C (9). Protein was dissolved in rehydration buffer containing 8 M urea, 2% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), and 25 mM DTT and estimated by Bradford's method (6).
(ii) 2-D PAGE.
Two-dimensional polyacrylamide gel electrophoresis (2-D PAGE) of proteins extracted from A. fumigatus was performed as described by Görg et al. (18). Proteins (600 μg) solubilized in 300 μl rehydration buffer (0.2% Bio-Lyte [pI range, 5 to 8] and 0.002% bromophenol blue) were applied to immobilized-pH-gradient (IPG) strips (17 cm; pI range, 5 to 8) (Bio-Rad, Hercules, CA). The first-dimensional separation was carried out with a Protean isoelectric focusing (IEF) cell system (Bio-Rad, Hercules, CA). IEF was performed at 20°C, using the following conditions: 250 V for 30 min, 10,000 V for 3 h, and 10,000 V for an additional 40,000 V·h. After IEF, the IPG strips were kept in equilibration buffer (6 M urea, 20% glycerol, 2% sodium dodecyl sulfate [SDS], 1.5 M Tris-HCl buffer, pH 8.8) with 2% DTT for 15 min and then in equilibration buffer with 2.5% iodoacetamide for 15 min (Bio-Rad protocol). The second-dimensional separation was carried out with 12% SDS-PAGE gels (20 by 20 cm2), initially at 70 V for 30 min and then at 150 V per gel, until the dye front reached the gel bottom. Proteins were visualized by Coomassie brilliant blue R-250 staining (Sigma, St. Louis, MO).
(iii) Gel image and image analysis.
Images of Coomassie-stained 2-D gels were acquired using a Fluor-S MultiImager (Bio-Rad, Hercules, CA) with a visible light source, and image analysis was carried out using PDQuest image analysis software, version 7.2. All images were taken under uniform settings, and three or four major spots in different parts of the gel were used for fixing the coordinates. 2-D gels were normalized for small variations in staining or protein loads by determining the total optical densities of the protein spots. The consensus protein spots present in 2-D gel replicates with spot intensity variations of <10% were considered for further analysis. The pooled spot intensities from the replicates were considered for determination of differentially expressed proteins (twofold up- or downregulated). These protein spots were then excised from the 2-D gel and subjected to trypsin digestion.
(iv) In situ tryptic digestion of proteins.
Protein spots of interest were excised from the gel, and in situ tryptic digestion was carried out as described earlier (10). In the final step, the lyophilized tryptic digestion product was reconstituted in 4 to 5 μl of 50% acetonitrile-0.1% trifluoroacetic acid to dissolve the extracted peptides for mass spectrometric (MS) analysis.
(v) MS analysis.
Peptide mass mapping was performed with 0.6 μl reconstituted extract with a 0.6-μl fresh α-cyano-4-hydroxycinnamic acid (CHCA) (Applied Biosystems, Framingham, MA) matrix on a 384-well matrix-assisted laser desorption ionization (MALDI) plate. The samples were analyzed with a MALDI-tandem time of flight (TOF-TOF) MS (model 4800 proteomics analyzer; Applied Biosystems, Framingham, MA) working in positive-ion-reflector mode. The instrument was calibrated to an accuracy level of <10 ppm by using a calibration mixture of known standard synthetic peptides with an m/z range of 800 to 4,000. Peptide mass fingerprints (PMFs) of the tryptic digests were acquired in automation mode. Then, PMF data were interrogated for protein identification with the NCBI database by using the Mascot search engine, and analysis was done with global proteomic solutions software (GPS Explorer, version 3.6; Applied Biosystems, Framingham, MA) automatically. The search parameters for the PMFs were set as follows. Mascot search engine version 1.9 was used to search the NCBI fungus database. Fixed and variable modifications were considered (Cys as an S-carboamidomethyl derivate and methionine as oxidized methionine), and one missed cleavage site was allowed. A Mascot score above 58 was taken as the cutoff, molecular mass was unrestricted, and sequence coverage was >25% (17). A mass tolerance of 50 ppm and a requirement of at least five peptides matched were also used for protein identification. Only those proteins that showed significant matches with A. fumigatus proteins were considered in the present study (17).
For confirmation of protein identification by PMF, the peptides were further subjected to fragmentation by tandem MS (MS-MS) in result-dependent analysis mode. The resulting MS-MS ion spectra were again interrogated with the NCBI database (fungi) for confirmation of the protein identifications. If a minimum of two peptides showed individual ion scores of ≥30, the identification was considered significant. The function and functional category of each differentially expressed protein on exposure to AMB were assigned from the ExPASy database for biochemical pathways (http://www.expasy.ch/tools/pathways/).
Microarray analysis. (i) Isolation of total RNA from A. fumigatus.
DNA-free total RNA was isolated from A. fumigatus by using RNeasy plant minikits (Qiagen, GmbH, Hilden, Germany). Fungal pellets stored at −70°C were crushed using a mortar and pestle separately in the presence of liquid nitrogen. The cell powder was resuspended in 2 ml of RLT lysis buffer provided with the RNeasy plant minikit and centrifuged at 10,000 rpm for 2 min, followed by further steps for isolation of total RNA, including treatment with RNase-free DNase, as described in the kit protocol. Gel electrophoresis of total RNA revealed intact 28S and 18S ribosomal bands and no genomic DNA contamination. RNA concentration was determined spectrophotometrically by measuring absorption at 260 nm with a Lambda Bio 20 UV-visible-light spectrophotometer (Perkin Elmer, Wellesley, MA).
(ii) Fluorescence labeling.
