Abstract
Marine microorganisms that consume one-carbon (C1) compounds are poorly described, despite their impact on global climate via an influence on aquatic and atmospheric chemistry. This study investigated marine bacterial communities involved in the metabolism of C1 compounds. These communities were of relevance to surface seawater and atmospheric chemistry in the context of a bloom that was dominated by phytoplankton known to produce dimethylsulfoniopropionate. In addition to using 16S rRNA gene fingerprinting and clone libraries to characterize samples taken from a bloom transect in July 2006, seawater samples from the phytoplankton bloom were incubated with 13C-labeled methanol, monomethylamine, dimethylamine, methyl bromide, and dimethyl sulfide to identify microbial populations involved in the turnover of C1 compounds, using DNA stable isotope probing. The [13C]DNA samples from a single time point were characterized and compared using denaturing gradient gel electrophoresis (DGGE), fingerprint cluster analysis, and 16S rRNA gene clone library analysis. Bacterial community DGGE fingerprints from 13C-labeled DNA were distinct from those obtained with the DNA of the nonlabeled community DNA and suggested some overlap in substrate utilization between active methylotroph populations growing on different C1 substrates. Active methylotrophs were affiliated with Methylophaga spp. and several clades of undescribed Gammaproteobacteria that utilized methanol, methylamines (both monomethylamine and dimethylamine), and dimethyl sulfide. rRNA gene sequences corresponding to populations assimilating 13C-labeled methyl bromide and other substrates were associated with members of the Alphaproteobacteria (e.g., the family Rhodobacteraceae), the Cytophaga-Flexibacter-Bacteroides group, and unknown taxa. This study expands the known diversity of marine methylotrophs in surface seawater and provides a comprehensive data set for focused cultivation and metagenomic analyses in the future.
Methylotrophic bacteria represent an important functional guild, contributing to the metabolism and assimilation of one-carbon (C1) compounds. Because the carbon sources that these bacteria depend on in the marine environment are present at low concentrations, characterizing marine methylotrophs has involved the use of enrichment and cultivation approaches with a variety of C1 substrates. The C1 substrates of relevance to the marine environment include methane, methanol, methylated amines, methyl halides, and methylated sulfur compounds. Methane is supersaturated in surface seawater, and several studies have isolated methanotrophs from the marine environment (14, 16, 28, 47). Methyl halides are produced by a number of phytoplankton species (42), and these ozone-depleting compounds have been used to isolate methylotrophic Alphaproteobacteria that belonged to the Roseobacter clade (43, 45). Methanol represents a marine C1 substrate derived from phytoplankton (13) and the atmosphere (7) which may be actively metabolized by marine methylotrophs (21). Methanol concentrations have been estimated at between 100 nM (48) and 300 nM (10) and were directly measured in one study, ranging between 50 and 250 nM in several tropical Atlantic samples (54). Enrichment and isolation studies using methanol as the sole carbon source have generated molecular fingerprint phylotypes and have characterized isolates of Methylophaga spp. (Gammaproteobacteria). Methylophaga spp. have also been isolated using dimethyl sulfide (DMS) (8, 44) and can grow on monomethylamine (23), both of which occur at nanomolar concentrations in surface seawater (11, 24). Together, these cultivation-based approaches have revealed the presence of organisms capable of C1 cycling in the marine environment. Their involvement in methylotrophic metabolism in situ can be experimentally addressed using stable isotope probing (SIP) (40).
DNA SIP recently identified Methylophaga-like organisms as active methylotrophs that assimilated methanol and methylamine in surface waters of the English Channel (36). That study also demonstrated that 16S rRNA gene sequences representing clades of uncultivated Gammaproteobacteria were also retrieved from the heavy DNA for each of these compounds that clustered close to Methylophaga. A DNA SIP experiment with methanol substrate dilution to concentrations anticipated to reflect those in situ (34) confirmed the involvement of Methylophaga spp. in methanol consumption and retrieved functional genes involved in methanol metabolism from these active methylotrophs using metagenomic libraries.
The goal of the current study was to extend our previous observations that were made under nonbloom conditions by studying methylotrophic populations in the context of a phytoplankton bloom dominated by Emiliania huxleyi and Karenia mikimotoi (formerly Gyrodinium aureolum). Both the coccolithophores (e.g., Emiliania) and the small dinoflagellates (e.g., Karenia) are associated with dimethylsulfoniopropionate production (22, 29), and phytoplankton blooms are known to produce relevant C1 compounds or their precursors, including methanol (13), methylated sulfur compounds (24), and methyl halides (42). As with our previous marine SIP studies (34, 36), seawater samples were incubated with methanol and methylamine, and in this investigation, SIP incubations were also carried out with 13C-labeled methane, dimethylamine, methyl bromide, and DMS to identify microbial populations that are actively involved in the cycling of these C1 compounds during phytoplankton blooms in situ.
MATERIALS AND METHODS
Bloom sampling.
A transect across a phytoplankton bloom dominated by E. huxleyi and K. mikimotoi (D. Schroeder, personal communication) was sampled in the English Channel bordering the southern coast of the United Kingdom. Surface seawater was taken from inside the bloom (49.3222 N, 5.1446 W to 49.5105 N, 5.1217 W), at the edge of the bloom (49.5472 N, 4.3966 W to 49.5523 N, 4.4045 W), and outside the bloom area (50.1158 N, 4.1998 W to 50.1053 N, 4.2062 W). The distances between the beginnings and ends of sampled areas differed for the three sampling stations and were 1.3 km, 0.8 km, and 20 km for the outside, the edge, and the inside of the bloom, respectively. All samples were taken between 1000 h and 2200 h on 26 July 2007. Water samples were returned to the laboratory, and aliquots were taken for filtration (for DNA extraction) and to establish SIP incubations on 27 July 2006. Multiple aliquots of approximately 1 liter were filtered through 0.2-μm Sterivex filters (Durapore, Millipore) and frozen at −80°C until processed for nucleic acid extraction.
Incubation with 13C-labeled substrates.
