Abstract
All 982 methicillin-resistant Staphylococcus aureus strains collected from August 2006 to December 2007 were tested for vancomycin susceptibility by using 3-μg/ml vancomycin brain heart infusion screening plates, a vancomycin Etest, and a vancomycin/teicoplanin macro Etest. Three vancomycin-intermediate Staphylococcus aureus (VISA) (0.3%) and two heterogeneous VISA (0.2%) isolates were identified. The screening method yielded 895 cases of ≤1 colony and 87 positive results (with growth of >1 colony after 48 h); further Etests showed 82/87 isolates with growth on screening plates to be false positive. Repeat testing showed a false-positivity rate of only 15 of the original 87 isolates by plate screening.
Because of the inability of routine antimicrobial susceptibility test methods to detect heterogeneous vancomycin-intermediate Staphylococcus aureus (hVISA) and VISA, the false perception that the incidences of these strains are rare exists (1, 4, 5, 6-10, 14-16). Walsh et al. (19, 21-24) have recommended an overnight vancomycin Etest to detect VISA and a macro Etest using a 2 McFarland standard, 48 h of incubation, brain heart infusion (BHI) agar, and vancomycin and teicoplanin strips to detect hVISA strains. Recently, Rybak et al. have indicated that the incidence of hVISA strains has increased over the past 22 years, for an overall incidence of 2.2% (11). Vancomycin-resistant S. aureus (VRSA) strains are currently very rare and thus far have not posed therapeutic problems (1, 17, 20).
We performed a prospective study to screen for the incidence and characteristics of vancomycin nonsusceptible phenotypes in all methicillin-resistant S. aureus (MRSA) strains isolated in the clinical laboratory at Hershey Medical Center from August 2006 to December 2007. We selected BHI agar because this medium is recommended and standardized by the CLSI for vancomycin agar screening plates (2, 3) and also for macro Etest (protocol EAS 003 on the AB Biodisk website) and is easily prepared in-house. On the basis of CDC-recommended screening for reduced vancomycin susceptibility (2, 3) and revised CLSI susceptible breakpoints (2, 3, 16), we decided upon a vancomycin concentration of 3 μg/ml in screening plates.
Strains were identified as Staphylococcus aureus by the Pastorex Staph-Plus method (Bio-Rad, Redmond, WA), and MRSA resistotype was defined using growth on 6-μg/ml oxacillin agar screening plates (Becton Dickinson, Inc. [BD], Sparks, MD) with a 30-μg cefoxitin disk (BD) and/or a PBP2 test (Oxoid, Ltd., Basingstoke, United Kingdom).
Only one isolate per patient visit or per day of hospitalization was stored. Additional isolates obtained from subsequent cultures were examined by multiple-locus variable-number tandem-repeat fingerprinting (MLVF; formerly multiple-locus variable-number tandem-repeat assay) typing (12, 13), and clones with profiles identical to that of the original MRSA isolate from the same patient were excluded. Strains were frozen at −70°C in double-strength skim milk (BD) until testing.
All strains were tested for (i) growth on BHI (BD) containing 3 μg/ml vancomycin by delivering 10 μl of a suspension with a 0.5 McFarland standard in saline onto the 3-μg/ml vancomycin plate and incubating the plate for 48 h at 35°C; (ii) vancomycin susceptibility by a conventional Etest methodology, using an inoculum with a 0.5 McFarland standard in saline plated onto Mueller-Hinton agar (BD) with MIC interpretation after 24 h of incubation at 35°C (protocol EAS 003 on AB Biodisk website); and (iii) susceptibility by a macro Etest, in which a 100-μl aliquot of a suspension with a 2 McFarland standard in Mueller-Hinton broth (BD) was streaked onto a BHI plate (BD). Vancomycin and teicoplanin Etest strips were placed on the plate and incubated at 35°C, and the results were interpreted at 24 and 48 h (protocol EAS 003 on AB Biodisk website). Controls run with each procedure included S. aureus strains ATCC 29213 (vancomycin susceptible), ATCC 700698 (hVISA; Mu3), and ATCC 700699 (VISA; Mu50). All strains with positive vancomycin and/or teicoplanin macro Etest results were considered possible hVISA strains and tested by the population analysis profile (PAP) method (18) in duplicate. PAP was the definitive criterion for defining the hVISA phenotype.