Total RNA (7 μg) from A. fumigatus alone or A. fumigatus treated with AMB (at the MIC50) was reverse transcribed separately using fluorescein-labeled dCTP or biotin-labeled dCTP from Micromax TSA labeling kits (Perkin Elmer, Wellesley, MA). Labeled cDNA was purified by isopropanol precipitation per the protocol of the Micromax TSA labeling kit. Each of the purified labeled cDNAs was dissolved in 20 μl of hybridization buffer (50% formamide, 5× sodium citrate-sodium chloride [SSC; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS, and 300 μg salmon sperm DNA. A dye swap experiment with two biological replicates was performed to ensure the reproducibility of each gene expression pattern.
(iii) Prehybridization of DNA slides.
The whole-genome A. fumigatus microarray slides (version 1) were obtained from the J. Craig Venter Institute (Rockville, MD). Each of the slides was prehybridized with prehybridization buffer (5× SSC, 0.1% SDS, 1% bovine serum albumin) for 1 h at 42°C. The slides were washed thrice with Milli-Q-purified water for 5 min, followed by 2 min of washing in isopropyl alcohol. The slides were then removed from isopropyl alcohol and immediately centrifuged at 700 rpm for 10 min at room temperature and subjected to a hybridization process (http://pfgrc.jcvi.org/index.php/microarray/protocols.html).
(iv) Hybridization and scanning of DNA slides.
Labeled cDNAs in hybridization buffer were mixed and heated at 94°C for 5 min before being loaded onto the array slides. The slides were covered with a 24- by 50-mm glass coverslip (Erie Scientific Company, Portsmouth, NH) and kept in hybridization chamber (Corning, St. Louis, MO) at 42°C for 16 to 18 h to prevent evaporation (http://pfgrc.jcvi.org/index.php/microarray/protocols.html). After hybridization, the slides were processed per the protocol of the Micromax TSA labeling kit and scanned in both the Cy3 and the Cy5 channels with a model 4200 Axon Instruments scanner with a 10-μm resolution. Each of the signals was converted into a resolution of 16 bits per pixel.
(v) Image analysis.
TIFF images were analyzed using GenePix Pro 6.0 software. The signal intensity value of each spot was reduced by the local background level for each spot. Total-intensity-based normalization by means was used as previously described by Foster and Huber (14). Gene expression ratios of ≥2 in both replicates (dye swap) were considered to be differentially expressed genes. Clustering of the two independent array data was carried out using Avadis microarray gene expression analysis software. The function and functional category of each differentially expressed gene on exposure to AMB were assigned from the ExPASy database for biochemical pathways (http://www.expasy.ch/tools/pathways/).
Real-time PCRs.
Total RNA was extracted from A. fumigatus alone or A. fumigatus treated with AMB (at the MIC50) as described above. cDNA was synthesized using total RNA after treatment with RNase-free DNase, using the Super Script III first-strand synthesis system for reverse transcription-PCR (RT-PCR; Invitrogen, CA). Primers were designed using AmplifX version 1.4.0 software, with a preference for binding of one primer of a pair to the exon-exon junction, to amplify amplicons of approximately 100 to 150 bp for the respective genes (see Table S1 in the supplemental material). The specificity of primers was examined by analyzing their cDNA amplification (PCR) product with agarose gel electrophoresis and later by analyzing the dissociation curve in the real-time RT-PCR.
Real-time RT-PCRs were performed using the ABI 7900 HT Fast real-time PCR system (Perkin-Elmer Applied Biosystems). Sybr green ER quantitative PCR supermix (Invitrogen, CA) was used to perform real-time PCRs to study relative changes in mRNA expression profile. In the pilot experiments with real-time RT-PCR for the GAPDH (glyceraldehyde-3-phosphate dehydrogenase), actin, and 18S rRNA genes in A. fumigatus alone or A. fumigatus treated with AMB (at the MIC50), GAPDH showed insignificant difference in expression. Therefore, it was included as a control. The results obtained for the target genes were normalized using the threshold cycles (CTs) obtained for the GAPDH (control gene) cDNA amplifications run on the same plate by using the ΔΔCT method (39). The RT-PCR thermal cycling conditions consisted of an initial step at 50°C for 2 min, followed by 95°C for 10 min. The next stage involved 40 cycles as follows: 95°C for 15 s, 58°C for 30 s, and 72°C for 30 s, followed by a dissociation step (40).
In all experiments, appropriate negative controls containing no template DNA or RNA were subjected to the same procedure to exclude or detect any possible contamination or carryover. The experiment was carried out with three biological replicates (performed in triplicate), and the average ΔΔCT values and standard deviations for the three biological replicates were calculated.
Nucleotide sequence accession number.
The microarray data have been made available in the public domain under GenBank accession number GSE10505 (http://www.ncbi.nlm.nih.gov/geo).
RESULTS
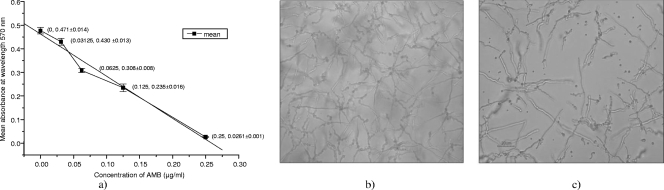
The studies of AMB response were carried out with the MIC50 of AMB against A. fumigatus in RPMI 1640, pH 7.4, which was found to be 0.125 ± 0.008 μg/ml at 37°C after 24 h (Fig. 1).
FIG. 1.
(a) Determination of MIC50s of AMB for A. fumigatus. (b, c) Microscopic images of A. fumigatus alone (b) and treated with AMB at 0.125 μg/ml (the MIC50) (c). The values in the graph (a) represent concentrations of AMB and mean absorbance values ± standard deviations. The MIC50 of AMB for A. fumigatus was determined to be 0.125 μg/ml. The microscopic images were taken at ×40 magnification on a Nikon Eclipse TE2000-S phase contrast inverted microscope (Nikon, Melville, New York, NY). Spores (1 × 106 per ml) of A. fumigatus were incubated for 24 h at 37°C in RPMI medium with or without different concentrations of AMB. An MTT assay was performed to study the inhibition of growth of A. fumigatus by AMB (at the MIC50) as described in Materials and Methods.
