Abstract
Pseudomonas aeruginosa strains isolated from patients with persistent lung infections and cystic fibrosis have been found to gradually develop aminoglycoside resistance over time. The aim of this study was to identify potential contributors to low-level aminoglycoside resistance, which may cause such graduated increases in resistance. The Harvard P. aeruginosa PA14 nonredundant library, consisting of approximately 5,800 mutants, was screened for twofold or greater increases in tobramycin resistance. Mutants carrying mutations in a total of 135 unique genes were identified and confirmed to have reduced susceptibility to tobramycin. Many of these genes were involved predominantly in energy metabolism; however, most of these mutants did not exhibit growth defects under the conditions tested, although some exhibited the small-colony phenotype and/or defects in growth under anaerobic conditions. Lipopolysaccharide mutants were also identified, and it was found that tobramycin had a reduced ability to permeabilize the outer membranes of these mutants. The results of this study emphasize the complexity of the interactions that tobramycin may have within the bacterial cell and introduce a large number of novel genes which may play a role in tobramycin resistance.
The majority of deaths of cystic fibrosis patients can be attributed to the progressive deterioration of lung function resulting from chronic infection by pathogens such as Pseudomonas aeruginosa (25). Antibiotic treatment of such chronic infections may temporarily suppress symptoms; however, it does not eradicate the pathogen. To overcome the inability of orally and parenterally administered antibiotics to adequately penetrate lung tissue and secretions, the aerosolized aminoglycoside tobramycin was formulated to directly target the site of infection. Clinical trials of aerosolized tobramycin revealed that long-term use of the agent against P. aeruginosa results in small, graduated increases in tobramycin MICs upon repeated isolation of the organism (21, 22), although the mechanisms contributing to this resistance have yet to be described.
Tobramycin is a bactericidal agent that targets the 30S ribosome and interferes with protein synthesis. Uptake occurs in three phases: an initial, reversible ionic-binding phase, a phase of slow energized uptake, and finally a phase of very rapid energy-dependent uptake (2, 4, 9). Despite the traditionally held view that antibiotic action can be simplified to interaction with a single target, it is evident that aminoglycosides exert pleiotropic effects on the cell (8, 9), as these effects may be antagonized by a variety of compounds known to affect cellular metabolic processes. Furthermore, the bactericidal nature of aminoglycosides such as tobramycin cannot be accounted for simply by protein synthesis inhibition or misreadings during translation because other protein synthesis inhibitors, such as chloramphenicol, and agents that promote misreading, including modified amino acids, are bacteriostatic. A limited number of cytochrome mutations leading to aminoglycoside resistance have been previously identified by selecting mutagenized P. aeruginosa strains on aminoglycoside-containing media (1, 3). As aminoglycosides exert a number of effects on the bacterial cell, we believe the potential sites for development of resistance are numerous. The purpose of this study was to identify the extent of the tobramycin “resistome,” i.e., potential genetic contributors to the graduated development of aminoglycoside resistance in P. aeruginosa, using a comprehensive mutant library for a tobramycin resistance screen.
MATERIALS AND METHODS
Bacterial strains.
The P. aeruginosa PA14NR set described by Liberati et al. (15) was used for screening genes related to tobramycin resistance. This mutant library comprises approximately 5,800 mutants representing around 4,600 genes. It was constructed using a mariner-based transposon, MAR2xT7, containing the resistance cassette aacC1, which confers resistance to the aminoglycosides gentamicin, astromicin, and sisomicin, but not to tobramycin. In addition, two PAO1 mutant libraries, the mini-Tn5-luxCDABE mutant library described by Lewenza et al. (14) and the University of Washington transposon mutant library (11), were used for verification and cross-referencing purposes.
Tobramycin resistance screening.
P. aeruginosa PA14 mutants were inoculated into 100 μl of Mueller-Hinton broth (MHB) in 96-well plates and incubated at 37°C overnight. Overnight cultures were diluted 1:100 into MHB and were replica plated onto MH agar containing 0.5 μg/ml tobramycin (the MIC of the wild-type strain under these conditions). Growth was assessed at 24 and 48 h. Resistance was defined as a twofold or greater increase in the MIC compared to that for the parent strain, PA14. MICs at 24 and 48 h were determined at least in triplicate, using broth microdilution in cation-adjusted MHB (CAMHB), according to CLSI guidelines (6).
