Abstract
PSI-6130 (β-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine) is a selective inhibitor of hepatitis C virus (HCV) replication that targets the NS5B polymerase. R7128, the prodrug of PSI-6130, has shown antiviral efficacy in patients chronically infected with HCV genotype 1a (GT-1a) and GT-1b. We observed that the compound exhibited potent in vitro activity against laboratory-optimized HCV replicons as well as against a panel of replicons containing NS5B HCV polymerases derived from GT-1a and GT-1b clinical isolates. We used the HCV replicon cell system to examine the emergence of variants with reduced sensitivity to PSI-6130. Short-term treatment of cells harboring the HCV subgenomic replicon with PSI-6130 cleared the replicon without generating resistant variants. Long-term culture of the cells under the compound selection generated the S282T substitution in a complex pattern with other amino acid substitutions in the NS5B polymerase. The presence of the coselected substitutions did not increase the moderate three- to sixfold loss of sensitivity to PSI-6130 mediated by the S282T substitution; however, their presence enhanced the replication capacity compared to the replication levels seen with the S282T substitution alone. We also observed a lack of cross-resistance between PSI-6130 and R1479 and demonstrated that long-term culture selection with PSI-6130 in replicon cells harboring preexisting mutations resistant to R1479 (S96T/N142T) results in the emergence of the S282T substitution and the reversion of S96T to wild-type serine. In conclusion, PSI-6130 presents a high barrier to resistance selection in vitro, selects for variants exhibiting only low-level resistance, and lacks cross-resistance with R1479, supporting the continued development of the prodrug R7128 as a therapeutic agent for the treatment of HCV infection.
Hepatitis C virus (HCV) is a positive-strand RNA virus and a member of the Hepacivirus genus in the Flaviviridae family. Acute infection with HCV progresses into chronic infection for approximately 80% of infected patients and is a major cause of liver cirrhosis and hepatocellular carcinoma. The current use of pegylated alpha interferon (IFN-α) in combination with ribavirin for chronic HCV infection treatment (10, 14, 16) results in a 37 to 50% sustained viral response in patients infected with HCV genotype 1 (GT-1) (15). This suboptimal sustained viral response, combined with the high prevalence of undesirable side effects, has prompted focused efforts to develop specifically targeted antiviral therapies for HCV infections. The HCV NS5B RNA-dependent RNA polymerase (RdRp) is a critical enzyme for viral RNA replication and presents an attractive therapeutic target. Specific inhibitors for the HCV polymerase, including nucleoside analog inhibitors (7, 14, 21, 22, 30) and nonnucleoside analog inhibitors (2-6, 8, 11, 17, 18, 19, 47), have been generated, and some have entered clinical development.
Specific nucleoside inhibitors having the 2′ and/or 4′ modification, including 2′-C-methylcytidine (NM107) (36, 37), 2′-C-methyladenosine (30), 2′-C-methylguanosine (30), 2′-C-methyl-7-deaza-adenosine (MK-0608) (35), 4′-azidocytidine (R1479) (22), 2′-deoxy-2′-β-fluoro-4′-azidocytidine (RO-0622), and 2′-deoxy-2′-β-hydroxy-4′-azidocytidine (RO-9187) (21), have shown specific inhibition of HCV replication in vitro. Moreover, a number of these nucleoside analog inhibitors, including R1626 (prodrug of R1479) (40, 42), NM283 (prodrug of NM107) (1), and MK-0608 (34), have demonstrated clinical efficacy for HCV-infected patients or for HCV-infected chimpanzees.
β-d-2′-Deoxy-2′-fluoro-2′-C-methylcytidine (PSI-6130) (32, 44) is a potent nucleoside analog inhibitor of the HCV NS5B polymerase. R7128, a prodrug of PSI-6130, has demonstrated efficacy in a phase 1b clinical study, with a mean HCV RNA reduction of 2.7 log10 IU/ml when administered at 1,500 mg twice daily for 2 weeks to treatment-experienced patients infected with HCV GT-1a or GT-1b (41). In combination with poyethylene glycol-IFN/ribavirin, administration of R7128 at 1,500 mg twice daily demonstrated a decline of 5.12 log10 IU/ml in HCV RNA levels, correlating with an 85% rapid virologic response (reduction to less than 15 IU/ml). PSI-6130 inhibits the replication of HCV through formation of its 5′ triphosphate form, which functions as an alternative substrate for the viral polymerase, competitively inhibiting viral RNA synthesis by preventing further extension after incorporation (44). In similarity to natural nucleotides and other nucleoside analog inhibitors, PSI-6130 undergoes uptake by the cells and phosphorylation to the 5′ triphosphate derivative (32). A recent study characterizing the metabolic activation of PSI-6130 in human primary hepatocytes identified a novel UTP metabolite that displayed inhibitory activity against the NS5B polymerase in vitro, indicating that PSI-6130 is metabolized into two pharmacologically active species in the cells (28).
An early understanding of the nature of resistant HCV variants likely to emerge under conditions of drug treatment may allow improved drug design and, ultimately, the selection of inhibitors with unique resistance profiles to be used in the clinic in combination treatment regimens of greater efficacy. Nucleoside analog inhibitors with 2′-C-methyl modification, including 2′-C-methylcytidine (NM107), 2′-C-methyl-7′-deaza-adenosine (MK-0608), and 2′-C-methyladenosine, were shown to select for the S282T substitution in the active site of the polymerase in HCV replicon studies (23, 30, 34, 35). Confirming the replicon study results, the S282T substitution emerged in HCV-infected chimpanzees that were treated with MK-0608 (34). In the replicon studies, the mutation conferred a significant loss of sensitivity to these inhibitors that ranged from approximately 20- to over 100-fold, depending on the particular analog (30). PSI-6130 has both a 2′-fluoro and a 2′-C-methyl modification on the ribose sugar moiety (Fig. 1), thus differing from other modified nucleoside analogs which have only the 2′-C-methyl modification. The additional modification may alter the molecular interactions between PSI-6130 and the wild type (WT) as well as the polymerase with the S282T substitution. The S282T substitution mediated only a moderate loss of activity for PSI-6130 (32), suggesting that PSI-6130 may have a different resistance profile compared to those of the other 2′-C-methyl-modified nucleoside analogs. The in vitro resistance profile emerging in the NS5B polymerase under selection with PSI-6130 has not been previously studied. Our goals were to examine the emerging amino acid substitutions following the treatment of replicon cells with PSI-6130 and to characterize the contribution of these mutations in conferring resistance to the compound.
FIG. 1.
Chemical structures of HCV inhibitors used in the study. PSI-6130, β-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine; R7128, PSI-6130 prodrug; RO2433, PSI-6130 uridine metabolite; NM107, 2′-C-methylcytidine; R1479, 4′-azidocytidine; NNI-1, thiophene-2 caboxylic acid.
MATERIALS AND METHODS
Compounds.
PSI-6130 was synthesized at Pharmasset Inc. R7128 (a prodrug of PSI-6130), R1479 (4′-azidocytidine), NM107 (2′-C-methylcytidine), and NNI-1 (thiophene-2-carboxylic acid) (Fig. 1) were synthesized at Roche Palo Alto LLC. Recombinant human IFN-α-2a (Roferon) was prepared at Hoffmann-La Roche (Basel, Switzerland). PSI-6130-TP, RO2433-TP, and NM107-TP were synthesized by TriLink BioTechnologies. 3′-Deoxycytidine-5′-triphosphate (3′-dCTP) was purchased from TriLink BioTechnologies (San Diego, CA).
Cell lines.
Cured or Lunet Huh7 cells (obtained from R. Bartenschlager) were cultured at 37°C in a humidified atmosphere with 5% CO2 in Dulbecco's modified Eagle medium supplemented with Glutamax and 100 mg/ml sodium pyruvate. The medium was further supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum and 1% (vol/vol) penicillin-streptomycin. All reagents were from Invitrogen (Carlsbad, CA).
The subgenomic replicon cell lines harboring stable WT GT-1b Con1 (2209-23) or WT GT-1a (pRLucH771b75S/I) were maintained under the same conditions as described above, with the addition of 0.5 mg/ml G418 (Geneticin; Invitrogen) (26, 43). The replicons in these cells express the Renilla luciferase as a reporter gene. All replicon cell lines in this study were maintained at subconfluence to ensure constant levels of replicon RNA (38, 39).
Determination of replicon inhibitory activity by use of the transient Firefly luciferase replicon, the stable Renilla replicon, and the stable qPCR assays.
Assays were performed as previously described (22, 23). Briefly, cells harboring replicons were treated with inhibitors for 72 h, and Firefly luciferase reporter signal, Renilla luciferase reporter signal, or replicon RNA levels were measured in the transient assay, the stable Renilla replicon assay, or the quantitative PCR (qPCR) assay, respectively.
The replication capacity in the transient assay was determined at 96 h following transfection, standardized to the 4-h luciferase signal following transfection, and in turn normalized against the WT replication levels. The lower limit for replication giving reliable 50% effective concentration (EC50) results was determined to be at an approximate signal-to-background ratio of 10 at 96 h following transfection.
Compound cytotoxicity was determined by using the WST-1 assay as described previously (23). Protein-adjusted shift results were determined by using the qPCR assay and cells cultured with the inhibitors in the presence of 40% (vol/vol) human serum (Serological Corporation, Toronto, Canada) (23).
Site-directed mutagenesis.
Mutations were introduced in the NS5B coding region of the transient subgenomic GT-1b replicon expressing the Firefly luciferase reporter gene by using either a QuikChange I or a QuikChange II site-directed mutagenesis kit and following the procedures of the manufacturer (Stratagene, La Jolla, CA). All introduced mutations were confirmed by double-strand DNA sequencing.
Generation of transient GT-1b and GT-1a shuttle replicons.
HCV subgenomic GT-1b and GT-1a shuttle replicons used in the transient replicon assay were prepared by adapting the repPI-luc/ET (obtained from R. Bartenschlager), by replacing the pBR322 backbone with the pUC18, and by including two restriction sites flanking the 5′ (AsiSI) and the 3′ (RsrII or SacII) ends of the NS5B gene. The transient replicon containing the GT-1a H77 sequence was adapted from the GT-1b transient replicon by replacing the nonstructural region of the Con1 strain with the sequence of the H77 strain (except the first 75 amino acids of NS3 of the Con1 origin that remained). For better replication efficiency, three adaptive mutations were introduced (48).
Generation of NS5B clinical isolates in the transient replicon.