Proteomic analysis.
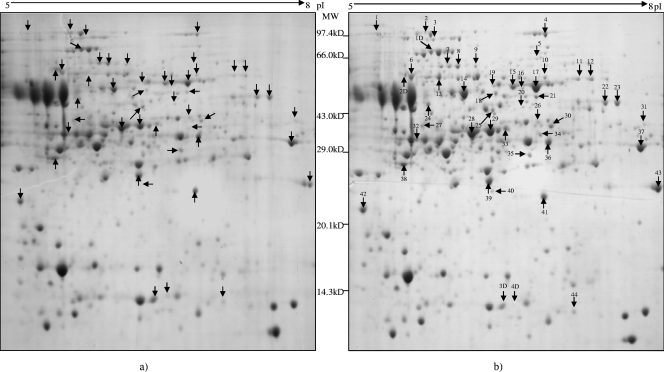
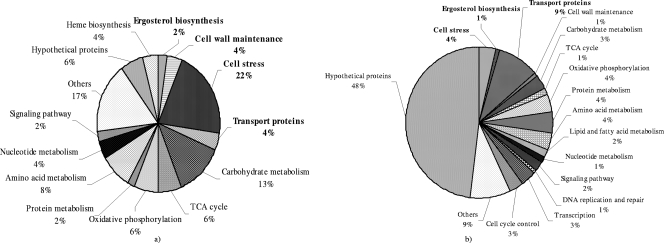
We used a 2-D PAGE-MS proteomics approach for the analysis of proteins from A. fumigatus with or without treatment with AMB. The experimental conditions used in protein profiling to clarify the differential expression levels induced by the drug allowed access to about 500 proteins of the fungus (according to the number of spots seen on the gel), of which 85 were differentially expressed (Fig. 2). Seventy-six of them were found to be upregulated and nine downregulated. PMF followed by MS-MS analysis of selected peptides led to the identification of 48 proteins (44 upregulated and 4 downregulated) (Fig. 2; also see Tables S2 and S3 in the supplemental material). Identification of the remaining differentially regulated protein spots was limited by mass signals that were weak, presumably due to the chemistry of the peptides and/or posttranslational modification such as glycosylation (37). The differentially expressed proteins belonged to various functional categories, such as those involving the ergosterol pathway, cell wall maintenance, cell stress, transporter proteins, carbohydrate metabolism, and amino acid metabolism, etc. (Fig. 3a). Proteins that may be of special regulatory importance include ERG13 (a 3-hydroxy-3-methylglutaryl-coenzyme A synthase involved in ergosterol biosynthesis), HEM13 (a coproporphyrinogen III oxidase involved in sterol synthesis and regulation), conidial hydrophobin protein B (a cell wall protein), Rho-GDP dissociation inhibitor (Rho-GDI), and secretory-pathway GDI (which inhibits Rho family-dependent cell functions, such as vesicular trafficking, cell shape, and cytokinesis). Proteins implicated in cell stress, such as manganese-superoxide dismutase (Mn-SOD), catalase, LsfA, and heat shock proteins (HSP30 and mitochondrial HSP70), were also among the differentially expressed proteins.
FIG. 2.
2-D PAGE of proteins extracted from A. fumigatus alone (a) and treated with AMB at 0.125 μg/ml (the MIC50) (b). Proteins (600 μg) were separated by IEF on an IPG strip (17 cm; pI range, 5 to 8) and SDS-PAGE on 12.0% Laemmli gels and stained with Coomassie blue R 250. 2-D gel images were compared to identify differential expression levels of A. fumigatus proteins on exposure to AMB, using PDQuest software. Differentially expressed proteins were then subjected to MALDI-TOF and MALDI-TOF-TOF analysis, which resulted in identification of a total of 48 proteins (marked with arrows). Forty-four proteins were observed to be upregulated, and four proteins were downregulated (b). The functions and functional categories of the differentially expressed proteins identified are given in Table 2 and Table S2 in the supplemental material. MW, molecular mass.
FIG. 3.
Pie chart grouping responsive proteins (n = 48) (a) and genes (n = 295) (b) of A. fumigatus with 2.0-fold changes or more on exposure to AMB. According to the proteomic study, 22% of the identified proteins are associated with cell stress, 4% are cell wall maintenance proteins, 4% of the responsive proteins belong to transport proteins, 2% are associated with the ergosterol biosynthesis pathway, and 6% of the responsive proteins are hypothetical proteins (a). According to the microarray study, 48% of the responsive genes encode hypothetical proteins, 4% of the genes are associated with cell stress, 1% are associated with cell wall maintenance or ergosterol biosynthesis, and 9% of the responsive genes are involved in transport (b). Differentially expressed proteins and genes belonging to various functional categories were annotated from the ExPASy database (http://expasy.org/uniprot). TCA, tricarboxylic acid.
Microarray analysis.
From two independent dye swap experiments (see Materials and Methods), differentially expressed genes appearing in both the channels were considered for analysis and clustering. A total of 295 out of 10,003 genes of A. fumigatus were differentially expressed on exposure to AMB. Of these, 165 genes were observed to be upregulated and 130 downregulated (see Table S4 in the supplemental material). The group of genes related to hypothetical proteins was the largest (n = 142; 90 upregulated and 52 downregulated). Other differentially expressed genes were classified into groups such as those involving the ergosterol biosynthesis pathway, cell wall maintenance, cell stress, transporter proteins, lipid and fatty acid metabolism, and carbohydrate and protein metabolism, etc. (Fig. 3b). Some of these genes of special functional significance correspond to enzymes involved in ergosterol biosynthesis (ERG11 [lanosterol 14-alpha-demethylase] and ERG6 [delta-24-sterol C-methyltransferase]), Rho-GDI, secretory-pathway GDI, GTPase-activating protein, multidrug resistance proteins, plasma membrane H+ ATPase, multidrug and toxin extrusion (MATE) efflux family protein, and Sit1p/Arn1p.