Tobramycin kill curves.
Kill curves were performed in triplicate, in 96-well plates to simulate the MIC conditions, using selected mutants representing the major gene class functions identified in the screen. Cultures were grown in CAMHB at 37°C to an optical density at 600 nm (OD600) of 0.4, and 1.25 ml was harvested by centrifugation. Pellets were resuspended in 1 ml CAMHB and then diluted 1:50. Fifty microliters of each sample was inoculated into the wells of 96-well plates, each containing 50 μl of a 4-μg/ml tobramycin solution, yielding a starting inoculum containing approximately 3 × 106 to 4 × 106 CFU/ml in 2 μg/ml tobramycin. Separate 96-well plates were inoculated for each time point, and aliquots were plated at 0, 10, 20, 30, 40, 50, and 60 min. Plates were incubated at 37°C overnight, and colony counts were performed to obtain numbers of CFU/ml at each time point.
Determination of growth rate.
Growth rates were determined for all tobramycin-resistant mutants related to energy metabolism. These mutants were inoculated and grown overnight in 3 ml CAMHB at 37°C. Two microliters of each overnight culture was inoculated in 200 μl CAMHB, and growth at 37°C was monitored using a Tecan Spectrafluor Plus by measuring the OD620 every 20 min.
Assessment of aerobic and anaerobic growth defects.
Cell suspensions were prepared to a 0.5 McFarland standard in LB broth and diluted 1:20 in 200 μl each of LB broth (aerobic) and LB broth containing 15 mM KNO3 (anaerobic) in 96-well microtiter plates. Anaerobic plates were placed in an anaerobic jar containing a GasPak Plus hydrogen and carbon dioxide generator envelope. All plates and jars were incubated at 37°C for 24 h. The OD600 of the mutants was measured in the 96-well plates and compared to the OD600 of PA14.
Tobramycin outer membrane interaction studies.
The hydrophobic fluorescent probe 1-N-phenylnaphthylamine (NPN) was used as described by Loh et al. to study the interaction of tobramycin with the outer membranes of the tobramycin-resistant lipopolysaccharide (LPS) mutants compared to its interaction with that of PA14 (16). Briefly, a 50-ml sample of mid-log-phase cells was harvested by centrifugation at 3,000 × g for 10 min and resuspended in 5 mM sodium HEPES buffer (pH 7.2), with 5 μM carbonyl cyanide m-chlorophenylhydrazone, at an OD600 of 0.5. NPN was added to a final concentration of 10 μM, and baseline fluorescence was measured at an excitation wavelength of 350 nm and an emission wavelength of 420 nm using a Perkin Elmer LS 50B fluorescent spectrophotometer. Tobramycin was added to final concentrations of 1, 5, 10, 20, 40, and 60 μg/ml, and increases in fluorescence were recorded.
Cloning of the PA5300 strain.
To complement the PAO1 PA5300::lux mutant strain and the PA14 PA5300 mutant strain, a PCR amplicon was generated using Phusion high-fidelity DNA polymerase (Finnzymes) from PAO1 genomic DNA using forward primer PA5300EcoRI and reverse primer PA5300BamHI. The amplicon was then ligated into the broad-host-range plasmid vector pUCP18 and transformed into Escherichia coli strain XL1-BlueMRF′. Plasmids were isolated (Qiagen) and transformed into both mutants as well as wild types PAO1 and PA14 by electroporation (5).
Cloning of radA.
In order to complement the PA14 radA mutant, the genomic DNA of P. aeruginosa PA14 was used as a template to amplify the whole radA gene using Phusion high-fidelity DNA polymerase (Finnzymes). The PCR fragment was subcloned using the Zero Blunt Topo kit (Invitrogen). The vector was cut with XbaI and HindIII (Invitrogen), and the radA fragment was cloned into the low-copy-number vector pBBR1MCS-1 (13) and transformed into E. coli Top10 (Invitrogen). Plasmids were isolated (Qiagen) and electroporated into the PA14 radA mutant as well as into wild-type PA14 (5).
RESULTS AND DISCUSSION
Tobramycin resistance screening and confirmation.