HCV RNA was extracted from the plasma of treatment-naïve GT-1a or GT-1b HCV-infected patients. The NS5B polymerase genes were reverse transcribed and amplified using primers that contained the restriction site sequences for AsiSI in the 5′ end and RsrII or SacII in the 3′ end of the NS5B gene. After restriction enzyme digestion, GT-1b-amplified products were cloned into the GT-1b shuttle replicon vector and GT-1a-amplified products were cloned into the GT-1a shuttle replicon vector. Ninety-six individual colonies were pooled for each clinical isolate. In vitro-transcribed RNA was then prepared using RiboMAX T7 Express (Promega, Madison, WI).
Replicon clearance assay.
Cells were cultured as previously described (23), with some modifications. HCV replicon cells were cultured with 10× the respective EC50 of PSI-6130, NM107, or NNI-1 in the absence of neomycin selection for 29 days (replicon clearance phase). Cells were passaged at a 1:3 ratio when confluent, and samples were taken for determination of replicon RNA levels. After 29 days, the inhibitor was removed and cells were cultured for a further 14 days in culture medium containing 0.25 mg of G418/ml. Levels of HCV RNA were determined by using the qPCR assay and were expressed as log RNA change compared to the HCV RNA level seen with untreated cells at day 0.
Long-term selection of drug-resistant replicon variants.
For the selection of replicon variants resistant to PSI-6130 or the prodrug R7128, cells harboring the WT GT-1b replicon were plated in the presence of increasing concentrations of the inhibitor. A sample without compound and a sample selected with NM107 were set up in parallel as controls. Culture medium was changed every 3 to 4 days, and/or cells were passaged at a 1:5 dilution upon reaching 95% confluence. Generally, the inhibitor concentrations were increased as follows: between 2.5 and 5 μM, at intervals of 0.5 μM; from 5 and 10 μM, at intervals of 1 μM; between 10 and 20 μM, at intervals of 2 to 4 μM; and from 20 and 50 μM, at intervals of 5 μM. Above 50 μM, the concentration was increased at intervals of 10 μM. In certain instances when excessive cell death was observed, the compound concentration was temporarily reduced to allow recovery and cell growth. Once growth resumed, the concentration of the compound was increased. Selected cells were tested for sensitivity to PSI-6130 or R7128 at several time points by using the qPCR assay, and the NS5B coding sequence was examined. cDNAs were prepared by using SuperScript III One-Step reverse transcription-PCR (Invitrogen, Carlsbad, CA). Clonal sequencing analysis of NS5B was carried out by cloning the NS5B coding sequences into a TOPO-TA vector (Invitrogen) and analyzing individual clones. Sequencing spanning the entire NS5B coding region was performed on either pooled populations or individual clones by using primers covering both strands. Sequencing was performed using an ABI 3730 xl DNA analyzer, and sequences were analyzed using VNTI software (Invitrogen).
HCV polymerase assay.
The inhibition potency of compounds with respect to the RdRp activity of recombinant NS5B570-BK, NS5B570-Con1, and NS5B570-H77 proteins was determined by measuring the incorporation of radiolabeled NMP into acid-insoluble RNA products by use of a complement strand of internal ribosomal entry site (cIRES) RNA template, as previously described (23), with some modifications. Briefly, 50% inhibitory concentration (IC50) determinations were carried out using 200 nM in vitro-transcribed cIRES RNA template, 1 μCi of tritiated UTP (42 Ci/mmol), 500 μM ATP, 500 μM GTP, 1 μM CTP, 1× TMDN buffer (40 mM Tris-HCl [pH 8.0], 4 mM MgCl2, 4 mM dithiothreitol, 40 mM NaCl), and 200 nM enzyme. The inhibition potency of compounds with respect to the RdRp activity of NS5B570-S282T-Con1 was determined under GT-1b assay conditions as previously described (22). NS5B570-BK and NS5B570-Con1 enzymes were used as controls. The final reaction volume was 50 μl under all assay conditions. All reactions contained a final 10% dimethyl sulfoxide. Km and Ki values were measured as described previously (23).
HCV replicase complex assay.
In vitro replicase assays using purified membrane fractions from stable HCV replicon cells were performed as previously described (29).
Compound crystallography.
Suitable crystals of NM107 and PSI-6130 were obtained by evaporation of a saturated solution with methanol as the solvent. Crystals were mounted in loops and cooled to 89°K in a nitrogen stream. Diffraction data were collected using a Swiss light source (beamline X10SA) and a MAR CCD225 detector with synchrotron radiation (0.70 Å), and data were processed with XDS software. The crystal structures were solved and refined with SheIXTL (Bruker AXS, Karlsruhe, Germany).
CCDC 671178 and 675351, deposited with the Cambridge Crystallographic Data Centre, contain supplementary crystallographic data for this paper. These data can be obtained free of charge via http://www.ccdc.cam.ac.uk/cgi-bin/catreq.cgi, by emailing data_request@ccdc.cam.ac.uk, or by contacting The Cambridge Crystallographic Data Centre, 12, Union Road, Cambridge CB2 1EZ, United Kingdom (fax, 44 1223 336033).
Nucleotide sequence accession numbers.
The novel NS5B sequences described in this work have been deposited in GenBank and are provided as the supplemental material. The GenBank accession numbers are FJ217353 to FJ217356.
RESULTS
PSI-6130 exhibits potent and specific inhibitory activity against HCV RNA replication mediated by the NS5B polymerase.
PSI-6130 exhibits potent inhibitory activity against the replication of GT-1b subgenomic replicons (44). In this study, the inhibitory effect of PSI-6130 was further characterized using laboratory-optimized GT-1a and GT-1b subgenomic replicons as well as a panel of replicons containing the NS5B coding region derived from GT-1a and GT-1b clinical isolates.
Using a cell line harboring a stable GT-1b (Con1 strain) subgenomic replicon expressing the Renilla luciferase and a transient (Con1) replicon system containing the Firefly reporter gene, PSI-6130 inhibited GT-1b subgenomic RNA replication with equivalent EC50 values in both replicon systems when measured by either kinetic qPCR or luciferase reporter expression levels (Table 1). The prodrug R7128, developed to improve in vivo pharmacokinetic properties, also demonstrated potent inhibition of the stable and transient replicons with EC50 values similar to those of PSI-6130, which is consistent with the presence of esterases in serum-containing culture media. Both PSI-6130 and R7128 inhibited HCV GT-1b (Con1 strain) and GT-1a (H77 strain) subgenomic RNA replication, with similar levels of potency (mean EC50 values, 0.51 and 0.30 μM, respectively; Table 1). The antiviral potency measurements obtained for the nucleoside analogs tested were independent of the method used for HCV replication quantification (quantitative reverse transcription-PCR or luciferase reporter gene quantification) and were similar when determined using stable or transient HCV replicon systems (Table 1).
TABLE 1.
PSI-6130 is a potent inhibitor of the GT-1b and GT-1a HCV replicona
| Compound | GT-1b (Con1) (μM [mean ± SEM])
|
GT-1a (H77) EC50 (transient Firefly luciferase) (μM [mean ± SEM]) | ||||
|---|---|---|---|---|---|---|
| EC50 (stable qPCR) | EC50 (stable Renilla luciferase) | EC50 (transient Firefly luciferase) | qPCR EC50 (40% human serum) | CC50c WST-1 | ||
| PSI-6130 | 0.46 ± 0.13 | 0.61 ± 0.04 | 0.51 ± 0.04 | 0.51 ± 0.18 | >300 | 0.30 ± 0.04 |
| NM107 | 1.13 ± 0.34 | 1.51 ± 0.17 | 1.1 ± 0.07 | 1.69 ± 0.44 | >100 | NDb |
| R7128 | 0.32 ± 0.09 | 0.70 ± 0.16 | 0.89 ± 0.07 | ND | >100 | ND |
SEM values represent standard errors of the means of the results of a minimum of three independent experiments.
ND, not determined.
CC50, 50% cytotoxicity concentration.
The effect of the presence of human serum on the activity of PSI-6130 was examined. The presence of 40% human serum did not modify the inhibitory activity of PSI-6130. The mean EC50 for PSI-6130 in the presence of 40% human serum was 0.51 μM, a value equivalent to the EC50 in the absence of human serum (Table 1). PSI-6130 did not exhibit any cytotoxic effects on Huh-7 cells at the highest concentration tested (300 μM) using the WST-1 cytotoxicity assay (Table 1), consistent with previously reported observations (44).
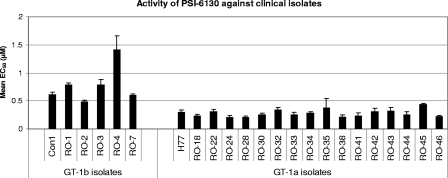
The inhibitory activity of PSI-6130 was evaluated using a panel of transient replicons containing the NS5B coding region derived from clinical isolates of treatment-naïve HCV-infected patients (Fig. 2). The mean EC50 values obtained for five GT-1b clinical isolates ranged from 0.60 to 1.41 μM (Fig. 2) and were comparable to the values obtained for the Con1 reference strain replicon (mean EC50 = 0.51 μM; Table 1). The mean EC50 values obtained for 16 GT-1a clinical isolates ranged from 0.20 to 0.43 μM and were comparable to the values obtained for the reference H77 strain replicon (mean EC50 = 0.30 μM; Table 1).
FIG. 2.
PSI-6130 exhibits potent activity against GT-1b and GT-1a NS5B clinical isolates. The inhibitory activity of PSI-6130 was evaluated using a panel of transient replicons containing the NS5B coding region derived from clinical isolates of treatment-naïve HCV GT-1a- or GT-1b-infected patients. After the amplified NS5B region was cloned from a patient serum into the corresponding genotype transient shuttle replicon, 96 individual molecular clones were pooled for each clinical isolate to mimic the natural polymorphism in the patient. The activity of PSI-6130 was assessed using the HCV transient replicon assay.
The inhibitory activity of the 5′ triphosphate form PSI-6130 (PSI-6130-TP) against the RdRp activity of NS5B was examined by using both GT-1b (NS5B570-BK and NS5B570-Con1) and GT-1a (NS5B570-H77) recombinant enzymes (Table 2). PSI-6130-TP exhibited equivalent levels of potency against the BK, Con1, and H77 NS5B polymerases (Table 2), based on IC50 values and inhibition constants (Ki). The inhibitory activity of PSI-6130-TP in the NS5B-dependent RNA synthesis assay was comparable to the inhibitory activity of NM107-TP within assay variability limits.
TABLE 2.
