Abstract
Retapamulin MICs of ≥2 μg/ml were noted for 6 of 5,676 S. aureus recent clinical isolates evaluated. The ABC proteins VgaAv and VgaA were found to be responsible for the reduced susceptibility to pleuromutilins exhibited by these six isolates.
Members of the pleuromutilin class of antibiotics selectively inhibit bacterial protein synthesis through interaction with prokaryotic ribosomes (11) and are structurally distinct from other classes of ribosome inhibitors. While the pleuromutilins tiamulin and valnemulin have found uses in veterinary applications, retapamulin (formerly SB-275833) is the only member of this class approved for use in humans (9). Currently, retapamulin is approved in countries including the United States for topical use to treat impetigo, a highly contagious skin infection typically caused by Staphylococcus aureus or Staphylococcus pyogenes.
The development of resistance to available antibiotics is a major medical issue that has spurred on the development of drugs such as retapamulin. Unfortunately, the development of resistance to any antibiotic is inevitable, thereby making an understanding of emerging resistance mechanisms important for the future development of antibiotics and diagnostic tools for detection. For these reasons, we have investigated the mechanism of decreased susceptibility to pleuromutilins among staphylococci in the GlaxoSmithKline strain collection, as well as isolates collected from retapamulin profiling of recent clinical isolates.
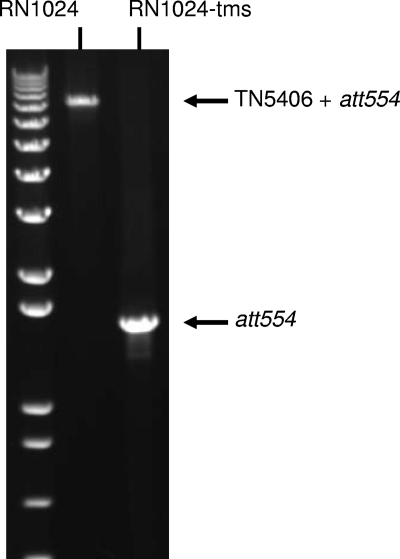
S. aureus strain RN1024 from the GlaxoSmithKline strain collection was identified during antimicrobial profiling of experimental compounds as being less susceptible to pleuromutilins. It was originally isolated in 1992 from a patient in northern France. A pleuromutilin-susceptible variant of this strain, RN1024-tms, was isolated, and subtractive-suppressive PCR (1) was carried out to generate a library enriched for DNA present in RN1024 but not RN1024-tms. Of 14 clones from the library that were sequenced, 10 were found to be identical to a fragment of the tnpA gene of the S. aureus transposon Tn5406 (5). Tn5406 is a small insertion element of 5,467 bp. It possesses four genes, tnpA, tnpB, tnpC, and vgaAv (5). Using PCR and gene-specific primers, we could detect each of the Tn5406-specific genes in RN1024 but not in RN1024-tms (data not shown). Tn5406 preferentially inserts into the chromosomal locus att554. We used primers that flank att554 to show that RN1024 carries a single copy of Tn5406 at this site, while RN1024-tms no longer carries the insertion (Fig. 1).
FIG. 1.
Confirmation of the presence of Tn5406 in S. aureus RN1024 but not RN1024-tms. Results of PCR using primers flanking the insertion site att554 or Tn5406.
Of the genes present on Tn5406, vgaAv is the only one not associated with transposon function, implicating it as the locus responsible for the decreased pleuromutilin sensitivity of RN1024. The plasmid pCU1-vgaAv, which consists of vgaAv and 660 bp of upstream DNA cloned into the Escherichia coli/S. aureus shuttle vector pCU1 (2), was constructed to test whether VgaAv can affect pleuromutilin MICs. pCU1-vgaAv was introduced into the laboratory strain S. aureus RN4220 (2, 6), and MICs were measured. Retapamulin and tiamulin MICs are considerably higher in RN4220 (pCU1-vgaAv) than in the control strain, RN4220 (pCU1) (see Table 2). Based on the findings that RN1024-tms lacks Tn5406 and that pCU1-vgaAv decreases pleuromutilin sensitivity, we conclude that vgaAv is responsible for the decreased pleuromutilin sensitivity of RN1024. Pleuromutilin MICs are significantly higher for RN4220 (pCU1-vgaAv) than for RN1024. This is likely due to copy number, as RN1024 carries a single copy of vgaAv while RN4220 (pCU1-vgaAv) carries multiple copies.
TABLE 2.