Screening of a preexisting transposon library allowed for the identification of a large number of potential genetic contributors to tobramycin resistance that have not been identified using the traditional method of using randomly mutagenized P. aeruginosa strains selected on antibiotic media. This tobramycin resistance screen identified a total of 348 mutants with reduced susceptibility compared to the PA14 parent strain. MICs determined using broth microdilution confirmed that 186 of these mutants have an MIC at least twofold greater than the MIC of the PA14 parent strain. These 186 mutants represented a total of 135 genes, as depicted in Fig. 1. A major benefit of this type of screen is that even small changes in resistance may be detected. Here the highest MIC found in the screen was an eightfold increase compared to that of PA14 for the mutL mutant (MIC of 4 μg/ml versus 0.5 μg/ml for the wild type). Additionally, 21 of the mutants identified had a fourfold increase in the MIC, while the remaining 165 mutants had a twofold increase in the MIC. Although twofold changes in MICs are typically considered within the acceptable range of error, we were able to confirm the increased tobramycin MIC in nearly all of these mutants in at least duplicate experiments, either by the redundancy of the PA14 mutant library or by the availability of corresponding mutants in either the mini-Tn5-luxCDABE library or the University of Washington PAO1 library. Of the 135 genes found in the PA14 screen, 44 genes had redundant mutants within the PA14 library, 31 genes were available in corresponding mutants in the PAO1 mini-Tn5-luxCDABE library, and 4 genes had corresponding mutants in the University of Washington PAO1 library. In total, 70 genes were confirmed to have increased tobramycin resistance in at least two independent mutants. An additional 60 genes that were not represented by more than one mutant belonged to operons with which other mutants had confirmed resistance. Only six genes neither were confirmed in duplicate experiments nor belonged to an operon. In addition, we were able to successfully restore wild-type tobramycin susceptibility in both the PAO1 and PA14 PA5300 mutants.
FIG. 1.
Distribution of genes identified in the tobramycin resistance screen around the PAO1 genome. This genome image was generated by using CGView.
Classification of tobramycin-resistant mutants using pseudoCAP.
Although twofold changes in MICs are typically considered within the acceptable range of error for this assay, the study of El'Garch et al. not only confirmed the reliability of twofold changes but indicated that independent mutants could act in a combinatorial fashion (7). Furthermore, slight increases in the MIC not only may tip the MIC above the clinical breakpoint but also may be indicative of a genotype with enhanced adaptability to stress conditions, such as exposure to antibiotics.
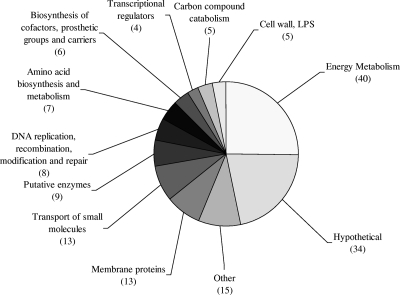
This enhanced adaptability has been observed for cystic fibrosis infections, for which the isolation of antibiotic-resistant strains with a hypermutator phenotype due to the presence of mutations in DNA mismatch repair systems, particularly in mutL, mutS, or uvrD, has previously been reported (18, 19). A number of isolates with other unknown genotypes have also been found to exhibit this hypermutator phenotype. Similarly, mutations in regulatory genes are known to lead to a cascade of effects ultimately causing adaptive resistance (20). When the mutants exhibiting reduced susceptibility to tobramycin were categorized according to their PseudoCAP functional class, according to the Pseudomonas Genome Database (www.pseudomonas.com) (Fig. 2), seven regulatory genes were identified along with eight distinct mutations known to be involved in DNA replication and repair processes, including mutations in mutL, mutS, mutY, micA, mfd, nth, uvrA, and radA.
FIG. 2.
Distribution of the 135 genes found to be involved in tobramycin resistance, grouped by PseudoCAP functional class. NB: the numbers of genes represented do not add up to 135, as many genes are represented in more than one functional group.