Samples taken from the edge of the bloom were chosen to set up SIP incubations with several 13C-labeled C1 substrates. Seawater sample aliquots of 750 ml were added to 1-liter serum bottles with the addition of 0.1% (750 μl) marine ammonium mineral salts medium (modified from that described in reference 12) and substrate. A total of 75 μmol of 13C-labeled substrate was added to bottles for methanol, monomethylamine, dimethylamine, methyl bromide, and methane (final concentration of 100 μM, assuming complete dissolution). For DMS, 187.5 μmol of substrate was added to make up a final concentration of 250 μM. All serum bottles were crimp sealed with butyl rubber bungs to prevent the loss of volatile substrates. All 13C-labeled compounds had a purity of 99% or higher and were obtained from Cambridge Isotope Laboratories (Hook Hampshire, United Kingdom), except methylated amines (Sigma, Gillingham, United Kingdom). [13C2]DMS was prepared by a method adapted from that described for labeled dimethyl sulfoxide synthesis (5). Sodium sulfide nonahydrate (6.5 g) was dissolved in 6.5 ml of sterile deionized water in a glass test tube and cooled to 0°C in an ice-water bath, with vigorous stirring. Subsequently, 5 g of [13C]methyl iodide (Cambridge Isotope Laboratories LTD, Andover, MA) was added dropwise over a period of 30 min prior to incubating the reaction mixture at 0°C for 5 h with stirring. Five milliliters each of 2 M sodium hydroxide solution and 1 M sodium thiosulfate solution was added, the reaction mixture vessel was then connected to a receiving tube held at −170°C in liquid nitrogen, and the reaction mixture was allowed to warm to 40°C in a water bath. The [13C2]DMS was distilled from the reaction mixture for 90 min and then redistilled into a sterile receiving vessel for 1 h. Sterile deionized water was added to the receiving vessel to dissolve the [13C2]DMS, and the resulting solution was transferred, with washings, to a sterile 1-liter serum vial which was then sealed with a butyl rubber bung. The concentration and purity of the [13C2]DMS solution were assessed by gas chromatography with a flame ionization detector. A total of 250 ml of a 7 mM solution of pure [13C2]DMS was obtained.
For all substrates, parallel incubations were set up as 12C-unlabeled controls and 13C- and 12C-labeled substrate incubations were harvested at a single time point. With the exception of monomethylamine and dimethylamine, substrate utilization was monitored by gas chromatography using a flame ionization detector. Measurement of DMS and methyl bromide concentrations in sterile seawater controls confirmed that the degradation observed for SIP incubations was due to biological processes and not to chemical decomposition. The concentrations of the methylamines were assumed to mirror those of methanol; recovery of [13C]DNA from methylamine and dimethylamine incubations confirmed that methylated amines had been assimilated. Following substrate depletion, SIP incubations were filtered through 0.22-μm Sterivex filters and frozen at −80°C until processed for nucleic acid extraction.
DNA extraction, SIP gradient centrifugation, and fractionation.
Total nucleic acids were extracted directly from Sterivex filters according to a previously described protocol (36). Briefly, lysozyme, proteinase K, and sodium dodecyl sulfate were used to lyse cells, and lysates were transferred to 15-ml phase lock tubes (Qiagen, West Sussex, United Kingdom) for phenol-chloroform and chloroform extractions. Purified DNA was quantified on a 1% (wt/vol) agarose gel. Aliquots (1 to 5 μg) of DNA extracts from each of the SIP incubations were added to cesium chloride (CsCl) solution (average density of ∼1.725 g ml−1) and transferred to an ultracentrifuge gradient tube for centrifugation and fractionation as previously described (37). Briefly, tubes were added to a Vti 65.2 rotor (Beckman Coulter, Fullerton, CA) and centrifuged at 44,100 rpm (177,000 × gav [average]) for 40 h at 20°C. Gradients were fractionated from bottom (fraction 1, highest density) to top (fraction 12, lowest density) into 425-μl fractions. DNA was purified from CsCl and quantitatively recovered by precipitation with glycogen (20 μg) and polyethylene glycol (30% PEG 6000 and 1.6 M NaCl). Purified DNA was suspended in 30 μl of sterile LoTE buffer (3 mM Tris [pH 8], 0.2 mM EDTA), and 5-μl aliquots were run on a 1% (wt/vol) agarose gel for quantification and to identify the distribution of [13C]DNA relative to background, unlabeled [12C]DNA (see Fig. S1 in the supplemental material). These data indicated that the 13C labeling of DNA was very high for methanol, monomethylamine, and DMS incubations; most of the DNA for these 13C-incubated samples eluted in heavy fractions (fractions 7 and 8, ∼1.725 to 1.730 g ml−1). The detection of 13C-labeled DNA confirmed that the substrate was incorporated into microbial biomass. For dimethylamine and methyl bromide incubations, the extent of DNA labeling was less pronounced. For methyl bromide incubations, there was almost no difference between the smears of DNA across gradients associated with 12C- and 13C-methyl bromide SIP incubations (see Fig. S1 in the supplemental material).
DGGE.
For each fractionated gradient, two fractions were selected for analysis of the “heavy” [13C]DNA (fractions 7 and 8, ∼1.725 to 1.730 g ml−1), and one fraction was selected for the characterization of “light” [12C]DNA (either fraction 11 or 12, ∼1.710 to 1.705 g ml−1). One-microliter aliquots of gradient fractions were used as the template for PCR to obtain 16S rRNA gene fragments suitable for denaturing gradient gel electrophoresis (DGGE) analysis. Each 50-μl reaction mixture consisted of 25 pmol of primers 341f-GC and 534r (32), 1× (NH4)2SO4 buffer (Fermentas, York, United Kingdom), 1.5 mM MgCl, 33.6 μg nonacetylated bovine serum albumin (Sigma, Gillingham, United Kingdom), 40 nmol of each deoxynucleoside triphosphate, 1.25 U Taq polymerase (Fermentas). The reaction mixture tubes were loaded directly into the block at 95°C (simplified hot start), followed by an initial denaturation at 95°C for 5 min and 30 cycles of 94°C for 1 min (denaturing), 55°C for 1 min (annealing), and 72°C for 1 min (extension). A final extension at 72°C for 7 min was followed by a holding step at 10°C. Five-microliter aliquots were quantified on a 1% (wt/vol) agarose gel.