Effects of vga genes on S. aureus antimicrobial susceptibility
| Strain | MIC (μg/ml) of:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Retapamulin | Tiamulin | Dalfopristin | Quinupristin | Quinupristin-Dalfopristin | Linco-mycin | Clindamycin | Levofloxacin | Linezolid | Mupirocin | Chloram-phenicol | Tetra-cycline | Rifampin (rifampicin) | Erythro-mycin | Genta-micin | Trimeth-oprim | |
| RN4220 | ≤0.06 | 0.5 | 4 | 4 | 0.125 | 0.5 | ≤0.06 | 0.125 | 2 | ≤0.06 | 8 | 0.25 | ≤0.06 | 0.25 | 1 | 2 |
| RN4220 (pVGA) | 32 | >64 | 32 | 4 | 0.5 | 32 | 4 | 0.125 | 2 | ≤0.06 | 8 | 0.25 | ≤0.06 | 0.25 | 0.5 | 2 |
| RN4220 (pCU1) | ≤0.06 | 0.25 | 4 | 4 | 0.125 | 0.5 | ≤0.06 | 0.125 | 2 | ≤0.06 | >64 | 0.25 | ≤0.06 | 0.25 | 0.5 | 2 |
| RN4220 (pCU1-vgaAv) | 32 | >64 | >64 | 1 | 1 | 32 | 2 | 0.125 | 1 | ≤0.06 | >64 | 0.125 | ≤0.06 | 0.125 | 0.5 | 2 |
| RN1024 | 2 | 16 | 64 | 4 | 0.5 | 8 | 0.25 | 16 | 2 | 0.25 | 8 | 64 | >64 | 0.25 | >64 | 2 |
| RN1024-tms | ≤0.06 | 0.5 | 8 | 4 | 0.25 | 1 | ≤0.06 | 32 | 2 | 0.25 | 8 | 64 | >64 | 0.25 | >64 | 2 |
| ATCC 29213 | ≤0.06 | 1 | 8 | 4 | 0.25 | 2 | 0.125 | 0.125 | 4 | 0.25 | 16 | 0.5 | <0.06 | 0.5 | 0.5 | 2 |
As part of the evaluations conducted during preclinical development, the potency of retapamulin was measured against multiple laboratory collections of recent clinical isolates (10). These combined studies identified six S. aureus strains with retapamulin MICs of ≥2 μg/ml. In five of these strains, the MICs ranged from 2 to 4 μg/ml. The MIC of the remaining strain was 64 μg/ml (Table 1). Using vgaAv-specific primers, vgaAv was detected in the five strains with retapamulin MICs between 2 and 4 μg/ml. The results of PCR and sequence analysis indicated that, like RN1024, all five strains carried Tn5406 inserted into the att554 site (data not shown).
TABLE 1.
Analysis of recent clinical S. aureus isolates with reduced pleuromutilin sensitivity
| Isolate | Origin | MIC (μg/ml) ofa:
|
Gene presentb
|
Genetic element | ||
|---|---|---|---|---|---|---|
| Tiamulin | Retapamulin | vgaA | vgaAv | |||
| S. aureus IV20204009 | France | 32 | 2 | − | + | Tn5406 |
| S. aureus IV20205004 | France | 16 | 2 | − | + | Tn5406 |
| S. aureus IV20217008 | Portugal | 32 | 4 | − | + | Tn5406 |
| S. aureus IV20217033 | Portugal | >64 | 64 | + | − | pVGA |
| S. aureus IV20222009 | Ireland | 16 | 4 | − | + | Tn5406 |
| S. aureus IHMA127277 | United States | ND | 2 | − | + | Tn5406 |
ND, not determined.
Determined by PCR using gene-specific primers.
vgaAv was not detected in S. aureus IV20217033 (retapamulin MIC, 64 μg/ml) but the homologous gene vgaA was detected by using vgaA-specific primers. Plasmid DNA from this strain transformed RN4220 to tiamulin resistance, indicating that the resistance determinant was plasmid borne. Suspecting that the plasmid encoded VgaA, we determined its complete sequence, named here pVGA (5.9 kb), and confirmed the presence of vgaA. In addition to vgaA, pVGA contains genes predicted to encode replication and mobilization functions and no other antibiotic resistance genes. pVGA-encoded VgaA varies by a single amino acid from a VgaA isoform (7, 8) so far only identified among coagulase-negative staphylococci. The higher retapamulin MIC of IV20217033 relative to those of RN1024 and the other isolates is likely due to the presence of vgaA in a higher copy number than the chromosomally encoded VgaAv. Consistent with this, the pleuromutilin MIC of RN4220 (pCU1-vgaAv), i.e., vgaAv in a high copy number, is comparable with that of RN4220 (pVGA) (Table 2).