All hVISA and VISA isolates were subjected to Panton-Valentin leukocidin (PVL)/mecA screening and staphylococcal cassette chromosome mec (SCCmec) and agr typing. The MLVF method (12, 13) was used for typing of all 26 VISA and VRSA isolates deposited in the Network on Antimicrobial Resistance in Staphylococcus aureus [NARSA] repository by December 2006. MLVF typing was also performed for all multiple strains isolated from the same patient.
We considered strains to be potential hVISA strains if they grew >1 colony on the 3-μg/ml screening plate or had macro Etest results at 48 h of ≥8 μg/ml for both vancomycin and teicoplanin or ≥12 μg/ml for teicoplanin alone (22, 24). All potential hVISA strains were tested by PAP (18) to confirm hVISA phenotype. Strains were considered VISA when vancomycin MICs of 3 to 8 μg/ml were obtained by a conventional Etest with incubation for 24 h (1, 3).
During the study period, 982 unique MRSA isolates were collected (Table 1). The results for the 3-μg/ml screen plate were as follows: no growth at 48 h, 843 strains; 1 colony at 48 h, 52 strains; 2 to 15 colonies at 48 h, 66 strains; slight growth (>15 colonies to 25% growth) at 48 h, 7 strains; moderate growth (25% to 75% growth) at 48 h, 13 strains; and confluent growth (75% or more growth) at 48 h, 1 strain. Upon repeat testing of the 87 strains with growth upon initial screening, an absence of growth was observed in 60 strains, 1 colony at 48 h in 12 strains, 2 to 15 colonies at 48 h in 12 strains, slight growth at 48 h in 1 strain, moderate growth at 48 h in 1 strain, and confluent growth at 48 h in 1 strain. Additionally, repeat testing of 10 vancomycin-susceptible strains with vancomycin MICs of 2 μg/ml yielded no growth on screens in eight strains, one colony in one strain, and two colonies in one strain.
TABLE 1.
Sources of isolates tested in the study
| Source of isolation | No. of isolates | % of isolates |
|---|---|---|
| Wound | 589 | 60.0 |
| Respiratory tract | 203 | 20.7 |
| Blood | 95 | 9.7 |
| Tissue | 32 | 3.3 |
| Fluid | 24 | 2.4 |
| Ear | 17 | 1.7 |
| Eye | 9 | 0.9 |
| Catheter tip | 3 | 0.3 |
| Bone | 3 | 0.3 |
| Genital tract | 3 | 0.3 |
| Gastric mucosa | 2 | 0.2 |
| Drainage | 2 | 0.2 |
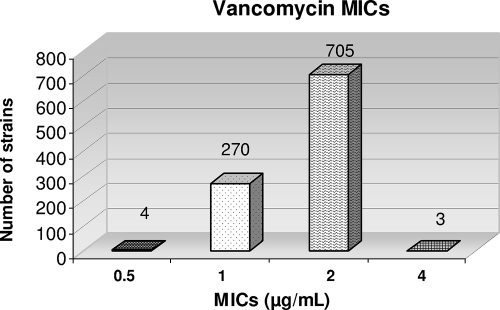
The conventional-vancomycin-Etest (24 h) MIC results for the 982 strains were as follows: for 4 strains, 0.5 μg/ml; for 270 strains, 1 μg/ml; for 705 strains, 2 μg/ml; and for 3 strains, 4 μg/ml (Fig. 1). There was clustering around the breakpoint, with 270 strains having vancomycin MICs of 1 μg/ml and 705 having vancomycin MICs of 2 μg/ml (1). Conventional-vancomycin-Etest MICs of 1.5 μg/ml (only available by Etest) were reported as 2 μg/ml. Of the 705 strains reported with MICs of 2 μg/ml, 632 strains actually had Etest results of 1.5 μg/ml. Eighty-two of the 87 isolates on initial testing and 15 isolates on repeat testing with growth were considered false positives, showing growth of >1 colony on the screening plate, a macro Etest MIC of ≤8 μg/ml for both vancomycin and teicoplanin or ≤12 μg/ml for teicoplanin or a negative PAP result, and a conventional-vancomycin-Etest MIC of ≤2 μg/ml.