For DGGE, 5-μl aliquots (100 to 300 ng) were run on a 10% polyacrylamide gel with a 30 to 70% denaturing gradient (100% denaturant is 7.0 M urea and 40% deionized formamide) according to the D-Code system instructions (Bio-Rad, Hercules, CA). Gels were run overnight (14 h) at 85 V and then stained for 1 h in SYBR Green I (Invitrogen, Paisley, United Kingdom). Gel images were captured with a FLA-5000 imaging system (Fujifilm, Tokyo, Japan). Bands selected for sequence analyses were sampled from the gel by means of sterile pipette tips and amplified from the gel, using the PCR conditions described above for DGGE. Sequencing was done with the 341f primer and a BigDye terminator version 3.1 kit (Applied Biosystems, Foster City), and the sequencing products were run on an ABI Prism 3130 genetic analyzer (Applied Biosystems) by the Molecular Biology Service, University of Warwick, Warwick, United Kingdom. DGGE band sequences were approximately 150 bases in length.
For determining the relatedness of the DGGE fingerprints, gels were imported into Gelcompar II (Applied Maths, Sint-Martens-Latem, Belgium) and normalized to ladder bands and additional internal standard bands. A dendrogram produced with the unweighted-pair group method using average linkages was generated by performing a Pearson correlation with background-subtracted densitometric curves, which takes band intensities into account. The output of the clustering analysis was independent of the input order of DGGE fingerprints.
16S rRNA gene libraries.
Clone libraries of bacterial 16S rRNA genes were generated from the original seawater samples (outside of bloom, edge of bloom, inside of bloom; 36 clones sequenced from each) and for the heavy DNA associated with the five substrates (methanol, monomethylamine, dimethylamine, methyl bromide, and DMS) that yielded 13C-labeled DNA (24 clones sequenced from each). The PCR to amplify the 16S rRNA gene used primers 27f and 1492r (25) and the same amplification reaction as that used for DGGE, except with an extension time of 1.5 min. Products were cloned into the TOPO-TA vector according to the manufacturer's protocol (Invitrogen). Screening was done as described previously (38), and cloned 16S rRNA gene inserts were sequenced at the Edinburgh node of the NERC Molecular Genetics Facility, using the 27f primer. Pintail (3) software was used to identify suspected chimeras and identified one heavy-band sequence which was likely chimeric in origin and several water library sequences that were likely chimeric; these sequences were excluded from further analyses. For seawater samples, classification of 16S rRNA gene sequences was done using an RDP-II classifier (53) after manually verifying base calls. For the 16S rRNA gene libraries constructed using [13C]DNA from SIP experiments, manually verified 16S rRNA gene sequences were compared to those in GenBank (6) to retrieve the three closest matches for each library sequence. Sequences were aligned within ARB (30) software, and an alignment was exported to MEGA4 (49). Evolutionary distances were computed using the maximum-composite likelihood method (50) and are presented in the units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There was a total of 466 nucleotide positions in the final data set. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches (9). The tree was drawn to scale, with branch lengths given in the same units as those of the evolutionary distances used to infer the phylogenetic tree. In the absence of cultivated methylotrophic organisms that fell within the groups of 16S rRNA genes derived from SIP experiments, clades were defined based on the consistent association with particular substrates, but a specific cutoff value was not used.
Nucleotide sequence accession numbers.
All sequences were deposited in GenBank for the marine samples taken from the edge (accession no. EU399242 to EU399272), the inside (accession no. EU399273 to EU399306), and the outside (accession no. EU399307to EU399340) of the phytoplankton bloom. The 16S rRNA gene clone library sequences from heavy DNA were deposited with the following accession numbers for SIP incubations with dimethylamine (accession no. EU399341 to EU399364), DMS (accession no. EU399365 to EU399386), methyl bromide (accession no. EU399387 to EU399407), monomethylamine (accession no. EU399408 to EU399428), and methanol (accession no. EU399429 to EU399451). DGGE band sequences from heavy DNA were deposited with the following accession numbers for SIP incubations with methanol (accession no. EU399452 to EU399457), monomethylamine (accession no. EU399458 to EU399464), dimethylamine (accession no. EU399465 to EU399469), DMS (accession no. EU399470 to EU399474), and methyl bromide (accession no. EU399475 to EU399477).
RESULTS
Phytoplankton bloom microbial community analysis.
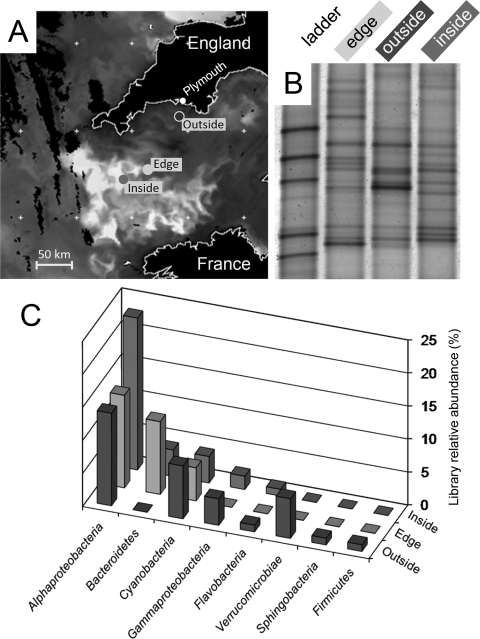
This study was conducted with samples from an extensive phytoplankton bloom with a predominance of both Emiliania huxleyi and Karenia mikimotoi (D. Schroeder, personal communication). Based on remotely sensed observations obtained from the day prior to sampling, three sampling stations within the western English Channel were selected to represent areas with varying chlorophyll concentrations (Fig. 1A), indicating regions internal to the bloom (“inside”), on the edge of the bloom (“edge”) and external to the bloom (“outside”). Prior to assessing the methylotrophs in the bloom (edge sample), we assessed the background bacterial community composition of the three water samples, using 16S rRNA gene fingerprinting (Fig. 1B) and clone libraries (Fig. 1C). The DGGE profiles indicate that the bacterial communities of these three water samples were represented by unique predominant band phylotypes, although several bands were shared among the three samples (Fig. 1B). Almost all sequences collected from the 16S rRNA gene clone libraries were most similar to GenBank sequences derived from other marine surface water samples, reflecting a composition similar to that described in previous studies (data not shown). All libraries were dominated by Alphaproteobacteria and Cyanobacteria, although Bacteroidetes organisms were also prevalent in the “inside” and “edge” libraries (Fig. 1C). Overall, the communities shared similar division-level compositions, but analysis also indicated that local sample heterogeneity existed across this relatively short bloom transect.