On the basis of the genes encoding the ergosterol biosynthesis pathway, cell stress, cell wall maintenance, and transporter proteins and showing differential expression levels on exposure to AMB, 12 representative target genes and a control gene, the GAPDH gene, were chosen for validation with real-time RT-PCR (Table 1). Nine of them showed expression profiles similar to those observed in the microarray data, reflecting that microarray and real-time RT-PCR data are largely consistent (Table 1). ERG6, observed to be downregulated in microarray analysis, did not show any significant change in gene expression profile in real-time RT-PCR analysis.
TABLE 1.
Comparison of ratios of gene expression for A. fumigatus alone and A. fumigatus treated with AMB, obtained with microarray hybridization and real-time RT-PCR
| Locus tag | Protein description | Fold increasea
|
|
|---|---|---|---|
| Microarray hybridization | Real-time RT-PCR | ||
| Afu7g03740 | 14-Alpha demethylase Cyp51A (ERG11) | 2.8 | 2.0 |
| Afu4g03630 | Sterol 24-C-methyltransferase (ERG6) | −2.9 | −0.76 |
| Afu6g07820 | Integral membrane protein, putative | 2.6 | 2.0 |
| Afu1g14550 | Mn-SOD | 2.3 | 3.1 |
| Afu3g07930 | Putative GST | 3.2 | 2.3 |
| Afu5g11380 | Rho-GDP dissociation inhibitor | 2.0 | 5.2 |
| Afu2g11150 | Secretory-pathway GDI | 2.0 | 4.0 |
| Afu6g12340 | GTPase-activating protein, putative | 2.5 | 4.5 |
| Afu7g04580 | TBC domain protein, putative | 2.0 | 1.9 |
| Afu3g03500 | Multidrug resistance protein 4, putative | 3.5 | 2.1 |
| Afu3g07640 | Plasma membrane H+ ATPase | 3.4 | 5.4 |
| Afu3g13670 | Siderochrome-iron transporter, putative | 4.4 | 1.9 |
Values shown are ratios for A. fumigatus treated with AMB for 24 h (at the MIC50) versus A. fumigatus cultured alone for 24 h. Changes in expression levels of target genes are averages for replicates, normalized to GAPDH mRNA expression.
To study the changes in the mRNA expression profiles of A. fumigatus at an early time point, we performed real-time RT-PCR with the representative genes after 3 h of exposure to AMB. Here, the Mn-SOD, plasma membrane H+ ATPase, and siderochrome transporter genes were observed to be downregulated, but no significant change was observed in the expression profiles of other genes of A. fumigatus (unpublished data).
DISCUSSION
The present study describes proteomic and transcriptomic analysis of differentially expressed genes and proteins of A. fumigatus in response to treatment with AMB. The differential expression information was extracted from microarray data with 10,003 genes. Hypergeometric probability analysis performed with the microarray data showed overrepresentation of the functional categories involving ergosterol biosynthesis, cell stress, transporter proteins, oxidative phosphorylation, nucleotide metabolism, cell cycle control, and protein metabolism (see Fig. S1 in the supplemental material). On the other hand, the proteomic data were from a set of about 500 protein spots separated in 2-D gel with a pI range of 5 to 8, with 48 of them differentially expressed and identified. This number did not permit comprehensive one-to-one comparisons of differentially expressed proteins with transcripts or hypergeometric probability analysis as done for microarray data. Nevertheless, the observation that at least some of the genes and proteins affected on AMB treatment were common in both proteomic and microarray analyses (Rho-GDI, secretory-pathway GDI, and Mn-SOD, etc.), with many of these differential genes and proteins representing common gene/protein functions, permits a discussion of dominant cellular processes targeted by AMB (Table 2 and Fig. 3a and b). We therefore highlight altered gene or protein expressions that are relevant to a given function and consistent with earlier literature on the mechanism of action of AMB, i.e., involving the ergosterol biosynthesis pathway, cell stress, cell wall maintenance, and transport proteins (1).
TABLE 2.