PSI-6130-TP is a potent inhibitor of GT-1a and GT-1b recombinant HCV NS5B polymerasesa
| Compound | Result for GT-1b (Con1) (μM [mean ± SEM])
|
Result for GT-1b (BK) (μM [mean ± SEM])
|
Result for GT-1a (H77) (μM [mean ± SEM])
|
|||
|---|---|---|---|---|---|---|
| IC50 | Ki | IC50 | Ki | IC50 | Ki | |
| PSI-6130-TP | 0.13 ± 0.01 | 0.034 ± 0.003 | 0.11 ± 0.03 | 0.04 ± 0.006 | 0.12 ± 0.03 | 0.032 ± 0.001 |
| NM107-TP | 0.09 ± 0.02 | 0.016 ± 0.004 | 0.18 ± 0.06 | 0.017 ± 0.01 | 0.25 ± 0.05 | 0.061 ± 0.012 |
SEM values represent standard errors of the means of the results of a minimum of three independent experiments.
Lack of emergence of replicon variants resistant to PSI-6130 in short-term in vitro selection.
In this study, we sought to determine the amino acid substitutions selected in the HCV replicon system conferring resistance to PSI-6130 and the level of activity reduction mediated by those substitutions. Several initial attempts to select replicons resistant to PSI-6130 in the presence of the selection agent G418 (five independent experiments, starting with ≥5× the EC50) and over short-term periods (less than 4 weeks) resulted in the clearance of the replicon and an inability to generate viable cells containing resistant variants (data not shown). To enhance the possibility of obtaining resistant variants, a replicon clearance assay was employed wherein the selection process was modified by separating the treatment with the inhibitor from the selection with G418, as previously described (23). In the first phase (the clearance phase) (Fig. 3), the cells were incubated in the presence of 10 times the EC50 of PSI-6130, NM107, or NNI-1 (Fig. 1) without the presence of G418 for a period of 29 days. In the second phase (rebound phase), the inhibitors were removed and the cells were cultured in the presence of G418 for a period of 14 days. As controls, cells were treated separately with either NM107 or NNI-1 (Fig. 1 and 3) as a nucleoside analog or as a nonnucleoside inhibitor, respectively.
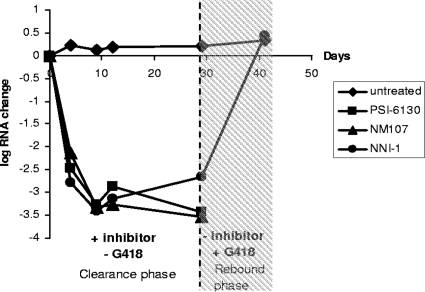
FIG. 3.
PSI-6130 and NM107 clear the HCV replicon. Cells harboring the stable HCV replicon were treated with 10× the EC50 of PSI-6130, NM107, or NNI-1 in the absence of G418 for 29 days (clearance phase). The inhibitors were then removed, and G418 was added for 14 days (rebound phase). HCV replicon levels were monitored using qPCR during the clearance and rebound phases.
In the control sample treated with NNI-1, multiple colonies formed after the course of treatment (Fig. 3 [representing the results of three experiments]). The presence of the replicon RNA in the cells was confirmed by qPCR analysis. Sequencing of the coding region for NS5B in the resulting colonies revealed the presence of the previously reported NNI-1 L419M resistance mutation (19). In contrast to the results of the treatment with NNI-1, no viable colonies formed in the samples that were treated with PSI-6130, suggesting the clearance of the replicon from the treated cells and the lack of emergence of replication-competent resistant variants under these conditions. Similarly, no colonies emerged in the samples treated with NM107 (Fig. 3) (23).
Long-term selection of replicon cells with PSI-6130 and generation of replicons with reduced sensitivity to the compound.
The inability to generate replicon variants resistant to PSI-6130 due to replicon clearance, as described above, prompted the use of a long-term culture approach. Cells harboring a GT-1b stable replicon were cultured over several months, with small increments in concentrations of PSI-6130 in the presence of the G418 selection reagent. In certain instances when excessive cell death occurred under conditions of selection, the concentration of PSI-6130 was reduced to encourage cell growth. Untreated replicon cells as well as cells treated with NM107 were cultured in parallel as controls. The sensitivity of the cells to PSI-6130 under conditions of selection was determined at frequent intervals, and the NS5B coding sequence was determined to examine the presence of amino acid substitutions (Table 3).
TABLE 3.
Amino acid substitutions and phenotypic changes observed in the resistance selection with PSI-6130 and R7128 in replicon cells
| Selection set | Passage no. | PSI-6130/R7128 concn (μM) | EC50 fold shifta | Amino acid substitution(s) in NS5B |
|---|---|---|---|---|
| Untreated | 35 | 0 | No shift | No substitutions |
| Untreated | 56 | 0 | No shift | No substitutions |
| 1 | 6 | 5 | 7 | Y586C |
| 1 | 28 | 50 | 32 | S282S/T, C575S, Y586C |
| 1 | 53 | 100 | 51 | S282T, K81R, S84S/P I239L, A300A/T, L320F/L/C, A421V, Y586C |
| 2 | 25 | 50 | NDb | S282S/T, I239V, A396G |
| 2 | 27 | 25 | 25 | S282T, A396G |
| 2 | 28 | 25 | ND | S282T, I239L, A396G, V485L |
| 2 | 32 | 25 | 13 | S282T, I239L, A396G |
| 3 | 21 | 30 | 120 | K72M, S282S/T |
| 3 | 26 | 30 | 107 | K72M, S282T, L564M |
Shift calculated as EC50 for selected cells divided by the EC50 for untreated cells.
ND, not determined.
In selection set 1 (Table 3), the cells were cultured in the presence of PSI-6130 for a total of 53 passages. The concentration of PSI-6130 was increased gradually, starting at 2.5 μM (equivalent to approximately 5× the EC50) and reaching 100 μM at passage 53. The sensitivity to PSI-6130 was examined at several passages. At passages 6, 28, and 53, EC50 increased 7-, 32-, and 51-fold, respectively (data representing selected passages are presented in Table 3). Examination of the cell population amino acid sequences of NS5B revealed that as the cells were continually passaged in the presence of PSI-6130, mutations accumulated in the polymerase. The mutations observed included Y586C at passage 6; S282T, C575S, and Y586C at passage 28; and K81R, I239L, S282T, L320F/L/C, A421V, and Y586C at passage 53 (Table 3). Some of the mutations emerged and persisted until the end of treatment, and others were transient (e.g., C575S; Table 3). In contrast, no mutations were observed in the NS5B region in the cells cultured in the absence of PSI-6130 (Table 3).
Two additional selection experiments were performed, one with PSI-6130 (set 2) and one with the prodrug R7128 (set 3) (Table 3). Similar patterns were observed in these two selection experiments; population sequencing revealed that mutations accumulated in the NS5B coding region under conditions of PSI-6130 treatment. In selection set 2, mutations I239L, I239V, S282T, A396G, and V485L were observed. In selection set 3, mutations K72M, S282T, and L564M were detected (Table 3). In similarity to the results seen with set 1, some mutations emerged and persisted until the end of treatment and others were transient (e.g., V485L and L564M; Table 3).
The S282T mutation, a substitution known to confer resistance to 2′-C-methyl-substituted nucleoside analogs, emerged in all three selection experiments, and it was accompanied by various other mutations. Generally, the emergence of the S282T substitution occurred after long-term passaging for more than 20 cell passages and the use of concentrations of PSI-6130 greater than 30 μM. For example, in selection set 1, the S282T mutation was first observed as a mix of threonine and WT serine at passage 24 under conditions of treatment with 45 μM PSI-6130 (data not shown), followed by a complete conversion to threonine at subsequent passages under conditions of treatment with 50 μM PSI-6130 (Table 3). Similar transitions were also observed in selection sets 2 and 3.
After several cell passages, the cells that were treated with NM107 exhibited a reduction in sensitivity to both PSI-6130 and NM107 accompanied by the emergence of the S282T substitution without additional substitutions being observed (data not shown).
Characterization of the S282T amino acid substitution observed in the PSI-6130-selected cells.
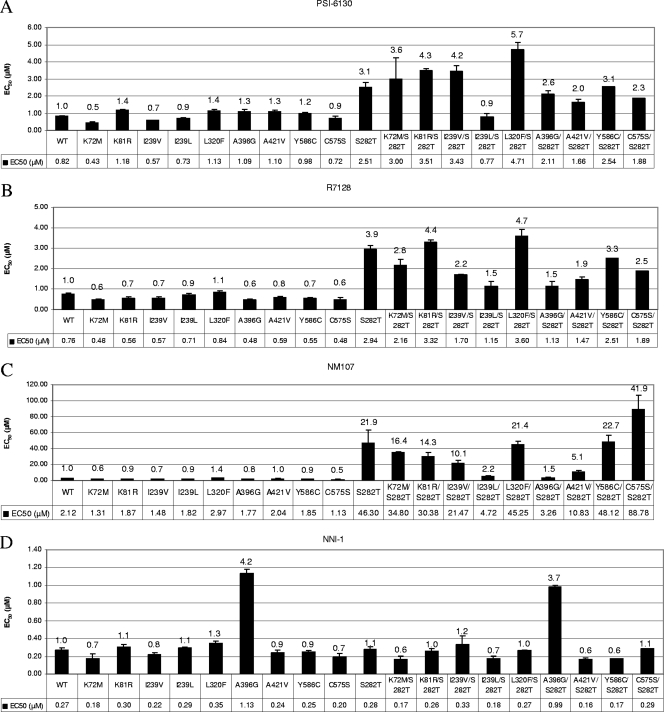
The S282T mutation was introduced into the replicon by site-directed mutagenesis and was tested in the transient replicon assay system. We observed that the S282T mutation reduced the replication capacity to 15% of the WT replication levels (Fig. 4), in agreement with previous observations (23, 30). We also observed that the S282T mutation conferred a moderate reduction in sensitivity to PSI-6130 and R7128. There was a three- to fourfold loss of sensitivity to PSI-6130 and R7128 compared to the results seen with WT replicons (Fig. 5A and B). The loss of sensitivity to NM107 with this substitution was appreciably higher (22-fold) (Fig. 5C) (23).
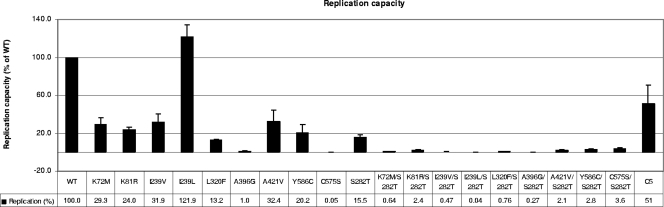
FIG. 4.