VgaA and VgaAv are members of the ATP-binding cassette (ABC) family of proteins and are likely involved in drug efflux (3). VgaA and VgaAv, which are 83% identical (5), have been shown to confer decreased sensitivity to two classes of protein synthesis inhibitors—lincosamides and streptogramin A compounds (4, 5). Streptogramins are mixtures of two unrelated classes of bacterial protein synthesis inhibitors, streptogramin A and streptogramin B, which act synergistically. As shown by the data in Table 2, of 16 antibiotics tested, the presence of pCU-vgaAv and pVGA in an isogenic background greatly increased the MICs of pleuromutilins, lincosamides (lincomycin and clindamycin), and a streptogramin A (dalfopristin). Quinupristin-dalfopristin (Synercid) MICs are also slightly higher in these strains, likely because of their higher dalfopristin MICs. Also shown by the data in Table 2, in addition to being more susceptible to pleuromutilins, RN1024-tms is more susceptible to lincosamides and dalfopristin than its Tn5406-carrying parent. These combined MIC data are consistent with our conclusion that the Vga proteins, in addition to conferring streptogramin A and lincosamide resistance, also confer reduced susceptibility to pleuromutilins.
Of 5,676 recent S. aureus clinical isolates evaluated (10), taken from pooled data from eight preclinical studies, one global surveillance study, and five phase III clinical trials, the six VgaA- or VgaAv-containing strains reported are the only isolates with retapamulin MICs of ≥2 μg/ml. In addition to demonstrating its low frequency, this suggests that reduced susceptibility to pleuromutilins will most commonly be due to the presence of a Vga protein.
Nucleotide sequence accession number.
The nucleotide sequence of pVGA has been submitted to the GenBank database and has been given the accession number FJ207465.
Footnotes
Published ahead of print on 6 October 2008.
REFERENCES
- 1.Akopyants, N. S., A. Fradkov, L. Diatchenko, J. E. Hill, P. D. Siebert, S. A. Lukyanov, E. D. Sverdlov, and D. E. Berg. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Augustin, J., R. Rosenstein, B. Wieland, U. Schneider, N. Schnell, G. Engelke, K. D. Entian, and F. Gotz. 1992. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur. J. Biochem. 204:1149-1154. [DOI] [PubMed] [Google Scholar]
- 3.Bouige, P., D. Laurent, L. Piloyan, and E. Dassa. 2002. Phylogenetic and functional classification of ATP-binding cassette (ABC) systems. Curr. Protein Pept. Sci. 3:541-559. [DOI] [PubMed] [Google Scholar]
- 4.Chesneau, O., H. Ligeret, N. Hosan-Aghaie, A. Morvan, and E. Dassa. 2005. Molecular analysis of resistance to streptogramin A compounds conferred by the Vga proteins of staphylococci. Antimicrob. Agents Chemother. 49:973-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haroche, J., J. Allignet, C. Buchrieser, and N. El Solh. 2000. Characterization of a variant of vga(A) conferring resistance to streptogramin A and related compounds. Antimicrob. Agents Chemother. 44:2271-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage 6. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 7.Novotna, G., and J. Janata. 2006. A new evolutionary variant of the streptogramin A resistance protein, Vga(A)LC, from Staphylococcus haemolyticus with shifted substrate specificity towards lincosamides. Antimicrob. Agents Chemother. 50:4070-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Novotna, G., J. Spizek, and J. Janata. 2007. In vitro activity of telithromycin and quinupristin/dalfopristin against methicillin-resistant coagulase-negative staphylococci with defined resistance genotypes Folia Microbiol. (Praha) 52:593-599. [DOI] [PubMed] [Google Scholar]
- 9.Parish, L. C., and J. L. Parish. 2008. Retapamulin: A new topical antibiotic for the treatment of uncomplicated skin infections. Drugs Today (Barcelona). 44:91-102. [DOI] [PubMed] [Google Scholar]
- 10.Scangarella, N., C. Jakielaszek, L. McCloskey, D. Gentry, S. Rittenhouse, F. Shawar, and D. Payne. 2007. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother., abstr. E-2089.
- 11.Yan, K., L. Madden, A. E. Choudhry, C. S. Voigt, R. A. Copeland, and R. R. Gontarek. 2006. Biochemical characterization of the interactions of the novel pleuromutilin derivative retapamulin with bacterial ribosomes. Antimicrob. Agents Chemother. 50:3875-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]