The most notable observation from the classification of the tobramycin-resistant mutants was, however, the large proportion of energy metabolism mutants. To rule out decreased growth as the reason for reduced tobramycin susceptibility, all 186 mutants were assessed for major growth deficiencies using an endpoint OD600 at 24 h as a measure of yield of growth. Of 186 mutants assayed, mutations in only 25 genes, comprising 10 individual genes and three operons (Table 1), were found to cause a significant reduction in growth yield under either aerobic or anaerobic conditions or both. Additionally, growth rates were determined for all energy metabolism mutants. With the exception of mutants belonging to the PA4429-PA4431 operon, for which mutants had a doubling time of approximately 80 min, all of the energy metabolism mutants exhibited normal growth in CAMHB compared to PA14, with doubling times of approximately 50 min. Finally, growth on solid media was assessed for the 57 energy metabolism mutants by observing colony morphology. Only 10 appeared as small-colony variants (of which 3 also had an observed growth defect in liquid media); all 10 of these had mutations in genes encoding cytochromes. Clinical isolates exhibiting this poorly understood small-colony phenotype have been shown to have increased antibiotic resistance compared to revertant colonies, and recovery of these isolates is strongly associated with daily inhalation of tobramycin or colistin (10).
TABLE 1.
Tobramycin resistance mutations causing defective growth in LB broth under aerobic and/or anaerobic conditions and small-colony variants
| Defect or variant type | PAO1 ortholog(s) of mutant gene | Gene name | Gene description |
|---|---|---|---|
| General growth defect | PA0432 | sahH | S-Adenosyl-l-homocysteine hydrolase |
| PA1547 | Putative membrane protein | ||
| PA1548 | Putative cytochrome oxidase maturation protein, cbb3 type | ||
| PA1588 | sucC | Succcinyl-CoA synthetase beta subunit | |
| PA4429 | Putative cytochrome c1 precursor | ||
| PA4430 | Putative cytochrome c reductase, iron-sulfur subunit | ||
| PA5070 | tatC | Sec-independent protein translocase TatC | |
| Aerobic growth defect | PA3818 | Inositol-1-monophosphatase | |
| PA3975 | Possible phosphomethylpyrimidine kinase | ||
| Anaerobic growth defect | PA1480 | ccmF | Cytochrome c-type biogenesis protein CcmF |
| PA2638-PA2648 | nuo | NADH dehydrogenase I | |
| PA2960 | pilZ | Type IV fimbrial biogenesis protein PilZ | |
| PA3233 | Putative signal transduction protein | ||
| PA4673 | Putative GTP-binding protein | ||
| PA4916 | Putative ADP-ribose pyrophosphatase | ||
| Small-colony variants (energy | PA1480 | ccmF | Cytochrome c-type biogenesis protein CcmF |
| metabolism mutations) | PA1483 | cycH | Cytochrome c-type biogenesis protein |
| PA1552 | Putative cytochrome c oxidase, cbb3 type, subunit III | ||
| PA1553 | Putative cytochrome c oxidase, cbb3 type, subunit II | ||
| PA1554 | ccoN | Cytochrome oxidase subunit, cbb3 type | |
| PA4429 | Putative cytochrome c1 precursor | ||
| PA4430 | Putative cytochrome b | ||
| PA4431 | Putative cytochrome c reductase, iron-sulfur subunit | ||
| PA5300 | cycB | Cytochrome c5 |
Several aminoglycoside-resistant cytochrome mutants were previously isolated by selective passaging on antibiotic media (1, 3); however, this study emphasizes the vast number of cytochrome genes which may impact aminoglycoside susceptibility. The definitive role that cytochromes play in aminoglycoside activity in P. aeruginosa is unknown. It is, however, known that the mechanism by which aminoglycosides cross the cytoplasmic membrane is an energy-dependent process which relies directly on the electron transport chain. Indeed, it has been suggested that a threshold membrane potential is required to support aminoglycoside uptake (17). Thus, for these energy metabolism mutants, we propose that resistance may result as a function of decreased uptake due to altered membrane potential. Nonetheless, cytochrome mutations were not the only energy metabolism mutations identified in the tobramycin screen. Of particular note is the identification of a number of genes related to NADH reduction, including the nuo and nqr operons, as well as the putative NADH:ubiquinone oxidoreductase encoded by PA3493. These results, along with the identification of several mutations involved in the tricarboxylic acid cycle and the cytochrome-electron transport chain mutations, are consistent with recent views suggesting that NADH depletion triggers the production of free radicals which ultimately contribute to antibiotic killing (12).