Integrated proteomic and microarray data showing differential expression levels of genes and proteins of A. fumigatus on exposure to AMBa
| Functional category and locus tag | Protein description | Function(s) | Fold increaseb
|
|
|---|---|---|---|---|
| Proteomic data | Microarray data | |||
| Ergosterol biosynthesis | ||||
| Afu3g10660 | Hydroxymethyl glutaryl-coenzyme A synthase (ERG13) | Ergosterol biosynthesis | 2 | |
| Afu7g03740 | 14-Alpha demethylase Cyp51A (ERG 11) | Ergosterol biosynthesis | 2.8 | |
| Afu4g03630 | Sterol 24-C-methyltransferase (ERG 6) | Ergosterol biosynthesis | −2.9 | |
| Heme biosynthesis | ||||
| Afu1g07480 | Coproporphyrinogen III oxidase | Porphyrin biosynthesis, heme biosynthesis | 2 | |
| Afu1g07480 | Coproporphyrinogen III oxidase | Porphyrin biosynthesis, heme biosynthesis | 2 | |
| Cell wall maintenance | ||||
| Afu1g17250 | Conidial hydrophobin RodB | Cell wall component | −2 | |
| Afu1g17250 | Conidial hydrophobin RodB | Cell wall component | −2 | |
| Afu6g07820 | Integral membrane protein, putative | Cell membrane protein | 2.6 | |
| Afu3g08110 | Cell wall tir-3 protein | Cell wall protein | −3.6 | |
| Afu1g14300 | Fasciclin domain family | Cell adhesion | −3.3 | |
| Cell stress | ||||
| Afu1g14550 | Mn-SOD | Antioxidant | 2 | 2.3 |
| Afu3g02270 | Mycelial catalase Cat1 | Oxygen and ROS metabolism; pathogenesis | 2 | |
| Afu4g08580 | Antioxidant protein LsfA | Regulation of cell redox homeostasis | 2 | |
| Afu7g01860 | Heat shock protein | Protein folding | 2 | |
| Afu3g14540 | 30-kDa heat shock protein | Molecular chaperone | 2 | |
| Afu3g14540 | 30-kDa heat shock protein | Molecular chaperone | 2 | |
| Afu2g09960 | Mitochondrial HSP70 | Molecular chaperone | −2 | |
| Afu3g07930 | Putative GST | Antioxidant | 3.2 | |
| Afu3g14970 | Thioredoxin | Self-defense | −3.4 | |
| Afu6g06470 | Heat shock protein Hsp88 | Allergen, chaperone | −2.6 | |
| Transport proteins | ||||
| Afu5g11380 | Rho-GDP dissociation inhibitor | Involved in transport | 2 | 2.0 |
| Afu2g11150 | Secretory-pathway GDI | Involved in transport | 2 | 2.0 |
| Afu6g12340 | GTPase-activating protein, putative | Endocytic pathway | 2.5 | |
| Afu4g06950 | Related to VAMP-associated protein | Endoplasmic reticulum type II integral membrane protein | 4.0 | |
| Afu7g04580 | TBC domain protein, putative | Regulation of plasma membrane and endosome trafficking | 2.0 | |
| Afu4g07700 | Clathrin, heavy polypeptide | Endocytosis | −2.1 | |
| Afu2g12980 | Protein transport protein (SEC31), putative | Protein transport | −2.1 | |
| Afu3g10740 | RAB GTPase Ypt51, putative | Regulation of endocytic pathway | −5.0 | |
| Afu3g03500 | Multidrug resistance protein 4, putative | Drug efflux | 3.5 | |
| Afu3g07640 | Plasma membrane H+ ATPase | Drug efflux | 3.4 | |
| Afu5g13170 | MATE efflux family protein subfamily, putative | Drug efflux | 2.0 | |
| Afu6g07280 | ABC transporter family protein, putative | Drug efflux | −2.0 | |
| Afu1g12620 | Efflux pump | Efflux | 2.9 | |
| Afu5g01520 | Major facilitator superfamily protein | Probably involved in iron-siderophore transport | −3.3 | |
| Afu3g13670 | Siderochrome-iron transporter, putative | Iron transport | 4.4 | |
| Afu7g06570 | Zinc/cadmium resistance protein | Uptake of zinc and cadmium ions | −3.0 | |
The list shows those altered gene and protein expressions of A. fumigatus which are related to the mechanism of action of AMB. Lists of all genes showing differential expression are given in Tables S2 and S3 in the supplemental material.
Values shown are ratios for A. fumigatus treated with AMB (at the MIC50) for 24 h versus A. fumigatus cultured alone for 24 h.
Ergosterol biosynthesis pathway.
Microarray analysis indicated upregulation of ERG11 and downregulation of ERG6 genes of the ergosterol biosynthesis pathway. Real-time RT-PCR analysis also showed upregulation of ERG11; however, no significant change in mRNA expression was observed for ERG6. ERG11 has been proposed to be a direct target of azole drugs, and its upregulation may reflect a response for overcoming the drug effect (1, 24). Proteomic analysis showed upregulation of ERG13 and two isoforms of an enzyme of the heme biosynthesis pathway, HEM13, which is consistent with other studies (1, 24). Cofactor heme is required for enzymatic activities of Erg3p (C-5 sterol desaturase) and Erg5p (C-22,23 desaturase) and regulation of expression of Erg11p by heme (4). The above observations suggest that the genes/proteins directly or indirectly involved in ergosterol biosynthesis are upregulated to overcome the stress due to AMB. However, ERG11 mutant (Cyp51A) and ERG3 knockout (single and double mutants of ERG3A and -3B) genes encoding C-5 sterol desaturase did not show alteration in sensitivity of A. fumigatus to AMB or other antifungal drugs (2, 26) and need to be explored further.
Similarly, ERG6 was reported to be downregulated in response to voriconazole in A. fumigatus (11). Deletion of ERG6 in clinical isolates of Candida lusitaniae conferred resistance to AMB on these isolates. ERG6 has a role in synthesis of secondary sterols, and its downregulation suggests that A. fumigatus downregulates synthesis of secondary sterols in response to AMB or voriconazole.
Cell wall proteins.
Proteomic analysis showed downregulation of conidial hydrophobin B (RodBp), a cell wall structural protein involved in the building of the conidial outer cell wall and uniquely present in filamentous fungi. Microarray analysis showed differential expression levels for other proteins/enzymes of the fungal cell wall, such as integral membrane proteins, cell wall tir-3 protein, and chitinase. These structural proteins could be involved in maintenance of fungal cell wall integrity while challenged with AMB. Chitinase is involved in morphogenetic changes such as conidial germination, mycelial formation by plasticization, and cell wall expansion (5), and its upregulation in response to AMB is reported here for the first time. These observations are in coherence with an A. fumigatus response to voriconazole (1, 11, 36).
Cell stress.