FIG. 1.
Comparison of overnight conventional vancomycin Etest MICs.
Three VISA strains, SA770, SA1287, and SA1984, as well as two hVISA strains, SA618 and SA873, were identified. Two strains, SA662 and SA2089, were identified as potential hVISA strains. Strain SA662 showed moderate growth on the 3-μg/ml vancomycin screen and a teicoplanin macro Etest result of 12 μg/ml at 48 h. Strain SA2089 had no growth on the vancomycin screen but a teicoplanin macro Etest result of 12 μg/ml. However, both of these strains were negative by PAP, so their phenotypic significance remains unclear (Table 2).
TABLE 2.
Phenotypic characteristics of VISA and hVISAs isolates
| Isolate | MIC (μg/ml) for indicated Etest
|
PAP result | ||
|---|---|---|---|---|
| Vancomycin | Macro
|
|||
| Vancomycinc | Teicoplaninc | |||
| VISA (SA770) | 4 | 6 | 24 | NTa |
| VISA (SA1287) | 4 | 4 | 8 | NT |
| VISA (SA1984) | 4 | 6 | 8 | NT |
| hVISA (SA618) | 2 | 3 | 24 | Confirmed presence of subpopulation |
| hVISA (SA873) | 2 | 3 | 12 | Confirmed presence of subpopulation |
| VSSAb (SA662) | 2 | 4 | 12 | No subpopulation, VSSA phenotype |
| VSSA (SA2089) | 1 | 3 | 12 | No subpopulation, VSSA phenotype |
NT, not tested.
VSSA, vancomycin-susceptible S. aureus.
Cutoffs rather than real MICs.
All VISA and hVISA strains were MRSA and PVL negative with agr type II (three isolates) or I (one hVISA isolate). SCCmec typing showed that one VISA isolate and all potential hVISA isolates had SCCmec type II and that one VISA strain had SCCmec type IV (Table 3). Strains that are SCCmec type II, PVL negative, and primarily agr type II are characteristic of hospital-acquired MRSA. Also, agr type II is associated with reduced susceptibility of S. aureus to glycopeptides. One VISA strain (SA770) and the Hershey VRSA strain (VRS2) were genetically identical (Fig. 2). No other genetic correlation among the two hVISA strains or the three VISA strains isolated at Hershey Medical Center and any of the VRSA or VISA strains currently deposited in NARSA was found.
TABLE 3.
Molecular analysis of VISA and potential hVISA isolates
| Isolate | MLVF type | mec/PVL result | SCCmec type | agr type |
|---|---|---|---|---|
| VISA (SA770) | A | +/− | II | II |
| VISA (SA1287) | B | +/− | IV | II |
| VISA (SA1984) | C | +/− | II | II |
| hVISA (SA618) | D | +/− | II | I |
| hVISA (SA873) | E | +/− | II | II |
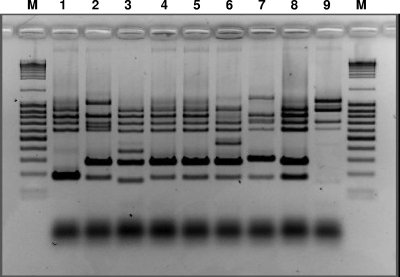
FIG. 2.
