Abstract
Future treatments for chronic hepatitis C virus (HCV) infection are likely to include agents that target viral components directly. Here, the preclinical characteristics of ITMN-191, a peptidomimetic inhibitor of the NS3/4A protease of HCV, are described. ITMN-191 inhibited a reference genotype 1 NS3/4A protein in a time-dependent fashion, a hallmark of an inhibitor with a two-step binding mechanism and a low dissociation rate. Under preequilibrium conditions, 290 pM ITMN-191 half-maximally inhibited the reference NS3/4A protease, but a 35,000-fold-higher concentration did not appreciably inhibit a panel of 79 proteases, ion channels, transporters, and cell surface receptors. Subnanomolar biochemical potency was maintained against NS3/4A derived from HCV genotypes 4, 5, and 6, while single-digit nanomolar potency was observed against NS3/4A from genotypes 2b and 3a. Dilution of a preformed enzyme inhibitor complex indicated ITMN-191 remained bound to and inhibited NS3/4A for more than 5 h after its initial association. In cell-based potency assays, half-maximal reduction of genotype 1b HCV replicon RNA was afforded by 1.8 nM; 45 nM eliminated the HCV replicon from cells. Peginterferon alfa-2a displayed a significant degree of antiviral synergy with ITMN-191 and reduced the concentration of ITMN-191 required for HCV replicon elimination. A 30-mg/kg of body weight oral dose administered to rats or monkeys yielded liver concentrations 12 h after dosing that exceeded the ITMN-191 concentration required to eliminate replicon RNA from cells. These preclinical characteristics compare favorably to those of other inhibitors of NS3/4A in clinical development and therefore support the clinical investigation of ITMN-191 for the treatment of chronic hepatitis C.
Chronic hepatitis C virus (HCV) infection afflicts more than 170 million people worldwide and is the leading cause of liver transplantation in the United States (23, 39). The standard of care for the treatment of chronic hepatitis C is weekly injection of pegylated alfa interferon (peginterferon alfa) and twice-daily oral administration of ribavirin. This combination achieves the clinically relevant endpoint of durable clearance of virus from serum, or sustained virologic response (SVR), in approximately half of treated patients (10, 22). Poorer response rates are observed in certain subpopulations, including individuals harboring genotype 1 virus or a high viral load, cirrhotic patients, and African Americans (10, 22). Thus, novel therapeutic approaches that enhance SVR rates are needed to better treat this prevalent and serious disease.
The term “specifically targeted antiviral therapy for HCV,” or STAT-C, has been coined to describe regimens targeting essential HCV-encoded enzymes. Inhibitors of the protease activity of nonstructural protein 3/4A (NS3/4A) and the viral polymerase NS5B are considered attractive STAT-C components (20, 24, 25, 45). The NS3 protein is a chymotrypsin-like serine protease that is activated by association with NS4A. Following translation of the HCV RNA genome, the NS3/4A protease cleaves four sites that demarcate five proteins proximal to the carboxy terminus of the HCV polyprotein. Thus, NS3/4A liberates the functional form of the viral polymerase and other viral proteins required for HCV replication. In addition, the proteolytic activity of NS3/4A has recently been shown to dampen cellular sensing of viral components and in doing so to reduce type 1 interferon production (11, 44). Thus, inhibitors of NS3/4A may disrupt two separate processes relevant to the suppression of HCV.
Several inhibitors of NS3/4A have shown potent antiviral activity in early clinical trials, highlighting the significant potential of this class of compounds. In landmark studies, ciluprevir (BILN-2061) was found to reduce the average plasma concentration of genotype 1 HCV by approximately 3.0 log10 units following twice-daily dosing of 200 mg for 2 days (14, 19). Despite the impressive virologic response promoted by this compound, further clinical development was placed on hold due to severe cardiac toxicity in rhesus monkeys receiving ciluprevir for 4 weeks (32). Another macrocyclic inhibitor, TMC435350, has recently been reported to promote a maximal decline in circulating HCV of 3.9 log10 units following a 5-day course of once-daily administration of 200 mg (49). However, at that dose, TMC435350 accumulated in healthy volunteers from days 1 to 5 and showed an even more pronounced increase in day 5 exposure in HCV patients, leaving the steady-state level of the compound undefined (47, 49).
Two additional compounds represent a distinct class of linear tetrapeptide inhibitors that act as mechanism-based covalent traps of the catalytic serine of NS3/4A. Boceprevir (SCH 503034) at a dose of 400 mg every 8 h promoted a mean maximum decline in HCV RNA of 1.6 log10 units in patients who previously did not respond to interferon-based therapy (36). A higher dose of the compound is reported to be under continued clinical study. Telaprevir (VX-950) has been subjected to the most extensive clinical testing program among the NS3/4A protease inhibitors. Administration of telaprevir as a monotherapy at various doses and schedules in genotype 1 patients results in serum HCV RNA reductions after 2 days similar to those reported for ciluprevir and a median reduction in the serum HCV RNA of 4.0 and 4.4 log10 units after 14 days of dosing with the optimal regimen of 750 mg every 8 h (8, 14, 19, 31). A significant number of patients experienced viral rebound when administered telaprevir monotherapy due to the emergence of viruses encoding NS3 proteases with reduced sensitivity to the drug (8, 31, 35), but the rate of viral escape was dramatically reduced when a standard dose of peginterferon alfa-2a was coadministered (8). Longer-duration clinical studies of telaprevir in combination with peginterferon alfa-2a and ribavirin continue to demonstrate that regimens including protease inhibitors can significantly improve treatment responses, but the side effect profile of this particular compound in combination with the current standard of care may compromise its clinical effectiveness (13, 15).
ITMN-191 is a novel NS3/4A protease inhibitor with potential utility as an adjuvant to the current standard of care or as a component of all oral STAT-C regimens. Here, the preclinical characteristics of ITMN-191 are described and compared to those of other NS3/4A inhibitors that have demonstrated antiviral effects in humans.
MATERIALS AND METHODS
Peptides, proteins, inhibitors, and replicons.
The NS4A peptide fragment was obtained from Midwest Bio-Tech (Fishers, IN). Full-length NS3 coding sequences derived from HCV genotype 1b HCV replicon K2040 (GenBank accession no. FJ031985) (41) and clinical isolates of HCV genotypes 1 to 6 (GenBank accession no. FJ024486 to FJ024492) were kind gifts from Michael Gale (University of Texas Southwestern). Full-length NS3 (genotype 1b amino acids 1 to 631) was expressed as an N-terminal hexahistidine fusion from recombinant baculoviruses generated using the Baculogold System (BD Biosciences, San Jose, CA) in High Five cells. Proteins were purified to >90% homogeneity via Ni affinity immobilization, followed by either gradient chromatography on poly(U) Sepharose and gel filtration (Superdex 200) or gel filtration alone.
Patient-derived HCV genotype 1b HCV replicon K2040 (41) harbored in Huh7 cells and HP luciferase HCV replicon plasmid were also kind gifts from M. Gale.
ITMN-191, telaprevir, boceprevir, and ciluprevir were synthesized by InterMune or a commissioned contractor.
Biochemical assays.
Protease activity for K2040 and genotype 1 to 6 NS3 proteins was followed in a continuous fluorescent resonance energy transfer (FRET)-based assay. The assay buffer contained 25 μM NS4A peptide, 50 mM Tris-HCl, pH 7.5, 15% (vol/vol) glycerol, 0.6 mM lauryldimethylamine N-oxide, 10 mM dithiothreitol, and 0.5 μM fluorescein/QXL520-labeled FRET substrate {Ac-DE-Dap(QXL520)-EE-Abu-ψ-[COO]-AS-Cys(5-FAMsp)-NH2; AnaSpec Inc., San Jose, CA}. Typically, 50 pM K2040 enzyme was added to initiate the reaction. Reactions were set up in black 96-well plates (Corning Inc., Corning, NY), and fluorescence data were collected using a SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA). Control reactions lacking inhibitors and enzyme were included. Initial rates were calculated from the linear phase of the reaction (up to 1 h) and were used to generate dose-response curves that were fitted to a four-parameter logistic function to obtain 50% inhibitory concentrations (IC50s) (XLfit; IDBS Inc., Guildford, United Kingdom). IC50s are reported as mean ± standard deviation and are the averages of at least three independent determinations. Recovery of activity from preformed ITMN-191·NS3/4A complex was assessed by preincubating 10 nM NS3/4A with a twofold excess of ITMN-191 in 1× assay buffer for 15 min, followed by a rapid 200-fold dilution of the preformed complex into assay buffer containing substrate. A control reaction with the same final conditions without preincubation of NS3/4A and ITMN-191 was initiated by the addition of enzyme to an otherwise-complete reaction mixture. Additional control reactions lacked either ITMN-191 or NS3. The progress of the reactions was followed over 5 h. Longer reaction times were not pursued due to potential loss of enzyme activity and possible substrate depletion.