It is reported that AMB promotes oxidative damage of cell membranes through generation of ROS (reviewed in references 27 and 44), and as anticipated, we have observed overexpression of various enzymes that act as antioxidants in response to the drug, which is in agreement with previous studies (24, 31, 46). Both proteomic and microarray analyses showed upregulation of Mn-SOD. In addition, proteomic analysis also showed upregulation of other enzymes that act as antioxidants, such as catalase and LsfA. Microarray analysis showed upregulation of antioxidants such as glutathione S-transferase (GST) and thioredoxin. Earlier studies had shown upregulation of Mn-SOD and catalase upon exposure to AMB in S. cerevisiae (46) and C. albicans (1). Upregulation of antioxidants such as GST, thioredoxin, and LsfA observed in the present study may be specific to the A. fumigatus response to AMB and may provide enhanced intracellular oxidative detoxification to the fungus for its survival. Heat shock proteins HSP30, mitochondrial HSP70 (in proteomics analysis), and HSP88 (in microarray analysis) were observed to be upregulated. Their overexpression may support A. fumigatus survival under the stress conditions induced by AMB. These antioxidants may also be relevant in conferring resistance to AMB on A. fumigatus by reducing oxidative stress (27, 28).
Transporter proteins.
Among transporter proteins, Rho-GDI and secretory-pathway GDP dissociation inhibitor were observed to be upregulated in both proteomic and microarray analyses (16). Upregulation of genes encoding other proteins, such as the Tre-2/Bub2/Cdc16 domain (carrying GTPase-activating proteins for Rab family G proteins), and downregulation of genes encoding clathrin, Sec 31 (a subunit of the exocyst complex), and Rab GTPase Ypt51 (a key regulator of vesicular trafficking events between various subcellular compartments within the eukaryotic cell) have not been reported earlier and may represent a specific response of A. fumigatus to AMB. The differential expression levels of some of the genes, such as those encoding multidrug resistance proteins, plasma membrane H+ ATPase, and drug efflux pumps, also reflected mechanisms of resistance to antifungal drugs through their overexpression, as reported previously (8, 32, 41, 45). However, differential expression levels for other genes in response to AMB in A. fumigatus, such as those encoding Rho-GDI, secretory-pathway GDI, GTPase-activating protein, and MATE efflux family protein (12, 16, 21), are reported here for the first time and may be explored further.
Other genes encoding transport proteins upregulated in microarray analysis are implicated in transportation of iron, zinc, cadmium, arsenite, phosphate, malic acid, carboxylic acid, and uracil, as reported earlier (1, 29, 30, 33, 34, 46). Iron transport and siderophore uptake by Sit1p/Arn1p have been reported to be upregulated in response to AMB in S. cerevisiae (1) and voriconazole in A. fumigatus (11). Sit1p/Arn1p has also been implicated in virulence of Cryptococcus neoformans (20) and C. albicans (19). The above observations suggest that the fungus overexpresses genes and proteins involved in small-molecule and vesicular transport and drug efflux, which is one of the mechanisms adapted by the fungus to develop resistance and survive against antifungal drug.
Other important functional groups detected by proteomic and transcriptomic analysis include those for energy metabolism and amino acid metabolism and genes/proteins involved in transcription, translation, DNA repair, or signal transduction. Genes encoding hypothetical proteins were observed to be the largest group of A. fumigatus genes with differential expression in response to AMB in the microarray study, similar to what was found for other studies of fungal responses to AMB (1, 24). A BLASTp search of one of the hypothetical proteins (spot no. 1) in the proteomic study showed homology to H+/K+ ATPase alpha of Neosartorya fischeri and Aspergillus clavatus. Another hypothetical protein (spot no. 9) showed homology to UbiE/COQ5 family methyltransferase of Neosartorya fischeri and Aspergillus clavatus. Further understanding of the functions of these hypothetical proteins may add new information on the mechanisms of action of AMB.
The integrated proteomic and transcriptomic analyses of differential gene expression described here are largely consistent with the postulated mechanisms of AMB action and support the existence of these mechanisms in both filamentous and nonfilamentous fungi. They represent processes affected during sustained exposure to drug. The specific and primary molecular targets, however, may evolve from early changes in gene expression under the effect of the drug. We believe that the findings of the current study provide a basis for further understanding for development of new antifungal drug targets.
Supplementary Material
Acknowledgments
We are grateful to the Council of Scientific and Industrial Research and the Department of Science and Technology, Government of India, for financial support.
We are grateful to the Pathogen Functional Genomics Resource Center at the J. Craig Venter Institute (Rockville, MD) for providing A. fumigatus microarray slides. We are also thankful to the Centre for Genomic Application for providing the microarray facility and to Dev Ananthan, Centre for Genomic Application, for analysis work. We acknowledge D. W. Denning, School of Medicine, University of Manchester, Manchester, United Kingdom, for providing the Af293 strain. We sincerely thank M. Sridevi (biostatistician at Sristek, Hyderabad, India) for statistical analysis and Nitin C. Tupperwar (scientist at the Centre for Cellular and Molecular Biology) for real-time RT-PCR analysis. Jata Shankar and Poonam Gautam were recipients of senior research fellowships from the Council of Scientific and Industrial Research, Government of India.