FIG. 1.
Physical location and bacterial composition of samples. (A) A grayscale Aqua-MODIS satellite radiance image generated from a mixed-wavelength composite image (551-nm, 488-nm, and 443-nm wavebands) on 26 July 2006. Emiliania huxleyi coccolithophore reflectance is readily visible in this image. A chlorophyll a satellite image from the same day shows chlorophyll abundance corresponding to the Karenia bloom adjacent to the bloom on the north side of the English Channel (image not shown). Satellite data were received and processed by the NERC Earth Observation Data Acquisition and Analysis Service (NEODAAS) at Dundee University and Plymouth Marine Laboratory (www.neodaas.ac.uk). (B) Bacterial DGGE fingerprints of samples taken from the locations indicated in panel A. (C) Frequency of 16S rRNA gene clones belonging to major phylogenetic groups across the different gene libraries analyzed.
DNA SIP incubations.
Enrichment incubations with six C1 substrates were established on the day following sampling (day 0), with substrate concentrations of 100 μM (250 μM for DMS). Substrate had been depleted by day 3 in methanol incubations, and these were filtered for DNA extraction as were those containing methylamines. Approximately 83 μmol of [12C] and [13C]DMS were consumed by the fourth day (data not shown), and these incubations were subsequently sacrificed for DNA extraction. Methyl bromide incubations (with 12C and 13C) had consumed >90% of the 75 μmol of substrate originally present by day 18 and were filtered for DNA extraction. Changes in headspace concentrations of methane (100 μM total in the bottle; ∼0.63% in the headspace) for seawater incubations with methane were unchanged for several months (data not shown), and these incubations were not analyzed further.
16S rRNA gene fingerprinting of DNA from SIP experiments.
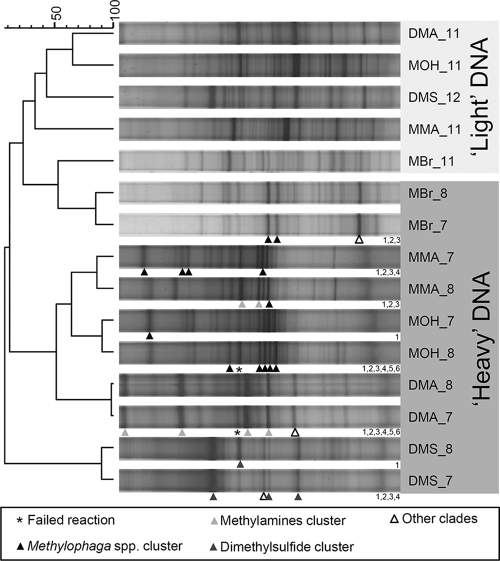
DGGE was used to profile the bacterial communities associated [“heavy”] and “light” fractions for both the 12C-labeled control samples and the 13C-labeled incubated samples. The 12C-labeled incubated samples showed no profile differences between the “heavy” and the “light” fractions (data not shown), whereas unique fingerprints were evident for all C1 substrate incubations (Fig. 2). As expected, the fingerprints for the light fractions of all tubes clustered together. The “background” bacterial communities in each SIP incubation were more similar to one another than to the “heavy” 13C-labeled fraction fingerprints of the same incubation. However, for the methyl bromide SIP incubation, the fingerprint of the “heavy” DNA was less clearly unique for the light DNA than for the other substrate incubations, reflecting the possibility that only a small amount of DNA was labeled and was just detectable above background [12C]DNA. All other DGGE fingerprints from heavy fractions (fractions 7 and 8) of 13C-labeled substrate incubations clustered in distinct clades apart from the “light” DNA, with monomethylamine and methanol fingerprints clustering closely, with some similarity to the dimethylamine fingerprints. Dimethylsulfide [13C]DNA fingerprints were distinct from all other patterns in this study, reflecting the unique composition of active methylotrophs enriched in these SIP incubations. Individual bands from fingerprints representing “heavy” DNA from 13C1 incubations were selected for PCR reamplification and sequencing. These sequences were used to assign band sequences to specific SIP-related 16S rRNA gene clades derived from this study and from a previous SIP study that was carried out under nonbloom conditions in the English Channel (36). The results indicate that Methylophaga spp. were associated with methyl bromide, methanol, and methylamine SIP incubations, whereas additional clades were affiliated with dimethylamine and DMS, likely contributing to their more distinct fingerprint profiles (Fig. 2).
FIG. 2.
DGGE fingerprint comparison of “light” and “heavy” DNA associated with DNA SIP incubations with different carbon sources (MMA, monomethylamine; DMA, dimethylamine; MOH, methanol; MBr, methyl bromide; and DMS). The dendrogram scale bar refers to the percent similarity of Pearson correlations between fingerprint densitometric curves. Shading of the triangle pointers indicates the phylogenetic affiliations of sequenced bands, most of which were associated with clades in Fig. 3. Numbers at the bottom right of the fingerprints correspond to sequenced bands submitted to GenBank. For example, the open triangle for fraction 7 of the [13C]methylbromide SIP (MBr_7) is labeled MBr_7_3 for the GenBank submission. Several bands were not associated with clades but were affiliated with sequences in Fig. 3, as follows: MBr_7_3 is identical to MBr_587_7; DMA_7_6 is closest to MBr_587_24; DMS_7_2 is identical to DMS_584_3.
16S rRNA gene clone libraries of [13C]DNA.