Biochemical selectivity screens.
ITMN-191, telaprevir, and ciluprevir were submitted to MDS Pharma Services (Taipei, Taiwan) for assessment of the inhibition of proteases and interference with receptors, transporters, and ion channels. Protease assays employed spectrophotometric or spectrofluorometric substrates and isolated enzymes. Interference with receptors, transporters, and ion channels was assessed in radioligand binding assays. The reported values are the averages of two assay points.
Cell-based assays.
Huh7 cells harboring HCV replicon were grown under standard conditions (43). Serially diluted ITMN-191 was added to K2040 replicon cells 1 day after cell plating. Final ITMN-191 concentrations ranged from 100 nM to 5 pM for antiviral assays and from 1 mM to 5.6 nM for cytotoxicity assays; ITMN-191 was prepared through threefold serial dilution for both assays. After a 48-h incubation, intracellular RNA was extracted (RNeasy kit; Qiagen, Valencia, CA), and the level of HCV replicon RNA was quantified by reverse transcription (RT)-PCR assay with primers (5′-CAC TCC CCT GTG AGG AAC TAC TG-3′ and 5′-AGG CTG CAC GAC ACT CAT ACT-3′) and a probe (5′-6-FAM-CTT CAC GCA GAA AGC GTC TAG CCA TGG-MGBNFQ-3′, where FAM is 6-carboxyfluorescin and MGBNFQ is molecular-groove binding non-fluorescence quencher) specific to the HCV 5′ untranslated region using an ABI Prism 7900 sequence detection system (Applied Biosystems, Foster City, CA). Single-tube reactions were performed using the TaqMan Gold RT-PCR kit according to the manufacturer's instructions (Applied Biosystems, Foster City, CA). Triplicate reactions for the RNA standards and samples were performed in 50 μl with 5 μl intracellular RNA (50 ng). RT was carried out at 48°C for 30 min, followed by 10 min at 95°C. The PCR was as follows: 15 s at 95°C and 1 min at 60°C for 40 cycles. Each RNA concentration was determined in triplicate. The absolute concentration of replicon RNA was calculated based on its signal relative to that of a standard curve generated by known concentrations of an in vitro-transcribed RNA corresponding to a genotype 1b 5′ untranslated region. Replicon levels in the presence of ITMN-191 were fitted to a four-parameter logistic function to obtain a 50% effective concentration (EC50).
The viability of Huh7 cells, human cardiac myocytes, and human cardiac fibroblasts was assessed following 72 h of exposure to ITMN-191 using a CellTiter-Glo Luminescent Cell Viability kit (Promega, Madison, WI). For these cell types, viability in the presence of ITMN-191 was fitted to a four-parameter logistic function to obtain a 50% cytotoxic concentration (CC50). Additionally, the CC50s were determined for six primary normal human cell types in stationary and proliferating (dividing) phases by Lonza, Inc. (Walkersville, MD). The cell types included normal human hepatocytes, microvascular endothelial cells, human skeletal muscle myoblasts, human articular chondrocytes, human lung fibroblasts, and renal proximal tubule epithelial cells. For the proliferating phase, ITMN-191 was added when cells reached 50% confluence. For the stationary phase, ITMN-191 was added 1 day after cells reached 100% confluence. Cell viability was quantified following 72 h of exposure to ITMN-191 using a ViaLight Plus kit on a microplate reader. CC50s were determined as described above.
For replicon clearance assays, plated K2040 HCV replicon cells were treated with ITMN-191 (final concentration, 45 nM, 30 nM, 15 nM, 7.5 nM, or 3.7 nM) or 0.1% (vol/vol) dimethyl sulfoxide in the absence of G418. Every 3 to 4 days, the cells were counted and split into fresh medium that contained ITMN-191 or 0.1% (vol/vol) dimethyl sulfoxide, and a cell sample was frozen. After 2 weeks, the cells were washed three times with phosphate-buffered saline, placed in fresh medium containing 0.5 mg/ml G418, and cultured for 4 weeks. The cells were counted and split when they reached 80% confluence, and a cell sample was frozen. HCV replicon RNA was quantified as described above. Experiments were performed in duplicate.
Analysis of antiviral synergy.
Huh7 cells harboring HCV replicon were grown under standard conditions (43), and HCV replicon levels were determined by quantification of a replicon-borne neomycin gene product (NPT II) using a commercially available enzyme-linked immunosorbent assay kit (Agdia, Elkhart, IN). Two experimental strategies were used to investigate the antiviral synergy of ITMN-191 with peginterferon alfa-2a. For Loewe additivity modeling (1, 2), dose-response curves were generated at fixed ratios of the two agents and for each agent separately. CalcuSyn (Biosoft, Ferguson, MO) was used to calculate the dose reduction index (DRI) and combination index (CI) and to plot isobolograms. The DRI was used to calculate the CI as follows: CI = 1/(DRI)drug 1 + 1/(DRI)drug 2. Synergy was taken as a CI of <1, strong synergy as a CI of <0.3, and antagonism as a CI of >1. For isobologram analysis, a conservative assumption of mutually nonexclusive drug interaction was used to generate a line of additive theoretical drug interaction.
Dose-response curves were generated using a checkerboard design in which drug ratios and concentrations were both varied. Bliss independence modeling was used to quantify areas where observed effects were significantly greater (synergy) or less (antagonism) than those predicted from single-drug control data. Triplicate data sets were assessed at the 95% confidence level with MacSynergy II (29) for evidence of minor synergy (>25 to <50 μM2), moderate synergy (>50 to <100 μM2; log volumes, >5 and <9), and strong synergy (>100 μM2; log volume, >9).
In vivo preclinical studies.
Pharmacokinetic properties were evaluated in rats and monkeys. Procedures were performed under protocols approved by the Institutional Animal Care and Use Committee of the test facility. Sprague-Dawley rats (three per time point) were administered a 30-mg/kg of body weight dose of ITMN-191 by oral gavage (a 6-mg/ml solution in water). Cynomolgus monkeys (two per time point; Charles River, Wooster, MA) were administered a 30-mg/kg dose of ITMN-191 by oral gavage (a 3-mg/ml solution in water). For each species, terminal blood samples and the entire perfused liver were collected 1, 4, 8, 12, and 24 h after dose administration. Blood samples were collected in EDTA, processed for plasma by centrifugation at 5°C, and stored at −20°C until analysis was performed. Liver samples were snap-frozen and stored at −70°C until analysis was performed. Blank, standard, and unknown plasma samples and homogenized liver containing an internal standard (ITMN-191 analog) were treated with acidified acetonitrile and centrifuged to remove precipitated proteins. The density of liver tissue was taken into account to allow concentrations in both compartments to be expressed as weight per unit volume. The cleared supernatants were diluted 1:1 into high-performance liquid chromatography grade water and analyzed on a 4000 Q-trap liquid chromatography-tandem mass spectrometer (Applied Biosystems, Foster City, CA) fitted with the Turbo-Ionspray source operating in negative-ion mode. Analytes and internal standards were monitored using multiple-reaction-monitoring scans and calibrated with ABI Analyst software, version 1.4.2. The calibration standards ranged from 0.0169 ng/ml to 37.0 ng/ml and from 7.47 ng/ml to 5,440 ng/ml for the quantification of plasma samples and liver homogenates, respectively. Quadratic fitting with 1/x weighting was utilized where an R2 value of >0.999 was achieved in both matrices.
RESULTS
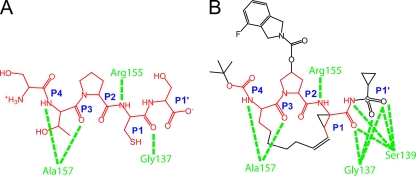
The identities and binding modes of NS3/4A protease substrates are known (Fig. 1A). ITMN-191 is an inhibitor of the HCV NS3/4A protease that was designed through a structure-informed drug discovery campaign to mimic and enhance contacts made by NS3/4A to its natural substrates (Fig. 1B). Specifically, ITMN-191 was designed to contain a rigid 15-member P1-P3 macrocyclic core that provided appropriate attachment points for acyl sulfonamide, fluoroisoindoline, and t-butyl moieties that mimicked P1′, P2, and P4 groups of the native substrates, respectively. Together, the appended groups and the macrocycle recapitulated and enhanced many of the interactions made by P1′, P1, P2, P3, and P4 groups of the natural substrates of NS3/4A and therefore promoted tight binding by ITMN-191.
FIG. 1.