Replication capacity of transient replicons containing the selected NS5B amino acid substitutions. The amino acid substitutions observed in the selection with PSI-6130 or R1728 were introduced into the GT-1b Con1 transient HCV replicon. In vitro-transcribed RNA was transfected into Huh-7 cells, and the replication capacity was calculated (error bars represent the standard errors of the means of the results of three to five independent experiments). Clone C5 was generated by cloning the NS5B coding sequence from the selected cells into the transient replicon and was tested as described above.
FIG. 5.
Characterization of the observed amino acid substitutions in NS5B in the transient replicon assay. The amino acid substitutions observed in the selection with PSI-6130 were introduced into the GT-1b Con1 transient HCV replicons by site-directed mutagenesis. In vitro-transcribed RNA was transfected into Huh-7 cells, and the sensitivity of the replicons to PSI-6130 (A), R7128 (B), NM107 (C), and NNI-1 (D) was measured at 72 h following treatment (error bars represent the standard errors of the means of the results of three to five independent experiments). A value representing a severalfold shift in EC50 compared to the WT replicon value (determined by dividing the EC50 mutant value by the EC50 WT value) is presented above each bar graph point.
The effect of the S282T substitution was also examined in the context of clinical isolates. The substitution was introduced into four GT-1b clinical isolates obtained by isolating the molecular clones representing the consensus NS5B coding sequence of RO-1, RO-2, RO-3 (Fig. 2), or RO-11 (data not shown) and cloned into the GT-1b transient replicon. The substitution resulted in a reduction in the replication capacity (6 to 26% of that seen with the parent unsubstituted replicon) which is comparable to the reduction observed in the Con1 replicon containing the S282T substitution (15% of that seen with the parent unsubstituted Con1) (Table 4 and Fig. 4). The S282T substitution also resulted in a reduction in sensitivity to PSI-6130 (3.5- to 5.4-fold) comparable to the reduction observed with the Con1 replicon (3.1-fold) (Table 4; Fig. 5A). The S282T substitution conferred a greater reduction in susceptibility to NM107, as previously observed for the S282T mutant in the GT-1b Con1 replicon (Table 4; Fig. 5C).
TABLE 4.
PSI-6130 activity against GT-1b clinical isolates with the S282T substitution
| Strain and mutationa | % Replication capacityb | EC50 (μM [mean ± SEMc])
|
|
|---|---|---|---|
| PSI-6130 | NM107 | ||
| Con1 | |||
| None | 100 | 0.82 ± 0.04 | 0.52 ± 0.07 |
| S282T | 15 | 2.51 ± 0.29 | >30d |
| RO-1-H06 | |||
| None | 100 | 0.28 ± 0.04 | 0.60 ± 0.13 |
| S282T | 15 | 1.07 ± 0.35 | >30d |
| RO-2-D09 | |||
| None | 100 | 0.25 ± 0.02 | 0.66 ± 0.06 |
| S282T | 26 | 0.90 ± 0.095 | >30d |
| RO-3-D10 | |||
| None | 100 | 0.26 ± 0.03 | 0.66 ± 0.11 |
| S282T | 6 | 1.41 ± 0.24 | >30d |
| RO-11-D07 | |||
| None | 100 | 0.30 ± 0.03 | 0.52 ± 0.06 |
| S282T | 25 | 1.04 ± 0.17 | >30d |
RO-1-H06, RO-2-D09, RO-3-D10, and RO-11-D07 are single molecular clones representing the consensus sequences of RO-1, RO-2, RO-3 (Fig. 2), and RO-11 (data not shown), respectively.
The replication capacity for each S282T mutant is expressed as the percentage of the corresponding parent sequence.
SEM values represent standard errors of the means of the results of a minimum of three independent experiments.
Maximum concentration of NM107 used in the assay.
We sought to further characterize the effects on the activity of PSI-6130, R7128, NM107, and NNI-1 mediated by the S282T substitution by using the stable HCV replicon system. The sensitivity of the HCV replicon containing the S282T substitution to PSI-6130 was examined in cells harboring the stable GT-1b HCV replicon by using the qPCR assay. The S282T substitution resulted in a moderate 2.4-fold reduction in sensitivity to PSI-6130 and a 3.4-fold reduction in sensitivity to R7128 (Table 5). In contrast, the S282T substitution resulted in a more pronounced reduction in sensitivity to NM107, with a 17-fold shift in the EC50 (Table 5). No change in sensitivity to NNI-1 was observed (Table 5).
TABLE 5.
PSI-6130 activity against WT and S282T stable HCV replicon
| Compound | qPCR EC50 (μM [mean ± SEMa])
|
EC50 fold shiftb | |
|---|---|---|---|
| WT | S282T | ||
| PSI-6130 | 0.31 ± 0.08 | 0.75 ± 0.22 | 2.4 |
| R7128 | 0.32 ± 0.09 | 1.08 ± 0.03 | 3.4 |
| NM107 | 1.55 ± 0.31 | 26.31 ± 3.77 | 17.0 |
| NNI-1 | 0.31 ± 0.04 | 0.22 ± 0.03 | 0.8 |
SEM values represent standard errors of the means of the results of a minimum of three independent experiments.
Shift calculated as EC50 for S282T mutants divided by EC50 for the WT.
The sensitivity to PSI-6130-TP and NM-107-TP of the GT-1b (Con1) recombinant NS5B enzyme containing the S282T substitution was examined using the in vitro polymerase enzymatic assay. Confirming the previously reported loss of sensitivity of the NS5B polymerase observed with the S282T mutation (32) and in agreement with the sensitivity profile observed in the replicon system, the S282T substitution resulted in a moderate reduction in NS5B sensitivity to PSI-6130-TP (5.4-fold shift in IC50 and 2-fold shift in Ki) (Table 6), which is in contrast to the more pronounced reduction in sensitivity to NM107-TP (111-fold shift in IC50 and 36-fold shift in Ki) (Table 6). Recently, an active intracellular uridine metabolite of PSI-6130, RO2433 (Fig. 1), was identified (28, 33). The activity of RO2433-TP was examined. The recombinant NS5B enzyme containing the S282T substitution exhibited a reduced sensitivity to RO2433-TP (20.2-fold shift in IC50 and 21.1-fold shift in Ki) (Table 6) (33). No change in sensitivity to the control 3′-dCTP was found.
TABLE 6.
PSI-6130-TP activity against WT and S282T recombinant HCV NS5B polymerases
| Compound | WT (μM [mean ± SEMa])
|
S282T (μM [mean ± SEMa])
|
Fold shiftb
|
|||
|---|---|---|---|---|---|---|
| IC50 | Ki | IC50 | Ki | IC50 | Ki | |
| PSI-6130-TP | 0.13 ± 0.01 | 0.023 ± 0.001c | 0.70 ± 0.12 | 0.044 ± 0.004 | 5.4 | 1.9 |
| NM107-TP | 0.09 ± 0.02 | 0.016 ± 0.001 | 10 ± 0.6 | 0.58 ± 0.035 | 111.1 | 36.5 |
| RO2433-TP | 0.52 ± 0.11 | 0.14 ± 0.03c | 11 ± 3 | 2.97 ± 0.81 | 20.2 | 21.1 |
| 3′-dCTP | 0.25 ± 0.09 | 0.045 ± 0.016 | 0.46 ± 0.19 | 0.027 ± 0.011 | 1.8 | 0.6 |
SEM values represent standard errors of the means of the results of a minimum of three independent experiments.
Shift calculated as value for S282T mutants divided by value for the WT.
From Ma et al. (28).
The effect of the S282T substitution on the activity of PSI-6130 and NM107 was further examined by using the replicase assay. Membrane fractions containing the replicase complexes were isolated from Huh-7 cells harboring either the WT replicon or the replicon with the S282T mutation and were tested for sensitivity to PSI-6130-TP and NM107-TP. The S282T mutation resulted in a less pronounced reduction in sensitivity to PSI-6130-TP (3.1-fold shift in IC50) than to NM107-TP (54.9-fold shift in IC50) (Table 7). These results confirm the results obtained in the replicon and the NS5B enzymatic assays (Table 5 and Table 6). No reduction in sensitivity to the control 3′-dCTP was observed (Table 7).
TABLE 7.
PSI-6130 activity against WT and S282T isolated replicase complexes
| Compound | Replicase IC50 (μM [mean ± SEMa])
|
IC50 fold shiftb | |
|---|---|---|---|
| WT | S282T | ||
| PSI-6130-TP | 0.34 ± 0.10 | 1.05 ± 0.18 | 3.1 |
| NM107-TP | 0.22 ± 0.17 | 12.07 ± 4.60 | 54.9 |
| 3′-dCTP | 0.78 ± 0.50 | 0.76 ± 0.12 | 0.97 |
SEM values represent standard errors of the means of the results of a minimum of three independent experiments.
Shift calculated as IC50 for S282T mutants divided by IC50 for the WT.
Taken together, results from this study, along with previously reported observations (32), indicate that the S282T substitution that emerged under conditions of PSI-6130 selection only moderately reduced the activity of the compound against HCV RNA replication mediated by the NS5B polymerase.
Individual and pair-wise characterization of amino acid substitutions emerging with the S282T substitution.
The amino acid substitutions that emerged and became persistently present in the three selection experiments with PSI-6130 or R7128 were tested for their effect on the replication capacity and for their effect on the sensitivity of the replicon to PSI-6130, R7128, and NM107. We characterized the K81R, K72M, I239V, I239L, L320F, A396G, A421V, C575S, and Y586C mutations (Table 3). These amino acid substitutions were introduced as single mutations by site-directed mutagenesis into a GT-1b transient replicon containing the Firefly luciferase reporter gene. All the substitutions, except I239L, reduced the replication capacity compared to the WT replicon results (Fig. 4). Certain substitutions had significantly lower replication capacity compared to the results seen with the WT replicon that included the A396G (1.0%) and the C575S (0.05%) substitutions. Other amino acid substitutions, including K72M, K81R, I239V, L320F, A421V, and Y586C, reduced the replication capacity to approximately 10 to 30% of WT replication levels (Fig. 4).
The effect of the observed amino acid substitutions on the sensitivity of the replicons was examined. We observed that the levels of sensitivity of the replicons containing these mutations were comparable to those observed with the WT replicons (Fig. 5A, B, and C). With the exception of the S282T substitution, none of the substitutions, when present alone, resulted in a noticeable change in the sensitivity to PSI-6130, R7128, or NM107.