As the diversity of active methylotrophs was anticipated to be relatively low, 24 clones were sequenced from each library associated with SIP incubations with each of the five substrates analyzed in this study. The results of the sequencing confirmed the relatively low diversity of methylotrophs within each SIP incubation, but across the different substrates applied indicated a broad diversity of active marine methylotrophs in this study.
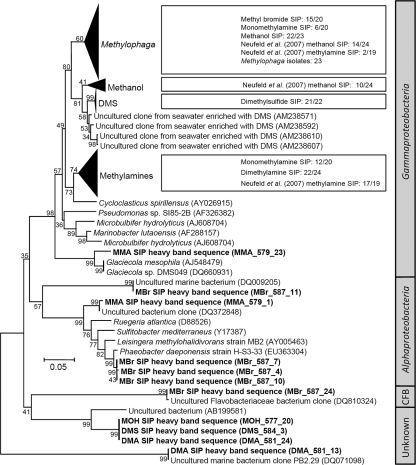
Methanol-assimilating phylotypes.
Phylogenetic analysis demonstrated that sequences associated with SIP incubations with methanol clustered in the Methylophaga species clade of the Gammaproteobacteria together with sequences from a previous SIP incubation (36) and with several characterized Methylophaga isolates (Fig. 3 and see Fig. S2 in the supplemental material). In addition, one cloned 16S rRNA gene sequence obtained from the methanol SIP was a member of a clade of unknown phylogenetic affiliation, which also contained one DMS and DMA-SIP-derived cloned 16S rRNA gene. The methanol clade identified in a previous methanol SIP experiment (Fig. 3; see Fig. S2 in the supplemental material) has the closest affiliation to Gammaproteobacteria sequences in GenBank that were retrieved from multiple marine Arctic surface sediments or detected on the surface of submerged artificial substrates incubated in marine water near China. This clade, however, was not detected in the current study.
FIG. 3.
Phylogenetic affiliations of 16S rRNA gene sequences obtained by 13C-labeled SIP incubations with methanol (MOH), monomethylamine (MMA), dimethylamine (DMA), methyl bromide (MBr), and DMS SIP incubations. Selected GenBank sequences from uncultivated clones and reference strains are included for comparison. Bootstrap values are included for all branch points on this neighbor-joining tree. GenBank accession numbers are shown in parentheses. The scale bar (in the tree) represents 5% sequence divergence. The collapsed clades are expanded in Fig. S2, S3, and S4 in the supplemental material. The division-level affiliation of sequences is indicated in the boxes at the right. CFB, Cytophaga-Flavobacterium-Bacteroides.
Phylotypes assimilating methylated amines.
As with the methanol SIP incubation, a previously characterized clade of sequences associated with a monomethylamine SIP incubation (36) was also represented by sequences from the monomethylamine SIP from the current study and also from the dimethylamine SIP incubation. In particular, 22 of the 24 sequences generated from the dimethylamine SIP incubation and most of the corresponding DGGE band sequences (Fig. 2) fell within this clade (Fig. 3 and see Fig. S3 in the supplemental material). This clade also contained several sequences isolated from arctic sediment (H. Li, Y. Yu, W. Luo, Y. Zeng, and B. Chen, unpublished), a mangrove ecosystem (27), and a deep-sea coral ecosystem (K. Penn, D. Wu, J. Eisen, and N. Ward, unpublished) and a strain isolated from the Yellow Sea (H. Kim and J.-C. Cho, unpublished; GenBank accession no. EF468718). Additional 16S rRNA gene sequences obtained from the monomethylamine SIP incubation belonged to the Methylophaga clade, many species of which can grow on methylated amines.
Phylotypes assimilating DMS.
Almost all 16S rRNA gene sequences derived from the DMS SIP “heavy” DNA were nearly identical and formed an additional clade with low relative diversity (Fig. 3; see Fig. S4 in the supplemental material). The DMS clade was most closely related to the methanol SIP clade associated with the class Gammaproteobacteria and identified in a previous study (35); it shared close similarity (96%) with sequences retrieved from clone libraries associated with DMS-enriched seawater samples from the Sargasso Sea (52) and was approximately 91% similar to the Methylophaga sp. clade, based on the percent similarity between sequences of DMS_584_22 and Methylophaga marina (GenBank accession no. X95459) over 722 bases. Another sequence from the “heavy” DNA of the DMS SIP was affiliated with a clade of unknown phylogeny.
DISCUSSION
The study site was chosen based on the mixed Emiliania and Karenia bloom that occurred in the English Channel in July 2006. The growth of phytoplankton in oceanic surface water has been associated with the direct or indirect production of methanol (13), methylamines (33), methyl halides (2, 4, 31, 42), methylated sulfur compounds (19, 20, 26), and methane, through decomposition (15, 39). In sampling from the edge of the bloom for SIP analysis (Fig. 1), the objective was to retrieve sequences of methylotrophs relevant to bloom C1 substrate production. Although the sample chosen was relevant to C1 metabolism, it is important to note that the substrate concentrations (100 μM) were far higher than those normally present in marine surface water samples. This was done because for a previous bloom in Bergen, Norway, the application of C1 substrates at low μM concentrations did not result in the detection of 13C-labeled DNA, possibly due to relatively high bacterial biomass associated with the bloom (J. D. Neufeld, R. Boden, H. Moussard, H. Schäfer, and J. C. Murrell, unpublished). In the present study, the objective was to identify phylotypes associated with the use of labeled C1 substrates, and the use of elevated substrate concentrations may have biased the results obtained. Typically, SIP experiments require substrate concentrations that exceed those found naturally, and the data may have to be interpreted with caution (35). Nonetheless, a comparison of near in situ substrate concentrations (1 μM) with a marine methanol SIP incubation detected the same Methylophaga species phylotypes as detected in the present study (34). As a result, for C1 substrates in the marine environment, the results may be consistent despite the range of substrate concentrations used. In all SIP incubations thus far, the incubation times were extended to days, and an addition of nutrients may have also selected for fast-growing species of methylotrophs. However, the uncultivated methylotrophs detected here are consistently present, which suggests that they do play an active role in C1 metabolism in coastal marine environments.