Tobramycin kill curves.
Tobramycin kill curves mimicking MIC conditions were performed for selected mutants of various functional classes, including six energy metabolism mutants (PA1320, PA1480, PA1551, PA2638, PA2644, and PA5300), two transcriptional regulator mutants (PA0149 and PA5438), one DNA replication and repair mutant (PA4609), two LPS mutants (PA5447 and PA5450), two small molecule transport mutants (PA3408 and PA5070), two membrane protein mutants (PA1767 and PA3115), and two hypothetical protein mutants (PA0338 and PA1588) (Fig. 3). All selected mutants showed a significant reduction (P < 0.05 by one-tailed Student's t test) in the extent of killing at 60 min by 2 μg/ml tobramycin compared to the wild type, with the exception of the PA3115 mutant (P = 0.068) (Fig. 4). As a number of mutants found in the tobramycin screen were impaired in DNA mismatch repair systems and many of these mutants are characterized by a hypermutator phenotype, it may be assumed that resistance in these mismatch repair mutants is due to a highly probable second-site mutation. We recently demonstrated that the radA mutant from the PA14 library shows an elevated mutation rate compared to PA14 and that the complemented strain has a mutation rate similar to that of the wild type (26). The kill curves performed here for the same radA mutant show that the tobramycin resistance phenotype is stable under the conditions tested. As these kill curves are performed over a short period of time, only 60 min, this indicates that a second-site mutation is not developing upon treatment. Tobramycin MICs were determined for the complemented radA strain to determine whether resistance in this mutant was due primarily to the radA mutation or to a preexisting second-site mutation. Tobramycin susceptibility was not restored in the complemented strain, indicating that resistance was due to a preexisting secondary mutation. In contrast, complementation of the PA5300 mutant, which does not exhibit a hypermutator phenotype, restored susceptibility to wild-type MIC levels.
FIG. 3.
Impact of PA1551 gene mutation on tobramycin killing of strain PA14. Error bars represent the standard deviations calculated from at least three separate experiments.
FIG. 4.
Influence of tobramycin resistance mutations on killing by tobramycin. Represented is the percent a survival after a 60-min exposure to 2 μg/ml tobramycin for selected tobramycin-resistant mutants compared to wild-type PA14. Error bars represent the standard deviations calculated from at least three separate experiments.
Interaction of tobramycin with the outer membranes of LPS mutants.
P. aeruginosa is capable of expressing two distinct forms of LPS, known as the A band and the B band (O antigen), which differ with respect to their O polysaccharides. The O polysaccharide of A-band LPS is a conserved poly-rhamnose molecule, and proteins involved in the assembly of the A-band LPS are encoded by the PA5447-PA5454 operon (23, 24). Several genes of this operon, namely, wbpZ, wbpY, wzt, wzm, and wbpW, were identified in the tobramycin resistance screen. Tobramycin crosses the outer membrane of the bacterial cell via the process of self-promoted uptake (9). It competitively binds LPS, displacing divalent cations and disrupting the integrity of the outer membrane, thus causing increased membrane permeability. NPN is a fluorescent probe that fluoresces weakly in aqueous solution but strongly in a nonpolar or hydrophobic environment. Under normal conditions, NPN is excluded from the membrane and does not fluoresce. Upon disruption of the outer membrane, e.g., by aminoglycosides (16), NPN partitions into the outer membrane interior and an increase in fluorescence can be observed. The initial rate of increase in fluorescence varies with the concentration of aminoglycoside and defines the extent of outer membrane permeabilization, which in turn relates to the extent of self-promoted uptake. The NPN assay was used to determine if the mutations in A-band LPS genes were in fact contributing to resistance, resulting in a reduced ability of tobramycin to interact with and permeabilize the outer membranes of these mutants. A maximum increase in fluorescence intensity was achieved using 40 μg/ml tobramycin for PA14 and 60 μg/ml for the six LPS mutants, indicating that higher concentrations of tobramycin were required to attain a similar level of disruption to the outer membrane. Differences in permeabilization were observed for all LPS mutants compared to the wild type at all concentrations assayed above 1 μg/ml, at which concentration no permeabilization was observed for any of the mutants or the wild type during the relatively short period that could be assessed in this assay. These findings reflect the substantially delayed killing (∼1 h) observed when using concentrations near the MIC. While a concentration of 5 μg/ml resulted in slight permeabilization of the wild-type outer membrane, this was not observed for any of the LPS mutants during the time frame observed. At 10 μg/ml tobramycin, the initial rate of fluorescence increase was approximately twofold higher in the PA14 parent strain than in any of the LPS mutants tested (Fig. 5), suggesting that the reduced ability of tobramycin to permeabilize the outer membranes of these mutants explains their reduced susceptibility.