Since the S282T substitution appeared to be the only mutation mediating a reduced sensitivity to PSI-6130, we investigated the effect of the combination of the S282T mutation in a pair-wise manner with the other observed amino acid substitutions on the replication capacity and on the sensitivity to PSI-6130, R7128, and NM107. The majority of the amino acid substitutions, when combined with the S282T mutation, caused a further reduction in the replication capacity compared to that of each substitution alone (Fig. 4). The replication capacities of all the double mutants were less than 5% of that seen with the WT replicon. Therefore, those amino acid substitutions in this pair-wise combination pattern did not generally confer a replication advantage.
The majority of the pair-wise combinations with S282T did not significantly influence the sensitivity of the replicons to PSI-6130, R7128, or NM107 beyond the reduction observed with the S282T substitution alone (Fig. 5A, B, and C). Substitutions C575S and L320F appeared to further reduce the sensitivity to NM107 and PSI-6130, respectively, but not sensitivity to the other analogs. In addition, the I239L, A396G, and A421V mutations appeared to partially compensate for the loss of the sensitivity induced by the S282T mutation. Despite the low replication capacity seen with the replicons with the C575S, L239L/S282T, and A396G/S282T substitutions, the replication levels were above the lower limit of the assay (see Materials and Methods) and the EC50s obtained were reproducible.
The amino acid substitutions, either individually or in pair-wise combinations with S282T, had no effect on the sensitivity of the replicons to NNI-1 (Fig. 5D), with the exception of the A396G substitution that conferred a three- to fourfold reduction in the sensitivity to NNI-1.
Multiple combined amino acid substitutions coselected with S282T enhance replication capacity.
The S282T substitution reduces the replication capacity to 15% of WT replication levels (Fig. 4). In selection set 1 at passage 53, the NS5B coding region in those selected cells contained the S282T substitution in a complex pattern together with other mutations, including K81R, S84S/P, I239L, A300A/T, L320F/L/C, A421V, and Y586C (Table 3). To examine the effect of having all mutations together, the NS5B coding sequence from selection set 1 at passage 53 (Table 3) was cloned into the HCV transient replicon cassette vector. A total of 15 individual clones were examined for NS5B sequences, and it was determined that all the substitutions were genetically linked and were present in the majority of the clones. Of the 15 clones, 9 contained all the major substitutions, including K81R, S84P, I239L, S282T, A300T, L320C/F, A421V, and Y586C. A total of 3 of the 15 clones were lacking either the S84P substitution or the L320C/F substitution. Another three clones were lacking either two, three, or four substitutions among the S84P, I239L, L320C/F, and the A300T. Substitutions K81R, S282T, A421V, and Y586C were present in all 15 clones.
A single clone, C5, containing only the major substitutions (including K81R, S84P, I239L, S282T, A300T, L320C, A421V, and Y586C) observed in the selected cells, was examined by transfection into Huh-7 cells and testing of replication levels as well as sensitivity to PSI-6130. When these substitutions coexisted, the replication capacity was 51% of the WT level (Fig. 4, last column), suggesting that it was enhanced compared to the replication in replicons containing the S282T substitution alone (Fig. 4). Furthermore, the generated replicon had a sensitivity to inhibition by PSI-6130 and NM107 (mean EC50s, 0.79 and 2.03 μM, respectively) similar to that seen with the WT replicon (mean EC50s, 0.66 and 1.98 μM, respectively). The nature of this change in sensitivity is not understood. The enhanced replication capacity suggests that the coemergence of a number of amino acid substitutions in a complex pattern together with the S282T adds a replication advantage to the HCV RNA by compensating for the hampered replication capacity mediated by the S282T mutation but may also be associated with reduced resistance.
Characterization of the sensitivity of the replicons generated in the selected cells.
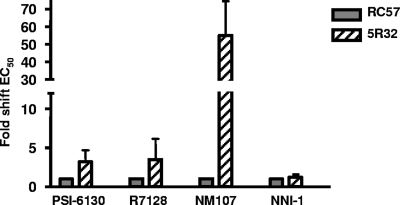
The reduced sensitivity observed in the PSI-6130- and R7128-selected cells, presented as shifts in EC50 values (Table 3), could be due to the emergence of resistant mutations in the replicon, to aberrant metabolism of PSI-6130 to form the active triphosphate in the cells, or to a combination of both factors. To isolate the effects attributable to changes in the replicon, whole cellular RNA was extracted from passage 32 of selection set 2 and passage 57 of the untreated cells (Table 3) and transfected into naïve Huh-7 cells to generate stable replicon cell lines 5R32 and RC57, respectively. The sequences of the NS5B coding region in the generated stable cells were confirmed to contain the mutations observed in the selected cells (Table 3). The sensitivity of the cells to PSI-6130, R7128, NM107, and NNI-1 was examined. 5R32 replicon cells exhibited a three- to fourfold reduction in sensitivity to both PSI-6130 and R7128 compared to the results seen with the RC57 control cell line (Fig. 6). The reduction in sensitivity (55-fold shift in EC50) to NM107 in these cells was more pronounced (Fig. 6). These results suggest that the emergence of mutations in the replicon as a result of selection with PSI-6130 contributes to a low level of change (three- to fourfold) in resistance. This reduction in sensitivity corresponds to that observed with the S282T substitution in the transient assay (Fig. 5A, B, and C).
FIG. 6.
Stable cells containing the replicons obtained from the cells selected with PSI-6130 show moderate reduced sensitivity to PSI-6130 compared to NM107 results. Whole cellular RNA was isolated from the cells selected with PSI-6130 and transfected into naïve Huh-7 cells. Stable cell lines were generated from selection set 2 (5R32) and from untreated cells (RC57). The generated stable cell lines were tested for sensitivity to PSI-6130, NM107, and NNI-1 by the use of qPCR (error bars represent the standard errors of the means of the results of three independent experiments). The shift values were calculated based on the untreated cell line value, which was set to 1.
Lack of cross-resistance between PSI-6130 and R1479.
R1479 (Fig. 1) is a potent nucleoside inhibitor of HCV NS5B RdRp. R1626, the prodrug of R1479, has demonstrated a potent antiviral activity in a phase 2 clinical study (40, 42).
As reported previously, selection of R1479 resistance replicons resulted in the identification of the NS5B mutation S96T, either alone or in combination with N142T, and the observation that this mutation was associated with R1479 resistance. S96T, alone or in combination with N142T, conferred a moderate three- to fourfold change in resistance to R1479 (23).
In this study, we examined whether there was cross-resistance between PSI-6130 and R1479. R1479 is known to exhibit slightly enhanced potency against the S282T mutant replicon compared to WT results (Table 8) (23). We tested the activity of PSI-6130 against replicons containing the S96T mutation. The mutation was introduced by site-directed mutagenesis, and the effect on activity was examined by using the transient replicon assay. We observed that PSI-6130 maintained full activity against the replicon with the S96T mutation in NS5B (EC50 of 0.42 μM compared to 0.82 μM for the WT) (Table 8). Similarly, PSI-6130 also maintained full activity against the S96T/N142T double-mutant replicon (data not shown). Therefore, each of the two compounds PSI-6130 and R1479 retains antiviral activity against replicons resistant to inhibition by the other compound, indicating the lack of cross-resistance.
TABLE 8.
Lack of cross-resistance between PSI-6130 and R1479 in the transient replicon assay
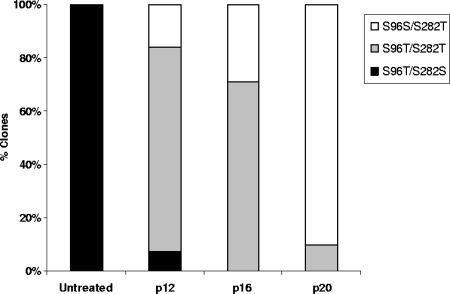
We next examined the feasibility of selecting replicons containing mutations conferring resistance to PSI-6130, starting with a replicon with preexisting mutations conferring resistance to R1479. Cells harboring a stable GT-1b replicon containing the S96T/N142T substitutions in NS5B that confer resistance to R1479 were treated with PSI-6130 (Table 9). The concentration of PSI-6130 was increased gradually with increasing cell passage numbers, and the coding sequence of NS5B was examined periodically. Sequencing of NS5B region from the pooled population of selected cells at passages 12, 16, and 20 indicated the emergence of the S282T mutation during incubation with PSI-6130 (mixture with WT at passage 12 and full mutation at passage 16) (Table 9). Notably, the emergence of the S282T mutation appeared to be accompanied by a reversion of the S96T mutation back to the WT serine (Table 9). To determine the proportions of each of the mutations in the replicon, we performed clonal sequence analysis. The NS5B coding sequences were amplified from the selected cells and then cloned into a recipient vector, and individual NS5B sequences were examined. The analysis confirmed that the emergence of the S282T substitution coincided with a reversion of the S96T substitution to the WT serine. At passage 12, the majority of the replicons contained the S282T substitution; however, it was present with a mixture of serine and threonine at position 96 (43 clones analyzed; 77% S96T and S282T, 16% S96S and S282T, 0% S96S and S282S, and 7% S96T and S282S) (Fig. 7). The percentage of clones carrying both S282T and the revertant S96S increased at passage 16 (45 clones analyzed; 71% S96T and S282T, 29% S96S and S282T, 0% S96S and S282S, and 0% S96T and S282S) (Fig. 7). Subsequently at passage 20, under conditions of treatment with 18 μM PSI-6130, the S282T substitution was present in 100% of the clones and S96T nearly fully reverted to S96S (21 clones analyzed; 10% S96T and S282T, 90% S96S and S282T, 0% S96S and S282S, and 0% S96T and S282S) (Fig. 7). No change was observed for the N142T substitution under the incubation conditions, and the mutation persisted until the end of the selection. This is in agreement with previous observations that the N142T substitution alone does not decrease the replication capacity and the sensitivity to R1479 (23).
TABLE 9.
Selection of PSI-6130 in cells harboring a replicon with the S96T/N142T substitutions in NS5B
| Cell category | PSI-6130 concn (μM) | Substitutions in NS5B population sequences |
|---|---|---|
| Untreated | 0 | S96T, N142T, S282S |
| p12 | 10 | S96S/T, N142T, S282S/T |
| p16 | 18 | S96S/T, N142T, S282T |
| p20 | 18 | S96S, N142T, S282T |
FIG. 7.
The R1479-resistant mutant S96T reverts to WT under conditions of treatment with PSI-6130. Cells harboring replicons with the S96T/N142T mutations were passaged in the presence of PSI-6130. Clonal sequencing analysis of NS5B was performed on samples collected at passages 12, 16, and 20.