Relatedness of VISA and hVISA isolates and VRSA and VISA isolates deposited in the NARSA repository. Lanes: 1, VRS1 (first VRSA strain isolated in Michigan, 2002); 2, VRS3 (third VRSA strain isolated in New York, 2004); 3, VRS5 (fifth VRSA strain isolated in Michigan, 2005); 4, VRS2 (second VRSA strain isolated in Pennsylvania [Hershey Medical Center], 2002); 5, SA770 VISA; 6, SA1984 VISA; 7, SA 1287 VISA; 8, SA873 hVISA; 9, SA618 hVISA. M, molecular mass marker.
Case reports for the five patients with VISA or hVISA are presented in the supplemental material.
The 3-μg/ml screenings yielded 87/982 and 15/982 positive results (growth of >1 colony after 48 h), which were 8.9% and 1.5% of the total isolates on the two separate screenings. Among the 87 strains with growth, 82 were false positives based on negative Etest results, while one isolate with a positive teicoplanin macro Etest result of 12 μg/ml had a negative PAP result. Repeat testing showed lower false-positivity rates for 15 of the 87 original strains. A total of 5/982 isolates (0.5%) were identified as either hVISA (positive macro Etest and PAP results) or VISA (conventional vancomycin Etest) strains. Colony counts varied from >1 to slight growth for hVISA strains and slight to confluent growth for VISA strains. Among all negative isolates (no growth on 3-μg/ml vancomycin plates), only one isolate was teicoplanin macro Etest positive (12 μg/ml), but PAP proved this isolate to be vancomycin susceptible. The reason for this discrepancy is unclear; in any event, accuracy in this screening method requires validation by testing as many other hVISA isolates as possible. Yusof et al. (25) in a very recent paper have reported accuracy for a new double-sided vancomycin-teicoplanin Etest strip in identification of hVISA and VISA.
It is clearly noted that, although the 3-μg/ml screening plate detected all five hVISA and VISA strains, a very high false-positivity rate of 94% (82 false-positive strains out of 87 strains with growth) was found upon initial testing. However, especially in developing countries without Etests, a screening test which does not miss hVISA or VISA but yields a high false-positivity rate is still of clinical use. Additionally, very few U.S. laboratories use the macro Etest and PAP methods, and our screening method would at least alert clinicians to positive strains which might otherwise not be detected by the methods used. Also, it is noteworthy that only a maximum of 87 tests on the first run required additional Etests, which seems easier to manage than routine Etesting of all MRSA isolates to look for hVISA or VISA. Also, if Etests are unavailable, it is easier to send only strains with positive screen results to another institution with more facilities.
We propose routine use of a three-step procedure to screen for hVISA/VISA phenotypes, as follows: (i) initial screening with 3-μg/ml vancomycin BHI agar plates is performed as described above, (ii) all isolates showing growth of >1 colony on screening plates after 48 h of incubation should be subjected to a conventional vancomycin Etest (to identify possible VISA) and a macro Etest using both teicoplanin and vancomycin cutoffs, and (iii) possible hVISA isolates (macro Etest results) should be discussed with the clinician to compare clinical responses for patients treated with vancomycin and the hVISA phenotype. The clinical use of PAP in the routine laboratory is debatable; if we had not included PAP, one vancomycin-susceptible MRSA strain would have been misidentified as hVISA. Many more hVISA strains are required before the above methods can be validated.
Our study confirms the efficacy of conventional and macro Etest in identification of a small number of hVISA and VISA strains. All five patients with the latter had predisposing conditions (see the supplemental material), previous vancomycin and/or daptomycin treatment, and similar physical conditions. The significance of organisms identified as hVISA by the macro Etest but having negative PAP results is unknown. Without inclusion of the regular and vancomycin and teicoplanin macro Etests, rates of hVISA and VISA will remain underreported. We propose the 3-μg/ml vancomycin screening method as an alternative for clinical laboratories without resources for routine Etests or for large-scale surveillance studies for VISA/hVISA. However, our reported method for screening out false-negatives and preliminarily identifying hVISA and VISA strains must be balanced against its false-positivity rate. Much more work on this subject is required.