This study represents a comprehensive survey of active methylotrophs in a marine surface water sample during a bloom of phytoplankton associated with the production of dimethylsulfoniopropionate. The methylotrophs detected in this survey are consistent with the results of our pilot study with only methanol and monomethylamine under nonbloom conditions obtained a year prior to the current sampling event (36); however, the use of a wider range of C1 substrates allowed the identification of a larger diversity of methylotrophs than that found previously, including populations that assimilate dimethylamine, DMS, and methyl bromide. The DMS SIP clones obtained were most closely related to clones obtained from DMS enrichments from Pensacola, FL, and the Sargasso Sea by Vila-Costa and colleagues (52), suggesting that the latter had similar metabolic activities and indeed represented DMS-degrading populations. Those sequences were classified as “uncultivated Methylophaga”; however, given the relatively low similarity of the 16S rRNA gene sequences of these cloned 16S rRNA gene sequences to those of Methylophaga isolates (around 92%) and their distinct clustering, supported by bootstrap analysis (see Fig. S2 and S4 in the supplemental material), it is also possible that these represent DMS-degrading populations belonging to a different genus. Conversely, none of the DMS SIP clones was closely related to previously isolated DMS-degrading Methylophaga isolates (44), which belonged to the Methylophaga clade detected with methanol, monomethylamine, and methyl bromide. This strongly suggests that populations closely related to the isolated strains may have a preference for other C1 substrates and/or are outcompeted by those represented by the DMS clade under the specific incubation conditions. The methyl bromide SIP sequences indicate that methyl bromide may be used by members of the genus Methylophaga and an organism with a 16S rRNA gene sequence most similar to that of Phaeobacter gallaeciensis (formerly Roseobacter) within the family Rhodobacteraceae. The notion that the Phaeobacter-related population degraded methyl halides would be supported by previous cultivation-based identification of marine methyl halide-degrading organisms which were closely related (43, 45, 46); however, screening of several Methylophaga isolates has failed to show their ability to degrade methyl halides (H. Schäfer, unpublished). The observation of Methylophaga-like sequences in the [13C]methyl bromide incubation could therefore be due to the slow hydrolytic conversion of methyl bromide to methanol (1) and subsequent utilization of the resulting methanol by these organisms. If Methylophaga populations in the methyl bromide incubations became labeled with 13C due to the uptake of methanol produced by conversion of methyl halides to methanol, this would further underline their ability to take up methanol at ambient concentrations and support using SIP incubations with elevated substrate concentrations to investigate substrate-responsive populations in seawater. Together, these data suggest that marine waters harbor a diverse suite of active methylotrophs that, apart from Methylophaga spp., have been unnoticed by previous cultivation studies (8, 17, 23, 43-45) and are almost completely without representation in marine clone libraries. The sequences represented here are important targets for directed cultivation and focused activity-based studies of marine methylotrophy.
Given the focus of past marine metagenomic studies on abundant community members, it is perhaps not surprising that few genes (phylogenetic or “functional”) have reflected the predominance of methylotrophic bacteria. Although formaldehyde oxidation genes were identified in the Sargasso Sea metagenomic libraries (51), genes for methane, methylamine, and methanol oxidation were not detected (18). Furthermore, the only presumed methylotroph 16S rRNA gene sequences identified in a marine metagenomic library was from Methylophilus spp., and these sequences occurred at ∼0.4% of the total 16S rRNA gene data set from the global ocean survey (41). The contribution of Methylophilus to marine C1 cycling remains unclear, and Methylophilus spp. have not been detected in [13C]DNA from the incubations carried out in this study. One possibility is that Methylophilus spp. represent K-selected organisms, which adapted to concentrations of carbon and nutrients that are lower than those used in this study. Cultivation-based approaches (17), enrichment cultures (44, 52), and SIP incubations (current study; 34, 36) have all demonstrated that Methylophaga spp. and related Gammaproteobacteria from multiple disparate marine samples (including estuary sediment; unpublished data) are present in the seawater samples and rapidly respond to the presence of C1 substrates. It is possible that these organisms may represent low-abundance and r-selected bacteria capable of opportunistic growth in the presence of relatively high concentrations of growth substrates during phytoplankton blooms, for example.
This study represents a comprehensive cultivation-independent survey of active marine methylotrophs and demonstrates that previously unrecognized bacterial groups are present in seawater, which are capable of responding to the presence of added C1 substrates. The presence of numerous clades of presumed substrate-specific methylotrophs presents a challenge to microbiologists to focus cultivation and quantitative molecular approaches to better understand the metabolism and distribution dynamics of these organisms with potentially enormous biogeochemical significance.
Supplementary Material
Acknowledgments
We thank Ian Joint, Clare Evans, Declan Schroeder, and Jack Gilbert for assistance with sampling. We also thank the NERC Earth Observation Data Acquisition and Analysis Service (NEODAAS) for supplying satellite data for this study.
This work was supported by funding from the Natural Environment Research Council (United Kingdom) Aquatic Microbial Metagenomics and Biogeochemical Cycles grant, reference NE/C001 923/1. H.S. was supported by a NERC postdoctoral fellowship (NE/B501404/1). J.D.N. acknowledges support from the Natural Sciences and Engineering Research Council (Canada).