Structure of ITMN-191 compared to that of a natural substrate of NS3/4A. (A) Structure of the NS4B/5A junction of the HCV polyprotein that is cleaved by NS3/4A. Peptide sites P1′, P1, P2, P3, and P4 are indicated. Groups directly transferred to ITMN-191 are in red. Suspected polar contacts with NS3/4A are shown in green, and the protein amino acids are indicated. (B) Structure of ITMN-191. Groups analogous to peptide sites P1′, P1, P2, P3, and P4 are indicated. Groups found in natural substrates of NS3/4A are shown in red. Suspected polar contacts with NS3/4A are shown in green, and the protein amino acids are indicated.
Biochemical inhibition of NS3/4A.
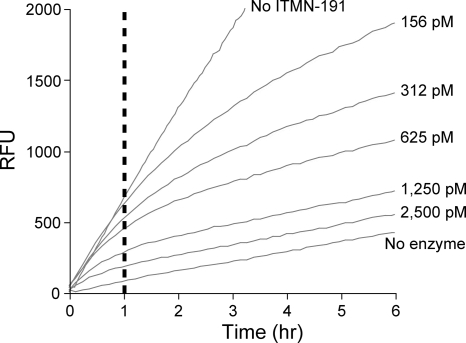
In peptide cleavage assays, ITMN-191 reduced genotype 1b NS3/4A (K2040) protease activity in a concentration-dependent fashion (Fig. 2). The magnitude of inhibition at any given concentration appeared approximately constant for an hour. The magnitude of inhibition increased upon prolonged observation, as evidenced by a progressive reduction in the slopes of progress curves (Fig. 2). Such biphasic progress curves in the presence but not the absence of inhibitor are a hallmark of a slow/tight binding mechanism. In this mechanism, the inhibitor first associates with its target in an initial complex that then rearranges into a more stable form (37, 42, 50, 51).
FIG. 2.
Progress curves describing biochemical inhibition of full-length NS3/4A. Representative progress curves demonstrating cleavage of a FRET-labeled NS3/4A substrate peptide over a 6-h period in the absence or presence of the indicated concentrations of ITMN-191 are shown. The reaction was performed without any preincubation of ITMN-191 and K2040 reference NS3/4A protein. RFU, relative fluorescence units. The dashed line at 1 hour indicates the time after which biphasic behavior becomes apparent.
In the above-mentioned two-step binding mechanism, the inhibition constants Ki and Ki* describe inhibition in the initial complex and in the full binding equilibrium, respectively, with the latter including the more stable and slowly dissociating complex. The IC50s of ITMN-191 and other NS3/4A protease inhibitors of known structure were determined under preequilibrium conditions analogous to those shown in Fig. 2 with data collection for 1 hour, which represented conditions that do not fully account for slow/tight binding. Under these conditions, ITMN-191, telaprevir, boceprevir, and ciluprevir displayed IC50s of 0.29 nM, 130 nM, 80 nM, and 0.73 nM, respectively, against a full-length genotype 1b NS3/4A reference protein (K2040) (Table 1). The values determined here were similar to those previously described as Ki values for these compounds against genotype 1 NS3/4A proteins (19, 26, 48). For inhibitors with slow/tight binding mechanisms, such as telapravir, boceprevir, and ITMN-191, the Ki* is more relevant, as it takes into account the more stable and slowly dissociating complex. The Ki* is 7 nM for telapravir (19, 26, 48), 14 nM for boceprevir (48), and 0.036 nM for ITMN-191 (30). Full characterization of the inhibition kinetics of ITMN-191, including derivation of Ki, Ki*, and koff (compound dissociation rate) from microscopic rate constants, will be presented elsewhere (P. T. R. Rajagopalan, S. D. Seiwert, and K. Kossen, unpublished data).
TABLE 1.
Biochemical potencies of ITMN-191 and other NS3/4A protease inhibitors
| Genotype | IC50 (nM)a
|
Fold shift from K2040 reference
|
||||||
|---|---|---|---|---|---|---|---|---|
| ITMN-191 | Telaprevir | Boceprevir | Ciluprevir | ITMN-191 | Telaprevir | Boceprevir | Ciluprevir | |
| 1b-(K2040) | 0.29 ± 0.07 | 130 ± 61 | 80 ± 15 | 0.73 ± 0.08 | ||||
| 1a | 0.20 ± 0.01 | 87 ± 5 | 80 ± 15 | 0.9 ± 0.2 | 0.69 | 0.67 | 1 | 1 |
| 1b | 0.23 ± 0.01 | 98 ± 2 | 83 ± 6 | 0.8 ± 0.1 | 0.79 | 0.75 | 1 | 1 |
| 2b | 1.6 ± 0.1 | 145 ± 5 | 76 ± 6 | 11 ± 1 | 5.5 | 1.1 | 1 | 15 |
| 3a | 3.5 ± 0.5 | 1590 ± 22 | 230 ± 33 | 11.5 ± 0.8 | 12 | 12 | 3 | 16 |
| 4 | 0.24 ± 0.02 | 470 ± 16 | 170 ± 10 | 0.6 ± 0.1 | 0.83 | 3.6 | 2 | 0.8 |
| 5 | 0.35 ± 0.01 | 130 ± 58 | 70 ± 10 | 0.8 ± 0.2 | 1.2 | 1.0 | 0.9 | 1 |
| 6 | 0.45 ± 0.01 | 36.9 ± 0.8 | 54 ± 6 | 1.0 ± 0.2 | 1.6 | 0.28 | 0.7 | 1.4 |
Values are reported as mean ± standard deviation based on a minimum of three independent experiments.
Potency was also determined against full-length NS3/4A sequences derived from clinical isolates representing all six genotypes of HCV, again under preequilibrium conditions. The IC50 of each inhibitor against genotype 1a, 1b, 5, or 6 NS3/4A was similar to its IC50 against the reference genotype 1b protein (Table 1). Against NS3/4A derived from genotype 2b, ciluprevir lost the most significant amount of potency, followed by ITMN-191. Telaprevir and boceprevir displayed similar potencies against genotype 1 and genotype 2b NS3/4A. All inhibitors except boceprevir showed slightly more than a 10-fold loss of potency against NS3/4A derived from genotype 3a. Both linear tetrapetide inhibitors additionally showed slightly reduced potency against genotype 4 NS3/4A, whereas the macrocyclic inhibitors ITMN-191 and ciluprevir did not.
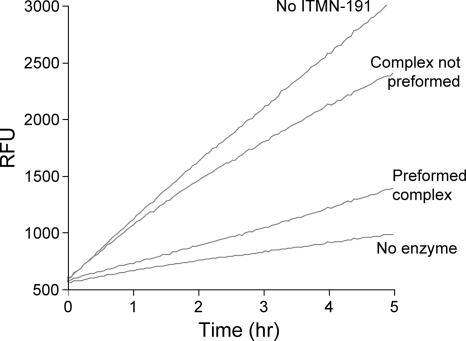
Kinetic principles associated with the time-dependent inhibition evidenced in Fig. 2 suggest that ITMN-191 may dissociate very slowly from NS3/4A and may therefore have a persistent inhibitory effect (37, 42, 50, 51). The persistence of inhibition was examined by monitoring NS3/4A activity following rapid dilution of a preformed complex of 20 nM ITMN-191 and genotype 1b NS3/4A (K2040) to a concentration of 100 pM ITMN-191. A final concentration of 100 pM would be expected to exhibit submaximal inhibition of NS3/4A if ITMN-191 participated in a rapid binding equilibrium, but if ITMN-191 bound NS3/4A through a slow/tight binding mechanism, it would dissociate slowly following its initial association at 20 nM and continue to significantly inhibit NS3/4A following dilution. In support of a slow/tight binding mechanism, suggested by Fig. 2, NS3/4A activity was substantially lower in samples subjected to preincubation with 20 nM ITMN-191 than in samples with the same final enzyme and inhibitor concentrations that were not preincubated (Fig. 3). Importantly, the extent of inhibition of NS3/4A preincubated with 20 nM ITMN-191 did not noticeably decrease during 5 h of measurement (i.e., upward curvature of reaction progress was not evident), and the amount of product formed was similar to that in the absence of NS3/4A, indicating that ITMN-191 remained stably bound to NS3/4A for at least 5 h. Thus, ITMN-191 disassociated from genotype 1b NS3/4A with a half-life on the order of several hours, as evidenced by the persistence of inhibition over the same time scale.
FIG. 3.
Protease activity following dilution of preformed ITMN-191·NS3/4A complex. Shown is a representative time course of cleavage of a FRET-labeled NS3/4A peptide substrate by K2040 reference NS3/4A protein over a 5-h period. The “Complex not preformed” reaction had 100 pM ITMN-191 and FRET-labeled substrate peptide added directly to 50 pM NS3/4A. “Preformed complex” had 20 nM ITMN-191 and 10 nM NS3/4A diluted 200-fold to the same final ITMN-191 and NS3/4A concentrations, with concomitant addition of FRET-labeled substrate peptide. Omission of NS3/4A or ITMN-191 determined the background and maximal rate of substrate cleavage, respectively. RFU, relative fluorescence units.