Supplementary Material
Acknowledgments
This study was supported by a grant from Pfizer, Inc., New York, NY.
Footnotes
Published ahead of print on 6 October 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Appelbaum, P. C. 2007. Reduced glycopeptide susceptibility in methicillin-resistant Staphylococcus aureus (MRSA). Int. J. Antimicrob. Agents 30:398-408. [DOI] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing, 16th informational supplement, M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A7, 7th ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- 4.de Lassence, A., N. Hidri, J. F. Timsit, M. L. Joly-Guillou, G. Thiery, A. Boyer, P. Lable, A. Blivet, H. Kalinowski, Y. Martin, J. P. Lajonchere, and D. Dreyfuss. 2006. Control and outcome of a large outbreak of colonization and infection with glycopeptide-intermediate Staphylococcus aureus in an intensive care unit. Clin. Infect. Dis. 42:170-178. [DOI] [PubMed] [Google Scholar]
- 5.Delgado, A., J. T. Riordan, R. Lamichhane-Khadka, D. C. Winnett, J. Jimenez, K. Robinson, F. G. O'Brien, S. A. Cantore, and J. E. Gustafson. 2007. Hetero-vancomycin-intermediate methicillin-resistant Staphylococcus aureus isolate from a medical center in Las Cruces, New Mexico. J. Clin. Microbiol. 45:1325-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garnier, F., D. Chainier, T. Walsh, A. Karlsson, A. Bolmström, C. Grelaud, M. Mounier, F. Denis, and M. C. Ploy. 2006. A 1 year surveillance study of glycopeptide-intermediate Staphylococcus aureus strains in a French hospital. J. Antimicrob. Chemother. 57:146-149. [DOI] [PubMed] [Google Scholar]
- 7.Julian, K., K. Kosowska-Shick, C. Whitener, M. Roos, H. Labischinski, A. Rubio, L. Parent, L. Ednie, L. Koeth, T. Bogdanovich, and P. C. Appelbaum. 2007. Characterization of daptomycin nonsusceptible vancomycin-intermediate Staphylococcus aureus in a patient with endocarditis. Antimicrob. Agents Chemother. 51:3445-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu, C., and H. F. Chambers. 2003. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob. Agents Chemother. 47:3040-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maor, Y., G. Rahav, N. Belausov, D. Ben-David, G. Smollan, and N. Keller. 2007. Prevalence and characteristics of heteroresistant vancomycin-intermediate Staphylococcus aureus bacteremia in a tertiary care center. J. Clin. Microbiol. 45:1511-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robert, J., R. Bismuth, and V. Jarlier. 2006. Decreased susceptibility to glycopeptides in methicillin-resistant Staphylococcus aureus: a 20 year study in a large French teaching hospital. J. Antimicrob. Chemother. 57:506-510. [DOI] [PubMed] [Google Scholar]
- 11.Rybak, M. J., S. N. Leonard, K. L. Rossi, C. M. Cheung, H. S. Sadar, and R. N. Jones. 2008. Characterization of vancomycin-heteroresistant Staphylococcus aureus from the metropolitan area of Detroit, Michigan, over a 22-year period (1986 to 2007). J. Clin. Microbiol. 46:2950-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabat, A., J. Krzyszton-Russjan, W. Strzalka, R. Filipek, K. Kosowska, W. Hryniewicz, J. Travis, and J. Potempa. 2003. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 41:1801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabat, A., N. Malachowa, J. Miedzobrodzki, and W. Hryniewicz. 2006. Comparison of PCR-based methods for typing Staphylococcus aureus isolates. J. Clin. Microbiol. 44:3804-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sancak, B., S. Ercis, D. Menemenlioglu, S. Colakoglu, and G. Hascelik. 2005. Methicillin-resistant Staphylococcusd aureus heterogeneously resistant to vancomycin in a Turkish university hospital. J. Antimicrob. Chemother. 56:519-523. [DOI] [PubMed] [Google Scholar]