Footnotes
Published ahead of print on 10 October 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abbattista Gentile, I., L. Ferraris, M. Sanguinetti, M. Tiprigan, and G. Fisichella. 1992. Methyl bromide in fresh waters: hydrolysis and volatilisation. Pestic. Sci. 34:297-301. [Google Scholar]
- 2.Anbar, A. D., Y. L. Yung, and F. R. Chavez. 1996. Methyl bromide: ocean sources, ocean sinks, and climate sensitivity. Global Biogeochem. Cycles 10:175-190. [DOI] [PubMed] [Google Scholar]
- 3.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, J. M., C. E. Reeves, P. D. Nightingale, S. A. Penkett, S. W. Gibb, and A. D. Hatton. 1999. Biological production of methyl bromide in the coastal waters of the North Sea and open ocean of the northeast Atlantic. Mar. Chem. 64:267-285. [Google Scholar]
- 5.Beerli, R., and H.-J. Borschberg. 1991. Preparation of [13C2]-DMSO. J. Labelled Comp. Radiopharm. 29:957-961. [Google Scholar]
- 6.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, B. A. Rapp, and D. L. Wheeler. 2000. GenBank. Nucleic Acids Res. 28:15-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpenter, L. J., A. C. Lewis, J. R. Hopkins, K. A. Read, I. D. Longley, and M. W. Gallagher. 2004. Uptake of methanol to the North Atlantic Ocean surface. Global Biogeochem. Cycles 18:GB4027. [Google Scholar]
- 8.de Zwart, J., P. Nelisse, and J. Kuenen. 1996. Isolation and characterization of Methylophaga sulfidovorans sp. nov.: an obligately methylotrophic, aerobic, dimethylsulfide oxidizing bacterium from a microbial mat. FEMS Microbiol. Ecol. 20:261-270. [Google Scholar]
- 9.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 10.Galbally, I. E., and W. Kirstine. 2002. The production of methanol by flowering plants and the global cycle of methanol. J. Atmos. Chem. 43:195-229. [Google Scholar]
- 11.Gibb, S., and A. Hatton. 2004. The occurrence and distribution of trimethylamine-N-oxide in Antarctic coastal waters. Mar. Chem. 91:65-75. [Google Scholar]
- 12.Goodwin, K. D., R. K. Varner, P. M. Crill, and R. S. Oremland. 2001. Consumption of tropospheric levels of methyl bromide by C1 compound-utilizing bacteria and comparison to saturation kinetics. Appl. Environ. Microbiol. 67:5437-5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heikes, B. G., W. Chang, M. E. Q. Pilson, E. Swift, H. B. Singh, A. Guenther, D. J. Jacob, B. D. Field, R. Fall, D. Riemer, and L. Brand. 2002. Atmospheric methanol budget and ocean implication. Global Biogeochem. Cycles 16:80-81. [Google Scholar]
- 14.Heyer, J., Y. Malashenko, U. Berger, and E. Budkova. 1984. Verbreitung methanotropher Bakterien. Z. Allg. Mikrobiol. 24:725-744. [Google Scholar]
- 15.Holmes, A. J., N. J. P. Owens, and J. C. Murrell. 1995. Detection of novel marine methanotrophs using phylogenetic and functional gene probes after methane enrichment. Microbiology 141:1947-1955. [DOI] [PubMed] [Google Scholar]
- 16.Holmes, A. J., N. J. P. Owens, and J. C. Murrell. 1996. Molecular analysis of enrichment cultures of marine methane oxidising bacteria. J. Exp. Mar. Biol. Ecol. 203:27-38. [Google Scholar]
- 17.Janvier, M., C. Frehel, F. Grimont, and F. Gasser. 1985. Methylophaga marina gen. nov., sp. nov. and Methylophaga thalassica sp. nov., marine methylotrophs. Int. J. Syst. Bacteriol. 35:131-139. [Google Scholar]
- 18.Kalyuzhnaya, M. G., O. Nercessian, A. Lapidus, and L. Chistoserdova. 2005. Fishing for biodiversity: novel methanopterin-linked C1 transfer genes deduced from the Sargasso Sea metagenome. Environ. Microbiol. 7:1909-1916. [DOI] [PubMed] [Google Scholar]
- 19.Kiene, R. P. 1990. Dimethyl sulfide production from dimethylsulfoniopropionate in coastal seawater samples and bacterial cultures. Appl. Environ. Microbiol. 56:3292-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiene, R. P. 1992. Dynamics of dimethyl sulfide and dimethylsulfoniopropionate in oceanic water samples. Mar. Chem. 37:29-52. [Google Scholar]
- 21.Kiene, R. P. 1993. Microbial sources and sinks for methylated sulfur compounds in the marine environment, p. 15-33. In D. P. Kelley and J. C. Murrell (ed.), Microbial growth on C1 compounds. Intercept Ltd., London, United Kingdom.
- 22.Kim, S. K., F. Rassoulzadegan, B. Krajka, B. C. Nguyen, N. Mihalopoulos, and P. Buat-Menard. 1990. Production of dimethylsulfonium propionate (DMSP) and dimethylsulfide (DMS) by a microbial food web. Limnol. Oceanogr. 35:1810-1821. [Google Scholar]
- 23.Kimura, T., I. Sugahara, and K. Hayashi. 1990. Use of short-chain amines and amino acids as sole sources of nitrogen in a marine methylotrophic bacterium, Methylophaga sp. AA-30. Agric. Biol. Chem. 54:1873-1874. [Google Scholar]
- 24.Kwint, R. L. J., and K. J. M. Kramer. 1995. Dimethylsulphide production by plankton communities. Mar. Ecol. Prog. Ser. 121:227-237. [Google Scholar]
- 25.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., Chichester, United Kingdom.
- 26.Levasseur, M., S. Michaud, J. Egge, G. Cantin, J. C. Nejstgaard, R. Sanders, E. Fernandez, P. T. Solberg, B. Heimdal, and M. Gosselin. 1996. Production of DMSP and DMS during a mesocosm study of an Emiliania huxleyi bloom: influence of bacteria and Calanus finmarchicus grazing. Mar. Biol. 126:609-618. [Google Scholar]
- 27.Liao, P.-C., B.-H. Huang, and S. Huang. 2007. Microbial community composition of the Danshui River estuary of northern Taiwan and the practicality of the phylogenetic method in microbial barcoding. Microb. Ecol. 54:497-507. [DOI] [PubMed] [Google Scholar]
- 28.Lidstrom, M. E. 1988. Isolation and characterization of marine methanotrophs. Antonie van Leeuwenhoek 54:189-199. [DOI] [PubMed] [Google Scholar]
- 29.Liss, P., G. Malin, and S. Turner. 1992. Production of DMS by marine phytoplankton, p. 1-14. In G. Restelli and G. Angeletti (ed.), Dimethylsulphide: oceans, atmosphere and climate. Kluwer Academic Publishing, Dordrecht, The Netherlands.