Footnotes
Published ahead of print on 6 October 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Agarwal, A. K., P. D. Rogers, S. R. Baerson, M. R. Jacob, K. S. Barker, J. D. Cleary, L. A. Walker, D. G. Nagle, and A. M. Clark. 2003. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cereviseae. J. Biol. Chem. 278:34998-35015. [DOI] [PubMed] [Google Scholar]
- 2.Alcazar-Fuoli, L., E. Mellado, G. Garcia-Effron, M. J. Buitrago, J. F. Lopez, J. O. Grimalt, J. M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2006. Aspergillus fumigatus C-5 sterol desaturases Erg3A and Erg3B: role in sterol biosynthesis and antifungal drug susceptibility. Antimicrob. Agents Chemother. 50:453-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arikan, S., M. Lozano-Chiu, V. Paetznick, S. Nangia, and J. H. Rex. 1999. Microdilution susceptibility testing of amphotericin B, itraconazole, and voriconazole against clinical isolates of Aspergillus and Fusarium species. J. Clin. Microbiol. 37:3946-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bammert, G. F., and J. M. Fostel. 2000. Genome-wide expression patterns in Saccharomyces cereviseae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob. Agents Chemother. 44:1255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard, M., and J. P. Latge. 2001. Aspergillus fumigatus cell wall: composition and biosynthesis. Med. Mycol. 39(Suppl. 1):9-17. [PubMed] [Google Scholar]
- 6.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Brakhage, A. A. 2005. Systemic fungal infections caused by Aspergillus species: epidemiology, infection process and virulence determinants. Curr. Drug Targets 6:875-886. [DOI] [PubMed] [Google Scholar]
- 8.Burghoorn, H. P., P. Soteropoulos, P. Paderu, R. Kashiwazaki, and D. S. Perlin. 2002. Molecular evaluation of the plasma membrane proton pump from Aspergillus fumigatus. Antimicrob. Agents Chemother. 46:615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow, L. P., S. L. Liu, C. J. Yu, H. K. Liao, J. J. Tsai, and T. K. Tang. 2000. Identification and expression of an allergen Asp f 13 from Aspergillus fumigatus and epitope mapping using human IgE antibodies and rabbit polyclonal antibodies. Biochem. J. 346:423-431. [PMC free article] [PubMed] [Google Scholar]
- 10.Chumbalkar, V. C., C. Subhashini, V. M. Dhople, C. S. Sundaram, M. V. Jagannadham, K. N. Kumar, P. N. Srinivas, R. Mythili, M. K. Rao, M. J. Kulkarni, S. Hegde, A. S. Hegde, C. Samual, V. Santosh, L. Singh, and R. Sirdeshmukh. 2005. Differential protein expression in human gliomas and molecular insights. Proteomics 5:1167-1177. [DOI] [PubMed] [Google Scholar]
- 11.da Silva Ferreira, M. E., I. Malavazi, M. Savoldi, A. A. Brakhage, M. H. Goldman, H. S. Kim, W. C. Nierman, and G. H. Goldman. 2006. Transcriptome analysis of Aspergillus fumigatus exposed to voriconazole. Curr. Genet. 50:32-44. [DOI] [PubMed] [Google Scholar]
- 12.Diener, A. C., R. A. Gaxiola, and G. R. Fink. 2001. Arabidopsis ALF5, a multidrug efflux transporter gene family member, confers resistance to toxins. Plant Cell 13:1625-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis, D. 2002. Amphotericin B: spectrum and resistance. J. Antimicrob. Chemother. 49(Suppl. 1):7-10. [DOI] [PubMed] [Google Scholar]
- 14.Foster, W. R., and R. M. Huber. 2002. Current themes in microarray experimental design and analysis. Drug Discov. Today 7:290-292. [DOI] [PubMed] [Google Scholar]
- 15.Gabrielskaa, J., M. G. J. Gubernatord, and W. I. Gruszeckic. 2006. Binding of antibiotic amphotericin B to lipid membranes: A 1H NMR study. FEBS Lett. 580:2677-2685. [DOI] [PubMed] [Google Scholar]
- 16.Garrett, M. D., J. E. Zahner, C. M. Cheney1, and P. J. Novick. 1994. GD11 encodes a GDP dissociation inhibitor that plays an essential role in the yeast secretory pathway. EMBO J. 13:1718-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gautam, P., C. S. Sundaram, T. Madan, W. N. Gade, A. Shah, R. Sirdeshmukh, and P. U. Sarma. 2007. Identification of novel allergens of Aspergillus fumigatus using immunoproteomics approach. Clin. Exp. Allergy 37:1239-1249. [DOI] [PubMed] [Google Scholar]
- 18.Görg, A., W. Postel, S. Günther, and C. Friedrich. 1988. Horizontal two-dimensional electrophoresis with immobilized pH gradients using PhastSystem. Electrophoresis 9:57-59. [DOI] [PubMed] [Google Scholar]
- 19.Heymann, P., M. Gerads, M. Schaller, F. Dromer, G. Winkelmann, and J. F. Ernst. 2002. The siderophore iron transporter of Candida albicans (Sit1p/Arn1p) mediates uptake of ferrichrome-type siderophores and is required for epithelial invasion. Infect. Immun. 70:5246-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard, D. H. 1999. Acquisition, transport, and storage of iron by pathogenic fungi. Clin. Microbiol. Rev. 12:394-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaatz, G. W., F. McAleese, and S. M. Seo. 2005. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 49:1857-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kontoyiannis, D. P., and R. E. Lewis. 2002. Antifungal drug resistance of pathogenic fungi. Lancet 359:1135-1144. [DOI] [PubMed] [Google Scholar]
- 23.Levitz, S. M., and R. D. Diamond. 1985. A rapid colorimetric assay of fungal viability with the tetrazolium salt MTT. J. Infect. Dis. 152:938-945. [DOI] [PubMed] [Google Scholar]