- 15.Sievert, D. M., J. T. Rudrik, J. B. Patel, L. C. McDonald, M. J. Wilkins, and J. C. Hageman. 2008. Vancomycin-resistant Staphylococcus aureus in the United States 2002-2006. Clin. Infect. Dis. 46:668-674. [DOI] [PubMed] [Google Scholar]
- 16.Tenover, F. C., and R. C. Moellering, Jr. 2007. The rationale for revising the Clinical and Laboratory Standards Institute vancomycin minimal inhibitory concentration interpretive criteria for Staphylococcus aureus. Clin. Infect. Dis. 44:1208-1215. [DOI] [PubMed] [Google Scholar]
- 17.Tenover, F. C., L. M. Weigel, P. C. Appelbaum, L. K. McDoμgal, J. M. Chaitram, S. K. McAllister, N. C. Clark, G. E. Killgore, C. M. O'Hara, L. Jevitt, J. B. Patel, and B. Bozdogan. 2004. Vancomycin-resistant Staphylococcus aureus isolate from a patient in Pennsylvania. Antimicrob. Agents Chemother. 48:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trakulsomboon, S., S. Danchaivijitr, Y. Rongrungruang, C. Dhiraputra, W. Susaemgrat, T. Ito, and K. Hiramatsu. 2001. First report of methicillin-resistant Staphylococcus aureus with reduced susceptibility to vancomycin in Thailand. J. Clin. Microbiol. 39:591-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walsh, T. R., A. Bolmström, A. Qwärnstrom, P. Ho, M. Wootton, R. A. Howe, A. MacGowan, and D. Diekema. 2001. Evaluation of current methods for detection of staphylococci with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 39:2439-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weigel, L. M., R. M. Donlan, D. H. Shin, B. Jensen, N. C. Clark, L. K. McDoμgal, W. Zhu, K. A. Musser, J. Thompson, D. Kohlerschmidt, N. Dumas, R. J. Limberger, and J. B. Patel. 2007. High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob. Agents Chemother. 51:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wootton, M., P. M. Bennett, A. P. MacGowan, and T. R. Walsh. 2005. Reduced expression of the atl autolysin gene and susceptibility to autolysis in clinical heterogeneous glycopeptide-intermediate Staphylococcus aureus (hGISA) and GISA strains. J. Antimicrob. Chemother. 56:944-947. [DOI] [PubMed] [Google Scholar]
- 22.Wootton, M., A. P. MacGowan, and T. R. Walsh. 2006. Comparative bactericidal activities of daptomycin and vancomycin against glycopeptide-intermediate Staphylococcus aureus (GISA) and heterogeneous GISA strains. Antimicrob. Agents Chemother. 50:4195-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wootton, M., A. P. MacGowan, T. R. Walsh, and R. A. Howe. 2007. A multicenter study evaluating the current strategies for isolating Staphylococcus aureus strains with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 45:329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wootton, M., T. R. Walsh, and A. P. MacGowan. 2005. Evidence for reduction in breakpoints used to determine vancomycin susceptibility in Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3982-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yusof, A., A. Englehardt, A. Karlsson, L. Bylund, P. Vidh, K. Mills, M. Wootton, and T. R. Walsh. 2008. Evaluation of a new Etest vancomycin-teicoplanin strip for detection of glycopeptide-intermediate Staphylococcus aureus (GISA), in particular, heterogenous GISA. J. Clin. Microbiol. 46:3042-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.