- 30.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonald, I. R., K. L. Warner, C. McAnulla, C. A. Woodall, R. S. Oremland, and J. C. Murrell. 2002. A review of bacterial methyl halide degradation: biochemistry, genetics and molecular ecology. Environ. Microbiol. 4:193-203. [DOI] [PubMed] [Google Scholar]
- 32.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naqvi, S. W. A., H. W. Bange, S. W. Gibb, C. Goyet, A. D. Hatton, and R. C. Upstill-Goddard. 2005. Biogeochemical ocean-atmosphere transfers in the Arabian Sea. Prog. Oceanogr. 65:116. [Google Scholar]
- 34.Neufeld, J. D., Y. Chen, M. G. Dumont, and J. C. Murrell. 2008. Marine methylotrophs revealed by stable-isotope probing, multiple displacement amplification and metagenomics. Environ. Microbiol. 10:1526-1535. [DOI] [PubMed] [Google Scholar]
- 35.Neufeld, J. D., M. G. Dumont, J. Vohra, and J. C. Murrell. 2007. Methodological considerations for the use of stable isotope probing in microbial ecology. Microb. Ecol. 53:435-442. [DOI] [PubMed] [Google Scholar]
- 36.Neufeld, J. D., H. Schäfer, M. J. Cox, R. Boden, I. R. McDonald, and J. C. Murrell. 2007. Stable-isotope probing implicates Methylophaga spp. and novel Gammaproteobacteria in marine methanol and methylamine metabolism. ISME J. 1:480-491. [DOI] [PubMed] [Google Scholar]
- 37.Neufeld, J. D., J. Vohra, M. G. Dumont, T. Lueders, M. Manefield, M. W. Friedrich, and J. C. Murrell. 2007. DNA stable-isotope probing. Nat. Protoc. 2:860-866. [DOI] [PubMed] [Google Scholar]
- 38.Neufeld, J. D., Z. Yu, W. Lam, and W. W. Mohn. 2004. Serial analysis of ribosomal sequence tags (SARST): a high-throughput method for profiling complex microbial communities. Environ. Microbiol. 6:131-144. [DOI] [PubMed] [Google Scholar]
- 39.Oremland, R. S. 1979. Methanogenic activity in plankton samples and fish intestines: a mechanism for in situ methanogenesis in oceanic subsurface waters. Limnol. Oceanogr. 24:1136-1141. [Google Scholar]
- 40.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 41.Rusch, D. B., A. L. Halpern, G. Sutton, K. B. Heidelberg, S. Williamson, S. Yooseph, D. Wu, J. A. Eisen, J. M. Hoffman, K. Remington, K. Beeson, B. Tran, H. Smith, H. Baden-Tillson, C. Stewart, J. Thorpe, J. Freeman, C. Andrews-Pfannkoch, J. E. Venter, K. Li, S. Kravitz, J. F. Heidelberg, T. Utterback, Y.-H. Rogers, L. I. Falcón, V. Souza, G. Bonilla-Rosso, L. E. Eguiarte, D. M. Karl, S. Sathyendranath, T. Platt, E. Bermingham, V. Gallardo, G. Tamayo-Castillo, M. R. Ferrari, R. L. Strausberg, K. Nealson, R. Friedman, M. Frazier, and J. C. Venter. 2007. The Sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 5:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sæmundsdóttir, S., and P. A. Matrai. 1998. Biological production of methyl bromide by cultures of marine phytoplankton. Limnol. Oceanogr. 43:81-87. [Google Scholar]
- 43.Schaefer, J. K., K. D. Goodwin, I. R. McDonald, J. C. Murrell, and R. S. Oremland. 2002. Leisingera methylohalidivorans gen. nov., sp. nov., a marine methylotroph that grows on methyl bromide. Int. J. Syst. Evol. Microbiol. 52:851-859. [DOI] [PubMed] [Google Scholar]
- 44.Schäfer, H. 2007. Isolation of Methylophaga spp. from marine dimethylsulfide-degrading enrichment cultures and identification of polypeptides induced during growth on dimethylsulfide. Appl. Environ. Microbiol. 73:2580-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schäfer, H., I. R. McDonald, P. D. Nightingale, and J. C. Murrell. 2005. Evidence for the presence of a CmuA methyltransferase pathway in novel marine methyl halide-oxidizing bacteria. Environ. Microbiol. 7:839-852. [DOI] [PubMed] [Google Scholar]
- 46.Schäfer, H., L. Miller, R. Oremland, and J. C. Murrell. 2007. Bacterial cycling of methyl halides. Adv. Appl. Microbiol. 61:307-346. [DOI] [PubMed] [Google Scholar]
- 47.Sieburth, J. N., P. W. Johnson, M. A. Eberhardt, M. E. Sieracki, M. Lidstrom, and D. Laux. 1987. The first methane-oxidizing bacterium from the upper mixing layer of the deep ocean: Methylomonas pelagica sp. nov. Curr. Microbiol. 14:285-293. [Google Scholar]
- 48.Singh, H. B., A. Tabazadeh, M. J. Evans, B. D. Field, D. J. Jacob, G. Sachse, J. H. Crawford, R. Shetter, and W. H. Brune. 2003. Oxygenated volatile organic chemicals in the oceans: inferences and implications based on atmospheric observations and air-sea exchange models. Geophys. Res. Lett. 30:1862. [Google Scholar]
- 49.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596. [DOI] [PubMed] [Google Scholar]
- 50.Tamura, K., M. Nei, and S. Kumar. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 101:11030-11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y.-H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 52.Vila-Costa, M., D. A. del Valle, J. M. Gonzalez, D. Slezak, R. P. Kiene, O. Sanchez, and R. Simo. 2006. Phylogenetic identification and metabolism of marine dimethylsulfide-consuming bacteria. Environ. Microbiol. 8:2189-2200. [DOI] [PubMed] [Google Scholar]
- 53.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams, J., R. Holzinger, V. Gros, X. Xu, E. Atlas, and D. W. R. Wallace. 2004. Measurements of organic species in air and seawater from the tropical Atlantic. Geophys. Res. Lett. 31:L23S06. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.