The sensitivity of the replicon cells selected with PSI-6130 was examined using the qPCR assay. Replicon cell samples from passage 20 were treated with NNI-1, PSI-6130, R7128, or NM107 and compared to untreated control cells (Table 10). The treatment with PSI-6130 resulted in a reduction in the sensitivity to the PSI-6130 compound, to the prodrug R7128, and to NM107 (change from 0.73 μM to 5.1 μM for PSI-6130, from 0.58 μM to 3.2 μM for R7128, and from 0.69 μM to 36.5 μM for NM107) (Table 10). The loss of sensitivity was as predicted, mainly due to the emergence of the S282T mutation (Table 9 and Fig. 7), although, as was observed previously (Table 3), cellular-factor-mediated resistance (cellular factors affecting the formation of the active triphosphate metabolites) may also have contributed to the overall observed loss of sensitivity. The sensitivity of the cells to R1479 increased (change from 15.9 μM to 2.13 μM; Table 10), which corresponded to the reversion of the S96T mutation to WT (Table 9 and Fig. 7). No change in sensitivity was observed for the control NNI-1 (Table 10).
TABLE 10.
Inhibitor sensitivity of S96T/N142T replicon cells selected with PSI-6130
| Compound | Replicon qPCR EC50 (mean ± SEM [μM])a
|
Fold shift (p20/untreated) | |
|---|---|---|---|
| Untreated | p20 | ||
| NNI-1 | 0.42 ± 0.01 | 0.33 ± 0.07 | 0.8 |
| PSI-6130 | 0.73 ± 0.34 | 5.1 ± 0.55 | 7.0 |
| R7128 | 0.58 ± 0.17 | 3.2 ± 0.75 | 5.5 |
| R1479 | 15.9 ± 0.9 | 2.13 ± 0.27 | 0.13 |
| NM-107 | 0.69 ± 0.34 | 36.5 ± 15.6 | 52.9 |
SEM values represent standard errors of the means of the results of a minimum of three independent experiments.
When present alone, the S96T or the S282T mutation significantly reduces the replication capacity of the replicon (4% or 15% of WT, respectively; Table 8) (23). To examine the effect of the combination of the two mutations when present in the same replicon, we generated replicons containing both mutations by site-directed mutagenesis and the replication capacity was examined by using the transient replicon assay. When combined, the S96T and the S282T mutations caused a drastic reduction in the replication capacity to 0.6% of WT (Table 8), indicating low viability for such replicons. The NS5B/S96T substitution reduces the sensitivity of the replicon to R1479 (Table 8) (23); however, the mutation did not reduce the sensitivity to PSI-6130 (EC50 of 0.42 μM compared to 0.82 μM for WT) (Table 8), suggesting the lack of cross-resistance. The sensitivity of replicons containing the combination of both S96T and S282T could not be determined due to the low replication capacity of the replicon (Table 8).
Therefore, treatment with PSI-6130 generates the predicted resistance mutation and reverts the preexisting resistance mutation for R1479 to WT, suggesting that the two compounds possess nonoverlapping, mutually exclusive mutations, likely due to the low replication capacity of the double-mutant S96T/S282T replicon.
DISCUSSION
PSI-6130 exhibited potent activity when examined with laboratory-optimized GT-1a and GT-1b stable and transient HCV replicons, as well as with the respective NS5B enzymes. The inhibitor also consistently demonstrated potent activity against replicons expressing GT-1a or GT-1b NS5B obtained from clinical isolates.
The HCV replicon system has been useful in predicting the emergence of resistant mutations in patients treated with polymerase or protease HCV inhibitors, for example, HCV-796 (46) or telaprevir (13, 20), respectively. We used the in vitro HCV replicon system in this study to identify the mutations that emerge under conditions of treatment with PSI-6130 and that confer reduced sensitivity to the compound. To date, no evidence of the presence of the S282T mutation has emerged in studies of patients exposed to monotherapy with R7128 for 14 days (25).
One explanation for the lack of detection of resistance mutations in the clinic to date is the apparent high barrier to the generation of resistance to PSI-6130, most probably due to the low replication fitness combined with only a low level of resistance gained from the mutant virus. A short-term (less than 4 week) direct selection method that has been used successfully to obtain resistant variants in vitro for other inhibitors (23, 24, 27, 31, 45) did not generate variants resistant to PSI-6130. In the replicon clearance assay, treatment of the cells with 10× the EC50 of PSI-6130 for 29 days resulted in the clearance of the replicon from the cells. This feature also seems to be shared with NM107 and R1479, which also cleared the replicon during the clearance process (23) This was in contrast to the results seen with treatment with NNI-1. Resistant variants containing the previously described L419M substitution (19) readily emerged under conditions of drug treatment. This suggests that the barrier to resistance to PSI-6130 and NM107 is higher than that to resistance to NNI-1.
Long-term culturing of the cells, accompanied by incremental increases in PSI-6130 concentrations, was required to obtain variants conferring reduced sensitivity to PSI-6130. Several months of culturing of the cells in the presence of PSI-6130 generated variants with reduced sensitivity to PSI-6130. This provides further evidence for a high barrier to the generation of variants resistant to PSI-6130 in vitro.
The higher hurdle to the generation of resistance to PSI-6130 may be explained by the fact that the S282T substitution causes only a moderate loss of sensitivity to PSI-6130, as demonstrated in the enzymatic assays and the replicon assays using laboratory-optimized as well as clinical isolate-derived replicons. However, the substitution also caused a drastic 85% loss of replication capacity when tested in the transient replicon system (Fig. 4) (23).
In the three selection sets, two using PSI-6130 and one using the R7128 prodrug, the S282T was the only substitution commonly selected in all three experiments; however, several other substitutions emerged along with the S282T substitution. The three sets were selected under identical conditions, but the coemerging mutations appeared to differ. The difference in coemerging substitutions is not predicted to be due to the difference between PSI-6130 and the prodrug R7128, as they are both known to convert to the same active species (28). We hypothesize that the seemingly higher level of resistance observed for the cells selected with the prodrug R7128 (120-fold) compared to the levels seen with cells selected with PSI-6130 (30- to 50-fold) is due to cellular effects on the metabolisms of both the drug and the prodrug to form the active triphosphate species. The effects attributed to the replicon itself were determined through retransfection of RNA from the selected cells into naïve Huh-7 cells and generation of stable cell lines. The resulting stable replicon cells showed only a low level of resistance (three- to fivefold), a level that is comparable to the resistance observed with the S282T substitution alone. Therefore, the different apparent resistance level in the original selected cells is not due to the replicon but rather to the host cells. Although the phenomenon of selecting host cells with a modified nucleoside metabolism may be specific to the tissue culture experimentation, it suggests that, in the clinic, individual differences in the metabolism of the nucleoside inhibitor are likely to play a role in determining the virological response to the treatment.
The reduced replication capacity appears to be rescued by the generation of other mutations in NS5B, thus providing a plausible explanation for the length of time required to generate variants with reduced sensitivity to PSI-6130. In the selection studies, a number of amino acid substitutions emerged under conditions of PSI-6130 treatment (Table 3). The presence of the other coselected mutations appeared to enhance the replication capacity. The replicon containing the mutations observed in selection set 1 at passage 53 (Table 3) had an replication capacity (51% of WT) that was improved in comparison to the replication capacity of the S282T mutant alone, which exhibited only a 15% replication capacity. Also, stable cell lines generated by the transfection of RNA from the PSI-6130-selected cells (for example, 5R32 of set 2) (Fig. 6 and data not shown) had an improved replication capacity based on replicon levels in the cells (range, 50 to 82% of the WT levels). These results suggest that the presence of the coselected mutations in NS5B in a complex combination pattern can compensate for the reduced replication capacity caused by the S282T mutation. It is possible that substitutions outside of the NS5B region (not analyzed in this study) could also contribute to the replication advantage. Further investigation is required to discern the exact combination of mutations required for the improved replication capacity.
PSI-6130 appears to possess a more favorable activity profile with respect to the NS5B polymerase containing the S282T substitution compared to other 2′-C-methyl-modified polymerase inhibitors. The reduction in sensitivity to PSI-6130 with the S282T substitution was moderate (range, two- to sixfold loss of activity). In contrast, the reduction of sensitivity to NM107 (range, 17- to 50-fold loss of activity) and to 2′-C-methyladenosine (range, 18- to 100-fold loss of activity) (30) was more pronounced. RO2433-TP is the active uridine metabolite for PSI-6130, which has 20-fold loss of inhibitory activity with S282T mutation polymerase. This activity and resistance profile suggests that this metabolite may also contribute to the emergence of the S282T substitution.
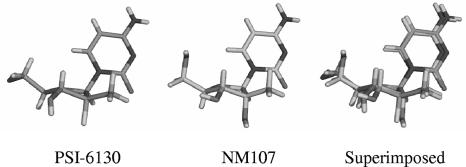
The reason for the higher activity of PSI-6130 against the NS5B polymerase containing the S282T substitution compared to that seen with other 2′-C-methyl-substituted nucleoside inhibitors is unclear. Molecular modeling suggested that the change from serine to threonine at position 282 causes a steric clash between the methyl group of the NM107 and the methyl group of the threonine, hence causing a significant loss in the affinity to NM107 (12). Shifts in Ki values indicated a more moderate loss of affinity to PSI-6130 than that seen with NM107 (Table 6). One possible explanation is that PSI-6130 may adopt a conformation that alleviates the steric hindrance, allowing more favorable interactions with the mutant polymerase. Alternatively, the weakening of the hydrogen bonding interaction between D225 and the ribose 2′-hydroxyl group by fluorine substitution may increase ribose structural flexibility in the NS5B active site, allowing PSI-6130 to avoid the S282T steric clash. In support of this hypothesis, it was recently shown that the capability of a hydrogen bonding interaction with D225 was not required for potent inhibition of HCV replication by nucleoside analogs (21). PSI-6130 is known to adopt the 3′-end conformation in the solid state (9, 12). We determined the crystal structures of NM107 and PSI-6130 (Fig. 8) and observed that both inhibitors adopt the 3′-end conformation at the ribose sugar moiety. However, many nucleosides differ in conformation in the solution phase from the solid state. For example, AZT is known to adopt the C-2′-endo conformation in the crystal state but exists as a 1:1 mixture of C-2′-endo and C-3′-endo in the solution phase. Therefore, it is possible that differences observed in the potency of NM107 and PSI-6130 against the S282T mutant might be due to differences in the conformations of these two structures in the solution phase.
FIG. 8.
PSI-6130 and NM107 adopt similar 3′-end conformations. X-ray crystal structures for PSI-6130 and NM107 are shown.