Biochemical specificities of ITMN-191, ciluprevir, and telaprevir.
In contrast to the highly potent inhibition of NS3/4A by ITMN-191, none of a panel of 53 proteases was inhibited more than 50% by a 10 μM screening concentration, indicating an IC50 higher than 10 μM against every protease in the panel (Table 2). Ciluprevir inhibited eight proteases and telaprevir inhibited nine proteases in the same panel at levels between 50% and 100%, which indicated that their IC50s against these proteases were 10 μM or less (Table 2; see Table S1 in the supplemental material). Neither ITMN-191 nor telaprevir showed appreciable activity against a broad panel of ion channels, receptors, and transporters, while ciluprevir inhibited human ERG (Table 2; see Table S2 in the supplemental material). From these data, the specificity index of ITMN-191, defined as the ratio of IC50s against nontarget enzymes (>10 μM) to its IC50 against full-length genotype 1b NS3/4A reference protein (K2040) (0.29 nM) (Table 1), was more than 35,000-fold. Calculated in a similar fashion, the biochemical specificity indexes of ciluprevir and telaprevir were less than 14,000-fold and less than 77-fold, respectively.
TABLE 2.
Biochemical specificities
| Panela | No. of proteins inhibited by 10 μM drugb
|
||
|---|---|---|---|
| ITMN-191 | Ciluprevir | Telaprevir | |
| Protease selectivity (53 proteins) | 0 | 8 | 9 |
| Broad ligand (26 proteins) | 0 | 1 | 0 |
Additional results for protease selectivity and broad ligand panels are shown in the supplemental material.
Proteins displaying ≥50% inhibition at a drug concentration of 10 μM, suggesting an IC50 of ≤10 μM.
Cellular potency against an HCV subgenomic replicon.
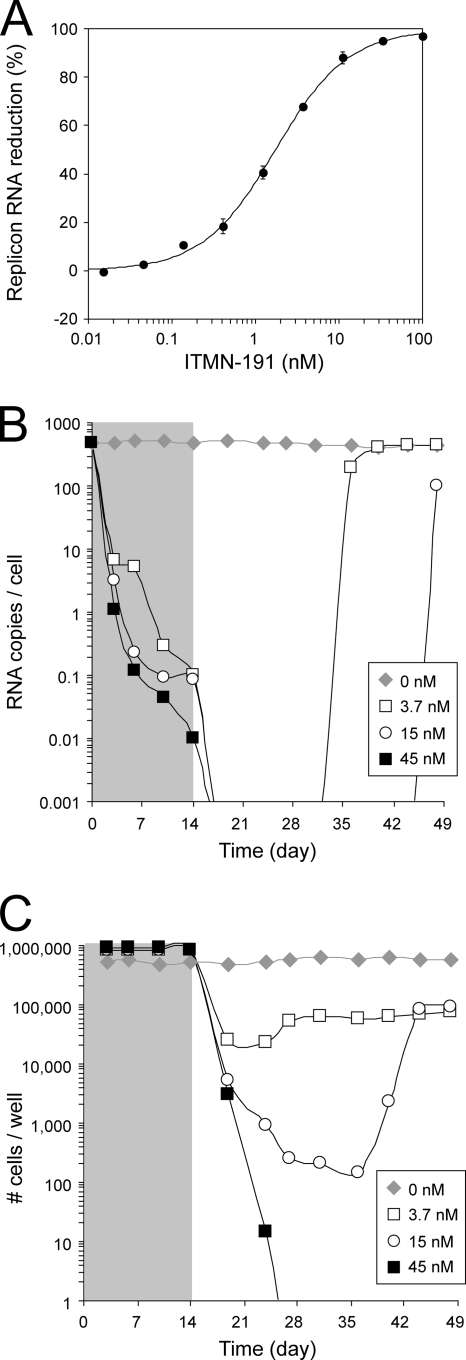
Dose-dependent reductions of a patient-derived HCV genotype 1b replicon harbored in hepatocyte-derived Huh7 cells were observed following 2-day incubation with ITMN-191 (Fig. 4A). The data were readily fitted to a four-parameter logistic equation to yield an EC50 of 1.8 nM with a slope of approximately 1.0. Calculation of the compound amount required for a 1 log10, 2 log10, or 3 log10 drop in replicon RNA (i.e., EC90, EC99, and EC99.9) yielded 14 nM, 160 nM, and 1,600 nM, respectively. Thus, ITMN-191 was a highly potent inhibitor of HCV replication in a cell-based system, as well as a highly potent inhibitor in biochemical assays.
FIG. 4.
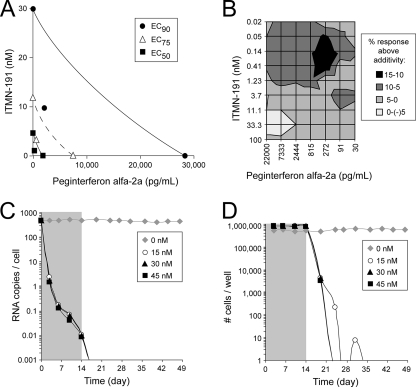
Cellular potency. (A) Average percent reduction in HCV replicon RNA in Huh7 cells promoted by the indicated concentrations of ITMN-191 as determined by RT-PCR from 18 independent experiments. The standard deviation between experiments is indicated. (B) HCV replicon RNA copies per cell as determined by RT-PCR during a 2-week treatment in the absence or presence of the indicated concentrations of ITMN-191 (shaded gray) and 4-week follow-up. Averaged data from two independent experiments are shown. The limit of detection by RT-PCR was 4.5 × 10−3 ± 1.9 × 10−4 copies/cell. (C) Number of cells per culture well during a 2-week treatment in the absence or presence of the indicated concentrations of ITMN-191 (shaded gray) and 4-week follow up. Averaged data from two independent experiments are shown.
Cytotoxicity of ITMN-191 upon 72 h of incubation was investigated with Huh7 cells and primary cultures of normal human hepatocytes, microvascular endothelial cells, human skeletal muscle myoblasts, human cardiac myocytes, human cardiac fibroblasts, human articular chondrocytes, human lung fibroblasts, and renal proximal tubule epithelial cells cultured under proliferating and nonproliferating conditions. CC50s ranged from 75 μM to 340 μM (data not shown), indicating a specificity index that minimally was approximately 41,000-fold relative to the cell-based potency of ITMN-191. Thus, ITMN-191 displayed a high degree of specificity for its intended target in both cell-based assays and biochemical assays.
ITMN-191 will be used clinically over durations longer than that employed in the 2-day determination of the EC50. Therefore, the antiviral activity of ITMN-191 was investigated following 14 days of exposure to replicon-bearing cells. ITMN-191 concentrations of 3.7 nM and 15 nM promoted a 3.7 log10 reduction in replicon levels upon 14 days of in vitro treatment (Fig. 4B) but did not clear HCV replicon from every cell, as judged by the selection of replicon-harboring cells during a 4-week follow-up period (Fig. 4C). Although no rebound in the replicon copy number was observed during ITMN-191 treatment, currently available data do not address whether the rebound observed following withdrawal of ITMN-191 reflected the emergence of drug-resistant NS3. In any case, treatment with 45 nM ITMN-191 (∼3 times its EC90) reduced HCV replicon RNA levels below the RT-PCR detection limit in a sustained fashion (Fig. 4B) and completely cleared replicon RNA, as judged by the inability to select for replicon-containing cells in a 4-week follow-up period (Fig. 4C).
Antiviral activity in combination with peginterferon alfa-2a.
To examine the combined antiviral effects of ITMN-191 and peginterferon alfa-2a, dose-response curves were generated for each agent at several fixed ratios. The EC50, EC75, and EC90 of each agent improved significantly when they were used in combination, indicating that their actions were either additive or synergistic (data not shown). Loewe additivity modeling (1, 2) provided CI values of <1 at EC50, EC75, and EC90 for each of the seven drug ratios tested, indicating synergy to strong synergy (Table 3).
TABLE 3.
ITMN-191/peginterferon alfa-2a CIsa
| ITMN-191 (nM)/peginterferon alfa-2a (pg/ml) ratio | CIb
|
||
|---|---|---|---|
| EC50 | EC75 | EC90 | |
| 3.7/22,000 | 0.45 ± 0.23 | 0.45 ± 0.17 | 0.45 ± 0.09 |
| 11/22,000 | 0.36 ± 0.13 | 0.36 ± 0.10 | 0.36 ± 0.07 |
| 33/22,000 | 0.6 ± 0.53 | 0.57 ± 0.45 | 0.56 ± 0.36 |
| 11/2,444 | 0.34 ± 0.11 | 0.36 ± 0.02 | 0.4 ± 0.11 |
| 100/7,333 | 0.43 ± 0.13 | 0.47 ± 0.09 | 0.53 ± 0.05 |
| 100/2,444 | 0.68 ± 0.40 | 0.71 ± 0.25 | 0.75 ± 0.06 |
| 100/815 | 0.39 ± 0.26 | 0.57 ± 0.20 | 0.84 ± 0.08 |
Using NPTII enzyme-linked immunosorbent assay and CalcSyn (2), the EC50s for ITMN-191 and peginterferon alfa-2a as single agents were 4.4 nM and 1,752 pg/ml, respectively.