- 24.Liu, T. T., R. E. Lee, K. S. Barker, R. E. Lee, L. Wei, R. Homayouni, and P. D. Rogers. 2005. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 49:2226-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meletiadis, J., C. Antachopoulos, T. Stergiopoulou, S. Pournaras, E. Roilides, and T. J. Walsh. 2007. Differential fungicidal activities of amphotericin B and voriconazole against Aspergillus species determined by microbroth methodology. Antimicrob. Agents Chemother. 51:3329-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellado, E., G. Garcia-Effron, M. J. Buitrago, L. Alcazar-Fuoli, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2005. Targeted gene disruption of the 14-α sterol demethylase (cyp51A) in Aspergillus fumigatus and its role in azole drug susceptibility. Antimicrob. Agents Chemother. 49:2536-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore, C. B., N. Sayers, J. Mosquera, J. Slaven, and D. W. Denning. 2000. Antifungal drug resistance in Aspergillus. J. Infect. 41:203-220. [DOI] [PubMed] [Google Scholar]
- 28.Moore, C. B., C. M. Walls, and D. W. Denning. 2000. In vitro activity of the new triazole BMS-207147 against Aspergillus species in comparison with itraconazole and amphotericin B. Antimicrob. Agents Chemother. 44:441-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreno, M. A., J. Amich, R. Vicentefranqueira, F. Leal, and J. A. Calera. 2007. Culture conditions for zinc- and pH-regulated gene expression studies in Aspergillus fumigatus. Int. Microbiol. 10:187-192. [PubMed] [Google Scholar]
- 30.Moreno, M. A., O. Ibrahim-Granet, R. Vicentefranqueira, J. Amich, P. Ave, F. Leal, J. P. Latgé, and J. A. Calera. 2007. The regulation of zinc homeostasis by the ZafA transcriptional activator is essential for Aspergillus fumigatus virulence. Mol. Microbiol. 64:1182-1197. [DOI] [PubMed] [Google Scholar]
- 31.Mousavi, S. A. A., and D. Geoffrey. 2004. Oxidative and amphotericin B-mediated cell death in the opportunistic pathogen Aspergillus fumigatus is associated with an apoptotic-like phenotype. Microbiology 150:1937-1945. [DOI] [PubMed] [Google Scholar]
- 32.Nascimento, A. M., G. H. Goldman, S. Park, S. A. Marras, G. Delmas, U. Oza, K. Lolans, M. N. Dudley, P. A. Mann, and D. S. Perlin. 2003. Multiple resistance mechanisms among Aspergillus fumigatus mutants with high-level resistance to itraconazole. Antimicrob. Agents Chemother. 47:1719-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oberegger, H., M. Schoeser, I. Zadra, M. Schrettl, W. Parson, and H. Haas. 2002. Regulation of freA, acoA, lysF, and cycA expression by iron availability in Aspergillus nidulans. Appl. Environ. Microbiol. 68:5769-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oberegger, H., I. Zadra, M. Schoeser, and H. Haas. 2000. Iron starvation leads to increased expression of Cu/Zn-superoxide dismutase in Aspergillus. FEBS Lett. 485:113-116. [DOI] [PubMed] [Google Scholar]
- 35.Olson, J. A., J. P. Adler-Moore, J. Schwartz, G. M. Jensen, and R. T. Proffitt. 2006. Comparative efficacies, toxicities, and tissue concentrations of amphotericin B lipid formulations in a murine pulmonary aspergillosis model. Antimicrob. Agents Chemother. 50:2122-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paris, S., J. P. Debeaupuis, R. Crameri, M. Carey, F. Charles, M. C. Prevost, C. Schmitt, B. Philippe, and J. P. Latge. 2003. Conidial hydrophobins of Aspergillus fumigatus. Appl. Environ. Microbiol. 69:1581-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng, X., X. Ye, and S. Wang. 2004. Identification of novel immunogenic proteins of Shigella flexneri 2a by proteomic methodologies. Vaccine 22:2750-2756. [DOI] [PubMed] [Google Scholar]
- 38.Pfaller, M. A., P. G. Pappas, and J. R. Wingard. 2006. Invasive fungal pathogens: current epidemiological trends. Clin. Infect. Dis. 43:S3-S14. [Google Scholar]
- 39.Puckette, M. C., Y. Tang, and R. Mahalingam. 2008. Transcriptomic changes induced by acute ozone in resistant and sensitive Medicago truncatula accessions. BMC Plant Biol. 8:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Semighini, C. P., M. Marins, M. H. S. Goldman, and G. H. Goldman. 2002. Quantitative analysis of the relative transcript levels of ABC transporter Atr genes in Aspergillus nidulans by real-time reverse transcription-PCR assay. Appl. Environ. Microbiol. 68:1351-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serrano, R., M. C. Kielland-Brandt, and G. R. Fink. 1986. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+K+), K+- and Ca2+-ATPase. Nature 319:689-693. [DOI] [PubMed] [Google Scholar]
- 42.Singh, J., D. Rimek, and R. Kappe. 2006. Intrinsic in vitro susceptibility of primary clinical isolates of Aspergillus fumigatus, Aspergillus terreus, Aspergillus nidulans, Candida albicans and Candida lusitaniae against amphotericin B. Mycoses 49:96-103. [DOI] [PubMed] [Google Scholar]
- 43.Sokol-Anderson, M. L., J. Brajtburg, and G. Medoff. 1986. Amphotericin B-induced oxidative damage and killing of Candida albicans. J. Infect. Dis. 154:76-83. [DOI] [PubMed] [Google Scholar]
- 44.Sterling, T. R., and W. G. Merz. 1998. Resistance to amphotericin B: emerging clinical and microbiological patterns. Drug Resist. Updat. 1:161-165. [DOI] [PubMed] [Google Scholar]
- 45.Tobin, M. B., R. B. Peery, and P. L. Skatrud. 1997. Genes encoding multiple drug resistance-like proteins in Aspergillus fumigatus and Aspergillus flavus. Gene 200:11-23. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, L., Y. Zhang, Y. Zhou, S. An, Y. Zhou, and J. Cheng. 2002. Response of gene expression in Saccharomyces cereviseae to amphotericin B and nystatin measured by microarrays. J. Antimicrob. Chemother. 49:905-915. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.