PSI-6130 and R1479 select for different resistance mutations (S282T and S96T/N142T, respectively). When tested for cross-resistance, each of those two inhibitors did not lose activity against the resistant mutants of the other compound, indicating the lack of cross-resistance. In addition, we also observed that in replicon cells containing the S96T substitution, treatment with PSI-6130 induced a reversion of the S96T mutation to the WT serine accompanied by the emergence of the S282T substitution. The pressure for mutation reversion could be explained by the fact that the combination of the S96T and S282T mutations causes a dramatic reduction in the replication capacity, from 4% of WT for the S96T mutation and 15% for the S282T mutation to 0.6% for the combination of the two mutations. The incompatibility of the two mutations when present in the replicon offers a clinical advantage for a potential combination therapy. Viruses simultaneously resistant to both inhibitors under conditions of combined drug treatment are predicted to have severely reduced replication fitness.
The results from this study demonstrate that the generation of variants resistant to PSI-6130 in vitro requires long-term passage. The emergence of the S282T mutation, which mediates only a moderate three- to sixfold reduction in sensitivity to PSI-6130, is accompanied by a number of coselected mutations that appear to enhance the replication capacity. This indicates a high hurdle to the generation of mutations conferring reduced sensitivity to PSI-6130 in vitro. The clinical relevance and the potential benefit are yet to be demonstrated in the ongoing R7128 clinical trial, which is designed to observe the clinical response to the drug and the emerging resistance profile. Finally, PSI-6130 and R1479 lack cross-resistance, and the simultaneous presence of mutations resistant to both compounds drastically reduces the replication capacity, offering a promising potential for a future combination therapy.
Supplementary Material
Acknowledgments
We are grateful to Phil Furman and Michelle Berrey of Pharmasset, Inc., for reviewing the manuscript. We thank Juanita Bernal-Fussell, Sandra Clausen, Nixy Zutshi, and Sharon Jiang for assistance with this work.
Footnotes
Published ahead of print on 6 October 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Afdhal, N., M. Rodriguez-Torres, E. Lawitz, E. Godofsky, G. Chao, B. Fielman, S. Knox, and N. Brown. 2005. Enhanced antiviral efficacy for valopicitabine (NM283) plus Peg-interferon in hepatitis C patients with HCV genotype-1 infection: results of a phase IIa multicenter trial, abstr. LB03. Abstr. 40th Annu. Meet. Eur. Assoc. Study Liver, Paris, France, 13 to 17 April, 2005.
- 2.Beaulieu, P. L. 2006. Finger loop inhibitors of the HCV NS5B polymerase: discovery and prospects for new HCV therapy. Curr. Opin. Drug Discov. Devel. 9:618-626. [PubMed] [Google Scholar]
- 3.Beaulieu, P. L. 2007. Non-nucleoside inhibitors of the HCV NS5B polymerase: progress in the discovery and development of novel agents for the treatment of HCV infections. Curr. Opin. Investig. Drugs 8:614-634. [PubMed] [Google Scholar]
- 4.Beaulieu, P. L., M. Bos, Y. Bousquet, P. DeRoy, G. Fazal, J. Gauthier, J. Gillard, S. Goulet, G. McKercher, M. A. Poupart, S. Valois, and G. Kukolj. 2004. Non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase: discovery of benzimidazole 5-carboxylic amide derivatives with low-nanomolar potency. Bioorg. Med. Chem. Lett. 14:967-971. [DOI] [PubMed] [Google Scholar]
- 5.Beaulieu, P. L., J. Gillard, D. Bykowski, C. Brochu, N. Dansereau, J. S. Duceppe, B. Hache, A. Jakalian, L. Lagace, S. LaPlante, G. McKercher, E. Moreau, S. Perreault, T. Stammers, L. Thauvette, J. Warrington, and G. Kukolj. 2006. Improved replicon cellular activity of non-nucleoside allosteric inhibitors of HCV NS5B polymerase: from benzimidazole to indole scaffolds. Bioorg. Med. Chem. Lett. 16:4987-4993. [DOI] [PubMed] [Google Scholar]
- 6.Beaulieu, P. L., and Y. S. Tsantrizos. 2004. Inhibitors of the HCV NS5B polymerase: new hope for the treatment of hepatitis C infections. Curr. Opin. Investig. Drugs 5:838-850. [PubMed] [Google Scholar]
- 7.Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, F. R. De, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. [DOI] [PubMed] [Google Scholar]
- 8.Chan, L., S. K. Das, T. J. Reddy, C. Poisson, M. Proulx, O. Pereira, M. Courchesne, C. Roy, W. Wang, A. Siddiqui, C. G. Yannopoulos, N. Nguyen-Ba, D. Labrecque, R. Bethell, M. Hamel, P. Courtemanche-Asselin, L. L'Heureux, M. David, O. Nicolas, S. Brunette, D. Bilimoria, and J. Bedard. 2004. Discovery of thiophene-2-carboxylic acids as potent inhibitors of HCV NS5B polymerase and HCV subgenomic RNA replication. Part 1: sulfonamides. Bioorg. Med. Chem. Lett. 14:793-796. [DOI] [PubMed] [Google Scholar]
- 9.Clark, J. L., L. Hollecker, J. C. Mason, L. J. Stuyver, P. M. Tharnish, S. Lostia, T. R. McBrayer, R. F. Schinazi, K. A. Watanabe, M. J. Otto, P. A. Furman, W. J. Stec, S. E. Patterson, and K. W. Pankiewicz. 2005. Design, synthesis, and antiviral activity of 2′-deoxy-2′-fluoro-2′-C-methylcytidine, a potent inhibitor of hepatitis C virus replication. J. Med. Chem. 48:5504-5508. [DOI] [PubMed] [Google Scholar]
- 10.Cornberg, M., H. Wedemeyer, and M. P. Manns. 2001. Hepatitis C: therapeutic perspectives. Forum (Genoa) 11:154-162. [PubMed] [Google Scholar]
- 11.Dhanak, D., K. J. Duffy, V. K. Johnston, J. Lin-Goerke, M. Darcy, A. N. Shaw, B. Gu, C. Silverman, A. T. Gates, M. R. Nonnemacher, D. L. Earnshaw, D. J. Casper, A. Kaura, A. Baker, C. Greenwood, L. L. Gutshall, D. Maley, A. DelVecchio, R. Macarron, G. A. Hofmann, Z. Alnoah, H. Y. Cheng, G. Chan, S. Khandekar, R. M. Keenan, and R. T. Sarisky. 2002. Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:38322-38327. [DOI] [PubMed] [Google Scholar]
- 12.Dutartre, H., C. Bussetta, J. Boretto, and B. Canard. 2006. General catalytic deficiency of hepatitis C virus RNA polymerase with an S282T mutation and mutually exclusive resistance towards 2′-modified nucleotide analogues. Antimicrob. Agents Chemother. 50:4161-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forestier, N., H. W. Reesink, C. J. Weegink, L. McNair, T. L. Kieffer, H. M. Chu, S. Purdy, P. L. Jansen, and S. Zeuzem. 2007. Antiviral activity of telaprevir (VX-950) and peginterferon alfa-2a in patients with hepatitis C. Hepatology 46:640-648. [DOI] [PubMed] [Google Scholar]
- 14.Fried, M. W. 2001. Advances in therapy for chronic hepatitis C. Clin. Liver Dis. 5:1009-1023. [DOI] [PubMed] [Google Scholar]
- 15.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 16.Hadziyannis, S. J., and J. S. Koskinas. 2004. Differences in epidemiology, liver disease and treatment response among HCV genotypes. Hepatol. Res. 29:129-135. [DOI] [PubMed] [Google Scholar]
- 17.Harper, S., S. Avolio, B. Pacini, F. M. Di, S. Altamura, L. Tomei, G. Paonessa, M. S. Di, A. Carfi, C. Giuliano, J. Padron, F. Bonelli, G. Migliaccio, F. R. De, R. Laufer, M. Rowley, and F. Narjes. 2005. Potent inhibitors of subgenomic hepatitis C virus RNA replication through optimization of indole-N-acetamide allosteric inhibitors of the viral NS5B polymerase. J. Med. Chem. 48:4547-4557. [DOI] [PubMed] [Google Scholar]
- 18.Howe, A. Y., J. Bloom, C. J. Baldick, C. A. Benetatos, H. Cheng, J. S. Christensen, S. K. Chunduru, G. A. Coburn, B. Feld, A. Gopalsamy, W. P. Gorczyca, S. Herrmann, S. Johann, X. Jiang, M. L. Kimberland, G. Krisnamurthy, M. Olson, M. Orlowski, S. Swanberg, I. Thompson, M. Thorn, V. A. Del, D. C. Young, Z. M. van, J. W. Ellingboe, J. Upeslacis, M. Collett, T. S. Mansour, and J. F. O'Connell. 2004. Novel nonnucleoside inhibitor of hepatitis C virus RNA-dependent RNA polymerase. Antimicrob. Agents Chemother. 48:4813-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howe, A. Y., H. Cheng, I. Thompson, S. K. Chunduru, S. Herrmann, J. O'Connell, A. Agarwal, R. Chopra, and A. M. Del Vecchio. 2006. Molecular mechanism of a thumb domain hepatitis C virus nonnucleoside RNA-dependent RNA polymerase inhibitor. Antimicrob. Agents Chemother. 50:4103-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kieffer, T. L., C. Sarrazin, J. S. Miller, M. W. Welker, N. Forestier, H. W. Reesink, A. D. Kwong, and S. Zeuzem. 2007. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology 46:631-639. [DOI] [PubMed] [Google Scholar]
- 21.Klumpp, K., G. Kalayanov, H. Ma, P. S. Le, V. Leveque, W. R. Jiang, N. Inocencio, W. A. De, S. Rajyaguru, E. Tai, S. Chanda, M. R. Irwin, C. Sund, A. Winqist, T. Maltseva, S. Eriksson, E. Usova, M. Smith, A. Alker, I. Najera, N. Cammack, J. A. Martin, N. G. Johansson, and D. B. Smith. 2008. 2′-Deoxy-4′-azido nucleoside analogs are highly potent inhibitors of hepatitis C virus replication despite the lack of 2′-alpha-hydroxyl groups. J. Biol. Chem. 283:2167-2175. [DOI] [PubMed] [Google Scholar]
- 22.Klumpp, K., V. Leveque, P. S. Le, H. Ma, W. R. Jiang, H. Kang, C. Granycome, M. Singer, C. Laxton, J. Q. Hang, K. Sarma, D. B. Smith, D. Heindl, C. J. Hobbs, J. H. Merrett, J. Symons, N. Cammack, J. A. Martin, R. Devos, and I. Najera. 2006. The novel nucleoside analog R1479 (4′-azidocytidine) is a potent inhibitor of NS5B-dependent RNA synthesis and hepatitis C virus replication in cell culture. J. Biol. Chem. 281:3793-3799. [DOI] [PubMed] [Google Scholar]
- 23.Le Pogam, S., W. R. Jiang, V. Leveque, S. Rajyaguru, H. Ma, H. Kang, S. Jiang, M. Singer, S. Ali, K. Klumpp, D. Smith, J. Symons, N. Cammack, and I. Najera. 2006. In vitro selected Con1 subgenomic replicons resistant to 2′-C-methyl-cytidine or to R1479 show lack of cross resistance. Virology 351:349-359. [DOI] [PubMed] [Google Scholar]
- 24.Le Pogam, S., H. Kang, S. F. Harris, V. Leveque, A. M. Giannetti, S. Ali, W. R. Jiang, S. Rajyaguru, G. Tavares, C. Oshiro, T. Hendricks, K. Klumpp, J. Symons, M. F. Browner, N. Cammack, and I. Najera. 2006. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J. Virol. 80:6146-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Pogam, S., A. Seshaadri, A. Kosaka, S. Hu, A. Beard, J. Symons, N. Cammack, and I. Najera. 2008. Lack of viral resistance after 14-day monotherapy treatment with R7128 in treatment-experienced patients infected with HCV genotype 1, abstr. 26. Abstr. Conf. Hepatitis B & C Virus Resist. Antivir. Ther., Paris, France, 14 to 16 February, 2008.