Values are reported as mean ± standard deviation based on a minimum of three independent experiments.
Isobologram analysis, which graphically represents additive, synergistic, and antagonistic drug effects based on Loewe principles of additivity, was used to depict the antiviral activities promoted by the two agents at fixed dose ratios (1, 2). By this analysis, the combination of ITMN-191 and peginterferon alfa-2a clearly showed strong synergistic direct antiviral effects at EC50, EC75, and EC90 effect levels, since fixed dose ratio combinations fell below the line of theoretical additivity (Fig. 5A). The cytotoxicity of each agent was no worse in combination than when used alone (data not shown).
FIG. 5.
Antiviral activity in combination with peginterferon alfa-2a. (A) Isobologram analysis. The EC90, EC75, and EC50 are indicated for ITMN-191 alone (y axis), peginterferon alfa-2a alone (x axis), and the two agents at a fixed ratio of 100 nM ITMN-191 to 7,333 pg/ml peginterferon alfa-2a. The theoretical line of fixed ratio additivity assuming mutually nonexclusive antiviral effects connects single-agent EC90, EC75, and EC50. (B) Two-dimensional representation of combined antiviral effects of ITMN-191 and peginterferon alfa-2a as determined by Bliss independence modeling of variable drug ratio combinations. The log values associated with synergy (positive numbers) and antagonism (negative numbers) are indicated. Averaged data from three independent experiments are shown. (C) HCV replicon RNA copies per cell as determined by RT-PCR during a 2-week treatment in the absence or presence of the indicated concentrations of ITMN-191 and 2 ng/ml of peginterferon alfa-2a (shaded gray) and 4-week follow up. Note that 2 ng/ml corresponds to the human minimum plasma concentration of peginterferon alfa-2a as described in the package insert for this product. Averaged data from two independent experiments are shown. (D) Numbers of cells per well during a 2-week treatment in the absence or presence of the indicated concentrations of ITMN-191 and 2 ng/ml of peginterferon alfa-2a (shaded gray) and 4-week follow up. Averaged data from two independent experiments are shown.
Analysis of variable-ratio drug combinations by the Bliss independence model provided a nonparametric approach to independently quantify effects that are significantly enhanced (synergistic) or reduced (antagonistic) relative to the response predicted from single-drug effects. Assessment of drug interaction at the 95% confidence level indicated that ITMN-191 and peginterferon alfa-2a displayed significant synergy (log volume, >50 μM2%) (Fig. 5B). The synergy was most significant at low ITMN-191 concentrations. Thus, two orthogonal, formal analyses indicated that ITMN-191 and peginterferon alfa-2a display significant antiviral synergy.
To monitor the combined antiviral effects of ITMN-191 and peginterferon alfa-2a upon longer-term exposure, their combined anti-HCV replicon effects were investigated following 14 days of exposure. When the minimum human plasma concentration of peginterferon alfa-2a was added to the lowest concentration of ITMN-191 tested (15 nM), and to higher concentrations, replicon RNA levels were reduced below the limit of detection by RT-PCR (Fig. 5C), and no replicon-containing cells were selected in a 4-week follow-up period (Fig. 5D). When ITMN-191 was examined separately, higher concentrations were required to eliminate HCV replicon RNA from cells (Fig. 4B and C). Thus, not only did these two agents display formal antiviral synergy, peginterferon alfa-2a significantly enhanced the ability of ITMN-191 to clear HCV replicon from Huh7 cells.
Liver exposure in animal species.
Pharmacokinetic parameters were obtained following administration of a single oral dose to rats or cynomolgus monkeys by harvesting livers and sampling plasma at multiple time points (Table 4). Doses of 30 mg/kg were administered to rats and monkeys via oral gavage, which corresponded to a human equivalent dose of 290 mg or 580 mg, respectively.
TABLE 4.
Nonclinical pharmacokinetic performance
| Animal | Organ or ratio | AUCinfa (μg·h/ml) | Cmaxb (μg/ml) | C12 hb (μg/ml) | Replicon response supportedc (log10 reduction)
|
Elimination of HCV replicon supportedd
|
||
|---|---|---|---|---|---|---|---|---|
| Cmax | C12 h | Cmax | C12 h | |||||
| Rat | Liver | 90.8 | 12.7 ± 4.3 | 2.0 ± 1.3 | 4.0 | 3.2 | Yes | Yes |
| Plasma | 9.50 | 1.1 ± 0.3 | 0.16 ± 0.12 | 2.9 | 2.1 | Yes | Yes | |
| Ratio | 10 | 11 | 12 | |||||
| Primate | Liver | 7.61 | 1.61 | 0.138 | 3.1 | 2.0 | Yes | Yes |
| Plasma | 0.06 | 0.02 | 0.001 | 1.2 | 0.3 | No | No | |
| Ratio | 127 | 85 | 116 | |||||
Sacrifice of animal cohorts at various times prevented determination of average AUCinf.
Values are reported as mean ± standard deviation based on concentrations in three rodents or concentrations in two cynomolgus monkeys.
In vitro antiviral effect in 2-day assay supported by in vivo concentration. The values were calculated based on an EC50 of 1.8 nM in the 48-h replicon reduction assay and a four-parameter logistic fit of inhibition data and rounded to the nearest 0.1 log10 unit.
Describes the whether in vivo concentration exceeds the concentration required for elimination of HCV replicon from Huh7 cells upon 14 days of exposure.
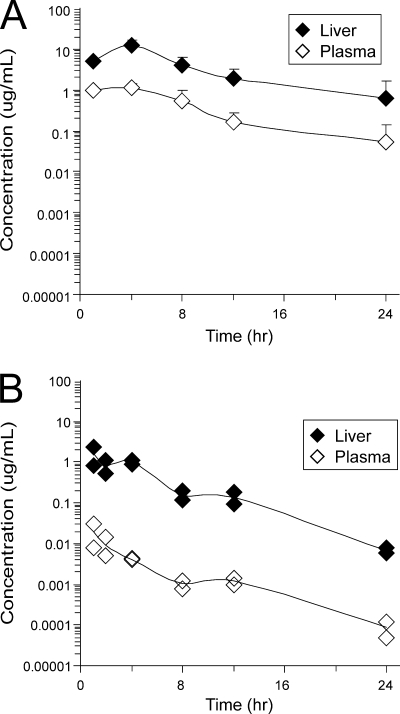
Importantly, the concentrations of ITMN-191 observed in the livers of both species were significantly above the compound's EC50, although concentrations in rats were higher than in monkeys (Table 4 and Fig. 6). In rats, the maximum concentration in the liver (Cmax) and the 12-h-postdose concentration in the liver (C12 h) were sufficient in vitro to reduce HCV replicon RNA levels by 4.0 log10 and 3.2 log10 units, respectively, in 2-day assays and to clear HCV replicon from cells in 14-day antiviral assays (Table 4). In monkey liver tissue, the Cmax and C12 h were sufficient to reduce HCV replicon RNA by 3.1 log10 and 2.0 log10 units, respectively, and also resulted in HCV replicon clearance from cells in 14-day antiviral assays (Table 4). While HCV is thought to replicate exclusively or nearly exclusively in the liver, significant reduction in HCV replicon RNA would also be supported by plasma concentrations (Table 4). Thus, although the exposure of ITMN-191 in monkeys is lower than that observed in rats, concentrations achieved in the livers and plasma of both species would be predicted to significantly impair viral replication.
FIG. 6.
Plasma and liver exposures of ITMN-191. The concentrations of ITMN-191 in liver and plasma were determined by liquid chromatography-tandem mass spectrometry following administration of a single oral dose of 30 mg/kg of ITMN-191 as an aqueous solution. (A) Average exposure in three rats. The standard deviation is indicated. (B) Exposure in each of two cynomolgus monkeys.
In each species, liver and plasma exposures roughly paralleled one another, but liver exposure was significantly greater than plasma exposure (Fig. 6). In rats, the liver-to-plasma ratios were roughly 10-fold, 11-fold, and 12-fold, based on total observed exposure (area under the observed concentration-time curve [AUCobs]), Cmax, and C12 h, respectively, while in monkeys, these ratios were 127-fold, 85-fold, and 116-fold, respectively (Table 4). Previously reported plasma and liver exposures in multiple-dose studies largely reflected exposures observed in single-dose studies (33).