FIG. 5.
Interaction of tobramycin with the outer membrane of the tobramycin-resistant wbpZ mutant (PA5450) compared to its interaction with wild-type PA14, using 10 μg/ml and 40 μg/ml tobramycin.
Concluding remarks.
The results presented here indicate that a wide variety of genetic determinants may affect aminoglycoside resistance. This finding is in contrast to the previously held belief, supported by direct in vitro selection studies of randomly mutagenized P. aeruginosa strains, that mutations leading to aminoglycoside resistance are relatively rare. In previous studies, aminoglycoside-resistant mutants were selected on media containing >12 μg/ml gentamicin from P. aeruginosa isolates subjected to ethyl methane sulfonate mutagenesis, while at a much lower selection concentration of 3.2 μg/ml gentamicin, nonmutatgenized cultures did not produce any aminoglycoside-resistant mutants (1, 3). The contrasting results demonstrated here are almost certainly because the ability to screen existing comprehensive libraries permitted us to identify many mutants with only twofold changes in resistance, which would be difficult to obtain through direct selection. Despite the large number of genes identified here, it is of importance to note that this type of screen has the limitation of identifying only nonessential genes and so may in fact underestimate the size of the tobramycin “resistome.” Nonetheless, we believe that this screen is a particularly sensitive method of detecting nonessential genes involved in resistance, as we have not only successfully detected mutations in two genetic backgrounds but have also independently identified other known resistance genes, such as the nuo (7) and cytochrome (1, 3) genes. Although the changes in the MICs of the mutants are modest, such modest changes have been shown to be real, and when double, triple, and quadruple mutants were generated for genes that when independently mutated lead to only twofold increases in the MICs, the mutations were shown to be capable of acting cumulatively to result in higher levels of resistance (7). By including such modest changes here, we have provided strong evidence for a very extensive aminoglycoside “resistome.” We have also related several phenotypes often associated with clinical aminoglycoside resistance to genes identified in this screen. Although the clinical relevance of the individual mutations found in this screen has yet to be elucidated, these results provide a large framework for future studies investigating the gradual development of aminoglycoside resistance due to the potential cumulative effects of genetic mutations. As well, they may provide insights into the actual mechanisms by which aminoglycosides act upon the cell.
Acknowledgments
This work was supported by grants from the Canadian Cystic Fibrosis Foundation and the Canadian Institutes of Health Research. K.N.S. holds a Natural Sciences and Engineering Council of Canada postgraduate scholarship and a Michael Smith Foundation for Health Research senior graduate studentship. R.E.W.H. holds a Canada Research Chair.
Footnotes
Published ahead of print on 29 September 2008.
REFERENCES
- 1.Bryan, L. E., and S. Kwan. 1981. Aminoglycoside-resistant mutants of Pseudomonas aeruginosa deficient in cytochrome d, nitrite reductase, and aerobic transport. Antimicrob. Agents Chemother. 19:958-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryan, L. E., and S. Kwan. 1983. Roles of ribosomal binding, membrane potential, and electron transport in bacterial uptake of streptomycin and gentamicin. Antimicrob. Agents Chemother. 23:835-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryan, L. E., T. Nicas, B. W. Holloway, and C. Crowther. 1980. Aminoglycoside-resistant mutation of Pseudomonas aeruginosa defective in cytochrome c552 and nitrate reductase. Antimicrob. Agents Chemother. 17:71-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Busse, H. J., C. Wostmann, and E. P. Bakker. 1992. The bactericidal action of streptomycin: membrane permeabilization caused by the insertion of mistranslated proteins into the cytoplasmic membrane of Escherichia coli and subsequent caging of the antibiotic inside the cells due to degradation of these proteins. J. Gen. Microbiol. 138:551-561. [DOI] [PubMed] [Google Scholar]
- 5.Choi, K.-H., A. Kumar, and H. P. Schweizer. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391-397. [DOI] [PubMed] [Google Scholar]
- 6.Clinical Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; 7th ed. Approved standard M7-A7. Clinical Laboratory Standards Institute, Wayne, PA.