- 26.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 27.Lu, L., H. Mo, T. J. Pilot-Matias, and A. Molla. 2007. Evolution of resistant M414T mutants among hepatitis C virus replicon cells treated with polymerase inhibitor A-782759. Antimicrob. Agents Chemother. 51:1889-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma, H., W. R. Jiang, N. Robledo, V. Leveque, S. Ali, T. Lara-Jaime, M. Masjedizadeh, D. B. Smith, N. Cammack, K. Klumpp, and J. Symons. 2007. Characterization of the metabolic activation of a HCV nucleoside inhibitor beta-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine (PSI-6130) and identification of a novel active 5′-triphosphate species. J. Biol. Chem. 282:29812-29820. [DOI] [PubMed] [Google Scholar]
- 29.Ma, H., V. Leveque, W. A. De, W. Li, T. Hendricks, S. M. Clausen, N. Cammack, and K. Klumpp. 2005. Inhibition of native hepatitis C virus replicase by nucleotide and non-nucleoside inhibitors. Virology 332:8-15. [DOI] [PubMed] [Google Scholar]
- 30.Migliaccio, G., J. E. Tomassini, S. S. Carroll, L. Tomei, S. Altamura, B. Bhat, L. Bartholomew, M. R. Bosserman, A. Ceccacci, L. F. Colwell, R. Cortese, F. R. De, A. B. Eldrup, K. L. Getty, X. S. Hou, R. L. LaFemina, S. W. Ludmerer, M. MacCoss, D. R. McMasters, M. W. Stahlhut, D. B. Olsen, D. J. Hazuda, and O. A. Flores. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164-49170. [DOI] [PubMed] [Google Scholar]
- 31.Mo, H., L. Lu, T. Pilot-Matias, R. Pithawalla, R. Mondal, S. Masse, T. Dekhtyar, T. Ng, G. Koev, V. Stoll, K. D. Stewart, J. Pratt, P. Donner, T. Rockway, C. Maring, and A. Molla. 2005. Mutations conferring resistance to a hepatitis C virus (HCV) RNA-dependent RNA polymerase inhibitor alone or in combination with an HCV serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 49:4305-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murakami, E., H. Bao, M. Ramesh, T. R. McBrayer, T. Whitaker, H. M. Micolochick Steuer, R. F. Schinazi, L. J. Stuyver, A. Obikhod, M. J. Otto, and P. A. Furman. 2007. Mechanism of activation of beta-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine and inhibition of hepatitis C virus NS5B RNA polymerase. Antimicrob. Agents Chemother. 51:503-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murakami, E., C. Niu, H. Bao, H. M. Steuer, T. Whitaker, T. Nachman, M. Sofia, P. Wang, M. J. Otto, and P. A. Furman. 2008. The mechanism of action of β-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine involves a second metabolic pathway leading to β-d-2′-deoxy-2′-fluoro-2′-C-methyluridine 5′-triphosphate, a potent inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob. Agents Chemother. 52:458-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olsen, D. B. 2006. The nucleoside inhibitor MK-0608 mediates suppression of HCV replication for greater than 30 days in chronically infected chimpanzees, abstr. 1159. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother., San Francisco, CA, 27 to 30 September, 2006.
- 35.Olsen, D. B., A. B. Eldrup, L. Bartholomew, B. Bhat, M. R. Bosserman, A. Ceccacci, L. F. Colwell, J. F. Fay, O. A. Flores, K. L. Getty, J. A. Grobler, R. L. LaFemina, E. J. Markel, G. Migliaccio, M. Prhavc, M. W. Stahlhut, J. E. Tomassini, M. MacCoss, D. J. Hazuda, and S. S. Carroll. 2004. A 7-deaza-adenosine analog is a potent and selective inhibitor of hepatitis C virus replication with excellent pharmacokinetic properties. Antimicrob. Agents Chemother. 48:3944-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pierra, C., A. Amador, S. Benzaria, E. Cretton-Scott, M. D'Amours, J. Mao, S. Mathieu, A. Moussa, E. G. Bridges, D. N. Standring, J. P. Sommadossi, R. Storer, and G. Gosselin. 2006. Synthesis and pharmacokinetics of valopicitabine (NM283), an efficient prodrug of the potent anti-HCV agent 2′-C-methylcytidine. J. Med. Chem. 49:6614-6620. [DOI] [PubMed] [Google Scholar]
- 37.Pierra, C., S. Benzaria, A. Amador, A. Moussa, S. Mathieu, R. Storer, and G. Gosselin. 2005. Nm 283, an efficient prodrug of the potent anti-HCV agent 2′-C-methylcytidine. Nucleosides Nucleotides Nucleic Acids 24:767-770. [DOI] [PubMed] [Google Scholar]
- 38.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pockros, P., D. Nelson, E. Godofsky, M. Rodriguez-Torres, G. T. Everson, M. W. Fried, R. H. Ghalib, S. Harrison, L. M. Nyberg, M. L. Shiffman, G. Hill, and A. Chan. 2007. Robust synergistic antiviral effect of R1626 in combination with Peginterferon alfa-2a (40KD), with or without ribavirin—interim analysis results of phase 2a study, abstr. 167. Abstr. 58th Annu. Meet. Am. Assoc. Study Liver Diseases, Boston, MA, 2 to 6 November, 2007.
- 41.Reddy, R., M. Rodriguez-Torres, E. Gane, R. Robson, J. Lalenzari, G. T. Everson, E. DeJesus, J. G. McHutchison, H. E. Vargas, A. Beard, C. A. Rodriguez, G. Z. Hill, W. T. Symonds, and M. M. Berry. 2007. Antiviral activity, pharmacokinetics, safety, and tolerability of R7128, a novel nucleoside HCV RNA polymerase inhibitor, following multiple, ascending, oral doses in patients with HCV genotype 1 infection who have failed prior interferon therapy, abstr. LB9. Abstr. 58th Annu. Meet. Am. Assoc. Study Liver Diseases, Boston, MA, 2 to 6 November, 2007.
- 42.Roberts, S., G. Cooksley, D. Shaw, H. K. Berns, M. T. Brandt, S. H. Fettner, G. Hill, D. Ipe, K. Klumpp, M. Mannino, E. O'Mara, Y. Tu, and C. B. Washington. 2006. Interim results of a multiple ascending dose study of R1626, a novel nucleoside analog targeting HCV polymerase in chronic HCV patients, abstr. 731. Abstr. 41th Annu. Meet. Eur. Assoc. Study Liver, Vienna, Austria, 26 to 30 April, 2006.
- 43.Sarrazin, C., R. Rouzier, F. Wagner, N. Forestier, D. Larrey, S. K. Gupta, M. Hussain, A. Shah, D. Cutler, J. Zhang, and S. Zeuzem. 2007. SCH 503034, a novel hepatitis C virus protease inhibitor, plus pegylated interferon alpha-2b for genotype 1 nonresponders. Gastroenterology 132:1270-1278. [DOI] [PubMed] [Google Scholar]
- 44.Stuyver, L. J., T. R. McBrayer, P. M. Tharnish, J. Clark, L. Hollecker, S. Lostia, T. Nachman, J. Grier, M. A. Bennett, M. Y. Xie, R. F. Schinazi, J. D. Morrey, J. L. Julander, P. A. Furman, and M. J. Otto. 2006. Inhibition of hepatitis C replicon RNA synthesis by beta-D-2′-deoxy-2′-fluoro-2′-C-methylcytidine: a specific inhibitor of hepatitis C virus replication. Antivir. Chem. Chemother. 17:79-87. [DOI] [PubMed] [Google Scholar]
- 45.Tomei, L., S. Altamura, L. Bartholomew, A. Biroccio, A. Ceccacci, L. Pacini, F. Narjes, N. Gennari, M. Bisbocci, I. Incitti, L. Orsatti, S. Harper, I. Stansfield, M. Rowley, F. R. De, and G. Migliaccio. 2003. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 77:13225-13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villano, S., A. Y. Howe, D. Raible, D. Harper, J. Speth, and G. Bichier. 2007. Analysis of HCV NS5B genetic variants following monotherapy with HCV-796, a non-nucleoside polymerase inhibitor, in treatment-naïve HCV-infected patients, abstr. 1302. Abstr. 58th Annu. Meet. Am. Assoc. Study Liver Diseases, Boston, MA, 2 to 6 November, 2007.
- 47.Wang, M., K. K. Ng, M. M. Cherney, L. Chan, C. G. Yannopoulos, J. Bedard, N. Morin, N. Nguyen-Ba, M. H. Alaoui-Ismaili, R. C. Bethell, and M. N. James. 2003. Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. Crystal structures and mechanism of inhibition. J. Biol. Chem. 278:9489-9495. [DOI] [PubMed] [Google Scholar]
- 48.Yi, M., and S. M. Lemon. 2004. Adaptive mutations producing efficient replication of genotype 1a hepatitis C virus RNA in normal Huh7 cells. J. Virol. 78:7904-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.