DISCUSSION
The current standard of care for chronic hepatitis C results in the clinically meaningful endpoint of durable clearance of circulating virus, or SVR, in approximately 50% of all patients. Clearly, new treatment modalities are needed both to improve response rates in treatment-naïve patients and to provide therapeutic options in patients for whom the standard of care has failed. Agents that inhibit essential viral components have shown promise in initial clinical studies and in combination with the current standard of care (8, 28, 31, 36). This report describes the preclinical profile of ITMN-191, a novel inhibitor of the HCV NS3/4A protease, which recently entered clinical development as a STAT-C component.
To gauge the potential utility of ITMN-191 as a therapeutic agent, it is useful to compare its preclinical profile to those of ciluprevir, telaprevir, boceprevir, and TMC435350, since these compounds have demonstrated antiviral effects in clinical studies and since robust preclinical efficacy models are not available to evaluate the in vivo antiviral effect of ITMN-191.
Enzyme kinetic analysis indicates that ITMN-191 associates with NS3/4A through a slow/tight binding mechanism that is associated with slow dissociation (Fig. 2). While ciluprevir also possesses a P1-P3 macrocycle, it dissociates quickly from NS3/4A (19), indicating that macrocyclic compounds can inhibit NS3/4A in qualitatively different fashions. Although both telaprevir (26) and boceprevir bind via a slow/tight mechanism, ITMN-191 does not contain a functional group designed to facilitate adduct formation, and X-ray crystallographic studies suggest that ITMN-191 does not form a covalent adduct with NS3/4A analogous to that observed with telaprevir (3, 26). Slow/tight binding mechanisms are also associated with noncovalent inhibitors and, as is the case with certain inhibitors of the human immunodeficiency virus protease, often involve a conformational rearrangement of the protein to “trap” inhibitor (4). The slow dissociation of ITMN-191 from NS3/4A evidenced in Fig. 3 may have important implications for the use of the compound in treating chronic hepatitis C patients, since drug delivered to hepatocytes at any given time will remain associated with NS3/4A and inhibit its activity long after unbound drug is cleared. The half-life of a complex of telaprevir and NS3/4A, which is estimated to be approximately 1 hour, has been invoked to partially explain that compound's effectiveness in clinical studies (26). These same benefits also may be captured by ITMN-191, since it also dissociates slowly from NS3/4A.
The biochemical potencies of telaprevir and ciluprevir against the K2040 NS3/4A protein used as a reference here are largely consistent with those previously determined as Ki values against genotype 1b (Table 1) (19, 26). The appreciable biochemical potency of ITMN-191 translates into significant antireplicon activity (Fig. 4), which is significantly greater than those of telaprevir (26) and boceprevir (48) and is 4-fold to 17-fold superior to TMC435350 (40). While the studies here suggest ITMN-191 and ciluprevir are equipotent against the replicon (19), other HCV genotype 1 replicon systems have indicated that ITMN-191 may be at least fivefold more potent than ciluprevir (12). Since activity in cell-based assays may be predictive of HCV clearance in liver tissue (26), the potency of ITMN-191 relative to inhibitors that have demonstrated significant virologic effects in HCV patients supports exploration of the antiviral activity of ITMN-191 in clinical studies.
The relative biochemical potencies of both macrocyclic inhibitors tested are roughly maintained across genotypes (Table 1). Both ITMN-191 and ciluprevir display reduced potency against genotype 3a NS3/4A. Relative to genotype 1 NS3/4A, genotype 3a carries a substitution at position 168 (D168Q), the same amino acid position subjected to substitution in genotype 1 HCV replicons that are resistant to ciluprevir and ITMN-191 (21, 38). Position 168 lies in the S2 region of NS3/4A, in close proximity to a site where the fluoroisoindolene moiety of ITMN-191 is found. Interestingly, clinical studies examining the antiviral efficacy of ciluprevir in a population comprised primarily of those harboring genotype 3 HCV indicated that the short-term virologic response of ciluprevir monotherapy is marked, albeit reduced relative to its effect against genotype 1 HCV (32). Interestingly, the biochemical activity of telaprevir is also compromised against genotype 3a NS3/4A, despite the compound being fully active against position 168 variants, such as D168V/A, that emerged in resistance selections with macrocyclic inhibitors (21). Consequently, provided that the relative biochemical potency of a compound against NS3/4A derived from different genotypes is related to the virologic effect in patients, ITMN-191 (as well as telaprevir) would be expected to have a difference in clinical activity in HCV genotype 3 patients relative to HCV genotype 1 patients that is similar to that displayed by ciluprevir.
Accumulating evidence suggests that the side effect profiles of certain HCV protease inhibitors can reduce treatment utility by increasing treatment discontinuations following longer-term exposure (13, 15). Off-target activity of the inhibitor is one potential source of drug-related side effects. Notably, the biochemical and cell-based specificity index of ITMN-191 compares favorably with those of telaprevir and ciluprevir (Table 2; see Tables S1 and S2 in the supplemental material) and those reported for TMC435350 and boceprevir (40, 48). While it remains to be determined how the in vitro specificities of the above-listed agents relate to their clinical side effect or toxicity profiles, the high degree of selectivity displayed by ITMN-191 and its preferential exposure in liver tissue compared to plasma (Fig. 6) are consistent with a favorable side effect profile.
Currently contemplated therapeutic regimens suggest the combination of a direct antiviral agent, such as a protease inhibitor, with a peginterferon and ribavirin. In part, peginterferon contributes to sustained virologic response by inducing direct antiviral effects in infected hepatocytes. Notably, peginterferon alfa-2a greatly enhanced the ability of ITMN-191 to eliminate HCV replicon from cells and, like other inhibitors of NS3/4A protease (18), displayed formal antiviral synergy with type I interferon. The combined use of an NS3/4A protease inhibitor and peginterferon alfa-2a reduces the emergence of NS3/4A escape mutations in vitro (46) and upon combined clinical dosing (8, 16, 17, 35). The in vitro antiviral synergy of ITMN-191 and peginterferon alfa-2a suggests that the clinical antiviral effect of ITMN-191 observed in a monotherapy study would likely significantly increase when used in combination with peginterferon alfa-2a. Thus, the potential exists to administer ITMN-191 at lower doses or with an improved schedule in combination with peginterferon alfa-2a relative to administration as a monotherapy.
Sufficient delivery to liver tissue is a key requirement for efficacy of a compound meant to disrupt HCV replication. Liver concentrations of ciluprevir, boceprevir, and TMC435350 have not been fully reported, which prevents direct comparison to these compounds. However, the animal liver concentrations of ITMN-191 reported here compare favorably with animal liver concentrations previously reported for telaprevir (26, 27). In terms of micrograms of compound per gram of tissue following administration of equal doses, the liver concentrations of ITMN-191 in rats are comparable to the liver concentrations of telaprevir in mice, rats, and dogs (Table 4) (26). The concentration of ITMN-191 in monkey liver is approximately 10-fold lower. However, liver concentrations of ITMN-191 in rats and monkeys at Cmax or C12 h afford clearance of an HCV replicon from cultured cells (Table 4), whereas animal liver concentrations of telaprevir at Cmax or C8 h do not (26, 27), owing to the higher potency of ITMN-191. Given the suggested importance of the drug concentration at trough in limiting viral escape and sustaining the second phase of viral decline (31), the favorable 12-h-postdose concentration of ITMN-191 in animals following oral dosing of an aqueous solution suggests exploration of twice-daily administration of ITMN-191.
In animals, plasma exposure of ITMN-191 is lower than liver exposure (Fig. 6). Plasma and liver compartments seem to be in rapid equilibrium, as the exposures in the two compartments parallel one another. The ratio of liver exposure to plasma exposure is animal species specific, making determination of liver exposure in humans based on plasma exposure difficult. Importantly, we note that the low plasma exposure of ITMN-191 in both species relative to other agents may reduce the potential for toxicities associated with higher systemic exposure; this same principle may relate to the differing toxicity profiles of various statins (6, 7).
When taken together, the potency, preclinical liver exposure, and preclinical safety profile (5) of ITMN-191 relative to equivalent parameters for other HCV NS3/4A protease inhibitors support exploration of its safety and efficacy in patients with chronic HCV infection. Recently, twice-daily and three-times-daily administration of ITMN-191 at a total daily dose of up to 600 mg were reported to result in robust decline of circulating HCV RNA and to display a favorable side effect profile in a 14-day proof-of-concept study in treatment-naïve chronic HCV patients (9, 34). Based on these initial data, ITMN-191 will be studied in combination with peginterferon alfa-2a and ribavirin and in combination with various inhibitors of NS5B. Thus, ongoing clinical studies will soon determine if the different preclinical profiles of the agents described here impact their relative clinical utility in patients with chronic hepatitis C.