- 7.El'Garch, F., K. Jeannot, D. Hocquet, C. Llanes-Barakat, and P. Plésiat. 2007. Cumulative effects of several nonenzymatic mechanisms on the resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 51:1016-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock, R. E. W. 1981. Aminoglycoside uptake and mode of action—with special reference to streptomycin and gentamicin. I. Antagonists and mutants. J. Antimicrob. Chemother. 8:249-276. [DOI] [PubMed] [Google Scholar]
- 9.Hancock, R. E. W. 1981. Aminoglycoside uptake and mode of action—with special reference to streptomycin and gentamicin. II. Effects of aminoglycosides on cells. J. Antimicrob. Chemother. 8:429-445. [DOI] [PubMed] [Google Scholar]
- 10.Haussler, S., B. Tummler, H. Weissbrodt, M. Rohde, and I. Steinmetz. 1999. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin. Infect. Dis. 29:621-625. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohanski, M. A., D. J. Dwyer, B. Hayete, C. A. Lawrence, and J. J. Collins. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797-810. [DOI] [PubMed] [Google Scholar]
- 13.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 14.Lewenza, S., R. K. Falsafi, G. Winsor, W. J. Gooderham, J. B. McPhee, F. S. Brinkman, and R. E. W. Hancock. 2005. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 15:583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liberati, N. T., J. M. Urbach, S. Miyata, D. G. Lee, E. Drenkard, G. Wu, J. Villanueva, T. Wei, and F. M. Ausubel. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 103:2833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loh, B., C. Grant, and R. E. W. Hancock. 1984. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 26:546-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mates, S. M., E. S. Eisenberg, L. J. Mandel, L. Patel, H. R. Kaback, and M. H. Miller. 1982. Membrane potential and gentamicin uptake in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 79:6693-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver, A., F. Baquero, and J. Blazquez. 2002. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol. Microbiol. 43:1641-1650. [DOI] [PubMed] [Google Scholar]
- 19.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 20.Overhage, J., M. Bains, M. D. Brazas, and R. E. W. Hancock. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J. Bacteriol. 190:2671-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pai, V. B., and M. C. Nahata. 2001. Efficacy and safety of aerosolized tobramycin in cystic fibrosis. Pediatr. Pulmonol. 32:314-327. [DOI] [PubMed] [Google Scholar]
- 22.Ramsey, B. W., M. S. Pepe, J. M. Quan, K. L. Otto, A. B. Montgomery, J. Williams-Warren, K. M. Vasiljev, D. Borowitz, C. M. Bowman, B. C. Marshall, S. Marshall, A. L. Smith, et al. 1999. Intermittent administration of inhaled tobramycin in patients with cystic fibrosis. N. Engl. J. Med. 340:23-30. [DOI] [PubMed] [Google Scholar]
- 23.Rocchetta, H. L., L. L. Burrows, J. C. Pacan, and J. S. Lam. 1998. Three rhamnosyltransferases responsible for assembly of the A-band d-rhamnan polysaccharide in Pseudomonas aeruginosa: a fourth transferase, WbpL, is required for the initiation of both A-band and B-band lipopolysaccharide synthesis. Mol. Microbiol. 28:1103-1119. [DOI] [PubMed] [Google Scholar]
- 24.Rocchetta, H. L., and J. S. Lam. 1997. Identification and functional characterization of an ABC transport system involved in polysaccharide export of A-band lipopolysaccharide in Pseudomonas aeruginosa. J. Bacteriol. 179:4713-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welsh, M. J., and R. B. Fick. 1987. Cystic fibrosis. J. Clin. Investig. 80:1523-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiegand, I., A. K. Marr, E. B. Breidenstein, K. N. Schurek, P. Taylor, and R. E. W. Hancock. 2008. Mutator genes giving rise to decreased antibiotic susceptibility in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52:3810-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]