Supplementary Material
Acknowledgments
This work was funded by InterMune, Inc., and conducted by employees of InterMune, Inc., and Array Biopharma, Inc. All authors are employees of either InterMune, Inc., or Array Biopharma, Inc.
We thank all members of the NS3/4A research team of InterMune, Inc., and Array BioPharma, Inc., for their valuable research contributions to the program and comments on the manuscript.
Footnotes
Published ahead of print on 29 September 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Chang, T. T., and T. C. Chou. 2000. Rational approach to the clinical protocol design for drug combinations: a review. Acta Paediatr. Taiwan 41:294-302. [PubMed] [Google Scholar]
- 2.Chou, T. C., and P. Talalay. 1984. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 22:27-55. [DOI] [PubMed] [Google Scholar]
- 3.Condroski, K. R., H. Zhang, S. D. Seiwert, J. A. Ballard, B. A. Bernat, B. Brandhuber, H. Colwell, D. Smith, G. Vigers, S. W. Andrews, A. L. Kennedy, Y. Jiang, S. M. Wenglowsky, J. A. Josey, and L. M. Blatt. 2006. Structure-based design of novel isoindolene inhibitors of HCV NS3/4A protease and binding mode analysis of ITMN 191 by X-ray crystallography. Gastroenterology 130:A-835.
- 4.Copeland, R. A. 2005. Evaluation of enzyme inhibitors in drug discovery, 1st ed. Wiley and Sons, Hoboken, NJ.
- 5.Dearlove, G. E., B. J. Marafino, Jr., J. T. Secrest, E. A. Smith, S. D. Seiwert, and L. M. Blatt. 2007. The safety evaluation of ITMN-191, a novel orally available hepatitis C serine protease inhibitor. Toxicologist 96:S1. [Google Scholar]
- 6.Evans, M., and A. Rees. 2002. Effects of HMG-CoA reductase inhibitors on skeletal muscle: are all statins the same? Drug Saf. 25:649-663. [DOI] [PubMed] [Google Scholar]
- 7.Evans, M., and A. Rees. 2002. The myotoxicity of statins. Curr. Opin. Lipidol. 13:415-420. [DOI] [PubMed] [Google Scholar]
- 8.Forestier, N., H. W. Reesink, C. J. Weegink, L. McNair, T. L. Kieffer, H. M. Chu, S. Purdy, P. L. Jansen, and S. Zeuzem. 2007. Antiviral activity of telaprevir (VX-950) and peginterferon alfa-2a in patients with hepatitis C. Hepatology 46:640-648. [DOI] [PubMed] [Google Scholar]
- 9.Forestier, N., D. Larrey, D. Guyader, P. Marcellin, R. Rouzier, A. Patat, W. Bradford, S. Porter, and S. Zeuzem. 2008. Treatment of chronic hepatitis C virus (HCV) genotype 1 patients with the NS3/4A protease inhibitor ITMN-191 leads to rapid reductions in plasma HCV RNA: results of a phase 1b multiple ascending dose (MAD) study. Hepatology 48:1132A. [DOI] [PubMed] [Google Scholar]
- 10.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 11.Gale, M., Jr., and E. M. Foy. 2005. Evasion of intracellular host defense by hepatitis C virus. Nature 436:939-945. [DOI] [PubMed] [Google Scholar]
- 12.He, Y., M. S. King, D. J. Kempf, L. Lu, H. B. Lim, P. Krishnan, W. Kati, T. Middleton, and A. Molla. 2008. Relative replication capacity and selective advantage profiles of protease inhibitor-resistant hepatitis C virus (HCV) NS3 protease mutants in the HCV genotype 1b replicon system. Antimicrob. Agents Chemother. 52:1101-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hezode, C., P. Ferenci, G. Dusheiko, S. Pol, T. Goeser, J.-P. Bronowicki, S. Gharakhanian, D. Devonish, R. Kauffman, J. Alam, J.-M. Pawlotsky, and S. Zeuzem. 2007. PROVE2: phase II study of VX-950 (telaprevir) in combination with peginterferon alfa 2A with or without ribavirin in subjects with chronic hepatitis C, first interim analysis. Hepatology 46:268A-269A. [Google Scholar]
- 14.Hinrichsen, H., Y. Benhamou, H. Wedemeyer, M. Reiser, R. E. Sentjens, J. L. Calleja, X. Forns, A. Erhardt, J. Cronlein, R. L. Chaves, C. L. Yong, G. Nehmiz, and G. G. Steinmann. 2004. Short-term antiviral efficacy of BILN 2061, a hepatitis C virus serine protease inhibitor, in hepatitis C genotype 1 patients. Gastroenterology 127:1347-1355. [DOI] [PubMed] [Google Scholar]
- 15.Jacobson, I. M., G. T. Everson, S. C. Gordon, R. Kauffman, L. McNair, A. Muir, and J. G. McHutchison. 2007. Interim analysis results from a phase 2 study of telaprevir with peginterferon alfa-2A and ribavirin in treatment-naïve subjects with hepatitis C. Hepatology 46:315A-316A.17654600 [Google Scholar]
- 16.Kieffer, T., C. Sarrazin, J. Miller, S. Traver, Y. Zhou, D. Bartels, B. Hanzelka, U. Müh, C. Chao Lin, H. Reesink, A. A. Kwong, and S. Zeuzem. 2006. Combination of telaprevir (VX-950) and PEG-IFNalfa suppresses both wild-type virus and resistance variants in HCV genotype 1-infected patients in a 14-day phase 1b study. Hepatology 44:63A. [Google Scholar]
- 17.Kieffer, T. L., C. Sarrazin, J. S. Miller, M. W. Welker, N. Forestier, H. W. Reesink, A. D. Kwong, and S. Zeuzem. 2007. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology 46:631-639. [DOI] [PubMed] [Google Scholar]
- 18.Koev, G., T. Dekhtyar, L. Han, P. Yan, T. I. Ng, C. T. Lin, H. Mo, and A. Molla. 2007. Antiviral interactions of an HCV polymerase inhibitor with an HCV protease inhibitor or interferon in vitro. Antivir. Res. 73:78-83. [DOI] [PubMed] [Google Scholar]
- 19.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 20.Lemon, S. M., M. Yi, and K. Li. 2005. “Strong reasons make strong actions”—the antiviral efficacy of NS3/4A protease inhibitors. Hepatology 41:671-674. [DOI] [PubMed] [Google Scholar]
- 21.Lin, C., C. A. Gates, B. G. Rao, D. L. Brennan, J. R. Fulghum, Y. P. Luong, J. D. Frantz, K. Lin, S. Ma, Y. Y. Wei, R. B. Perni, and A. D. Kwong. 2005. In vitro studies of cross-resistance mutations against two hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J. Biol. Chem. 280:36784-36791. [DOI] [PubMed] [Google Scholar]
- 22.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 23.National Institutes of Health. 2002. NIH consensus statement on management of hepatitis C: 2002. NIH Consens. State Sci. Statements 19:1-46. [PubMed] [Google Scholar]
- 24.Ni, Z. J., and A. S. Wagman. 2004. Progress and development of small molecule HCV antivirals. Curr. Opin. Drug Discov. Dev. 7:446-459. [PubMed] [Google Scholar]
- 25.Pawlotsky, J. M. 2004. Hepatitis C: it's a long way to new therapy, it's a long way to go. Gastroenterology 127:1629-1632. [DOI] [PubMed] [Google Scholar]
- 26.Perni, R. B., S. J. Almquist, R. A. Byrn, G. Chandorkar, P. R. Chaturvedi, L. F. Courtney, C. J. Decker, K. Dinehart, C. A. Gates, S. L. Harbeson, A. Heiser, G. Kalkeri, E. Kolaczkowski, K. Lin, Y. P. Luong, B. G. Rao, W. P. Taylor, J. A. Thomson, R. D. Tung, Y. Wei, A. D. Kwong, and C. Lin. 2006. Preclinical profile of VX-950, a potent, selective, and orally bioavailable inhibitor of hepatitis C virus NS3-4A serine protease. Antimicrob. Agents Chemother. 50:899-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perni, R. B., G. Chandorkar, P. R. Chaturvedi, L. F. Courtney, C. J. Decker, C. A. Gates, S. L. Harbeson, A. D. Kwong, C. Lin, Y.-P. Luong, W. Markland, B. G. Rao, R. D. Tung, and J. A. Thomson. 2003. VX-950: the discovery of an inhibitor of the hepatitis C NS3·4A protease and a potential hepatitis C virus therapeutic. Hepatology 38:624A. [Google Scholar]
- 28.Pockros, P. J., D. Nelson, E. Godofsky, M. Rodriguez-Torres, G. Everson, M. W. Fried, R. H. Ghalib, S. A. Harrison, L. M. Nyberg, M. L. Shiffman, G. Z. Hill, and A. Chan. 2007. Robust synergistic antiviral effect of R1626 in combination with peginterferon alfa-2a (40KD), with or without ribavirin: interim analysis results of phase 2a study. Hepatology 46:311A. [Google Scholar]
- 29.Prichard, M. N., and C. Shipman, Jr. 1990. A three-dimensional model to analyze drug-drug interactions. Antivir. Res. 14:181-205. [DOI] [PubMed] [Google Scholar]
- 30.Rajagopalan, P. T. R., S. S. Misialek, L. M. Blatt, S. D. Seiwert, and K. Kossen. 2007. Characterization of HCV NA3/4a protease inhibition by ITMN 191 reveals picomolar potency and slow dissociation: implications for the use of ITMN 191 in chronic HCV treatment. Gastroenterology 132:A-782. [Google Scholar]
- 31.Reesink, H. W., S. Zeuzem, C. J. Weegink, N. Forestier, A. van Vliet, J. van de Wetering de Rooij, L. McNair, S. Purdy, R. Kauffman, J. Alam, and P. L. Jansen. 2006. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase Ib, placebo-controlled, randomized study. Gastroenterology 131:997-1002. [DOI] [PubMed] [Google Scholar]
- 32.Reiser, M., H. Hinrichsen, Y. Benhamou, H. W. Reesink, H. Wedemeyer, C. Avendano, N. Riba, C. L. Yong, G. Nehmiz, and G. G. Steinmann. 2005. Antiviral efficacy of NS3-serine protease inhibitor BILN-2061 in patients with chronic genotype 2 and 3 hepatitis C. Hepatology 41:832-835. [DOI] [PubMed] [Google Scholar]
- 33.Rieger, R. A., P. A. Lee, S. D. Seiwert, S. W. Andrews, T. K. Pope, J. Pheneger, N. N. Neitzel, R. B. Franklin, J. A. Josey, and L. M. Blatt. 2006. Pharmacokinetic analysis and liver concentrations of a series of macrocyclic peptidomimetic inhibitors of HCV NS3/4A protease: identification of ITMN-191, a potent NS3/4A protease inhibitor with high liver exposure across multiple species. Gastroenterology 130:A-835. [Google Scholar]
- 34.Rubino, C., W. Bradford, A. Forrest, S. Porter, L. Blatt, S. Seiwert, and S. Zeuzem. 2008. Pharmacokinetic-pharmacodynamic (PK-PD) relationships for ITMN-191 in a phase 1 multiple ascending dose trial in patients with genotype 1 chronic hepatitis C (CHC) infection. Hepatology 48:1140A. [Google Scholar]
- 35.Sarrazin, C., T. L. Kieffer, D. Bartels, B. Hanzelka, U. Muh, M. Welker, D. Wincheringer, Y. Zhou, H. M. Chu, C. Lin, C. Weegink, H. Reesink, S. Zeuzem, and A. D. Kwong. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767-1777. [DOI] [PubMed] [Google Scholar]
- 36.Sarrazin, C., R. Rouzier, F. Wagner, N. Forestier, D. Larrey, S. K. Gupta, M. Hussain, A. Shah, D. Cutler, J. Zhang, and S. Zeuzem. 2007. SCH 503034, a novel hepatitis C virus protease inhibitor, plus pegylated interferon alpha-2b for genotype 1 nonresponders. Gastroenterology 132:1270-1278. [DOI] [PubMed] [Google Scholar]
- 37.Sculley, M. J., J. F. Morrison, and W. W. Cleland. 1996. Slow-binding inhibition: the general case. Biochim. Biophys. Acta 1298:78-86. [DOI] [PubMed] [Google Scholar]
- 38.Seiwert, S. D., S. W. Andrews, H. Tan, K. R. Condroski, J. A. Ballard, B. A. Bernat, J. A. Josey, and L. M. Blatt. 2006. Generation and characterization of HCV replicons with reduced sensitivity to ITMN 191, a macrocyclic inhibitor of NS3/4A. Gastroenterology 130:A-754. [Google Scholar]
- 39.Shepard, C. W., L. Finelli, and M. J. Alter. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558-567. [DOI] [PubMed] [Google Scholar]
- 40.Simmen, K., O. Lenz, T. Lin, G. Fanning, P. Raboisson, H. de Kock, G. van't Klooster, Å. Rosenquist, M. Edlund, M. Nilsson, L. Vrang, and B. Samuelsson. 2007. In vitro activity and preclinical pharmacokinetics of the HCV protease inhibitor, TMC435350. Hepatology 46:857A. [Google Scholar]
- 41.Sumpter, R., Jr., C. Wang, E. Foy, Y. M. Loo, and M. Gale, Jr. 2004. Viral evolution and interferon resistance of hepatitis C virus RNA replication in a cell culture model. J. Virol. 78:11591-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szedlacsek, S. E., and R. G. Duggleby. 1995. Kinetics of slow and tight-binding inhibitors. Methods Enzymol. 249:144-180. [DOI] [PubMed] [Google Scholar]
- 43.Tan, H., J. Derrick, J. Hong, C. Sanda, W. M. Grosse, H. J. Edenberg, M. Taylor, S. Seiwert, and L. M. Blatt. 2005. Global transcriptional profiling demonstrates the combination of type I and type II interferon enhances antiviral and immune responses at clinically relevant doses. J. Interferon Cytokine Res. 25:632-649. [DOI] [PubMed] [Google Scholar]
- 44.Thimme, R., V. Lohmann, and F. Weber. 2006. A target on the move: innate and adaptive immune escape strategies of hepatitis C virus. Antivir. Res. 69:129-141. [DOI] [PubMed] [Google Scholar]
- 45.Thomson, J. A., and R. B. Perni. 2006. Hepatitis C virus NS3-4A protease inhibitors: countering viral subversion in vitro and showing promise in the clinic. Curr. Opin. Drug Discov. Dev. 9:606-617. [PubMed] [Google Scholar]
- 46.Tong, X., R. Chase, A. Skelton, T. Chen, J. Wright-Minogue, and B. A. Malcolm. 2006. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antivir. Res. 70:28-38. [DOI] [PubMed] [Google Scholar]
- 47.van 't Klooster, G. A. E., I. Vanwelkenhuysen, R. Hooijmaijers, K. Bol, M. Voets, J. Van Houdt, R. A. L. Verloes, F. Aharchi, K. Marien, P. Van Remoortere, F. Broeckaert, H. de Kock, and K. A. Simmen. 2008. Once daily regimens of the HCV NS3/4A-protease inhibitor TMC435350 are predicted to provide therapeutic exposure in plasma and liver. J. Hepatol 48:S321. [Google Scholar]
- 48.Venkatraman, S., S. L. Bogen, A. Arasappan, F. Bennett, K. Chen, E. Jao, Y. T. Liu, R. Lovey, S. Hendrata, Y. Huang, W. Pan, T. Parekh, P. Pinto, V. Popov, R. Pike, S. Ruan, B. Santhanam, B. Vibulbhan, W. Wu, W. Yang, J. Kong, X. Liang, J. Wong, R. Liu, N. Butkiewicz, R. Chase, A. Hart, S. Agrawal, P. Ingravallo, J. Pichardo, R. Kong, B. Baroudy, B. Malcolm, Z. Guo, A. Prongay, V. Madison, L. Broske, X. Cui, K. C. Cheng, Y. Hsieh, J. M. Brisson, D. Prelusky, W. Korfmacher, R. White, S. Bogdanowich-Knipp, A. Pavlovsky, P. Bradley, A. K. Saksena, A. Ganguly, J. Piwinski, V. Girijavallabhan, and F. G. Njoroge. 2006. Discovery of (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3-[2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3- dimethyl-1-oxobutyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (SCH 503034), a selective, potent, orally bioavailable hepatitis C virus NS3 protease inhibitor: a potential therapeutic agent for the treatment of hepatitis C infection. J. Med. Chem. 49:6074-6086. [DOI] [PubMed] [Google Scholar]
- 49.Verloes, R., K. A. Farha, A. van Vliet, G. van′t Klooster, F. Aharchi, K. Marien, H. de Kock, and K. Kenneth Simmen. 2007. Results of a phase 1 placebo-controlled trial in healthy volunteers to examine the safety, tolerability and pharmacokinetics of the HCV protease inhibitor TMC435350 after single and repeated dosing. Hepatology 46:823A.17680645 [Google Scholar]
- 50.Williams, J. W., and J. F. Morrison. 1979. The kinetics of reversible tight-binding inhibition. Methods Enzymol. 63:437-467. [DOI] [PubMed] [Google Scholar]
- 51.Wolfenden, R. 1976. Transition state analog inhibitors and enzyme catalysis. Annu. Rev. Biophys. Bioeng. 5:271-306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.